SacB-SacR Gene Cassette As the Negative Selection Marker to Suppress Agrobacterium Overgrowth in Agrobacterium-Mediated Plant Transformation
- 1Department of Crop and Soil Environmental Science, Virginia Tech, Blacksburg, VA, USA
- 2Chinese Academy of Tropical Agricultural Sciences/Key Laboratory of Crop Gene Resources and Germplasm Enhancement in Southern China, Tropical Crops Genetic Resources Institute, Ministry of Agriculture, Danzhou, China
- 3Department of Horticulture, Virginia Tech, Blacksburg, VA, USA
- 4Department of Biology, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA
- 5USDA-ARS-Appalachian Fruit Research Station, Kearneysville, WV, USA
Agrobacterium overgrowth is a common problem in Agrobacterium-mediated plant transformation. To suppress the Agrobacterium overgrowth, various antibiotics have been used during plant tissue culture steps. The antibiotics are expensive and may adversely affect plant cell differentiation and reduce plant transformation efficiency. The SacB-SacR proteins are toxic to most Agrobacterium tumefaciens strains when they are grown on culture medium supplemented with sucrose. Therefore, SacB-SacR genes can be used as negative selection markers to suppress the overgrowth of A. tumefaciens in the plant tissue culture process. We generated a mutant A. tumefaciens strain GV2260 (recA-SacB/R) that has the SacB-SacR cassette inserted into the bacterial genome at the recA gene locus. The mutant Agrobacterium strain is sensitive to sucrose but maintains its ability to transform plant cells in both transient and stable transformation assays. We demonstrated that the mutant strain GV2260 (recA-SacB/R) can be inhibited by sucrose that reduces the overgrowth of Agrobacterium and therefore improves the plant transformation efficiency. We employed GV2260 (recA-SacB/R) to generate stable transgenic N. benthamiana plants expressing a CRISPR-Cas9 for knocking out a WRKY transcription factor.
Introduction
Agrobacterium-mediated genetic transformation is one of the most popular techniques used for the generation of transgenic plants (Gelvin, 2000; Tzfira and Citovsky, 2006). Efficient Agrobacterium-mediated transformation protocols have been developed for various plant species (Hiei et al., 1994; Ishida et al., 1996; Cheng et al., 1997; Tingay et al., 1997; Zhao et al., 2000). In general, the Agrobacterium-mediated transformation involves the generation of a sterile explant that can be co-cultured with Agrobacterium. Subsequently, the infected Agrobacterium cells are eliminated or suppressed by using various antibiotics, and the transgenic plant cells are selected by using antibiotics or other chemicals (Jones et al., 2005; Tsuda et al., 2012). During the Agrobacterium-mediated transformation process, one major problem is the overgrowth of Agrobacterium that could significantly reduce the plant transformation efficiency. To eliminate or inhibit the Agrobacterium overgrowth, different antibiotics such as carbenicillin, Timentin™, Augement, Clavamox, and Cefotaxime are used during the plant tissue culture selection steps (Bhau and Wakhlu, 2001; Tereso et al., 2006; Zang et al., 2009; Li and Qu, 2011; Ren et al., 2012). For example, carbenicillin is a semi-synthetic penicillin antibiotic that interferes with cell wall mucopeptide biosynthesis of gram-negative bacteria (Silva and Fukai, 2001). Under selection pressure, Agrobacterium cells could gain mutations that are resistant to the carbenicillin, which results in the overgrowth of Agrobacterium. Most antibiotics are generally expensive and may negatively affect plant cell differentiation (Ellis et al., 1989; Yu et al., 2001; Li and Qu, 2011). Therefore, a more reliable and cost-effective method to inhibit the overgrowth of Agrobacterium tumefaciens is highly desirable.
The SacB-SacR genes were originally isolated from Bacillus subtilis and encode levansucrase, an enzyme involved in both the hydrolysis of sucrose and the biosynthesis of levan (Chambert and Petitglatron, 1989; Quandt and Hynes, 1993; Traore and Zhao, 2011). Levan cannot be metabolized by most gram-negative bacteria including A. tumefaciens and is therefore toxic to this group of organisms (Gay et al., 1985; Schweizer, 1992). The SacB-SacR gene cassette, driven by its native promoter, has been used as a negative selectable marker for many gram-negative bacteria, and works by preventing the transformed bacterial cells from growing on culture medium supplemented with sucrose (Ried and Collmer, 1987). Our previous work demonstrated that the SacB-SacR genes can be used as negative selection markers to inhibit the growth of Agrobacterium strain GV2260 on Luria Broth agar medium supplemented with 5% sucrose (Traore and Zhao, 2011). Sucrose has been frequently used as the carbon source in synthetic plant tissue culture medium (Yaseen et al., 2013), although other sugars such as maltose, fructose, and sorbitol have also been used. It is interesting to test if growth of an Agrobacterium strain carrying the SacB-SacR gene cassette can be inhibited on plant tissue medium supplemented with sucrose.
The recA gene was originally identified as a conserved gene involved in homologous DNA recombination in various bacterial species (Clark and Margulies, 1965; Brendel et al., 1997; Song et al., 2003). RecA-dependent recombination was initially identified through analysis of conjugational recombination (Clark and Margulies, 1965; Bi and Liu, 1994), where recA can promote homologous pairing of DNA molecules and catalyzes the strand exchange reaction leading to the formation of hetero-duplex DNA in vitro (West, 1992; Bi and Liu, 1994). Deletion of the recA gene in the bacterial genome can reduce the rate of homologous recombination, and therefore increase plasmid DNA stability. Deletion of the recA gene in E. coli has no obvious deleterious effect on bacterial growth (Kurnit, 1989; Lovett et al., 1993). Several Agrobacterium strains with deletion of the recA gene were also developed (Farrand et al., 1989).
