Learning-dependent gene expression of CREB1 isoforms in the molluscan brain
- 1 Laboratory of Functional Biology, Kagawa School of Pharmaceutical Sciences, Tokushima Bunri University, Sanuki, Japan
- 2 School of Medicine and Health Sciences, Monash University Sunway Campus, Bandar Sunway, Malaysia
- 3 Department of System Neuroscience, School of Medicine, Sapporo Medical University, Sapporo, Japan
Cyclic AMP-responsive element binding protein1 (CREB1) has multiple functions in gene regulation. Various studies have reported that CREB1-dependent gene induction is necessary for memory formation and long-lasting behavioral changes in both vertebrates and invertebrates. In the present study, we characterized Lymnaea CREB1 (LymCREB1) mRNA isoforms of spliced variants in the central nervous system (CNS) of the pond snail Lymnaea stagnalis. Among these spliced variants, the three isoforms that code a whole LymCREB1 protein are considered to be the activators for gene regulation. The other four isoforms, which code truncated LymCREB1 proteins with no kinase inducible domain, are the repressors. For a better understanding of the possible roles of different LymCREB1 isoforms, the expression level of these isoform mRNAs was investigated by a real-time quantitative RT-PCR method. Further, we examined the changes in gene expression for all the isoforms in the CNS after conditioned taste aversion (CTA) learning or backward conditioning as a control. The results showed that CTA learning increased LymCREB1 gene expression, but it did not change the activator/repressor ratio. Our findings showed that the repressor isoforms, as well as the activator ones, are expressed in large amounts in the CNS, and the gene expression of CREB1 isoforms appeared to be specific for the given stimulus. This was the first quantitative analysis of the expression patterns of CREB1 isoforms at the mRNA level and their association with learning behavior.
Introduction
Cyclic AMP-responsive element binding protein (CREB) is a transcription factor regulating a remarkable spectrum of cellular responses involved in developmental events, cell survival, neuronal differentiation, and memory formation in the brains of animal species (Lonze and Ginty, 2002 ). CREB can activate the expression of target genes in a phosphorylation-dependent manner via dimerization by binding to a cyclic AMP-responsive element (CRE) in their promoter regions (Montminy and Bilezikjian, 1987 ).
The CREB family gene is also known to be expressed in a variety of spliced isoforms, giving rise to functionally different CREB family proteins with the potential to either activate or repress target gene expression (Ruppert et al., 1992 ; Mayr and Montminy, 2001 ; Blöcher et al., 2003 ). CREB activator isoforms contain the three functional domains, phosphorylation box (P-box) or kinase-inducible domain containing several phosphorylation sites for various kinases, transactivation domain (Q-domain) interacting with a component of the TFIID complex for basal transcription, and leucine zipper (bZIP) domain for DNA binding and dimerization. CREB repressor isoforms, lacking each one of P-box or Q-domain, have inhibitory effects on gene expression (Bartsch et al., 1998 ; Behr and Weinbauer, 2000 ; Blöcher et al., 2003 , 2005 ; Poels and Vanden Broeck, 2004 ;).
Spliced isoforms of CREB also have opposite effects on learning behavior. For example, mutant mice lacking the activator isoforms of CREB have deficiencies in long-term but not in short-term memory (Bourtchuladze et al., 1994 ). In Drosophila, seven spliced isoforms of CREB protein were identified: dCREB2-a, -b, -c, -d, -q, -r, and -s (Yin et al., 1995b ). Olfactory memory formation was enhanced by overexpression of transcription activator dCREB2-a (Yin et al., 1995a ), and impaired by overexpression of transcription repressor dCREB2-b lacking Q-domain (Yin et al., 1994 ; Perazzona et al., 2004 ). In Aplysia, at least three different CREB isoforms played competing roles in long-term memory formation (Bartsch et al., 1998 ; Upadhya et al., 2004 ). Recently, Mohamed et al. (2005) and Liu et al. (2008) reported that inducing CREB1 gene expression by a direct CREB1 feedback loop leads to increased transcriptional activity for long-term synaptic facilitation in Aplysia. These studies clearly showed that spliced isoforms of CREB regulate the gene induction for memory formation, and thus the ratio between the levels of CREB activator and repressor isoforms is important for memory formation. To date, however, there has been no absolute quantitative comparison of the expression levels of CREB isoforms in relation to learning and memory formation.
We previously cloned Lymnaea CREB1 (LymCREB1) cDNA, which is a homolog of mammalian CREB, in the central nervous system (CNS) of mollusk L. stagnalis, and confirmed its expression and function for synaptic facilitation (Sadamoto et al., 2004a ,b ). In the present study, we focused on the LymCREB1 mRNA, and isolated its activator and repressor isoforms. We then quantified the expression levels of these isoforms in Lymnaea CNS by real-time RT-PCR. Finally, we examined their expression levels after conditioned taste aversion (CTA) learning. Our present findings suggest that the spliced isoforms of LymCREB1, including both the activator and repressor isoforms, were constitutively produced, and that their expression levels changed in a learning-dependent manner.
Materials and Methods
Animals
Specimens of L. stagnalis with a 20-mm shell (young adults: Sadamoto et al., 2000) were maintained in tap water and fed on lettuce under a 12:12-light–dark cycle at 20°C. All snails were anesthetized with 25% Listerine® before dissection (Kojima et al., 1997 ). For RNA extraction, the isolated CNSs were frozen in liquid nitrogen.
Identification Of LymCREB1 Isoforms
Standard molecular biological techniques were performed according to Sambrook et al. (1989) . For identifying the spliced isoforms of LymCREB1, a Lambda Zap II phage library for Lymnaea CNS was screened by low-stringency hybridization with an ApCREB1a cDNA fragment (Sadamoto et al., 2004a ). PCR was also carried out using a sequence specific primer (5′-TCATGCATCTTTTTGACAGT-3′) for the terminal part of the LymCREB1-coding region and the vector-specific SK primer (5′-TCTAGAACTAGTGGATC-3′).
Genomic PCR and Southern Blotting
For determination of the LymCREB1 gene structure, the genomic DNA was isolated from Lymnaea ovotestis and genomic PCR was performed with the primers for the 5′- and 3′-ends of the coding region. To identify intron–exon junctions, the amplified products were further used as the template for secondary PCR with the primers for the intron–exon gaps predicted from the Aplysia CREB1 genomic sequence (Bartsch et al., 1998 ). For Southern blot analysis, genomic DNA (10 ζg) was digested with restriction enzymes (XhoI, SacI, and PstI, 30 U each), run on 1× TBE gel with 0.8% agar and transferred to a nylon membrane. The DNA fragment (465 bp) for intron B (see Figure 1 ) was a hexamer labeled with [α-32P]dCTP, and hybridized to the filter. The filter was washed in 1× SSC, 0.1% SDS at 55°C and autoradiographed.
