Genetic variations in the serotonergic system contribute to amygdala volume in humans
- 1State Key Laboratory of Cognitive Neuroscience and Learning, IDG/McGovern Institute for Brain Research, Beijing Normal University, Beijing, China
- 2Brainnetome Center, Institute of Automation, Chinese Academy of Sciences, Beijing, China
- 3National Laboratory of Pattern Recognition, Institute of Automation, Chinese Academy of Sciences, Beijing, China
- 4Center for Collaboration and Innovation in Brain and Learning Sciences, Beijing Normal University, Beijing, China
- 5Department of Psychology and Social Behavior, University of California, Irvine, Irvine, CA, USA
- 6Key Laboratory of Behavioral Science, Institute of Psychology, Chinese Academy of Sciences, Beijing, China
- 7Department of Biological Chemistry, University of California, Irvine, Irvine, CA, USA
- 8Institute of Genomics and Bioinformatics, University of California, Irvine, Irvine, CA, USA
The amygdala plays a critical role in emotion processing and psychiatric disorders associated with emotion dysfunction. Accumulating evidence suggests that amygdala structure is modulated by serotonin-related genes. However, there is a gap between the small contributions of single loci (less than 1%) and the reported 63–65% heritability of amygdala structure. To understand the “missing heritability,” we systematically explored the contribution of serotonin genes on amygdala structure at the gene set level. The present study of 417 healthy Chinese volunteers examined 129 representative polymorphisms in genes from multiple biological mechanisms in the regulation of serotonin neurotransmission. A system-level approach using multiple regression analyses identified that nine SNPs collectively accounted for approximately 8% of the variance in amygdala volume. Permutation analyses showed that the probability of obtaining these findings by chance was low (p = 0.043, permuted for 1000 times). Findings showed that serotonin genes contribute moderately to individual differences in amygdala volume in a healthy Chinese sample. These results indicate that the system-level approach can help us to understand the genetic basis of a complex trait such as amygdala structure.
Introduction
The amygdala, an almond-shaped brain structure which resides in the medial temporal lobe of the brain (Whalen and Phelps, 2009), is key in emotion processing (Sergerie et al., 2008). Lesion studies suggest that the amygdala plays a central role in the perception of emotional stimuli (Campanella et al., 2014), and fMRI studies show that the amygdala activates in response to emotional stimuli (Habel et al., 2007). Accordingly, the volume of the amygdala is a widely used index of emotional processing in both animal and human studies (Yang et al., 2008; Hartley et al., 2011). Rodent studies have found an association between amygdala volume and variation in emotion learning (Yang et al., 2008), while accumulating clinical studies have discovered volume abnormalities of the amygdala in patients with depression (Rubinow et al., 2014), bipolar disorder (Lisy et al., 2011), and borderline personality disorder (Ruocco et al., 2012). For instance, patients with personality disorder have a 13% smaller amygdala than healthy controls (Ruocco et al., 2012). Given these findings, amygdala volume could be a promising endophenotype in regards to emotional behavior and related psychiatric diseases.
Moderate to high heritability for amygdala volume (Hulshoff Pol et al., 2006; Kremen et al., 2010) suggest a significant genetic basis for this trait. The amygdala is densely innervated by serotonergic fibers (Bauman and Amaral, 2005), and the influence of synaptic serotonin on amygdala responsiveness has been identified (Fisher et al., 2006; Rhodes et al., 2007). Accumulating imaging genetics studies have linked individual differences in amygdala volume to genes affecting serotonergic signal. These genes encode proteins involved in serotonin synthesis (Inoue et al., 2010), reuptake (Frodl et al., 2008; Stjepanovic et al., 2013), metabolic degradation (Meyer-Lindenberg et al., 2006), and receptors (Zetzsche et al., 2008). However, the variance of the amygdala structure explained by a single genetic locus is small, no more than 1% (Hibar et al., 2015), far less than the reported 63–66% heritability (Kremen et al., 2010) from twin studies. This gap might be explained as the amygdala size, like many complex quantitative traits, is influenced by multiple genes, each with a small effect. Therefore, a gene set based model is needed to assess the additive effects of a group of functionally related genes that mediate a particular biological process (i.e., serotonin functioning) and potential interactions.
In the present study, we applied the system-level approach developed by our research group (Chen et al., 2015) to evaluate the overall contribution of the serotonin system genes on individual differences in amygdala morphology. A large sample of 417 Han Chinese adults was recruited, and 129 polymorphic loci within the serotonin system were genotyped to cover a substantial portion (by LD) of the common variations in serotonin system genes, including biosynthesis, vesicular release, active reuptake, metabolic degradation, and presynaptic and postsynaptic receptors. We hypothesized that the genes along the specified pathway would contribute greatly to variation in amygdala volume.
Materials and Methods
Participants
Our 417 participants (mean age 20.4 years, SD = 0.9; 179 males and 238 females) were a subset of a larger study of 480 healthy Chinese college students (mean age = 19.9 years, SD = 0.9; 208 males and 272 females) from Beijing Normal University, Beijing, China (Li et al., 2011), for whom structural imaging data was available. All participants were Han Chinese and were free of neurological and psychiatric disorders. This study was approved by the IRB of the State Key Laboratory of Cognitive Neuroscience and Learning at Beijing Normal University, China. All experiments were performed in accordance with approved guidelines and regulations. Written informed consent was obtained from each participant.
Gene Selection
We selected genes using the serotonin pathway defined in the Kyoto Encyclopedia of Genes and Genomes database, a collection of pathway maps widely used in gene-set analysis. Genes in the following four serotonin subsystems were selected: (1) the serotonin synthesis subsystem, which converts hydroxylation (by TPH) to 5-HT: TPH1, TPH2; (2) the degradation subsystem, which directly breaks down released 5-HT at the synapse into inactive metabolites (MAOA, MAOB); (3) the transportation subsystem, which pumps serotonin from synaptic spaces into presynaptic neurons [SLC6A4(also known as 5-HTT)] or integrates the membrane of intracellular vesicles of presynaptic neurons and transported monoamines into the synaptic vesicles [SLC18A1(also known as VMAT1), SLC18A2(also known as VMAT2)]; (4) the serotonin receptor subsystem (HTR1A, HTR1B, HTR1D, HTR1F, HTR2A, HTR2B, HTR2C, HTR3A, HTR3B, HTR3C, HTR3D, HTR3E, HTR4, HTR5A, HTR5B, HTR6, HTR7). Together, the selected genes represent all major genes involved in the four serotonin subsystems in humans (Chen et al., 2015). Several tag SNPs (tSNPs) defined by the HapMap project1 [Phase 3] (Frazer et al., 2007) were selected to sample the genetic diversity of these genes. Details of these genes and the selected loci (129 polymorphisms, including 127 SNPs and 2 VNTR polymorphisms) are shown in Supplementary Table S1.