In this study, we attempted to integrate the SacB-SacR gene cassette at the recA gene locus in the genome of A. tumefaciens strain GV2260. The derived mutant strain GV2260 (recA-SacB/R) was used to transform Nicotiana benthamiana (N. benthamiana) plant cells in both transient assays and stable transformation. We demonstrated that the mutant strain GV2260 (recA-SacB/R) maintains its capacity of transforming plant cells, and its growth can be efficiently inhibited by regular tobacco tissue culture medium supplemented with 3% sucrose. Stable transgenic plants carrying a CRISPR-Cas9 construct for knocking out a WRKY transcription factor were successfully recovered after transformation with the mutant Agrobacterium strain. Therefore, the mutant A. tumefaciens strain should have great value for large scale, high-throughput plant transformation applications in the future.
Materials and Methods
Bacteria Strains
Escherichia coli (E. coli) DH5α [F−endA glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG Φ80dlacZΔM15Δ(lacZYA-argF)U169, hsdR17(), λ−], A. tumefaciens (GV2260) [C58 background, rifampicin-resistant with the Ti plasmid (pTiB6s3)], and Escherichia coli (E. coli) helper P600 (Traore and Zhao, 2011).
Plant Materials
N. benthamiana (PI 555478) plants were propagated in a growth chamber programmed for 16 h light (140 μmol m−2s−1 cool white fluorescent irradiance) at 28°C and 8 h dark at 24°C Agrobacterium-mediated transient assays were conducted on three- to 4-week-old plants.
Cloning of the Agrobacterium recA Gene Fragment
A 780 bp DNA fragment with deletion of the N and C-terminus of the recA gene was amplified from the genomic DNA of GV2260 using primers: recA For, 5′-caccatcgatcatgaagctcggt-3′ and recA Rev, 5′-gcgccggacttctcgacgat-3′. The PCR reaction as performed using the iProof™ high fidelity Taq DNA polymerase (Bio-Rad, Hercules, CA). The PCR program consisted of 1 cycle at 98°C (2 min), followed by 30 cycles at 98°C (30 s), 55°C (45 s), and 72°C (1 min), and finished with a 1 cycle extension at 72°C (7 min). The PCR product was separated on a 0.8% agarose gel, stained with 0.01% ethidium bromide solution, and visualized using the Gel-Document Image System™ under UV light (Bio-Rad).
The PCR product was purified using the AccuPrep™ Gel Purification Kit (Bioneer, Alameda, CA) and cloned into the TopoEntr/D™ vector (Invitrogen, Carlsbad, CA) following the instructions of the user manual. The derived plasmid vector was designated as TopoEntr-recA and has been sequenced at the core facility of the Virginia Bioinformatics Institute (Blacksburg, VA).
Development of an Integrational Construct Carrying recA Fragment and the SacB-SacR Gene Cassette
The suicide vector pLVC18L (Zhao et al., 2011) was modified by insertion of the SacB/R and the ccdB gene cassettes. The Npt2 promoter-SacB/R fragment was amplified through overlap PCR from pEG101-SacB/R and pDSK519-GFP (Matthysse et al., 1996; Traore and Zhao, 2011) using primers: 1846pLvc18 XbaNpt2 Infusion For1, 5′-TGC CATTGCTGCAGGTCGACTCTAGAGATATCACATGGCGATAGCTAGACT G-3′; 1777Npt2Pro_SacB/R Rv Rev1, 5′-GTG ATGGGTTAAAAAGGATCGATCCGCGCCATCAGATCC TTG-3′; 1778Npt2Pro_SacB/R Rv For2, 5′-CAA GGATCTGATGGCGCGGATCGATCCTTTTTAACCCAT CAC-3′; 1847pLvc18 XbaSacB Infusion Rev2, 5′-CTC GGTACCCGGGGATCCTCTAGAGATATCTTATTTGTTAACTGTTAATTG TCCT-3′.
The PCR product was cloned into the XhoI site of pLVC18L using a Gibson cloning kit (New England BioLabs Inc., Ipswich, MA). The derived construct was designated as pLVC18L-Npt2-SacB/R. A ccdB gene cassette (frame B) (Invitrogen) was further cloned into the SmaI site of pLVC18L-Npt2-SacB/R to generate pLVC18L-Npt2-SacB/R-DesB. The recA gene fragment from TopoEntr-recA was subcloned into pLVC18L-Npt2-SacB/R-DesB using a Gateway®; LR cloning kit (Invitrogen) following the instructions of the user manual. The derived plasmid construct was named pLVC18L-Npt2-SacB/R-recA, and has been confirmed by sequencing at the core facility of the Virginia Bioinformatics Institute (Blacksburg, VA).
Integration of the pLVC18L-Npt2-SacB/R-recA Construct into the Genome of Agrobacterium tumefaciens Strain GV2260
The suicide vector pLVC18L-Npt2-SacB/R-recA was integrated to the genome of A. tumefaciens strain GV2260 by tri-parental conjugation and was selected on LB medium supplemented with tetracycline (10 μg/mL) as previously described (Traore and Zhao, 2011). A mutant GV2260 strain carrying the Npt2-SacB/R cassette was confirmed by PCR amplification of the tetracycline resistance gene and SacB/R gene using primers: tetracycline For, 5′-atgaaatctaacaatgcg ctcat-3′; tetracycline Rev, 5′-tacgagttgcatgataaagaa gaca-3′, and SacB-SacR For, 5′-cagcatatcatggcgtgt aatatg-3′; SacB-SacR Rev, 5′-ctcggtacccggggatcctctagagat atcttatttgttaactgttaattgtcct-3′. The derived mutant strain was designated as GV2260-SacB/R.