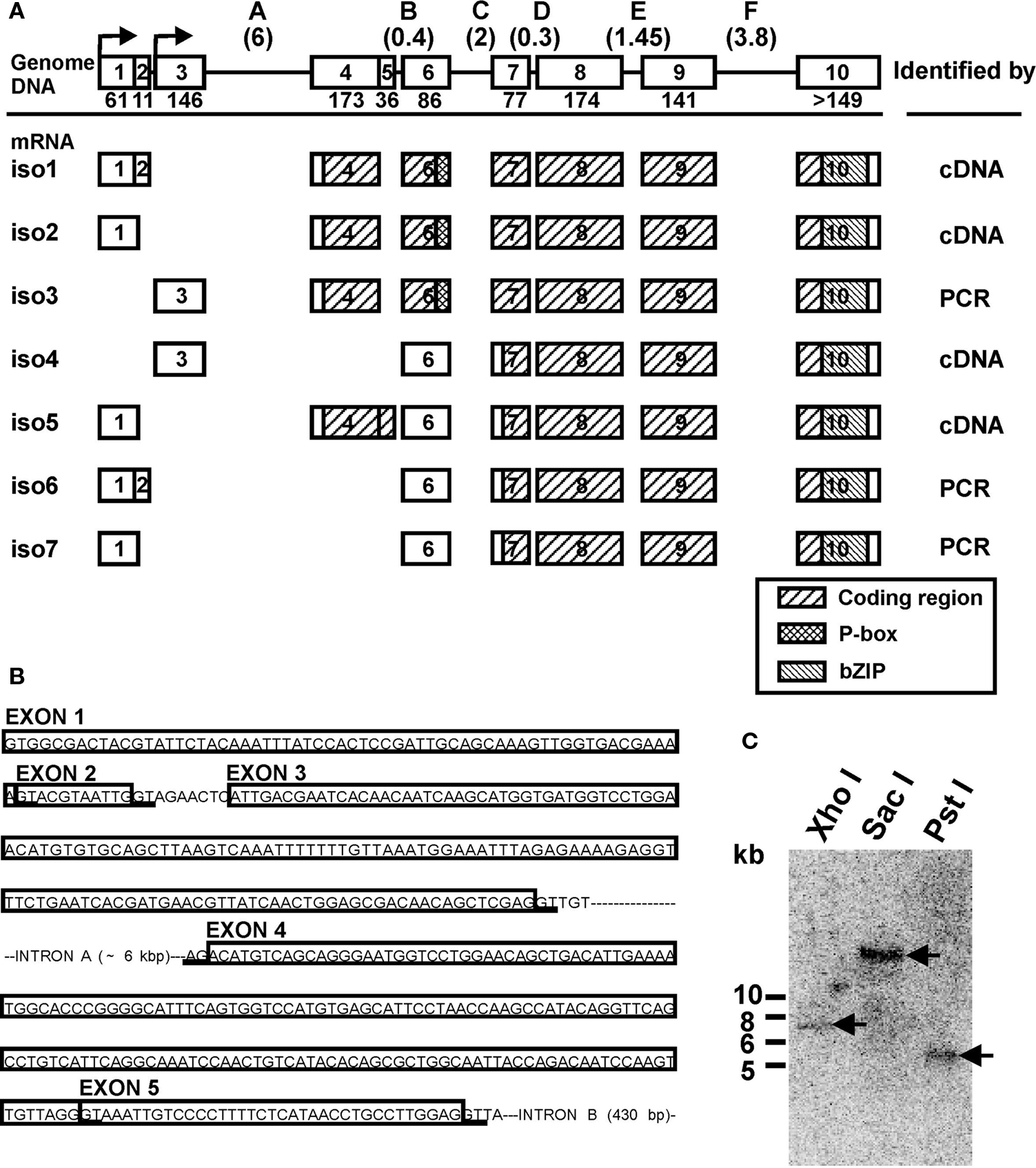
Figure 1. Genomic organization of the LymCREB1 gene and its spliced mRNA isoforms. (A) The genomic DNA and seven identified isoforms produced by alternative splicing are shown as iso1 to iso7. Above: Squares represent exons 1–10 and lines represent introns A–F. The numbers below the exons indicate the exon lengths (bp). The numbers in the parentheses above the introns indicate the intron lengths (kb). Below: Open boxes indicate non-coding regions; hatched boxes indicate coding regions. The identifying methodologies are shown on the right. (B) Intron and exon organization of the regions for alternative splicing in the LymCREB1 gene. The boxed sequences are the exons. The consensus sequence (GT/AG) for alternative splicing is underlined. (C) Southern blot analysis of the LymCREB1 gene. A 465-bp fragment labeled with 32P was hybridized to XhoI-, SacI- or PstI-digested genomic DNAs (10 μg per lane). A single fragment of approximately 7, 19, or 5.5 kb in size was detected in the XhoI-, SacI-, and PstI-digested DNA, respectively. Numbers at the left of the figure represent the DNA size markers (kb).
RNA Extraction and cDNA Synthesis
The animals were anesthetized and the CNSs were dissected and immediately frozen in liquid nitrogen. Total RNA was isolated with TRIzol Reagent (Gibco BRL, Gaithersburg, MD, USA). After DNase I treatment (Toyobo, Osaka, Japan), total RNA samples were extracted by phenol chloroform extraction followed by ethanol precipitation, and dissolved in RNase-free water. The quantity and the purity of RNA were determined by measuring the absorbance at 260 and 280 nm. To minimize the risk of contamination of other ganglia or loss of samples, we ensured that the concentration of samples remained unchanged between collections (30–100 ng/ζl). For cDNA synthesis, the total RNA sample (300 ng) was reverse-transcribed in a 30-ζl mixture containing 1× RT buffer (50 mM KCl, 10 mM Tris–HCl, pH 8.3), 5.5 mM MgCl2, 0.5 mM dNTPs, 0.2 ζM LymCREB1specific RT primer, 24 U RNase inhibitor and 100 U Multiscribe Reverse Transcriptase (PE Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions.
Quantitative mRNA Analysis by Real-Time PCR
After reverse transcription, the mRNA levels of LymCREB1 isoforms were determined by real-time PCR. The sequences of the primers were as follows:
isoform 1 sense 5′-GTTGGTGACGAAAAGTACGTAATTG-3′;
antisense, 5′-CTCACATGGACCACTGAAATGC-3′;
isoform 2 sense, 5′-AAGTTGGTGACGAAAAACAT-3′;
antisense, 5′-ACCGCAGCAACCCTAA-3′;
isoform 3 sense, 5′-TTATCAACTGGAGCGACAAC-3′;
antisense, 5′-ACCGCAGCAACCCTAA-3′;
isoform 4 sense, 5′-AGCGACAACAGCTCGAGGTT-3′;
antisense, 5′-GCTGTCAGCTGCAATGTTCCT-3′;
isoform 5 sense, 5′-GACGAAAAACATGTCAGCAG-3′;
antisense, 5′-CTCCAAGGCAGGTTATGAG-3′;
isoform 6 sense, 5′-GGTGACGAAAAGTACGTAATTGGTT-3′;
antisense, 5′-TGTGCTGTCAGCTGCAATGTT-3′;
isoform 7 sense, 5′-CAAAGTTGGTGACGAAAAGTTGCT-3′;
antisense, 5′-TGGCTCTGACTACTGTTCGAATCA-3′.