Genotyping Techniques
Genotyping was conducted as previously described (Li et al., 2011). Briefly, 4 ml venous blood sample was collected from each subject, and then genomic DNA was extracted according to standard methods. SNPs were genotyped using the Illumina GoldenGate Genotyping protocol (see Illumina GoldenGate Assay Protocol for details2). In addition, two genetic markers (5-HTTLPR, MAOA VNTR) were ascertained by standard PCR procedures (Chen et al., 2015).
Gene Data Preprocessing
Quality control of the genetic data was carried out based on the larger sample of 480 participants. Two subjects met the criteria of over 10% null genotyping, and were thus excluded from subsequent analyses. Of the 60228 genotypes (126 SNPs by 478 subjects), 120 genotypes (0.2%) were excluded because of low GenCall (<0.25). If any SNP had fewer than 10 (2%) heterozygotes or minor homozygotes, these two genotype groups were combined. If the combined group still had fewer than 10 subjects, that SNP was excluded from further analysis. We found that five SNPS showed significant Hardy–Weinberg disequilibrium (p < 0.01) based on a df of 1 (for SNPs located on X chromosome, only females were included in HWE calculation since males have only one X chromosome). However, these SNPs were retained because the HW disequilibrium here did not seem to result from genotyping error but rather reflected the characteristics of college students due to social selection (i.e., overrepresentation of alleles linked to school achievement and motivation; Chen et al., 2013). Because both tag SNPs and additional SNPs in regions detected in recent selection (Hawks et al., 2007) were selected in the current study, there was high LD among some SNPs. Thirty SNPs were excluded from multiple regression analysis because of their high LD with adjacent SNPs [r2 > 0.8, Plink calculated (Purcell et al., 2007)]. Genetic relatedness among subjects was checked following Anderson et al. (2010) protocol by Plink. All genotyped unrelated autosome SNPs (n = 240, r2 < 0.8) were used and the threshold was set at 0.95 (personal communication with Dr. Anderson and Dr. Zondervan). No pair of subjects showed high relatedness (all PI_HAT smaller than 0.5).
Four-hundred and seventy-eight subjects (99.6%) and 99 polymorphisms (77%) passed all of the aforementioned quality control procedures. Of these 478 subjects, 417 had structural imaging data and were thus included in the subsequent analyses of the 99 polymorphisms. The information for all 129 loci (127 SNPs and 2 VNTRs) is shown in Supplementary Table S1, including location (rs number, chromosome, position), gene, serotonin subsystem, allele polymorphism and frequency, HWE, LD, and whether they were included in the main analyses.
MRI Data Collection
MRI scans were performed in a 3.0T Siemens Magnetom Trio scanner equipped with a standard head coil at Beijing Normal University Brain Imaging Center. Structural MRI data were acquired with the T1-weighted MPRAGE pulse sequence (TE = 3.75 ms, TR = 2,530 ms, flip angle = 7°, FOV = 256 mm × 256 mm, voxel size = 1 mm × 1 mm × 1.33 mm, number of partitions = 128).
MRI Data Processing
Cortical surface reconstruction and volumetric segmentation were performed with the FreeSurfer software3 (Version 4.5.0). Each subject’s average T1-weighted image was segmented into gray matter volumes for seven subcortical regions relying upon variations in voxel signal intensities, probabilistic atlas location and local spatial relationships between the structures (Fischl et al., 2002). Quality control of scan images and segmentation was assured by visual inspection of the whole cortex of each subject, and any inaccuracies in Talairach-transformation, skull stripping and segmentation were manually corrected and re-inspected. High correlations between the automatic measures and manual measures in vivo and ex vivo have been demonstrated (Desikan et al., 2006). The amygdala volumes obtained from the Freesurfer procedure have been reported to be significantly correlated with manual parcellation (Stjepanovic et al., 2013).
Volumes of bilateral amygdala were retracted from the standard output of the FreeSurfer analysis. Then the mean bilateral amygdala volume was calculated. A preliminary analysis showed that both gender and ICV were significantly associated with amygdala size (for gender, F(1,415) = 48.06, p = 1.59 × 10-11; for ICV, F(1,415) = 114.23, p = 1.03 × 10-23). Therefore, to control for the confounding effects of gender and ICV, a regression analysis was conducted, with gender and ICV as independent variables, and amygdala size as the dependent variable. The residual for each subject was normally distributed (skewness = 0.456, kurtosis = 0.747, Kolmogorov–Smirnov test = 0.039, p = 0.136) and was used as an index of amygdala volume in subsequent association analyses. To avoid the possible confounding effect of emotion state on amygdala structure, the associations between Beck Anxiety Inventory (BAI) and Beck Depression Inventory (BDI) scores with amygdala size were tested separately. The associations did not reach significance [for BAI, F(1,415) = 0.74, p = 0.39; for BDI, F(1,415) = 2.67, p = 0.10]. Therefore, the BAI and BDI scores were not considered in subsequent analyses.
Statistical Analysis
The statistical procedure was comprised of three major analyses. First, analysis of variance (ANOVA) was conducted for each of these loci to detect variants which met the inclusion criterion (p < 0.05, uncorrected, to control for Type II error). Second, these loci were then entered into a regression model to estimate their overall contribution to amygdala size. In the regression model, all loci with significant main effects based on the ANOVA results were included with a forward stepwise method. In this step, all SNPs were coded in a linear way, i.e., the major homozygote, heterozygote, and minor homozygote were coded into 1, 2, and 3, respectively (SNPs on X chromosome were coded as 1 and 3 for major and minor allele homozygote, and also 3 for female heterozygotes). In addition, the MAOA VNTR was coded as 1 for the 3 repeat and 3 for the 4 repeat in males and 1 for 3 repeat homozygotes and 3 for others in females. Finally, the regression model was verified by permutation. Permutation tests were done 1000 times by shuffling amygdala volume data across subjects. In each iteration, selection of significant snps from the ANOVA tests, regression model estimation with a forward stepwise method, and r2 calculation were carried out on the shuffled data. The probability of getting a larger r2 in the shuffled data than in the real data was defined as p-value of the model.