Development of the Plasmid Vector pEarleygate101-YFP-HA
The YFP gene open reading frame plus the HA epitope tag was amplified from vector pEarleygate101 (Earley et al., 2006) with primers 2702pEG101-yfpHA For, 5′-ATTTGGAGAGGACACGctcgagAtgAGCAAGGGCGAGGAGCTGTTC ACCG-3′; 2703pEG101-yfpHA Rev, 5′-TCGACTGCAGAATTCGAAGCTTGAGctcgagATCTGAG-3′. The PCR product was gel purified and cloned into pEarleygate101 that had been digested with XhoI. The derived construct was designated as pEarleygate101-YFP-HA.
Development of a CRISPR-Cas9 Construct pgRNA-NbWRKY70 for Knocking Out Tobacco Transcription Factor WRKY70
A putative tobacco WRKY transcription factor NbWRKY70 was identified from GenBank (accession number AF421157). To knock out NbWRKY70, we identified a guiding RNA (GCAATCGACGGGTTAATTCGCGG) targeting the NbWRKY70 gene. An Arabidopsis U6 promoter, NbWRKY70 guiding RNA, and the PAM terminator were amplified through overlap-PCR using primers 2284AtU6gRNA common For1, CAGCAACTCATTACAACTTGTTTaagctttcgttgaacaacgga; 2285AtU6gRNA common Rev1, CGACTCTAGACACGGGGTGGTTTaaaaaaagcaccgactcggtgcc; 2846NbWRKY70 gRNA For, GCAATCGACGGGTTAATTCGgttttagagctagaaatag; 2847NbWRKY70 gRNA Rev, CGAATTAACCCGTCGATTGCaatcactacttcgactcta.
The PCR product was gel purified and cloned into the PmeI site of pM3UT-Cas9 using a Gibson cloning kit (New England BioLabs), where it contains a Cas9 gene driven by the Arabidopsis Ubiquitin 10 promoter. The Cas9 gene was originally codon optimized and synthesized based on the Arabidopsis genes (Zachary Nimchuk, unpublished data). The derived construct was designated as pgRNA-NbWRKY70.
Conjugation of pEarleygate101-YFP-HA and pgRNA-NbWRKY70 into Agrobacterium tumefaciens Strain GVV2260 and GV2260-SacB/R
The plasmid vectors pEarleygate101-YFP-HA and pgRNA-NbWRKY70 were conjugated into A. tumefaciens strain GVV2260 and GV2260-SacB/R using tri-parental conjugation, selected on LB medium supplemented with rifampin 100 μg/mL, and Kanamycin 50 μg/mL or Spectinomycin 50 μg/mL as previously described (Traore and Zhao, 2011).
Agrobacterium-Mediated Transient Assays in N. benthamiana Plants
Agrobacterium-mediated transient assays in N. benthamiana plants were performed as described previously (Wydro et al., 2006). In brief, the Agrobacterium strains were streaked on Yeast Extract Tryptone (YT) media supplemented with rifampicin 100 μg/mL, tetracycline 10 μg/mL, and kanamycin 50 μg/mL and incubated at 28°C for 2 days. Bacterial cells were harvested and re-suspended in induction buffer composed of 10 mM MgCl2, 10 mM MES (pH 5.6), and 100 μM acetosyringone and incubated for 3 h at room temperature. The bacterial inoculums were adjusted to OD600 nm = 0.6 and infiltrated into the stomata of the fully expanded N. benthamiana leaves using a 1-mL blunt-end syringe without a needle. The inoculated plants were incubated at room temperature under continuous light for 20–48 h before the detection of expressed proteins. The fluorescent signal of YFP-HA fusion protein was monitored 24 h after inoculation by fluorescent microscopy (Zeiss Axio Observer.A1, Carl Zeiss MicroImaging, Inc., Thornwood, NY).
Generation of Transgenic Tobacco Plants Using Either Wild Type or Mutant Agrobacterium Strains
Agrobacterium strain GV2260 or GV2260-SacB/R carrying plasmid pgRNA-NbWRKY70 were used for tobacco transformation following a previously described protocol (Horsch et al., 1989). In brief, the fully expanded leaf from a 4-week-old N. benthamiana plant was collected and sterilized in 10% bleach for 20 min. The leaf was cut into 1 cm2 leaf disks that were infected with Agrobacterium culture diluted to OD600 = 0.1. The infected leaf disks were co-cultured on MS medium supplemented with 6-BA (1 mg/L) and NAA (0.1 mg/L) and 3% maltose at 25°C in the dark for 2 days. The infected leaf disks were soaked in liquid MS medium for 5 min and then rinsed one time with liquid MS medium. The leaf disks were then blotted dry and transferred to a selection medium (MS medium supplemented with kanamycin 300 mg/L, cefotaxime 150 mg/L and 3 or 5% sucrose). Each treatment has at least 100 leaf disks with three replicates.
The selection mediums were incubated at 25°C under continuous light for 25–30 days for shoot regeneration. The transgenic shoots were transferred to rooting medium (MS medium supplemented with Kanamycin 100 mg/L and 3% sucrose).
The putative transgenic tobacco plants were confirmed by PCR with primers: PMOA36-TBS PmeI For, 5′-TGATAGAGTAGTTCATAT GGA-3′ and PMOA36-TBS PmeI Rev 5′-GCTTCC CAACCTTACCAGAG-3′. The PCR products were gel purified and sequenced at the core facility at Virginia Bioinformatics Institute.
Bacterial Genomic DNA, Plasmid DNA, and Plant Genomic DNA Isolation
Bacterial genomic DNAs were isolated using a ZR Fungal/Bacterial DNA MiniPrep™ (Zymo Research Corporation, Irvine, CA). Plasmid DNAs were isolated using an AccuPrep™ Plasmid Extraction Kit (Bioneer Corporation, Alameda, CA). Plant genomic DNAs were isolated by using the CTAB method as previously described (Zhang et al., 2013).