The fluorescent probes (PE Applied Biosystems) were labeled with the reporter dye 6-carboxyfluorescein (FAM) at the 5′ end and with the quencher dye 6-carboxytetramethylrodamine (TAMRA) at the 3′ end (probe 1 for isoforms 1, 2, 3, and 5, 5′-TTTTCAATGTCAGCTGTTCCAGGACCAT-3′ probe 2 for isoforms 4, 6 and 7, 5′-CTGAATGATCTCTCATCTCCTGTTAAAATGGA-3′).
The real-time PCR was performed using a GeneAmp 5700 Sequence Detection System (PE Applied Biosystems) in 12.5 ζl of reaction mixture (1× TaqMan Buffer A, 0.2 mM dNTPs, 0.31 U TaqMan Gold, 0.1 ζM each of the forward and reverse primer, 2 ζM fluorescent probe, 3 ζl of cDNA sample). PCR reaction was carried out at 95°C for 10 min, followed for 45 cycles of incubation at 95°C for 15 s and at annealing temperature for each primer set for 1 min (isoform 1, 62°C; isoform 2, 56°C; isoform 3, 56°C; isoform 4, 56°C; isoform 5, 56°C; isoform 6, 59°C; isoform 7, 59°C). At each PCR cycle, accumulation of PCR products was detected by monitoring the increase in fluorescence of the reporter dye from the fluorescent probes. Cycle threshold detection was converted into the number of gene copies in the starting material, and a standard curve was constructed using the known amounts of standard cDNA.
CTA and Backward Conditioning
Animals were conditioned by a CTA conditioning procedure that was previously described (Sugai et al., 2006 ). Briefly, the conditioned stimulus (CS) and unconditioned stimulus (US) were a 10-mM sucrose solution and a 10-mM KCl solution, respectively, which were poured into the experimental dish for 15 s with a 15-s inter-stimulus interval. The pairing of the CS and the US was repeated 10 times with a 10-min inter-trial interval. After the conditioning session, the CS was applied to the lips and washed out by distilled water. Then, the feeding response was determined for 1 min. A naive (distilled water only) cohort was employed, and a backward conditioning (US–CS) cohort was also employed as an example of “improper” conditioning. The conditioning procedure was performed in a blind manner. Thirty minutes after conditioning, the CNSs were dissected and immediately frozen in liquid nitrogen for RNA extraction.
Statistics
The data are expressed as the mean ± SEM. Statistical significance was examined by one-way ANOVA followed by the post hoc Scheffé test. In all analyses, values of P < 0.05 were considered to indicate statistical significance.
Results
LymCREB1 Isoforms in the Lymnaea CNS
We first screened a Lambda Zap II phage library for Lymnaea CNS to identify the LymCREB1 mRNA isoforms. PCR was also performed on the cDNA library with the vector specific primer (sense) and the sequence specific primer (antisense). Four kinds of clones were isolated from the cDNA library and three other kinds were further isolated by PCR. These seven identified clones were referred to as isoforms 1–7 (Figure 1 A).
Sequence characterization of the LymCREB1 isoforms allowed their grouping into two classes: (I) isoforms 1, 2 and 3, which encode the whole LymCREB1 protein containing both a phosphorylation site (P-box) and a leucine zipper (bZIP) domain (264 amino acids); and (II) isoforms 4, 5, 6, and 7, which encode a bZIP domain but not a P-box (167 amino acids). The P-box is the target region affected by several different kinase cascades and is required for induction of gene expression by a transcription factor. The bZIP domain is necessary for DNA binding and protein dimerization. On the other hand, all isoforms contain an intact Q2-domain corresponding to those of mammalian CREB1 proteins (exon 6–10, residues 108–221). Following the previous reports (Bartsch et al., 1998 ; Mayr and Montminy, 2001 ; Blöcher et al., 2003 , 2005 ), the class (I) isoforms are referred to as an activator group for gene induction, and can evoke dimerization, binding to target DNA and phosphorylation, whereas the class (II) isoforms make up a repressor group, with the potential to form a heterodimer with the activator protein, resulting in interference with gene induction due to the lack of a P-box.
To determine the intron–exon structure of the LymCREB1 gene, long-genomic PCR was performed with the primers for the LymCREB1-coding region. The 8-kb length of the obtained product was further used as the template for the determination of intron–exon junctions. Investigation of the LymCREB1gene by genomic PCR revealed that the 5′UTR and protein-coding regions were fragmented into 10 alternatively spliced exons (Figure 1 A). All of the 5′ and 3′ splice sites at the intron–exon junction conformed to the consensus splice site sequence (GT/AG).
The alternative splicing of LymCREB1 pre-mRNA transcripts resulted in the induction of transcriptional activators and repressors due to the selective inclusion or exclusion of a portion of exon 4 in which the AUG initiation codon for translation is located. In contrast to this alternatively spliced portion of the isoforms, the use of exons 7–10 appears to be a common feature in all the LymCREB1 isoforms (Figure 1 A). The sequence comparison of spliced isoforms showed that they alternate the transcription start site beginning at exon 1 or exon 3. Only isoform 5 contains another open reading frame that codes the N-terminal part (64 amino acids) of the whole LymCREB1 protein. Isoform 5 also includes an insert of exon 5 (36 bp) resulting in a shift of the codon reading frame with the appearance of an early stop codon (Figure 1 B; note that Figure 1 B is written as the gene structure). In addition, the initiation codon AUG was found in exon 7 in isoforms 4–7.
To estimate the copy number of the LymCREB1 gene, Southern blotting of the digested Lymnaea genomic DNA was performed (Figure 1 C). A single fragment of approximately 7, 19, or 5.5 kb in size was detected in the XhoI-, SacI-, and PstI-digested DNA, respectively. Together, these data indicate that all the LymCREB1 isoforms come from a single-copy CREB1 gene in Lymnaea CNS.
Quantitative mRNA Analysis of LymCREB1 Isoforms by Real-Time PCR
Next, we examined the expression levels of LymCREB1 isoforms in Lymnaea CNS by real-time PCR (Wagatsuma et al., 2005 ). Because of the large homologous regions among the members of LymCREB1 isoforms, the primer sets were chosen in specific regions of each isoform to avoid cross-reactions with the other isoforms. The location of the primer sets used for PCR analysis of the LymCREB1 gene is shown in Figure 2 A.