Results
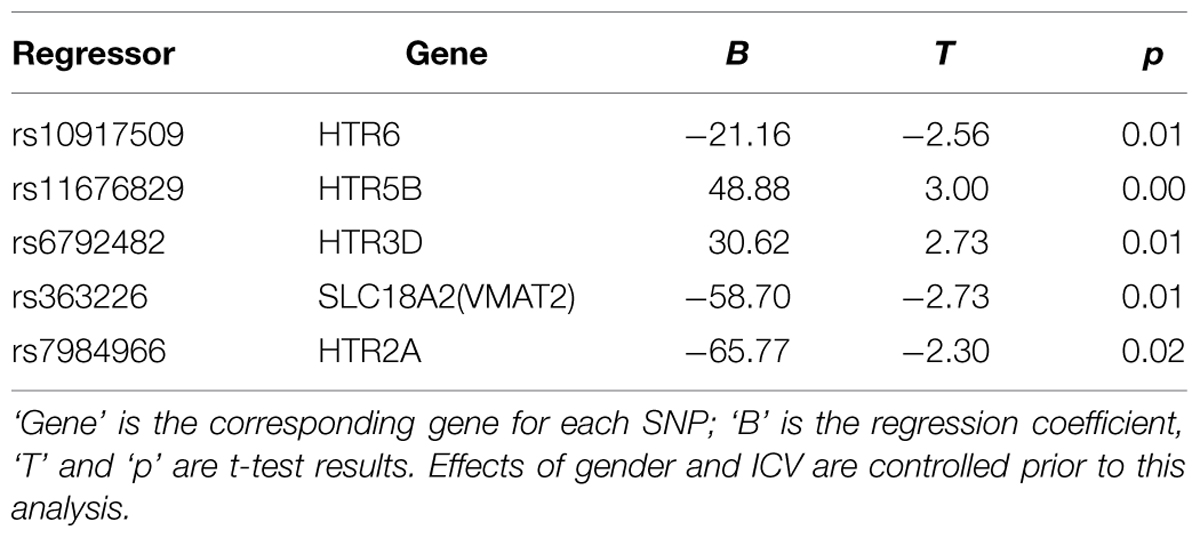
The mean bilateral amygdala volume across subjects was 1700.5 mm3 (SD = 177.7 mm3). ANOVA was used to screen the 99 loci that passed quality control procedures for associations with the amygdala size. Nine SNPs showed main effects on amygdala volume with uncorrected p < 0.05. Specifically, individuals who were major allele homozygotes for rs7997012 (HTR2A), rs7984966 (HTR2A), rs939334 (HTR3D), rs10917509 (HTR6), or rs363226 (SLC18A2), or minor allele homozygotes for rs1487275 (TPH2), rs6792482 (HTR3D), rs11676829 (HTR5B), or rs12249377 (HTR7), tended to have larger amygdala size than the remaining groups. (For details, see Table 1, and online Supplementary Table S2 ).
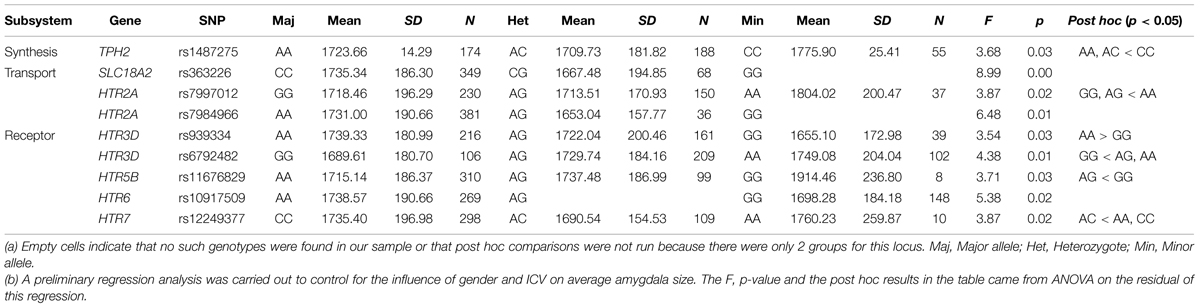
TABLE 1. Means and standard deviations of amygdala volume for each polymorphism in nine significant SNPs, and post hoc comparisons of each locus.
These nine SNPs were added to a regression model using the forward stepwise procedure to estimate their overall contribution to amygdala size. five of them made significant and unique contributions to the final model, while the other four SNPs were not included because of collinearity with other SNPs (see Table 2). The regression model accounted for 8.2% (6.8% adjusted) of the variance in amygdala size [F(5,417) = 7.33, p = 1.3 × 10-6]. The confidence interval of R2 estimated by bootstrap for 1000 times was 0.04–0.16, and that for adjusted R2 was 0.03–0.15.
Finally Monte Carlo permutation analyses were carried out to test the model. Figure 1 shows the permutation results. Based on 1000 permutation tests, the probability of attaining the R2 or adjusted R2 found in the model reached significance (p = 0.043 and 0.044, respectively). These results indicate that genes in the serotonin system contribute substantially to individual variance in amygdala volume.
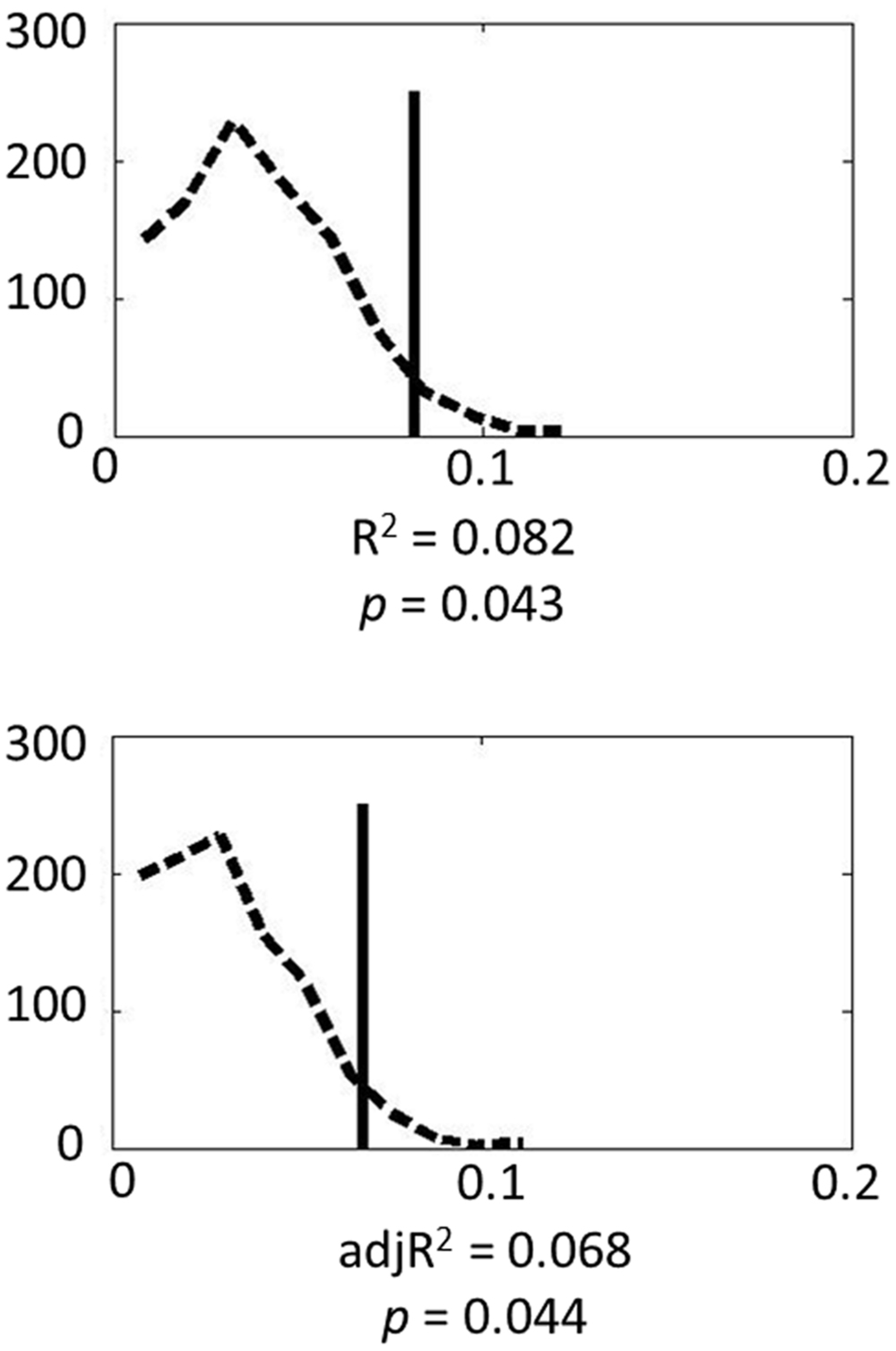
FIGURE 1. Permutation results for the genetic model: the dashed line represents the empirical distribution of R2 obtained from the randomized data, and the solid vertical line represents R2 obtained from the actual data.