Results and Discussion
Development of a Sucrose-Sensitive Mutant Agrobacterium Strain GV2260-SacB/R
To generate a mutant Agrobacterium strain that is sensitive to sucrose, a suicide vector pLVC18L carrying the SacB-SacR gene cassette was integrated into the recA gene locus in the genome of A. tumefaciens strain GV2260 through marker-exchange mutagenesis. The mutant strain was named GV2260-SacB/R. The integration of the SacB-SacR gene cassette in GV2260-SacB/R was confirmed by PCR amplification. As shown in Figure 1A, the plasmid DNA of pLVC18L-Npt2-SacB/R-recA and the genomic DNA of GV2260-SacB/R, but not the wild type strain GV2260, can amplify the SacB-SacR gene and a tetracycline resistance gene located on the suicide vector pLVC18L-Npt2-SacB/R-recA. All three DNAs can amplify the recA gene fragment. These results suggest that GV2260-SacB/R is carrying the SacB-SacR gene cassette. The mutant strain GV2260-SacB/R is expected to carry a non-functional recA gene (Lovett et al., 1993; Bi and Liu, 1994). In the future, it will be interesting to test the stability of plasmids maintained in GV2260-SacB/R.
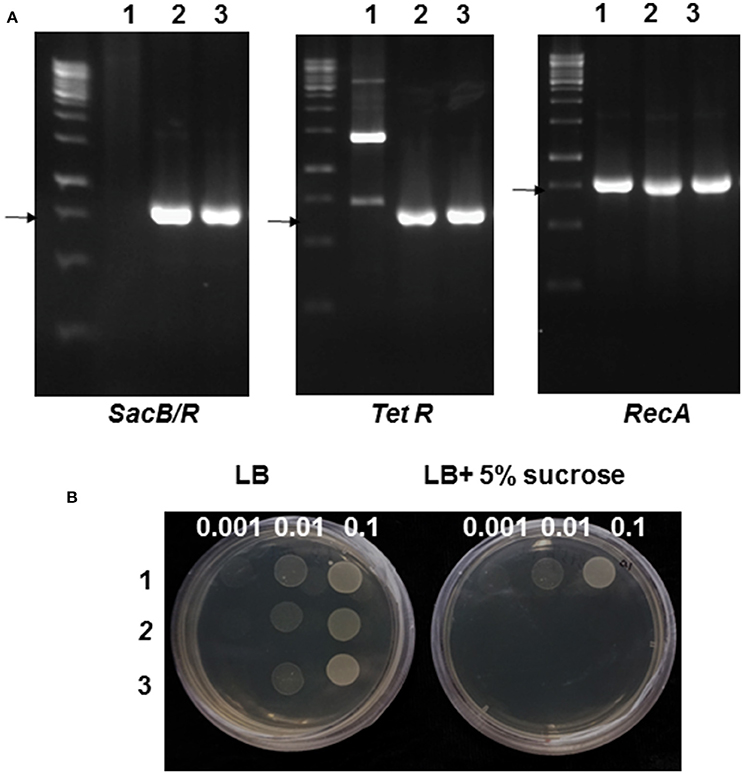
Figure 1. Genotype and phenotype validation of the mutant Agrobacterium strain GV2260-SacB/R. (A) PCR analysis of GV2260 (1), E. coli carrying pLVC18L-Npt2-SacB/R-recA (2), and mutant Agrobacterium strain GV2260-SacB/R (3) with primers for detecting the tetracycline resistance gene, SacB-SacR and recA genes. The arrows highlight the specifically amplified DNA fragments. (B) Testing for the inhibition of GV2260 (1), E. coli carrying pLVC18L-Npt2-SacB/R-recA (2), and mutant Agrobacterium strain GV2260-SacB/R (3) on LB media supplemented with 5% sucrose. Three bacterial dilutions, OD600 = 0.1, 0.01, and 0.001, have been used for testing on the LB medium.
To test the sucrose-sensitivity of GV2260-SacB/R, the mutant strain along with the wild type strain GV2260, and E. coli strain DH5α carrying pLVC18L-Npt2-SacB/R-recA were grown on LB agar medium supplemented with or without 5% sucrose. The wild type A. tumefaciens strain GV2260 grew equally well on LB medium with or without 5% sucrose, which suggests that sucrose in LB agar medium has no inhibitory effect on A. tumefaciens GV2260 (Figure 1B). In contrast, the E. coli strain DH5α carrying pLVC18L-Npt2-SacB/R-recA and GV2260-SacB/R grew well on LB agar medium without 5% sucrose, but showed almost no growth on LB agar medium with 5% sucrose. This result suggests that E. coli carrying pLVC18L-Npt2-SacB/R-recA and GV2260-SacB/R containing the SacB-SacR gene cassette can be effectively inhibited by the 5% sucrose presented in the culture medium.
Mutant Agrobacterium Strain GV2260-SacB/R Maintains Its Ability of Transforming Tobacco Plant Cells
To examine if GV2260-SacB/R can be used for plant cell transformation, the GV2260 and GV2260-SacB/R strains carrying plasmid construct pEarleygate101-YFP-HA were infiltrated into the leaves of N. benthamiana. In this construct, the YFP gene was cloned behind the CaMV 35S promoter, and it can be expressed in the transformed tobacco plant cells. Strong YFP fluorescence signals were detected from leaves inoculated with either GV2260 or GV2260-SacB/R carrying the YFP gene (Figure 2), which suggests that both strains can successfully transform the N. benthamiana plant cells.