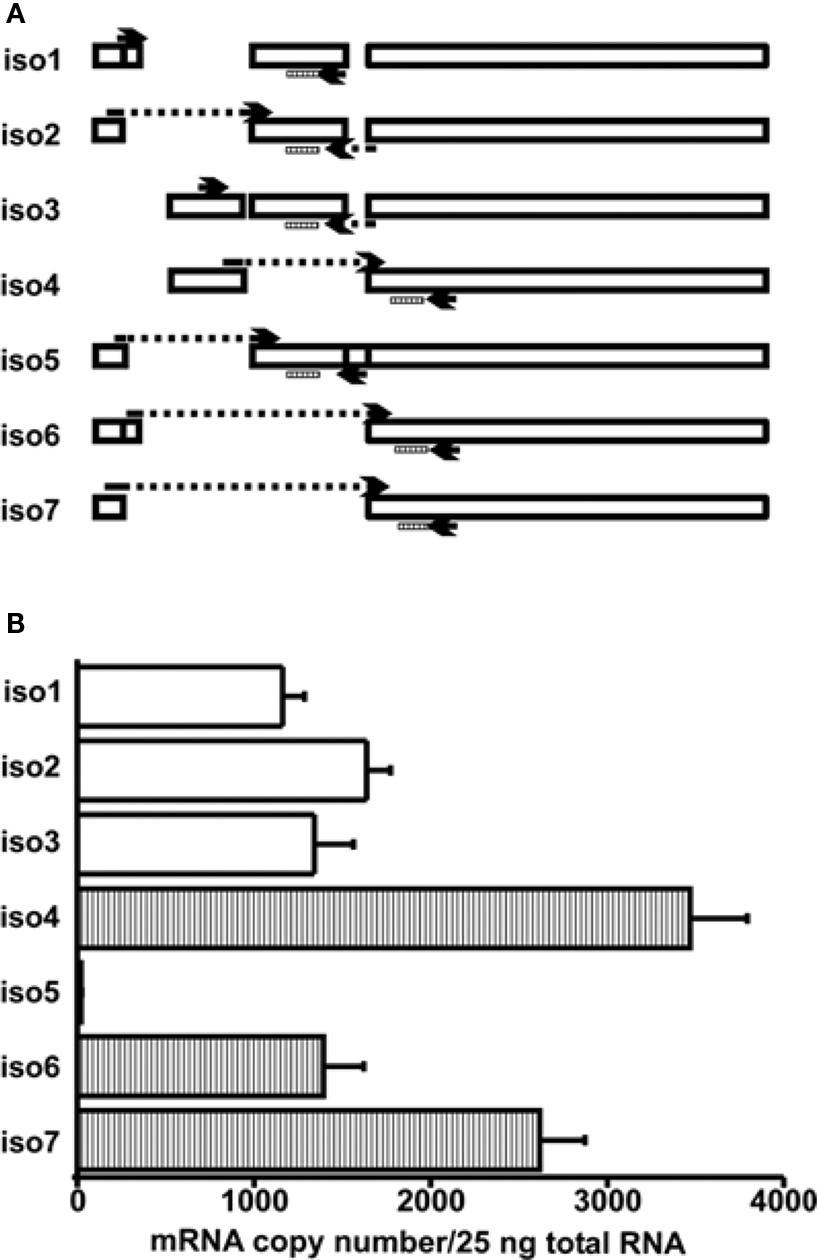
Figure 2. Determination of the mRNA copy numbers for LymCREB1 isoforms. (A) Schematic presentation of the LymCREB1 genes and location of the primer sets used for PCR analysis of the LymCREB1 gene. (B) Messenger RNA quantification of LymCREB1 isoforms. Twenty-five nanograms of total RNA for each isoform was used for the reverse transcription and quantification (mean ± SEM, n = 6 each). The copy numbers of the activator isoforms are indicated by white bars and those of the repressor isoforms by hatched bars.
To minimize the variability of reagents, all samples and standards were simultaneously reverse-transcribed in the solutions prepared from the same master mix. The amplification efficiency was determined for each primer-probe set as previously described (Bustin, 2000 ). Cycle threshold (Ct) values were determined for a series of 10-fold dilutions of standard templates, and the results were plotted as the log of the initial target copy number versus Ct. All sets produced slopes close to the theoretical ideal of −3.3 (−3.48 to −3.80). In this study, the specificity of PCR was determined in the 5′ end of primer sequence at exon–intron junction, and the lengths of amplicon were quite similar between isoforms as shown in Figure 2 A. Thus, to confirm the specificity of primer sets, we examined the cross-reactivity of isoform-specific primers with improper targets using the standard cDNA samples. The concentrations of the cross-reactants were consistently below one-thousandth of the concentrations of the target PCR product (data not shown).
We next summarized the patterns of the mRNA levels for the LymCREB1 isoforms in Lymnaea CNS (Figure 2 A). The isoforms 1–3 coding the activator proteins were expressed at a similar level. The isoforms 4, 6, and 7 coding the repressor proteins were expressed at a relatively high level in comparison with the activator isoforms 1–3. Only isoform 5 was expressed at a low level (21 ± 3), showing that this is a minor isoform in Lymnaea CNS. At the mRNA level, the ratio between activator isoforms and repressor isoforms for LymCREB1 was about 5:9 (the total number of isoforms 1–3 to that of isoforms 4–7) in Lymnaea CNS. The number of repressor isoforms was clearly greater than that of activator isoforms.
Change in the Copy Numbers of LymCREB1 Isoforms after Conditioned Taste Aversion Learning
To investigate the expression of LymCREB1 isoforms in association with learning and memory formation, we further examined the changes in the levels of LymCREB1 isoforms after CTA learning in Lymnaea (Kojima et al., 1997 ). The conditioning procedure is shown in Figure 3 A.
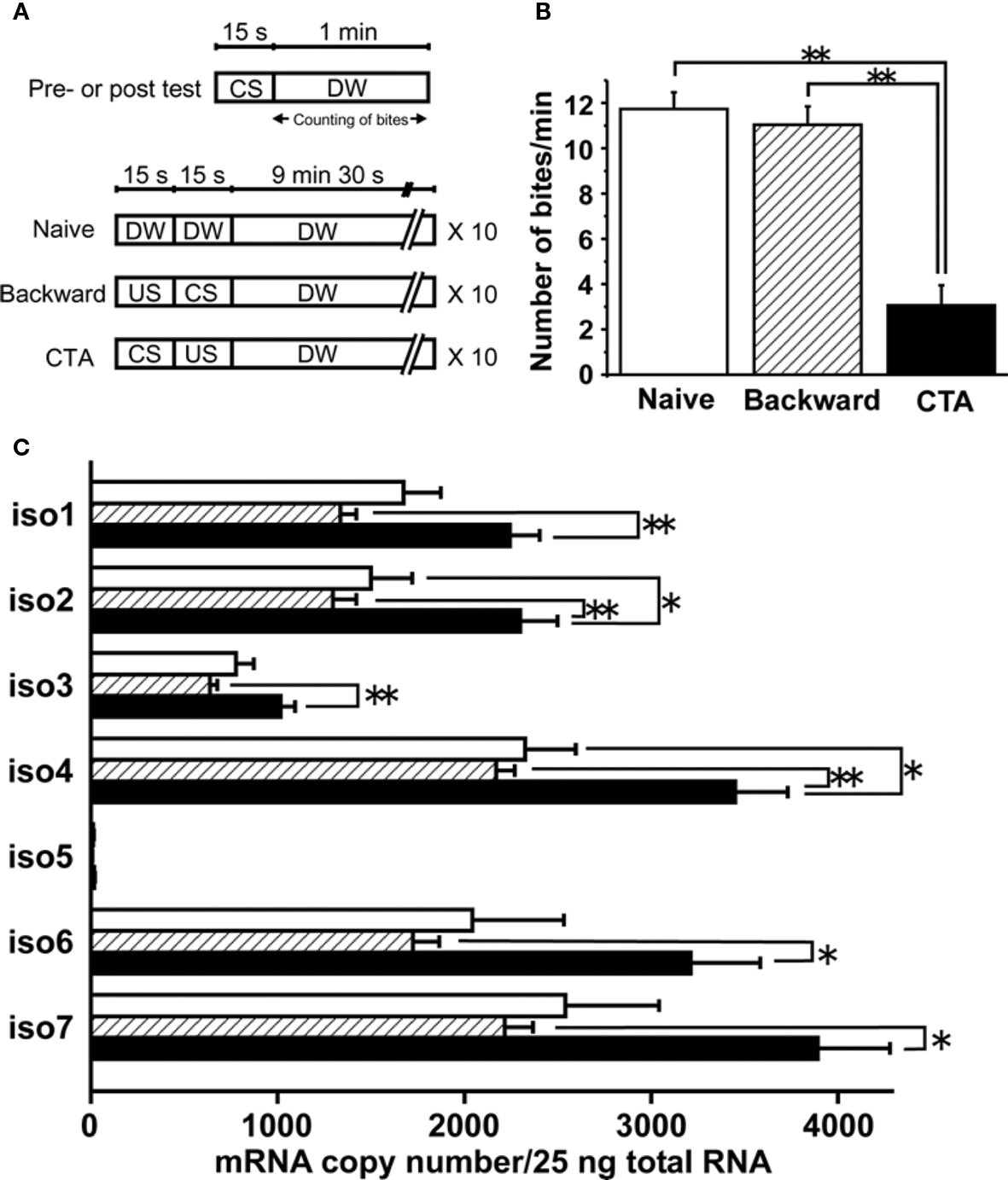
Figure 3. Quantification of mRNA copy numbers for LymCREB1 isoforms after CTA learning or backward conditioning. (A) Training paradigm. (B) Behavioral change after taste aversion learning or backward conditioning (mean ± SEM; **P < 0.01; n = 30 naive animals, n = 30 backward-conditioned animals, and n = 30 CTA-trained animals). (C) Messenger RNA quantification of LymCREB1 isoforms. Values are expressed as the mRNA copy number of cDNA per 25 ng input of total RNA (mean ± SEM; *P < 0.05; **P < 0.01; n = 8 naive animals, n = 10 backward-conditioned animals, and n = 17 CTA-trained animals). The copy numbers for naive control animals are indicated by the white bars, those for backward-conditioned animals by the hatched bars and those for CTA-trained animals by the black bars.