Discussion
The current study combined the advantages of both the candidate gene approach and dense genotyping technology. Our theory-driven method detected a group of biological relevant genes based on prior knowledge and thus avoided the heavy comparison correction necessary in GWAS. However, unlike candidate gene studies on single genes, our system level approach took into account the polygenic nature of amygdala structure. Given the innervation of serotonergic fibers in the amygdala (Bauman and Amaral, 2005) and the role of serotonergic genes on amygdala structure (Meyer-Lindenberg et al., 2006; Frodl et al., 2008; Zetzsche et al., 2008), we used dense gene chips to cover all tag SNPs of the serotonergic biological pathway in order to test the additive effect of potential genes. Results suggest that such a system level approach could bridge the gap between the small contributions of single genes and the considerable heritability of amygdala volume revealed by twin studies. Specifically, serotonergic genes collectively accounted for 8.2% of variance in amygdala volume. Although associations between these specific SNPs and amygdala structure have not been reported before, direct and indirect evidence have linked these genes to amygdala structure and amygdala-related psychological disorders. In the following paragraphs, we discuss each of these genes.
The TPH2 gene encodes TPH protein which is involved in the rate-limiting biosynthesis of serotonin. Postmortem studies have revealed the expression of TPH2 mRNA in the amygdala (Zill et al., 2007). Raphe neurons of Tph2 knockout mice were completely devoid of 5-HT, indicating that brain 5-HT synthesis across the lifespan is exclusively maintained by TPH2 (Gutknecht et al., 2009). Previous imaging genetic studies also found associations between several TPH SNPs and the amygdala, such as rs4570625 with the structure (Inoue et al., 2010) and function (Furmark et al., 2009) of the amygdala, and rs17110563 with bipolar disorder (Cichon et al., 2008). Thus far, however, no study has linked rs1487275 genotype to any emotion-related behavior or psychiatric disease, although its effect on amygdala structure was identified in the current study. Therefore, future studies should explore such potential associations in healthy or clinical samples.
SLC18A2 encodes VMAT2 that transports free serotonin from cellular cytosol into synaptic vesicles (Eiden et al., 2004). Rodent studies have reported early expression of VMAT2 in amygdala (Lebrand et al., 1998) and that mice lacking one copy of the VMAT2 gene develop with significantly reduced serotonin (Fon et al., 1997). Convergent studies have linked VMAT2 gene to brain development and amygdala-related psychiatric diseases. For instance, an increase in cell death in the superficial layers of the cingulate and retrosplenial cortices during early postnatal life in Vmat2 knockout mice (Stankovski et al., 2007) and a delayed maturation of the upper cortical layers in the Vmat2(sert-cre) and Tph2(-/-) mice (Narboux-Nême et al., 2013) were reported. Moreover, VMAT2 heterozygous mice exhibit ‘depression-like’ phenotype (Fukui et al., 2007). In human studies, patients with bipolar disorder showed higher binding of VMAT2 (Zubieta et al., 2001), patients with major depression also showed elevated VMAT2 density (Zucker et al., 2002) and structural change of VMAT2 (Zalsman et al., 2011) in platelets. Our finding of an association between SLC18A2 variation and the structure of the amygdala seems in accordance with the above previous results.
HTR2A, HTR3D, HTR5B, HTR6, and HTR7 encode different serotonin receptors. Studies indicated the expression of HTR2A (McDonald and Mascagni, 2007), HTR3 (Morales et al., 1998), HTR6 (Marazziti et al., 2012) in the amygdala. Their effects on amygdala structure might be partly accounted for by the distribution of serotonin receptors in the amygdala, and the modulation effect of serotonin receptors on different developmental processes (Gaspar et al., 2003), such as neurogenesis, apoptosis, axon branching, and dendritogenesis. Accumulating pharmacological studies have linked these receptors to the function of the amygdala and related psychiatric diseases. For example, HTR2 agonist has been found to increase neuronal firing of the amygdala (Stein et al., 2000) and to increase anxiety-like behavior (Pockros-Burgess et al., 2014); HTR2A antagonist can have an antidepressant-like effect (Quesseveur et al., 2013); HTR3 agonist attenuates antidepressants’ effect (Nakagawa et al., 1998), whereas HTR3 antagonist as well as HTR6 and HTR7 have an antidepressant effect (Wesolowska and Nikiforuk, 2007; Mnie-Filali et al., 2011; Gupta et al., 2014). A recent study also showed that social isolation stress could result in up-regulation of HTR5B, suggesting a close link between HTR5B and emotion and its neural substrates such as the amygdala (Maekawa et al., 2010). In addition to pharmacological studies, at least one molecular genetic study found a significant association between rs7997012 variation (HTR2A) and the therapeutic response to antidepressant treatments in major depression patients (Lin et al., 2014). In sum, previous studies have consistently shown that the above serotonin receptors play a major role in mood disorders, which are likely related to amygdala dysfunction. Moreover, HTR6 was indicated to mediate brain development in MAOA-deficient mouse embryos (Wang et al., 2014) and HTR7 signaling was reported to regulate neuronal morphology (Kobe et al., 2012). These documented genetic effects on neural development may have a permanent impact on the size of the amygdala. Given that the formation of serotonergic neurons and fiber distribution are not impaired (Gutknecht et al., 2008) inTph2 knockout mice (that are completely deficient in brain serotonin synthesis), we inferred that the effect of these genes on amygdala size was not through serotonergic neurons but through serotonin level and its effect on brain development. In the developing nervous system, an excess of serotonin affects interneuron migration (Riccio et al., 2009) and neocortical pyramidal neuron migration (Riccio et al., 2011). High levels of serotonin are also suggested to have neuroprotective effects on cortical neurons (Stankovski et al., 2007), while lack of brain serotonin is suggested to affect postnatal development and serotonergic neuronal circuitry formation (Migliarini et al., 2013). In summary, the association that we found between serotonergic genes and amygdala structure might result from the developmental role of serotonin.