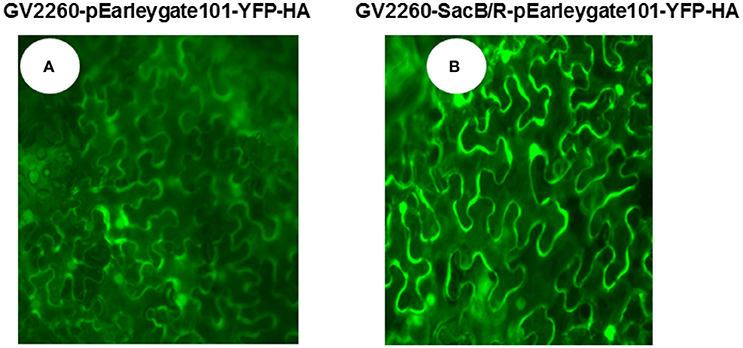
Figure 2. Agrobacterium-mediated transient expression of YFP in the leaves of N. benthamiana. (A) N. benthamiana leaf inoculated with Agrobacterium strain GV2260 carrying pEarleygate101-YFP-HA, (B) N. benthamiana leaf inoculated with Agrobacterium strain GV2260-SacB/R carrying pEarleygate101-YFP-HA.
Generation of Stable Transgenic N. benthamiana Plants Using the Mutant Agrobacterium Strain GV2260-SacB/R
To test the transformation efficiency of GV2260-SacB/R, N. benthamiana tissue culture and transformation was conducted with Agrobacterium strain GV2260 or GV2260-SacB/R carrying plasmid pgRNA-NbWRKY70. The construct pgRNA-NbWRKY70 carries a synthesized Cas9 gene driven by the Arabidopsis Ubiquitin 10 promoter. The expression of a guiding RNA targeting on the N. benthamiana WRKY70 gene was driven by the Arabidopsis U6 promoter. We modified the N. benthamiana leaf disk transformation protocol (An, 1985), where the leaf disks infected with Agrobacterium strains were only slightly washed, which usually can cause Agrobacterium overgrowth problems during the selection of transformed plant cells. The infected N. benthamiana leaf disks were cultured on selection medium supplemented with either 3 or 5% sucrose. The leaf-disk contamination caused by the overgrowth of Agrobacterium was recorded after 4 weeks of culture on the selection medium (Figure 3A). Under the test conditions, the contamination rates caused by GV2260-SacB/R on medium supplemented with 5 and 3% sucrose were 13.0 and 26.9% respectively, which are significant lower than the contamination rate of >80% caused by GV2260 (Figure 3B). Therefore, GV2260-SacB/R can be efficiently inhibited by the sucrose presented in the N. benthamiana tissue culture medium. However, under our testing conditions, there were still quite high numbers of contaminated leaf disks when in infections with GV2260-SacB/R. We speculate that the slightly rinsed leaf disks carried relatively high numbers of Agrobacterium cells, which may develop mutations on the SacB-SacR genes during the tissue culture process. It will be interesting to further test with different wash conditions, which may reduce the carry-on bacterium cells, and allow for further reduction of the contamination ratio. It will also be interesting to test if we can reduce or even eliminate antibiotic during the tissue culture process.
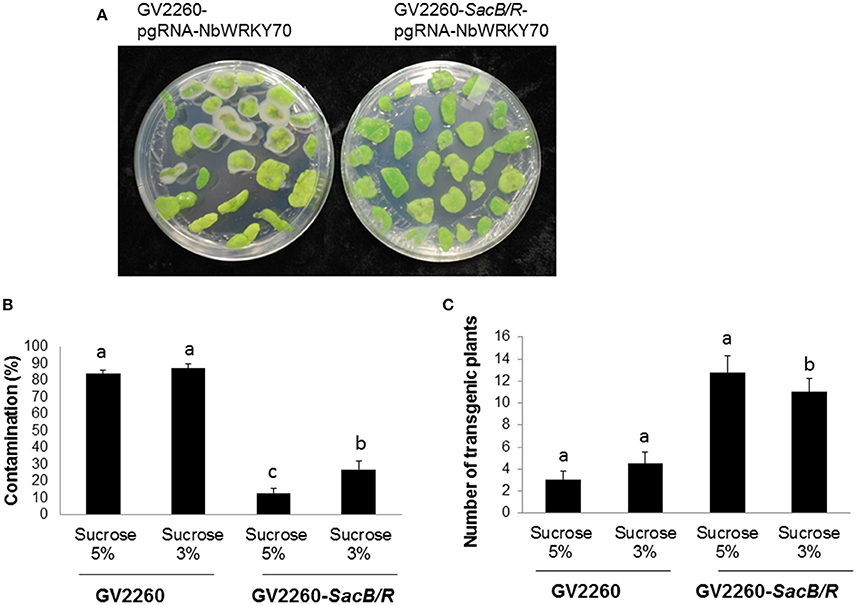
Figure 3. Comparison of the contamination ratio and transformation efficiency between GV2260 and mutant Agrobacterium strain GV2260-SacB/R during N. benthamiana tissue culture and transformation. (A) Leaf disks with Agrobacterium overgrowth. (B) Contamination ratio of GV2260 and mutant Agrobacterium strain GV2260-SacB/R during N. benthamiana transformation on tissue culture medium supplemented with 5 and 3% sucrose (Tukey HSD, P < 0.05). (C) Number of transgenic plants generated from GV2260 and GV2260-SacB/R (Tukey HSD, P < 0.05). Difference letters indicate statistical significant difference.