Animals were exposed to 10 mM sucrose (CS), followed by an exposure to 10 mM KCl (US). The CS–US pairing for CTA learning was performed 10 times. We also applied backward conditioning in which the CS followed the US as a behavioral control, and prepared naive animals. Ten minutes after the conditioning, the animals were given a memory test session (i.e., presentation of the CS alone). We found that the feeding response elicited by the CS in the test session was significantly reduced (P < 0.01) in comparison with the feeding response in the test session given to the naive and backward-conditioned cohort (Figure 3 B).
Thirty minutes after conditioning, the cerebral ganglia were dissected and used for mRNA quantification of each LymCREB1 isoform. The cerebral ganglia in Lymnaea are known to play a critical role in feeding behavior, and to be candidate loci for long-term memory in CTA learning (Kojima et al., 1997 ; Sadamoto et al., 2004a ,b ). After RNA extraction and reverse transcription, the cDNA from 25 ng total RNA was subjected to PCR analysis using the isoform-specific primer oligonucleotides. The results are summarized in Figure 3 C. The mRNA copy number in isoform 5 for backward conditioning was too low for reliable quantification (<10 copies).
A marked increase of LymCREB1 mRNA expression in each isoform was observed in CTA-trained animals compared with that of backward-conditioned animals (isoforms 1–4, P < 0.01; isoforms 6 and 7, P < 0.05). In isoforms 2 and 4, there was a significant difference between the CTA-trained and naive animals (P < 0.05, respectively). On the other hand, in backward-conditioned animals, no significant changes in the expression of LymCREB1 mRNA were found in comparison with naive animals.
Changes in LymCREB1 Gene Transcription and the Ratio between Activator and Repressor Isoforms after CTA Learning
To examine the effect of CTA learning on LymCREB1 gene expression, the total gene expression of LymCREB1 was calculated by adding together the mRNA copy numbers for all isoforms, and the results for each animal group are summarized in Figure 4 A.
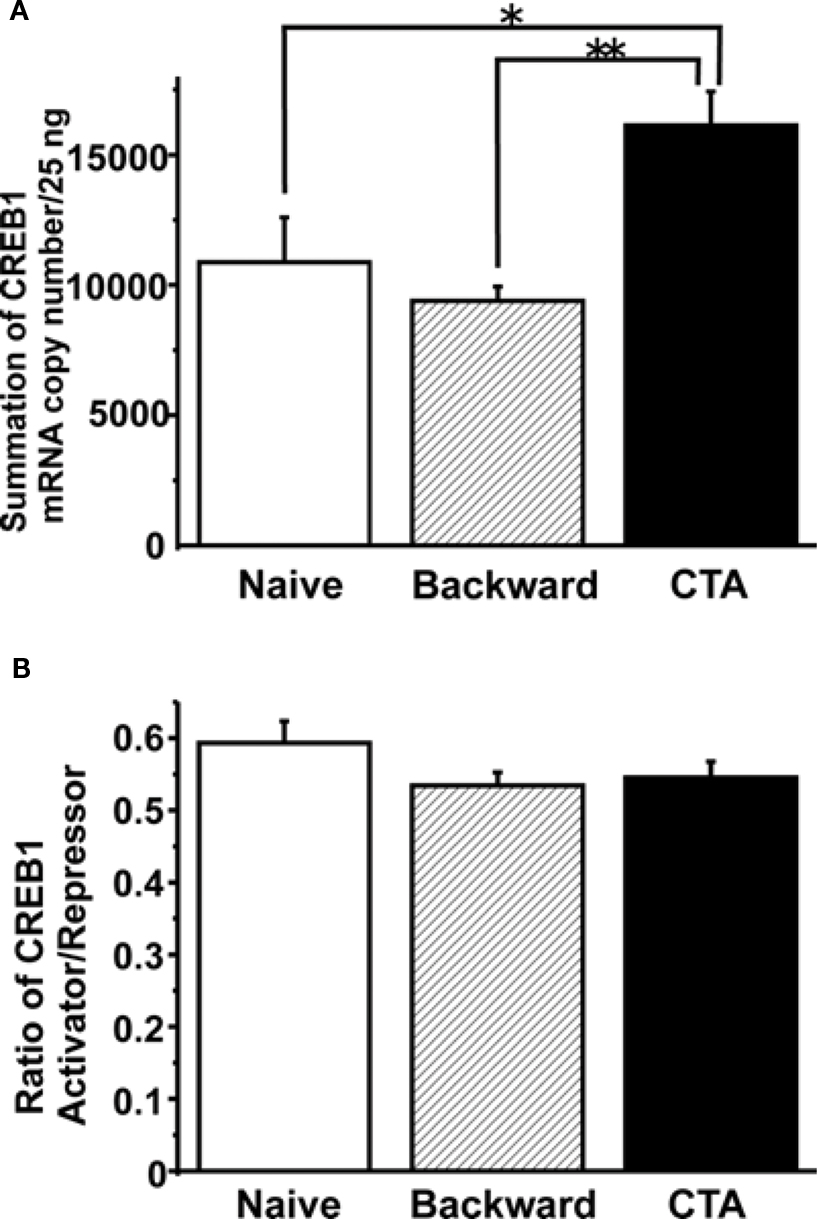
Figure 4. Summary of the LymCREB1 gene expression in Lymnaea CNSs after CTA learning and backward conditioning. Values are expressed as the mRNA copy number of cDNA per 25 ng input of total RNA (mean ± SEM; *P < 0.05; **P < 0.01; n = 8 for naive animals, n = 10 for backward-conditioned animals, and n = 17 for CTA-trained animals). The copy numbers for naive control animals are indicated by white bars, those for backward-conditioned animals by hatched bars and those for CTA-trained animals by black bars. (A) The total mRNA copy number for all LymCREB1 isoforms after CTA learning or backward conditioning. The mRNA copy numbers of backward-conditioned animals were significantly lower than those of CTA-trained animals (P < 0.01). The LymCREB1 gene expression after backward conditioning was significantly lower than that of CTA-trained animals. (B) The ratio between the mRNA copy numbers of activator and repressor isoforms. The ratio between the copy numbers of activator and repressor isoforms was not significantly different among the three groups.