Further research is required to support our findings for several reasons. First, some serotonergic genes, which have been found in several studies to impact amygdala structure, were not found to impact amygdala morphology in the present study (e.g., HTR1A). One possible explanation is that HTR1A may be important in amygdala structure and function in Caucasians (for whom most previous studies were on), but not Chinese. We were not surprised at the negative result in our Han Chinese sample, as several studies have reported that the same genetic variation can result in divergent psychological outcomes, depending on the population (Long et al., 2013; Wang et al., 2013). For example, a recent study genotyped the HTR1A polymorphism in European Americans and Koreans, and reported a significant interaction between HTR1A genotype and culture in the locus of attention (Kim et al., 2010). Moreover, the association between the HTR1A gene polymorphism (rs6295) and bipolar disorder in the Caucasian sample (Sullivan et al., 2009) was not found in the Korean population (Kim et al., 2014). Therefore, our sample of pure Han Chinese helps to prevent the confounding effect of population, but we should be cautious as it also limits the generalization of our results to other populations. Another possible explanation is that we may have missed the causative SNPs by using only tag SNPs to sample the genetic diversity of these genes. Further studies are required to test this association in other populations and to genotype more SNPs.
Second, in the current study, we focused on the role of serotonergic genes in healthy young adults to avoid the confounding effects of neurological diseases and age (Filippini et al., 2009). However, it should be noted that both developmental mechanisms and adult chronic disease may affect amygdala size, but through different mechanisms. Studies involving larger sample sizes and older adults (to explore possible effects of aging) and subjects with chronic disease are needed. Third, considering that amygdala dysfunction accompanying structural abnormality also underlies emotion related psychiatric disorders (Ubl et al., 2015) and reflects the effect of serotonergic genes (Hariri et al., 2002), further studies are needed to test the functional indices of the amygdala. Also, studies on amygdala subregions using higher resolution images could provide more information regarding the effects of serotonergic genes on the amygdala. Fourth, the current study could not identify the specific serotonin receptor(s) that transduced the effects. More direct biological evidence is required in further studies to elucidate the relationship between the serotonergic receptors and the downstream amygdala structural change.
Last but not the least, all associated SNPs in our study were located in non-coding regions. This result is consistent with the view that non-coding regions which were once labeled as “junk DNA” actually may play important functional roles (Birney et al., 2007). Some studies indicate that intron variants are involved in gene expression (Zhang et al., 2007) or mRNA secondary structure formation (Nackley et al., 2006). To explore the specific roles of the SNPs screened in our study, more systematic studies using animal models and other techniques (e.g., optogenetics) are required. Third, although genes from the serotonin system accounted for 8.2% of the variance in amygdala volume, there is still much more “missing heritability” (8.2% vs. 63–65% heritability) to be accounted for. Future studies regarding amygdala structure should incorporate other genetic systems, environmental factors, genetic epistasis, and gene-environmental interactions.
Conclusion
Our system-level approach indicated that several genes within the serotonin system had small effects on amygdala structure, and these genes together accounted for a sizable portion of the missing heritability of amygdala volume. The system-level analysis may enhance our understanding of the genetic basis of human amygdala structure and amygdala-related emotional behaviors and psychiatric diseases.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the Research Fund for the Doctoral Program of Higher Education (20110003120001), the 111 Project (B07008) of the Ministry of Education of China, the Fundamental Research Funds for the Central Universities (2012LYB05), the National Natural Science Foundation of China (31300934, 31100807, 31221003, and 31571150), and the Open Research Fund of the State Key Laboratory of Cognitive Neuroscience and Learning (Grant No. CNLYB1313). We thank all graduate research assistants who helped with data collection.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fnana.2015.00129
Abbreviations
5-HT, 5-hydroxytryptamine; 5-HTT, serotonin transporter; 5-HTTLPR, serotonin transporter gene linked polymorphic region; GWAS, Genome-Wide Association Study; HTR, serotonin receptor; ICV, intracranial volume; LD, linkage disequilibrium; MAOA, Monoamine oxidase A; MAOB, Monoamine oxidase B; SNP, Single Nucleotide Polymorphism; TPH, tryptophan hydroxylase; VMAT, Vesicular monoamine transporter; VNTR, variable number tandem repeat.
Footnotes
References
Anderson, C. A., Pettersson, F. H., Clarke, G. M., Cardon, L. R., Morris, A. P., and Zondervan, K. T. (2010). Data quality control in genetic case-control association studies. Nat. Protoc. 5, 1564–1573. doi: 10.1038/nprot.2010.116
Bauman, M. D., and Amaral, D. G. (2005). The distribution of serotonergic fibers in the macaque monkey amygdala: an immunohistochemical study using antisera to 5-hydroxytryptamine. Neuroscience 136, 193–203. doi: 10.1016/j.neuroscience.2005.07.040
Birney, E., Stamatoyannopoulos, J. A., Dutta, A., Guigo, R., Gingeras, T. R., Margulies, E. H., et al. (2007). Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816. doi: 10.1038/nature05874
Campanella, F., Shallice, T., Ius, T., Fabbro, F., and Skrap, M. (2014). Impact of brain tumour location on emotion and personality: a voxel-based lesion-symptom mapping study on mentalization processes. Brain 137, 2532–2545. doi: 10.1093/brain/awu183