After 2 months of culture selection, putative transgenic N. benthamiana plants were generated. The number of transgenic plants generated from GV2260-SacB/R was significantly higher than those generated from GV2260 (Figure 3C) because of the lower contamination rate caused by the GV2260-SacB/R strain. Four transgenic plants generated by GV2260-SacB/R were genotyped, showing the presence of the Cas9 gene (Figure 4A). To examine if the pgRNA-NbWRKY70 transgenic plants carry mutations in WRKY70, we amplified an NbWRKY70 DNA fragment carrying the guiding RNA targeting site. All four putative transgenic lines amplified an NbWRKY70 DNA fragment with similar size (Figure 4B). The PCR products were gel purified and sequenced. As shown in the chromatogram, the PCR product amplified from the wild type plants yields a clean sequence, while the PCR product from a transgenic plant (line 3) yields double peaks near the PAM site (CGG) (Figure 4C) (Li et al., 2011; Nekrasov et al., 2013). The double peak near the PAM site indicates the heterozygosity of template DNAs, which suggests there are Cas9 induced mutations at the WRKY70 gene. The PCR products were also cloned and individual clones were sequenced, which confirmed the presence of mutants (data not shown). The phenotype of transgenic plants will be further characterized in the future. Nevertheless, our result demonstrated that GV2260-SacB/R could successfully transform N. benthamiana plant cells to generate stable transgenic plants. It will be interesting to test the transformation capacity of GV2260-SacB/R in other plant species. In this study, we also confirmed that the CRISPR-Cas9 system is a powerful tool for introducing mutations on target genes in N. benthamiana (Nekrasov et al., 2013; Belhaj et al., 2015).
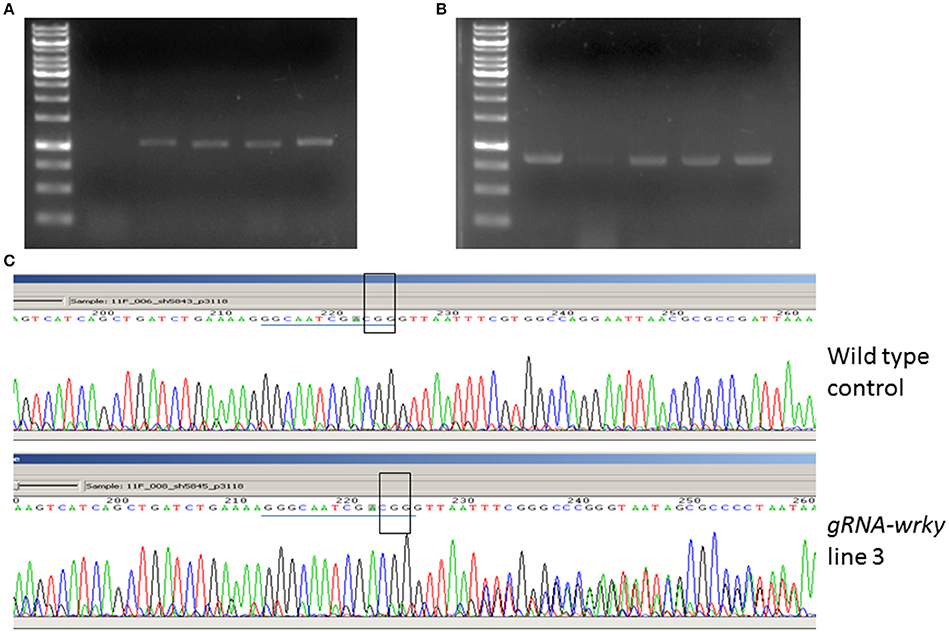
Figure 4. Detection of the pgRNA-NbWRKY construct and its induced mutation. (A) Amplification of the guiding RNA construct from transgenic and non-transgenic control. Lane 1, 1 Kb marker, lane 2, non-transgenic control, lanes 3–6, four transgenic plants. (B) Amplification of the NbWRKY DNA fragment carrying the guiding RNA targeting site. Lane 1, 1 Kb marker, lane 2, non-transgenic control, lanes 3–6, four transgenic plants. (C) Sequencing of the PCR product from the wild type and gRNA-NbWRKY transgenic line 3. The guiding RNA targeting sites are highlighted with a blue line, and the PAM sites are highlighted with an open box. The sequencing chromatograms showed mixed peak-signals after the PAM site in gRNA-NbWRKY transgenic line 3 but not in the non-transgenic control.
Conclusions
We generated a mutant Agrobacterium strain GV2260-SacB/R that is sensitive to sucrose. The mutant strain can be used for plant cell transformation as demonstrated by Agrobacterium-mediated transient assays and stable transformation. The overgrowth of mutant strain GV2260-SacB/R can be inhibited by 3–5% sucrose, a common carbon source used in plant tissue mediums. Therefore, GV2260-SacB/R can be a valuable tool for plant transformation research.
Author Contributions
BZ conceived the project and the cloning strategy. YML and JM performed the experiments. ST, DK, YL, ZN, and XZ contributed vectors and other reagents. YML, JM, ST, DK, YL, ZN, XZ, ZL, and BZ analyzed the data and wrote the draft manuscript. YML and BZ wrote the final manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank Dr. Guofu Hu for his technical assistance. The study was supported by Binational Agricultural Research and Development Fund (US-4216-09 to BZ), US National Science Foundation (IOS-0845283 to BZ), and the Virginia Agricultural Experiment Station (VA135872). The project was also partially supported by a grant from the program of Plant Feedstock Genomics for Bioenergy of the US Department of Energy (DE-SC0008338 to Kevin L. Childs, XZ, and BZ).