The results clearly showed that CTA learning significantly increased the total LymCREB1 transcription levels (P < 0.05 between CTA-trained and naive animals, P < 0.01 between CTA-trained and backward-conditioned animals). On the other hand, backward conditioning did not induce LymCREB1 gene expression even though the stimuli were the same as those used for CTA learning.
Finally, we examined the simple question of whether CTA learning changes the ratio between the activator and repressor isoforms. To calculate the effects of alternative splicing on CTA learning, we compared the ratios of the mRNA copy numbers of activator to repressor isoforms (Figure 4 B). The copy numbers of activator or repressor isoforms were calculated by adding together the mRNA copy numbers of isoforms 1–3 or those of isoforms 4–7, respectively. The ratio of activator/repressor isoforms was then calculated for the naive control, backward-conditioned and CTA-trained groups. No significant difference in the ratio was observed among the three groups (P = 0.28). These results showed that CTA learning increased LymCREB1 gene expression, but did not change the ratio between activator and repressor LymCREB1 isoforms, in the candidate loci for long-term memory.
Discussion
The transcription factor CREB is well known to play multiple roles in the gene regulation for numerous biological events. The complex function of CREB family proteins arises in part from the wide range of variation in their spliced isoforms, which allows them to interact with each other (Ruppert et al., 1992 ). Previous reports showed that alternatively spliced isoforms of CREB family protein have opposite functions in the induction of long-lasting learning behavior, indicating that the expression level of each isoform is critical for memory formation (Bourtchuladze et al., 1994 ; Yin et al., 1994 ; Blöcher et al., 2003 , 2005 ; Perazzona et al., 2004 ).
In the present study, we showed that (1) the activator and repressor isoforms of LymCREB1 are present in Lymnaea CNS; (2) the mRNA copy number of repressor isoforms is much higher than that of activator isoforms in Lymnaea CNS; (3) the CTA learning was accompanied with an increase of LymCREB1 gene expression; and (4) the ratio between activator and repressor isoforms was not changed after CTA learning or backward conditioning compared to that of the naive control group.
Expression of Activator and Repressor Isoforms of LymCREB1
We here identified seven different isoforms of LymCREB1 arising from the single-copy gene in the CNS of Lymnaea (Figure 1 ). Alternative splicing produces a variety of functionally distinct isoforms from a single gene by use of different combinations of splice junctions. In the CNS, all intron–exon junctions of LymCREB1 feature the consensus splice site sequences (GT/AG), and the genomic structure is quite similar to that of the Aplysia CREB1 gene (ApCREB1) (Bartsch et al., 1998 ). However, we did not identify any mRNA isoform without exon 7, which corresponds to ApCREB1β as a repressor isoform of ApCREB1. The PCR method used here is appropriate to identify this type of isoform. Thus, there is a difference between species in the splicing of the CREB1 gene.
Further, we succeeded in determining the expression levels of the spliced isoforms of LymCREB1 using quantitative PCR methodology (Figure 2 ). These results showed that the repressor isoforms are expressed more highly than the activators at the mRNA level, suggesting that the repressor isoforms also play a pivotal role for gene regulation due to the suppression of the function of activator isoforms via heterodimerization. There is clear evidence that differences in the activities or amounts of gene-specific splicing regulators in distinct species as well as during development or in different tissues might account for species-, tissue-, and developmental stage-specific differences in the patterns of splicing (Black, 2003 ). Thus, alternative splicing can regulate the ratio between the activator and repressor isoforms of LymCREB1, thereby determining the gene-induction ability of LymCREB1 proteins. More, spliced isoforms can also interact or compete with other CREB family protein, such as CREB2 (Sadamoto et al., 2004a ), which constitute the regulatory mechanism of CREB-dependent gene expression.
Whether the spliced isoforms of LymCREB1 actually display the differences of translation efficiency remains to be determined. However, protein analysis using the antibodies for CREB1 cannot allow us to examine the expression levels of proteins because of the similarity of amino acid sequences of the isoform proteins. Previous reports have also shown that the variable isoforms of CREB1 are also seen in other species (Ruppert et al., 1992 ; Bartsch et al., 1998 ; Eisenhardt et al., 2003 ), but the translation efficiency was not considered in these studies. Messenger RNA stability, protein translation, and protein modification, including phosphorylation, are important mechanisms for CREB1 function, and future research discussing these factors can advance the present knowledge.
Regulation Mechanism for the Expression Levels of LymCREB1 Isoforms
The transcriptional regulation of CREB1 genes appears to be another important mechanism for maintaining appropriate levels of CREB1 proteins. Previous studies have reported that transcription of the CREB1 gene is regulated by other transcription factors (Delfino and Walker, 1999 ; Shell et al., 2002 ), or by a feedback loop of CREB1 (Mohamed et al., 2005 ). In Lymnaea CNS, the expression levels of LymCREB1 isoforms did not show large variation between individuals (see the error bars in the data of Figures 2 B and 3 C), indicating the existence of mechanisms that regulate the expression of these isoforms. We also showed that the expression levels of all LymCREB1 isoforms were significantly increased after CTA learning but not after backward conditioning in candidate loci for memory formation of learning. These results enabled us to conclude that there exists a regulatory system of LymCREB1 expression that is sensitive to extracellular signals. More, the significant difference in LymCREB1 mRNA copy numbers was coincident with the difference in learning behavior between CTA-trained and backward-conditioned animals (Figures 3 B,C and 4 A). Such conditioning-specific regulation of CREB1 gene expression might also determine the function of CREB1 proteins in a learning-dependent manner. Finally, it is reasonable to assume that the change of CREB1 expression in this study is small because CREB1 quantity should not be changed drastically. Transcription factor CREB1 is also included in variable biological phenomena, such as cell differentiation and cancer development, and the change of CREB1 has critical effects on those biological events. Thus, the change of CREB1 should not be like those of immediate early genes, such as c-Fos or CCAAT/enhancer-binding protein (C/EBP). Supporting this idea, previous simulation with bifurcated analysis (Song et al., 2007 ) also showed an increase or a decrease of CREBs causes enormous effects on the continuous CREB-dependent gene regulation.