Chen, C., Chen, C., Moyzis, R. K., He, Q., Lei, X., Li, J., et al. (2013). Genotypes over-represented among college students are linked to better cognitive abilities and socioemotional adjustment. Cult. Brain 1, 47–63. doi: 10.1007/s40167-013-0003-3
Chen, C., Liu, C., Moyzis, R., Chen, W., and Dong, Q. (2015). Genetic variations in the serotoninergic system and environmental factors contribute to aggressive behavior in Chinese adolescents. Physiol. Behav. 138, 62–68. doi: 10.1016/j.physbeh.2014.09.005
Cichon, S., Winge, I., Mattheisen, M., Georgi, A., Karpushova, A., Freudenberg, J., et al. (2008). Brain-specific tryptophan hydroxylase 2 (TPH2): a functional Pro206Ser substitution and variation in the 5’-region are associated with bipolar affective disorder. Hum. Mol. Genet. 17, 87–97. doi: 10.1093/hmg/ddm286
Desikan, R. S., Segonne, F., Fischl, B., Quinn, B. T., Dickerson, B. C., Blacker, D., et al. (2006). An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31, 968–980. doi: 10.1016/j.neuroimage.2006.01.021
Eiden, L. E., Schafer, M. K., Weihe, E., and Schutz, B. (2004). The vesicular amine transporter family (SLC18): amine/proton antiporters required for vesicular accumulation and regulated exocytotic secretion of monoamines and acetylcholine. Pflugers Arch. 447, 636–640. doi: 10.1007/s00424-003-1100-5
Filippini, N., Macintosh, B. J., Hough, M. G., Goodwin, G. M., Frisoni, G. B., Smith, S. M., et al. (2009). Distinct patterns of brain activity in young carriers of the APOE-epsilon4 allele. Proc. Natl. Acad. Sci. U.S.A. 106, 7209–7214. doi: 10.1073/pnas.0811879106
Fischl, B., Salat, D. H., Busa, E., Albert, M., Dieterich, M., Haselgrove, C., et al. (2002). Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 33, 341–355. doi: 10.1016/S0896-6273(02)00569-X
Fisher, P. M., Meltzer, C. C., Ziolko, S. K., Price, J. C., Moses-Kolko, E. L., Berga, S. L., et al. (2006). Capacity for 5-HT1A-mediated autoregulation predicts amygdala reactivity. Nat. Neurosci. 9, 1362–1363. doi: 10.1038/nn1780
Fon, E. A., Pothos, E. N., Sun, B. C., Killeen, N., Sulzer, D., and Edwards, R. H. (1997). Vesicular transport regulates monoamine storage and release but is not essential for amphetamine action. Neuron 19, 1271–1283. doi: 10.1016/S0896-6273(00)80418-3
Frazer, K. A., Ballinger, D. G., Cox, D. R., Hinds, D. A., Stuve, L. L., Gibbs, R. A., et al. (2007). A second generation human haplotype map of over 3.1 million SNPs. Nature 449, 851–861. doi: 10.1038/nature06258
Frodl, T., Koutsouleris, N., Bottlender, R., Born, C., Jager, M., Morgenthaler, M., et al. (2008). Reduced gray matter brain volumes are associated with variants of the serotonin transporter gene in major depression. Mol. Psychiatry 13, 1093–1101. doi: 10.1038/mp.2008.62
Fukui, M., Rodriguiz, R. M., Zhou, J., Jiang, S. X., Phillips, L. E., Caron, M. G., et al. (2007). Vmat2 heterozygous mutant mice display a depressive-like phenotype. J. Neurosci. 27, 10520–10529. doi: 10.1523/JNEUROSCI.4388-06.2007
Furmark, T., Henningsson, S., Appel, L., Ahs, F., Linnman, C., Pissiota, A., et al. (2009). Genotype over-diagnosis in amygdala responsiveness: affective processing in social anxiety disorder. J. Psychiatry Neurosci. 34, 30–40.
Gaspar, P., Cases, O., and Maroteaux, L. (2003). The developmental role of serotonin: news from mouse molecular genetics. Nat. Rev. Neurosci. 4, 1002–1012. doi: 10.1038/nrn1256
Gupta, D., Radhakrishnan, M., Thangaraj, D., and Kurhe, Y. (2014). Antidepressant and anti-anxiety like effects of 4i (N-(3-chloro-2-methylphenyl) quinoxalin-2-carboxamide), a novel 5-HT3 receptor antagonist in acute and chronic neurobehavioral rodent models. Eur. J. Pharmacol. 735, 59–67. doi: 10.1016/j.ejphar.2014.04.008
Gutknecht, L., Kriegebaum, C., Waider, J., Schmitt, A., and Lesch, K.-P. (2009). Spatio-temporal expression of tryptophan hydroxylase isoforms in murine and human brain: convergent data from Tph2 knockout mice. Eur. Neuropsychopharmacol. 19, 266–282. doi: 10.1016/j.euroneuro.2008.12.005
Gutknecht, L., Waider, J., Kraft, S., Kriegebaum, C., Holtmann, B., Reif, A., et al. (2008). Deficiency of brain 5-HT synthesis but serotonergic neuron formation in Tph2 knockout mice. J. Neural Transm. 115, 1127–1132. doi: 10.1007/s00702-008-0096-6
Habel, U., Windischberger, C., Derntl, B., Robinson, S., Kryspin-Exner, I., Gur, R. C., et al. (2007). Amygdala activation and facial expressions: explicit emotion discrimination versus implicit emotion processing. Neuropsychologia 45, 2369–2377. doi: 10.1016/j.neuropsychologia.2007.01.023
Hariri, A. R., Mattay, V. S., Tessitore, A., Kolachana, B., Fera, F., Goldman, D., et al. (2002). Serotonin transporter genetic variation and the response of the human amygdala. Science 297, 400–403. doi: 10.1126/science.1071829
Hartley, C. A., Fischl, B., and Phelps, E. A. (2011). Brain structure correlates of individual differences in the acquisition and inhibition of conditioned fear. Cereb. Cortex 21, 1954–1962. doi: 10.1093/cercor/bhq253
Hawks, J., Wang, E. T., Cochran, G. M., Harpending, H. C., and Moyzis, R. K. (2007). Recent acceleration of human adaptive evolution. Proc. Natl. Acad. Sci. U.S.A. 104, 20753–20758. doi: 10.1073/pnas.0707650104
Hibar, D. P., Stein, J. L., Renteria, M. E., Arias-Vasquez, A., Desrivieres, S., Jahanshad, N., et al. (2015). Common genetic variants influence human subcortical brain structures. Nature 520, 224–229. doi: 10.1038/nature14101
Hulshoff Pol, H. E., Schnack, H. G., Posthuma, D., Mandl, R. C., Baare, W. F., Van Oel, C., et al. (2006). Genetic contributions to human brain morphology and intelligence. J. Neurosci. 26, 10235–10242. doi: 10.1523/JNEUROSCI.1312-06.2006
Inoue, H., Yamasue, H., Tochigi, M., Takei, K., Suga, M., Abe, O., et al. (2010). Effect of tryptophan hydroxylase-2 gene variants on amygdalar and hippocampal volumes. Brain Res. 1331, 51–57. doi: 10.1016/j.brainres.2010.03.057
Kim, H. S., Sherman, D. K., Taylor, S. E., Sasaki, J. Y., Chu, T. Q., Ryu, C., et al. (2010). Culture, serotonin receptor polymorphism and locus of attention. Soc. Cogn. Affect. Neurosci. 5, 212–218. doi: 10.1093/scan/nsp040