References
An, G. (1985). High efficiency transformation of cultured tobacco cells. Plant Physiol. 79, 568–570. doi: 10.1104/pp.79.2.568
Belhaj, K., Chaparro-Garcia, A., Kamoun, S., Patron, N. J., and Nekrasov, V. (2015). Editing plant genomes with CRISPR/Cas9. Curr. Opin. Biotechnol. 32, 76–84. doi: 10.1016/j.copbio.2014.11.007
Bhau, B. S., and Wakhlu, A. K. (2001). Effect of some antibiotics on the in vitro morphogenetic response from callus cultures of coryphantha elephantidens. Biol. Plant. 44, 19–24. doi: 10.1023/A:1017905917971
Bi, X., and Liu, L. F. (1994). RecA-independent and RecA-dependent intramolecular plasmid recombination: differential homology requirement and distance effect. J. Mol. Biol. 235, 414–423. doi: 10.1006/jmbi.1994.1002
Brendel, V., Brocchieri, L., Sandler, S. J., Clark, A. J., and Karlin, S. (1997). Evolutionary comparisons of RecA-like proteins across all major kingdoms of living organisms. J. Mol. Evol. 44, 528–541. doi: 10.1007/PL00006177
Chambert, R., and Petitglatron, M. (1989). Study of the effect of organic solvents on the synthesis of levan and the hydrolysis of sucrose by Bacillus subtilis levansucrase. Carbohydr. Res. 191, 117–123. doi: 10.1016/0008-6215(89)85051-7
Cheng, M., Fry, J. E., Pang, S., Zhou, H., Hironaka, C. M., Duncan, D. R., et al. (1997). Genetic transformation of wheat mediated by Agrobacterium tumefaciens. Plant Physiol. 115, 971–980. doi: 10.1104/pp.115.3.971
Clark, A. J., and Margulies, A. D. (1965). Isolation and characterization of recombination-deficient mutants of Escherichia coli K12. Proc. Natl. Acad. Sci. U.S.A. 53, 451–459. doi: 10.1073/pnas.53.2.451
Earley, K. W., Haag, J. R., Pontes, O., Opper, K., Juehne, T., Song, K., et al. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629. doi: 10.1111/j.1365-313X.2005.02617.x
Ellis, D. D., Lazaroff, W. R., Roberts, D. R., Flinn, B. S., and Webb, D. T. (1989). The effect of antibiotics on elongation and callus and bud formation from embryonic tissue of Piceaglauca. Can. J. For. Res. 19, 1343–1346. doi: 10.1139/x89-207
Farrand, S. K., O'Morchoe, S. P., and McCutchan, J. (1989). Construction of an Agrobacterium tumefaciens C58 recA mutant. J. Bacteriol. 171, 5314–5321.
Gay, P., Le coq, D., Steinmetz, M., Berkelman, T., and Kado, C. I. (1985). Positive selection procedure for entrapment of insertion-sequence elements in gram-negative bacteria. J. Bacteriol. 164, 918–921.
Gelvin, S. B. (2000). Agrobacterium and plant genes involved in T-DNA transfer and integration. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 223–256. doi: 10.1146/annurev.arplant.51.1.223
Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. doi: 10.1046/j.1365-313X.1994.6020271.x
Horsch, R., Fry, J., Hoffmann, N., Neidermeyer, J., Rogers, S., and Fraley, R. (1989). “Leaf disc transformation,” in Plant Molecular Biology Manual, eds S. Gelvin, R. Schilperoort, and D. Verma (Dordrecht: Springer Netherlands), 63–71.
Ishida, Y., Saito, H., Ohta, S., Hiei, Y., Komari, T., and Kumashiro, T. (1996). High efficiency transformation of maize (Zea mays L.) mediated by Agrobacterium tumefaciens. Nat. Biotechnol. 14, 745–750. doi: 10.1038/nbt0696-745
Jones, H. D., Doherty, A., and Wu, H. (2005). Review of methodologies and a protocol for the Agrobacterium-mediated transformation of wheat. Plant Methods 1, 1–9. doi: 10.1186/1746-4811-1-5
Kurnit, D. M. (1989). Escherichia coli recA deletion strains that are highly competent for transformation and for in vivo phage packaging. Gene 82, 313–315. doi: 10.1016/0378-1119(89)90056-5
Li, R., and Qu, R. (2011). High throughput Agrobacterium-mediated switchgrass transformation. Biomass Bioenergy 35, 1046–1054. doi: 10.1016/j.biombioe.2010.11.025
Li, Y., Mendiratta, S., Ehrhardt, K., Kashyap, N., White, M. A., and Bleris, L. (2011). Exploiting the CRISPR/Cas9 PAM constraint for single-nucleotide resolution interventions. PLoS ONE 11:e0144970. doi: 10.1371/journal.pone.0144970
Lovett, S. T., Drapkin, P. T., Sutera, V. A. Jr., and Gluckman-Peskind, T. J. (1993). A sister-strand exchange mechanism for recA-independent deletion of repeated DNA sequences in Escherichia coli. Genetics 135, 631–642.