Isoform 5 was dissimilar to the previously reported ApCREB1 isoforms (Bartsch et al., 1998 ), and the independent quantitative RT-PCR experiments showed that the expression level of isoform 5 was extremely low in Lymnaea CNS (Figures 2 B and 3 C). Therefore, isoform 5 seems to possess less potential for gene regulation in the Lymnaea CNS. One possible explanation for the low level gene expression of isoform 5 was that it was due to the mechanism of “nonsense-mediated mRNA decay (NMD),” which is responsible for the degradation of incorrectly spliced mRNAs with disrupted open reading frames (Conti and Izaurralde, 2005 ). Isoform 5 includes exon 5, which results in a shift of the codon reading frame with the appearance of an early stop codon (Figure 1 B), and appears to generate premature termination codons as the target of NMD (Conti and Izaurralde, 2005 ). More, exon 5 has a GT at its 5′-end (Figure 1 C), which is the splice donor site. Thus, isoform 5 mRNA is thought to arise from a failure in processing, and to be degraded by NMD immediately. These results indicate that the N-terminal part of LymCREB1 (64 amino acids) coded by isoform 5 is not translated into a functional protein.
CREB1 Function for Learning and Memory in Lymnaea
Various investigations have been carried out on CREB1 function in long-lasting behavioral changes due to CREB1-dependent gene induction for memory formation in vertebrates and invertebrates (Bourtchuladze et al., 1994 ; Perazzona et al., 2004 ). Lymnaea have provided us with insight into the richness of cellular correlation with learning and memory (Sadamoto et al., 1998 ; Ito et al., 1999 ; Kemenes et al., 2006 ; Parvez et al., 2006 ), and we have previously reported the localization and the function of LymCREB1 for long-lasting facilitation in the key neuron of CTA learning (Sadamoto et al., 2000, 2004a ,b ). In the present study, we found a significant difference in LymCREB1 gene expressions in the cerebral ganglia between CTA-trained and backward-conditioned groups, and also found a non-significant trend toward a difference between the CTA-trained and naive control groups (Figures 3 C and 4 A). Hence, we came to the conclusion that there is a possible pathway through which high expression of LymCREB1 is linked to the memory formation in CTA learning. Our observation is consistent with the previous reports in Aplysia that inducing CREB1 gene expression by a direct CREB1 feedback loop leads to increased transcriptional activity for long-term synaptic facilitation (Mohamed et al., 2005 ; Liu et al., 2008 ). Further, Ribeiro et al. (2003) reported that the appetitive learning of feeding behavior in Lymnaea increases phosphorylated-CREB1 in the cerebral ganglia which is the same ganglia as we observed in the present study. It is interesting that CREB1 plays a role in the same ganglia for memory formation even though the learning behavior is different.
In the present study, we also found a non-significant trend toward a decrease of LymCREB1 gene expression after backward conditioning. This means that, even though the same stimuli as in the CTA learning were applied, there was a significant difference in the gene expression between CTA-trained and backward-conditioning cohorts. We have previously reported that CTA learning was interfered after backward conditioning as a form of inhibitory learning (Sugai et al., 2007 ). A slight reduction in LymCREB1 gene expression may be one of the reasons for the inhibitory effect of backward conditioning. In support of this idea, Song et al. (2007) performed a bifurcation analysis using a computer simulation technique and showed that the number of CREB1 molecules reveals bimodal distribution in the bistability region. Thus, we now believe that backward-conditioning stimuli induce a shift to another stable state in the number of CREB1 molecules, and result in different learning behaviors, compared to CTA learning.
In contrast, the ratio between activator and repressor mRNA isoforms was not changed among the CTA-trained, backward-conditioning, and naive control groups (Figure 4 B). This means that the splicing pattern of LymCREB1 mRNA was not affected by CTA learning or backward conditioning. Thus, the LymCREB1-regulated transcriptional switch was not changed after CTA learning at the mRNA level. However, our present analysis also showed that the activator and repressor LymCREB1 were both increased after CTA learning. The increase of CREB1 activator molecules was presumed to result in an enhancement of the function of CREB.
On the other hand, it is notable that there are other mechanisms involved in the regulation of CREB1 protein, such as proteolysis. Upadhya et al. (2004) previously reported that the protein degradation of CREB1 repressor isoform occurs in neurons during induction of long-term facilitation. The final number of functional CREB1 molecules should be determined by several regulatory mechanisms, including gene regulation, translational regulation, and protein degradation.
Finally, we conclude that the procedure of CTA learning significantly increased the gene expression of both activator and repressor CREB1 isoforms in the candidate loci for memory formation, such as the cerebral ganglia. Our present study is the first to analyze the expression level of CREB1 isoforms quantitatively and accurately. Taken together with the previous results, our genetic analysis performed in the present study will enable us to design future studies to investigate the contribution of the CREB1 isoforms to the overall incidence of cellular or behavioral changes. Because of the conserved mechanism of CREB1-mediated gene regulation, our findings will further enhance understanding of the genetic bases for CREB1 functions in the tissues of a wide range of animal species.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the Japan Society for the Promotion of Science (Nos. 10765, 19770059 and 21770081 to Hisayo Sadamoto, Nos. 19370039 and 21657022 to Etsuro Ito) and by an OM Award from the Zoological Society of Japan (to Hisayo Sadamoto). The authors would like to thank Akiko Wagatsuma for her help with the quantitative RT-PCR analysis.
References
Bartsch, D., Casadio, A., Karl, K. A., Serodio, P., and Kandel, E. R. (1998). CREB1 encodes a nuclear activator, a repressor, and a cytoplasmic modulator that form a regulatory unit critical for long-term facilitation. Cell 95, 211–223.
Behr, R., and Weinbauer, G. F. (2000). CREM activator and repressor isoforms in human testis: sequence variations and inaccurate splicing during impaired spermatogenesis. Mol. Hum. Reprod. 6, 967–972.
Black, D. L. (2003). Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72, 291–336.
Blöcher, S., Behr, R., Weinbauer, G. F., Bergmann, M., and Steger, K. (2003). Different CREM-isoform gene expression between equine and human normal and impaired spermatogenesis. Theriogenology 60, 1357–1369.
Blöcher, S., Fink, L., Bohle, R. M., Bergmann, M., and Steger, K. (2005). CREM activator and repressor isoform expression in human male germ cells. Int. J. Androl. 28, 215–223.
Bourtchuladze, R., Frenguelli, B., Blendy, J., Cioffi, D., Schutz, G., and Silva, A. J. (1994). Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell 79, 59–68.
Bustin S. A. (2000). Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25, 169–193.
Conti, E., and Izaurralde, E. (2005). Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr. Opin. Cell. Biol. 17, 316–325.
Delfino, F. J., and Walker, W. H. (1999). NF-kappaB induces cAMP-response element-binding protein gene transcription in sertoli cells. J. Biol. Chem. 274, 35607–35613.
Eisenhardt, D., Friedrich, A., Stollhoff, N., Müller, U., Kress, H., and Menzel, R. (2003). The AmCREB gene is an ortholog of the mammalian CREB/CREM family of transcription factors and encodes several spliced variants in the honeybee brain. Insect Mol. Biol. 12, 373–382.