Kim, Y. K., Hwang, J. A., Lee, H. J., Lee, B. H., and Na, K. S. (2014). There is no association between the serotonin receptor gene and bipolar I disorder in the Korean population. Nord. J. Psychiatry 68, 488–493. doi: 10.3109/08039488.2013.877071
Kremen, W. S., Prom-Wormley, E., Panizzon, M. S., Eyler, L. T., Fischl, B., Neale, M. C., et al. (2010). Genetic and environmental influences on the size of specific brain regions in midlife: the VETSA MRI study. Neuroimage 49, 1213–1223. doi: 10.1016/j.neuroimage.2009.09.043
Kobe, F., Guseva, D., Jensen, T. P., Wirth, A., Renner, U., Hess, D., et al. (2012). 5-HT7R/G12 signaling regulates neuronal morphology and function in an age-dependent manner. J. Neurosci. 32, 2915–2930. doi: 10.1523/JNEUROSCI.2765-11.2012
Lebrand, C., Cases, O., Wehrlé, R., Blakely, R. D., Edwards, R. H., and Gaspar, P. (1998). Transient developmental expression of monoamine transporters in the rodent forebrain. J. Comp. Neurol. 401, 506–524. doi: 10.1002/(SICI)1096-9861(19981130)401:4<506::AID-CNE5>3.0.CO;2-#
Li, J., Chen, C., He, Q., Li, H., Moyzis, R. K., Xue, G., et al. (2011). Neurotensin receptor 1 gene (NTSR1) polymorphism is associated with working memory. PLoS ONE 6:e17365. doi: 10.1371/journal.pone.0017365
Lin, J. Y., Jiang, M. Y., Kan, Z. M., and Chu, Y. (2014). Influence of 5-HTR2A genetic polymorphisms on the efficacy of antidepressants in the treatment of major depressive disorder: a meta-analysis. J. Affect. Disord. 168, 430–438. doi: 10.1016/j.jad.2014.06.012
Lisy, M. E., Jarvis, K. B., Delbello, M. P., Mills, N. P., Weber, W. A., Fleck, D., et al. (2011). Progressive neurostructural changes in adolescent and adult patients with bipolar disorder. Bipolar Disord. 13, 396–405. doi: 10.1111/j.1399-5618.2011.00927.x
Long, H., Liu, B., Hou, B., Wang, C., Li, J., Qin, W., et al. (2013). The long rather than the short allele of 5-HTTLPR predisposes Han Chinese to anxiety and reduced connectivity between prefrontal cortex and amygdala. Neurosci. Bull. 29, 4–15. doi: 10.1007/s12264-013-1299-x
Maekawa, T., Kim, S., Nakai, D., Makino, C., Takagi, T., Ogura, H., et al. (2010). Social isolation stress induces ATF-7 phosphorylation and impairs silencing of the 5-HT 5B receptor gene. EMBO J. 29, 196–208. doi: 10.1038/emboj.2009.318
Marazziti, D., Baroni, S., Pirone, A., Giannaccini, G., Betti, L., Schmid, L., et al. (2012). Distribution of serotonin receptor of type 6 (5-HT6) in human brain post-mortem. A pharmacology, autoradiography and immunohistochemistry study. Neurochem. Res. 37, 920–927. doi: 10.1007/s11064-011-0684-y
McDonald, A. J., and Mascagni, F. (2007). Neuronal localization of 5-HT type 2A receptor immunoreactivity in the rat basolateral amygdala. Neuroscience 146, 306–320. doi: 10.1016/j.neuroscience.2007.01.047
Meyer-Lindenberg, A., Buckholtz, J. W., Kolachana, B., R Hariri, A., Pezawas, L., Blasi, G., et al. (2006). Neural mechanisms of genetic risk for impulsivity and violence in humans. Proc. Natl. Acad. Sci. U.S.A. 103, 6269–6274. doi: 10.1073/pnas.0511311103
Migliarini, S., Pacini, G., Pelosi, B., Lunardi, G., and Pasqualetti, M. (2013). Lack of brain serotonin affects postnatal development and serotonergic neuronal circuitry formation. Mol. Psychiatry 18, 1106–1118. doi: 10.1038/mp.2012.128
Mnie-Filali, O., Faure, C., Lambas-Senas, L., El Mansari, M., Belblidia, H., Gondard, E., et al. (2011). Pharmacological blockade of 5-HT7 receptors as a putative fast acting antidepressant strategy. Neuropsychopharmacology 36, 1275–1288. doi: 10.1038/npp.2011.13
Morales, M., Battenberg, E., and Bloom, F. E. (1998). Distribution of neurons expressing immunoreactivity for the 5HT3 receptor subtype in the rat brain and spinal cord. J. Comp. Neurol. 402, 385–401. doi: 10.1002/(SICI)1096-9861(19981221)402:3<385::AID-CNE7>3.0.CO;2-Q
Nackley, A. G., Shabalina, S. A., Tchivileva, I. E., Satterfield, K., Korchynskyi, O., Makarov, S. S., et al. (2006). Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science 314, 1930–1933. doi: 10.1126/science.1131262
Nakagawa, Y., Ishima, T., and Takashima, T. (1998). The 5-HT3 receptor agonist attenuates the action of antidepressants in the forced swim test in rats. Brain Res. 786, 189–193. doi: 10.1016/S0006-8993(97)01459-5
Narboux-Nême, N., Angenard, G., Mosienko, V., Klempin, F., Pitychoutis, P. M., Deneris, E., et al. (2013). Postnatal growth defects in mice with constitutive depletion of central serotonin. ACS Chem. Neurosci. 4, 171–181. doi: 10.1021/cn300165x
Pockros-Burgess, L. A., Pentkowski, N. S., Der-Ghazarian, T., and Neisewander, J. L. (2014). Effects of the 5-HT2C receptor agonist CP809101 in the amygdala on reinstatement of cocaine-seeking behavior and anxiety-like behavior. Int. J. Neuropsychopharmacol. 17, 1751–1762. doi: 10.1017/S1461145714000856
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi: 10.1086/519795
Quesseveur, G., Reperant, C., David, D. J., Gardier, A. M., Sanchez, C., and Guiard, B. P. (2013). 5-HT(2)A receptor inactivation potentiates the acute antidepressant-like activity of escitalopram: involvement of the noradrenergic system. Exp. Brain Res. 226, 285–295. doi: 10.1007/s00221-013-3434-3
Rhodes, R. A., Murthy, N. V., Dresner, M. A., Selvaraj, S., Stavrakakis, N., Babar, S., et al. (2007). Human 5-HT transporter availability predicts amygdala reactivity in vivo. J. Neurosci. 27, 9233–9237. doi: 10.1523/JNEUROSCI.1175-07.2007
Riccio, O., Jacobshagen, M., Golding, B., Vutskits, L., Jabaudon, D., Hornung, J. P., et al. (2011). Excess of serotonin affects neocortical pyramidal neuron migration. Transl. Psychiatry 1:e47. doi: 10.1038/tp.2011.49
Riccio, O., Potter, G., Walzer, C., Vallet, P., Szabó, G., Vutskits, L., et al. (2009). Excess of serotonin affects embryonic interneuron migration through activation of the serotonin receptor 6. Mol. Psychiatry 14, 280–290. doi: 10.1038/mp.2008.89
Rubinow, M. J., Mahajan, G., May, W., Overholser, J. C., Jurjus, G. J., Dieter, L., et al. (2014). Basolateral amygdala volume and cell numbers in major depressive disorder: a postmortem stereological study. Brain Struct. Funct. doi: 10.1007/s00429-014-0900-z [Epub ahead of print].