Matthysse, A. G., Stretton, S., Dandie, C., McClure, N. C., and Goodman, A. E. (1996). Construction of GFP vectors for use in gram-negative bacteria other than Escherichia coli. FEMS Microbiol. Lett. 145, 87–94. doi: 10.1111/j.1574-6968.1996.tb08561.x
Nekrasov, V., Staskawicz, B., Weigel, D., Jones, J. D., and Kamoun, S. (2013). Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31, 691–693. doi: 10.1038/nbt.2655
Quandt, J., and Hynes, M. (1993). Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127, 15–21. doi: 10.1016/0378-1119(93)90611-6
Ren, Y., Bang, H., Curtis, I., Gould, J., Patil, B., and Crosby, K. (2012). Agrobacterium-mediated transformation and shoot regeneration in elite breeding lines of western shipper cantaloupe and honeydew melons (Cucumis melo L.). Plant Cell Tissue Organ Cult. (PCTOC) 108, 147–158. doi: 10.1007/s11240-011-0024-6
Ried, J. L., and Collmer, A. (1987). An NptI-SacB-SacR cartridge for constructing directed, unmarked mutations in gram-negative bacteria by marker exchange-eviction mutagenesis. Gene 57, 239–246. doi: 10.1016/0378-1119(87)90127-2
Schweizer, H. (1992). Alielic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol. Microbiol. 6, 1195–1204. doi: 10.1111/j.1365-2958.1992.tb01558.x
Silva, J. A. T. D. Fukai, S. (2001). The impact of carbenicillin, cefotaxime and vancomycin on chrysanthemum and tobacco TCL morphogenesis and Agrobacterium growth. J. Appl. Horticult. 35, 71–77. Available online at: http://www.horticultureresearch.net/journal_pdf/20013-12.pdf
Song, J., Bradeen, J. M., Naess, S. K., Helgeson, J. P., and Jiang, J. (2003). BIBAC and TAC clones containing potato genomic DNA fragments larger than 100 kb are not stable in Agrobacterium. Theor. Appl. Genet. 107, 958–964. doi: 10.1007/s00122-003-1334-9
Tereso, S., Miguel, C., Maroco, J., and Oliveira, M. M. (2006). Susceptibility of embryogenic and organogenic tissues of maritime pine (Pinus pinaster) to antibiotics used in Agrobacterium-mediated genetic transformation. Plant Cell Tissue Organ Cult. 87, 33–40. doi: 10.1007/s11240-006-9130-2
Tingay, S., McElroy, D., Kalla, R., Fieg, S., Wang, M., Thornton, S., et al. (1997). Agrobacterium tumefaciens-mediated barley transformation. Plant J. 11, 1369–1376. doi: 10.1046/j.1365-313X.1997.11061369.x
Traore, S., and Zhao, B. (2011). A novel Gateway(R)-compatible binary vector allows direct selection of recombinant clones in Agrobacterium tumefaciens. Plant Methods 7:42. doi: 10.1186/1746-4811-7-42
Tsuda, K., Qi, Y., Nguyen, L. V., Bethke, G., Tsuda, Y., Glazebrook, J., et al. (2012). An efficient Agrobacterium-mediated transient transformation of Arabidopsis. Plant J. 69, 713–719. doi: 10.1111/j.1365-313X.2011.04819.x
Tzfira, T., and Citovsky, V. (2006). Agrobacterium-mediated genetic transformation of plants: biology and biotechnology. Curr. Opin. Biotechnol. 17, 147–154. doi: 10.1016/j.copbio.2006.01.009
West, S. C. (1992). Enzymes and molecular mechanisms of genetic recombination. Annu. Rev. Biochem. 61, 603–640. doi: 10.1146/annurev.bi.61.070192.003131
Wydro, M., Kozubek, E., and Lehmann, P. (2006). Optimization of transient Agrobacterium-mediated gene expression system in leaves of Nicotiana benthamiana. Acta Biochim. Pol. 53, 289–298. Available online at: http://www.actabp.pl/pdf/2_2006/289.pdf
Yaseen, M., Ahmad, T., Sablok, G., Standardi, A., and Hafiz, I. A. (2013). Review: role of carbon sources for in vitro plant growth and development. Mol. Biol. Rep. 40, 2837–2849. doi: 10.1007/s11033-012-2299-z
Yu, T.-A., Yeh, S.-D., and Yang, J.-S. (2001). Effects of carbenicillin and cefotaxime on callus growth and somatic embryogenesis from adventitious roots of papaya. Bot. Bull. Acad. Sin. 42, 281–286.
Zang, N., Zhai, H., Gao, S., Chen, W., He, S., and Liu, Q. (2009). Efficient production of transgenic plants using the bar gene for herbicide resistance in sweetpotato. Sci. Hortic. 122, 649–653. doi: 10.1016/j.scienta.2009.06.023
Zhang, X., Wang, L., and Shou, L. (2013). Modified CTAB method for extracting genomic DNA from wheat leaf. Agric. Sci. Technol. 14, 946–949. Available online at: http://search.proquest.com/openview/748d315d04b0db8ed3779e4549788799/1?pq-origsite=gscholar
Zhao, B., Dahlbeck, D., Krasileva, K. V., Fong, R. W., and Staskawicz, B. J. (2011). Computational and biochemical analysis of the Xanthomonas effector AvrBs2 and its role in the modulation of Xanthomonas type three effector delivery. PLoS Pathog 7:e1002408. doi: 10.1371/journal.ppat.1002408
Keywords: plant transformation, Agrobacterium overgrowth, CRISPR-Cas9, Nicotiana benthamiana, SacB-SacR gene cassette
Citation: Liu Y, Miao J, Traore S, Kong D, Liu Y, Zhang X, Nimchuk ZL, Liu Z and Zhao B (2016) SacB-SacR Gene Cassette As the Negative Selection Marker to Suppress Agrobacterium Overgrowth in Agrobacterium-Mediated Plant Transformation. Front. Mol. Biosci. 3:70. doi: 10.3389/fmolb.2016.00070
Received: 30 June 2016; Accepted: 07 October 2016;
Published: 26 October 2016.
Edited by:
Artur Burzyński, Polish Academy of Sciences, PolandReviewed by:
Benoit Lacroix, Stony Brook University, USALaura R. Jarboe, Iowa State University, USA
Copyright © 2016 Liu, Miao, Traore, Kong, Liu, Zhang, Nimchuk, Liu and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bingyu Zhao, bzhao07@vt.edu
 Yiming Liu
Yiming Liu Jiamin Miao
Jiamin Miao Sy Traore3
Sy Traore3  Yi Liu
Yi Liu Xunzhong Zhang
Xunzhong Zhang Bingyu Zhao
Bingyu Zhao