Ito, E., Kobayashi, S., Kojima, S., Sadamoto, H., and Hatakeyama, D. (1999). Associative learning in the pond snail, Lymnaea stagnalis. Zool. Sci. 16, 711–723.
Kemenes, I., Straub, V. A., Nikitin, E. S., Staras, K., O’Shea, M., Kemenes, G., and Benjamin, P. R. (2006). Role of delayed nonsynaptic neuronal plasticity in long-term associative memory. Curr. Biol. 16, 1269–1279.
Kojima, S., Nanakamura, H., Nagayama, S., Fujito, Y., and Ito, E. (1997). Enhancement of an inhibitory input to the feeding central pattern generator in Lymnaea stagnalis during conditioned taste-aversion learning. Neurosci. Lett. 230, 179–182.
Liu, R. Y., Fioravante, D., Shah, S., and Byrne, J. H. (2008). cAMP response element-binding protein 1 feedback loop is necessary for consolidation of long-term synaptic facilitation in Aplysia. J. Neurosci. 28, 1970–1976.
Lonze, B. E., and Ginty, D. D. (2002). Function and regulation of CREB family transcription factors in the nervous system. Neuron 35, 605–623.
Mayr, B., and Montminy, M. (2001). Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell. Biol. 2, 599–609.
Mohamed, H. A., Yao, W., Fioravante, D., Smolen, P. D., and Byrne, J. H. (2005). cAMP-response elements in Aplysia creb1, creb2, and Ap-uch promoters: implications for feedback loops modulating long term memory. J. Biol. Chem. 280, 27035–27043.
Montminy, M. R., and Bilezikjian, L. M. (1987). Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature 328, 175–178.
Parvez, K., Moisseev, V., and Lukowiak, K. (2006). A context-specific single contingent-reinforcing stimulus boosts intermediate-term memory into long-term memory. Eur. J. Neurosci. 24, 606–616.
Perazzona, B., Isabel, G., Preat, T., and Davis, R. L. (2004). The role of cAMP response element-binding protein in Drosophila long-term memory. J. Neurosci. 24, 8823–8828.
Poels, J., and Vanden, Broeck, J. (2004). Insect basic leucine zipper proteins and their role in cyclic AMP-dependent regulation of gene expression. Int. Rev. Cytol. 241, 277–309.
Ribeiro, M. J., Serfozo, Z., Papp, A., Kemenes, I., O’Shea, M., Yin, J. C., Benjamin, P. R., and Kemenes, G. (2003). Cyclic AMP response element-binding (CREB)-like proteins in a molluscan brain: cellular localization and learning-induced phosphorylation. Eur. J. Neurosci. 18, 1223–1234.
Ruppert, S., Cole, T. J., Boshart, M., Schmid, E., and Schutz, G. (1992). Multiple mRNA isoforms of the transcription activator protein CREB: generation by alternative splicing and specific expression in primary spermatocytes. EMBO J. 11, 1503–1512.
Sadamoto, H., Hatakeyama, D., Kojima, S., Fujito, Y., and Ito, E. (1998). Histochemical study on the relation between NO-generative neurons and central circuitry for feeding in the pond snail, Lymnaea stagnalis. Neurosci. Res. 32, 57–63.
Sadamoto, H., Sato, H., Kobayashi, S., Murakami, J., Aonuma, H., Ando, H., Fujito, Y., Hamano, K., Awaji, M., Lukowiak, K., Urano, A., and Ito, E. (2004a). CREB in the pond snail Lymnaea stagnalis cloning, gene expression and function in identifiable neurons of the central nervous system. J. Neurobiol. 58, 455–466.
Sadamoto, H., Azami, S., and Ito, E. (2004b). The expression pattern of CREB genes in the central nervous system of the pond snail Lymnaea stagnalis. Acta Biol. Hung. 55, 163–166.
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Shell, S. A., Fix, C., Olejniczak, D., Gram-Humphrey, N., and Walker, W. H. (2002). Regulation of cyclic adenosine 3′,5′-monophosphate response element binding protein (CREB) expression by Sp1 in the mammalian testis. Biol. Reprod. 66, 659–666.
Song, H., Smolen, P., Av-Ron, E., Baxter, D. A., and Byrne, J. H. (2007). Dynamics of a minimal model of interlocked positive and negative feedback loops of transcriptional regulation by cAMP-response element binding proteins. Biophys. J. 92, 3407–3424.
Sugai, R., Shiga, H., Azami, S., Watanabe, T., Sadamoto, H., Fujito, Y., Lukowiak, K., and Ito, E. (2006). Taste discrimination in conditioned taste aversion of the pond snail Lymnaea stagnalis. J. Exp. Biol. 209, 826–833.
Sugai, R., Azami, S., Shiga, H., Watanabe, T., Sadamoto, H., Kobayashi, S., Hatakeyama, D., Fujito, Y., Lukowiak, K., and Ito, E. (2007). One-trial conditioned taste aversion in Lymnaea: good and poor performers in long-term memory acquisition. J. Exp. Biol. 210, 1225–1237.
Upadhya, S. C., Smith, T. K., and Hegde, A. N. (2004). Ubiquitin-proteasome-mediated CREB repressor degradation during induction of long-term facilitation. J. Neurochem. 91, 210–219.
Wagatsuma, A., Sadamoto, H., Kitahashi, T., Lukowiak, K., Urano, A., and Ito, E. (2005). Determination of the exact copy numbers of particular mRNAs in a single cell by quantitative real-time RT-PCR. J. Exp. Biol. 208, 2389–2398.
Yin, J. C., Del Vecchio, M., Zhou, H., and Tully, T. (1995a). CREB as a memory modulator: induced expression of a dCREB2 activator isoform enhances long-term memory in Drosophila. Cell 81, 107–115.
Yin, J. C., Wallach, J. S., Wilder, E. L., Klingensmith, J., Dang, D., Perrimon, N., Zhou, H., Tully, T., and Quinn, W. G. (1995b). A Drosophila CREB/CREM homolog encodes multiple isoforms, including a cyclic AMP-dependent protein kinase-responsive transcriptional activator and antagonist. Mol. Cell. Biol. 15, 5123–5130.
Keywords: CREB1, gene expression, mollusk, learning, memory
Citation: Sadamoto H, Kitahashi T, Fujito Y and Ito E (2010) Learning-dependent gene expression of CREB1 isoforms in the molluscan brain. Front. Behav. Neurosci. 4:25. doi: 10.3389/fnbeh.2010.00025
Received: 26 February 2010;
Paper pending published: 19 March 2010;
Accepted: 30 April 2010;
Published online: 28 May 2010
Edited by:
Paul S. Katz, Georgia State University, USAReviewed by:
Károly Elekes, Balaton Limnological Research Institute of the Hungarian Academy of Sciences, HungaryJerry Yin, University of Wisconsin–Madison, USA
Copyright: © 2010 Sadamoto, Kitahashi, Fujito and Ito. This is an open-access article subject to an exclusive license agreement between the authors and the Frontiers Research Foundation, which permits unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are credited.
*Correspondence: Hisayo Sadamoto, Laboratory of Functional Biology, Kagawa School of Pharmaceutical Sciences, Tokushima Bunri University, 1314-1 Shido, Sanuki 769-2193, Japan. e-mail: sadamotoh@kph.bunri-u.ac.jp