Ruocco, A. C., Amirthavasagam, S., and Zakzanis, K. K. (2012). Amygdala and hippocampal volume reductions as candidate endophenotypes for borderline personality disorder: a meta-analysis of magnetic resonance imaging studies. Psychiatry Res. 201, 245–252. doi: 10.1016/j.pscychresns.2012.02.012
Sergerie, K., Chochol, C., and Armony, J. L. (2008). The role of the amygdala in emotional processing: a quantitative meta-analysis of functional neuroimaging studies. Neurosci. Biobehav. Rev. 32, 811–830. doi: 10.1016/j.neubiorev.2007.12.002
Stankovski, L., Alvarez, C., Ouimet, T., Vitalis, T., El-Hachimi, K. H., Price, D., et al. (2007). Developmental cell death is enhanced in the cerebral cortex of mice lacking the brain vesicular monoamine transporter. J. Neurosci. 27, 1315–1324. doi: 10.1523/JNEUROSCI.4395-06.2007
Stein, C., Davidowa, H., and Albrecht, D. (2000). 5-HT(1A) receptor-mediated inhibition and 5-HT(2) as well as 5-HT(3) receptor-mediated excitation in different subdivisions of the rat amygdala. Synapse 38, 328–337. doi: 10.1002/1098-2396(20001201)38:3<328::AID-SYN12>3.3.CO;2-K
Stjepanovic, D., Lorenzetti, V., Yucel, M., Hawi, Z., and Bellgrove, M. A. (2013). Human amygdala volume is predicted by common DNA variation in the stathmin and serotonin transporter genes. Transl. Psychiatry 3, e283. doi: 10.1038/tp.2013.41
Sullivan, G. M., Ogden, R. T., Oquendo, M. A., Kumar, J. S., Simpson, N., Huang, Y. Y., et al. (2009). Positron emission tomography quantification of serotonin-1A receptor binding in medication-free bipolar depression. Biol. Psychiatry 66, 223–230. doi: 10.1016/j.biopsych.2009.01.028
Ubl, B., Kuehner, C., Kirsch, P., Ruttorf, M., Flor, H., and Diener, C. (2015). Neural reward processing in individuals remitted from major depression. Psychol. Med. 1–10. doi: 10.1017/S0033291715001452 [Epub ahead of print].
Wang, Y., Li, J., Chen, C., Zhu, B., Moysis, R. K., Lei, X., et al. (2013). COMT rs4680 Met is not always the ‘smart allele’: Val allele is associated with better working memory and larger hippocampal volume in healthy Chinese. Genes Brain Behav. 12, 323–329. doi: 10.1111/gbb.12022
Wang, C. C., Man, G. C., Chu, C. Y., Borchert, A., Ugun-Klusek, A., Billett, E. E., et al. (2014). Serotonin receptor 6 mediates defective brain development in monoamine oxidase A-deficient mouse embryos. J. Biol. Chem. 289, 8252–8263. doi: 10.1074/jbc.M113.522094
Wesolowska, A., and Nikiforuk, A. (2007). Effects of the brain-penetrant and selective 5-HT6 receptor antagonist SB-399885 in animal models of anxiety and depression. Neuropharmacology 52, 1274–1283. doi: 10.1016/j.neuropharm.2007.01.007
Yang, R. J., Mozhui, K., Karlsson, R. M., Cameron, H. A., Williams, R. W., and Holmes, A. (2008). Variation in mouse basolateral amygdala volume is associated with differences in stress reactivity and fear learning. Neuropsychopharmacology 33, 2595–2604. doi: 10.1038/sj.npp.1301665
Zalsman, G., Rehavi, M., Roz, N., Laor, N., Weizman, A., and Toren, P. (2011). Altered affinity of the platelet vesicular monoamine transporter 2 to dihydrotetrabenazine in children with major depression. J. Neural Transm. 118, 1383–1387. doi: 10.1007/s00702-011-0643-4
Zetzsche, T., Preuss, U. W., Bondy, B., Frodl, T., Zill, P., Schmitt, G., et al. (2008). 5-HT1A receptor gene C -1019 G polymorphism and amygdala volume in borderline personality disorder. Genes Brain Behav. 7, 306–313. doi: 10.1111/j.1601-183X.2007.00353.x
Zhang, Y., Bertolino, A., Fazio, L., Blasi, G., Rampino, A., Romano, R., et al. (2007). Polymorphisms in human dopamine D2 receptor gene affect gene expression, splicing, and neuronal activity during working memory. Proc. Natl. Acad. Sci. U.S.A. 104, 20552–20557. doi: 10.1073/pnas.0707106104
Zill, P., Buttner, A., Eisenmenger, W., Moller, H. J., Ackenheil, M., and Bondy, B. (2007). Analysis of tryptophan hydroxylase I and II mRNA expression in the human brain: a post-mortem study. J. Psychiatr. Res. 41, 168–173. doi: 10.1016/j.jpsychires.2005.05.004
Zubieta, J. K., Taylor, S. F., Huguelet, P., Koeppe, R. A., Kilbourn, M. R., and Frey, K. A. (2001). Vesicular monoamine transporter concentrations in bipolar disorder type I, schizophrenia, and healthy subjects. Biol. Psychiatry 49, 110–116. doi: 10.1016/S0006-3223(00)00981-1
Keywords: serotonin, gene, amygdala, brain structure, missing heritability
Citation: Li J, Chen C, Wu K, Zhang M, Zhu B, Chen C, Moyzis RK and Dong Q (2015) Genetic variations in the serotonergic system contribute to amygdala volume in humans. Front. Neuroanat. 9:129. doi: 10.3389/fnana.2015.00129
Received: 13 July 2015; Accepted: 17 September 2015;
Published: 09 October 2015.
Edited by:
Yun-Qing Li, The Fourth Military Medical University, ChinaReviewed by:
Jean-Pierre Hornung, University of Lausanne, SwitzerlandKewei Chen, Banner Alzheimer’s Institute, USA
Copyright © 2015 Li, Chen, Wu, Zhang, Zhu, Chen, Moyzis and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuansheng Chen, Department of Psychology and Social Behavior, University of California, Irvine, Irvine, CA 92697, USA, chuansheng.chen@uci.edu; Qi Dong, State Key Laboratory of Cognitive Neuroscience and Learning, IDG/McGovern Institute for Brain Research, Beijing Normal University, Beijing 100875, China, dongqi@bnu.edu.cn
†These authors have contributed equally to this work.
 Jin Li
Jin Li Chunhui Chen1,4†
Chunhui Chen1,4†  Karen Wu
Karen Wu Chuansheng Chen
Chuansheng Chen Qi Dong
Qi Dong