Bypassing hazard of housekeeping genes: their evaluation in rat granule neurons treated with cerebrospinal fluid of multiple sclerosis subjects
- 1Department of Functional Biology, University of Valencia, Valencia, Spain
- 2Multiple Sclerosis Laboratory, Department of Biomedicine, Prince Felipe Research Center, Valencia, Spain
- 3Department of Biochemistry and Molecular Biology, University of Valencia and INCLIVA Biomedical Research Institute, Valencia, Spain
- 4CSUR-Esclerosi Múltiple, Hospital Universitari i Politècnic La Fe, Unitat Mixta d’Esclerosi Múltiple i Neurorregeneració de l’IIS-La Fe, València, Spain
- 5Hospital Clínico, Universitario de Valencia, Valencia, Spain
Gene expression studies employing real-time PCR has become an intrinsic part of biomedical research. Appropriate normalization of target gene transcript(s) based on stably expressed housekeeping genes is crucial in individual experimental conditions to obtain accurate results. In multiple sclerosis (MS), several gene expression studies have been undertaken, however, the suitability of housekeeping genes to express stably in this disease is not yet explored. Recent research suggests that their expression level may vary under different experimental conditions. Hence it is indispensible to evaluate their expression stability to accurately normalize target gene transcripts. The present study aims to evaluate the expression stability of seven housekeeping genes in rat granule neurons treated with cerebrospinal fluid of MS patients. The selected reference genes were quantified by real time PCR and their expression stability was assessed using GeNorm and NormFinder algorithms. GeNorm identified transferrin receptor (Tfrc) and microglobulin beta-2 (B2m) the most stable genes followed by ribosomal protein L19 (Rpl19) whereas β-actin (ActB) and glyceraldehyde-3-phosphate-dehydrogenase (Gapdh) the most fluctuated ones in these neurons. NormFinder identified Tfrc as the best invariable gene followed by B2m and Rpl19. ActB and Gapdh were the least stable genes as analyzed by NormFinder algorithm. Both methods reported Tfrc and B2m the most stably expressed genes and Gapdh the least stable one. Altogether our data demonstrate the significance of pre-validation of housekeeping genes for accurate normalization and indicates Tfrc and B2m as best endogenous controls in MS. ActB and Gapdh are not recommended in gene expression studies related to current one.
Introduction
Techniques employed for calibrating gene expression are paramount in studies directed toward accurate analysis of transcriptomic profiles. Quantitative real time PCR (qRT-PCR) has gained significant momentum over the past decade to quantify gene expression profiles. Considering the utmost sensitivity and reliability of qRT-PCR, a careful selection of a constitutively expressed gene is required to account for variation in the amount and quality of starting RNA and cDNA synthesis efficiency. In general, the expression of target gene transcripts is normalized with an internal control, often referred to as a housekeeping gene. Housekeeping (HK) genes are endogenous controls that are required for the primary function of a cell hence their expression should be constant in all conditions. However, recent research has indicated that their expression may not necessarily be stable in all cells/tissues. A gene showing consistent expression in one condition may show unstable expression in another. Invariable expression of the so-called housekeeping genes has been observed during cellular development (Al-Bader and Al-Sarraf, 2005) and under distinct experimental conditions (Zhong and Simons, 1999; Hamalainen et al., 2001; Deindl et al., 2002; Glare et al., 2002; Torres et al., 2003; Radonic et al., 2004; Toegel et al., 2007; Gubern et al., 2009). Therefore it is essential to pre-validate the expression stability of reference genes to accurately normalize the gene expression data. It is recommended that more than one stably expressed gene should be used for precise normalization procedure (Zhong and Simons, 1999; Tricarico et al., 2002; Vandesompele et al., 2002; Ohl et al., 2005).
In this context, we aimed to evaluate the expression stability of seven commonly used housekeeping genes in cerebellar granule neurons (CGNs) treated with cerebrospinal fluid (CSF) from multiple sclerosis (MS) and neuromyelitis optica (NMO) patients. Axonal damage is widely accepted as a major cause of persistent functional disability in MS. Therefore to study primary neuronal damage independent of secondary damage, resulting from demyelination, we used primary cultures of unmyelinated CGNs as a cellular model and exposed them to CSF derived from MS patients. Prior to comprehending mechanisms involved in axonal degeneration–regeneration, it was first necessary to identify best stably expressed housekeeping genes that can be used to normalize target mRNA transcripts in our experimental system. We therefore used a xenogeneic system comprising of primary rat CGN cultures incubated with CSF from patients with MS or controls and investigated the stability of reference genes in these rat neuronal cells. Previous studies in similar xenogeneic models showed that treatment with human CSF resulted in neurotoxicity in culture, although the molecular mechanisms remained unknown (Xiao et al., 1996; Alcazar et al., 2000). Recently, Vidaurre et al. (2014) reported that ceramides present in CSF from patients with MS disturb neuronal bioenergetics in rat neuronal cultures.
Primary cultures of rat CGNs represent an excellent model to study almost every aspect of neurobiology. While neuronal cell lines have been very useful in the study of neuronal cell cultures, there are certain drawbacks they exhibit. These cell lines are derived from neuronal tumors and hence will show many important physiological differences with the cell type from which they were derived. For instance, the human SH-SY5Y cell line, was derived by subcloning from the parental metastatic bone tumor biopsy cell line SK-N-SH (Biedler et al., 1973). Therefore, it is prudent to use primary cultures because they are not tumor-derived and hence are more likely to exhibit the properties of neuronal cells in vivo. Furthermore, CGNs are small and the most numerous unmyelinated neurons, therefore we used primary cultures of rat CGNs as a cellular model and exposed them to diseased CSF to comprehend the pathophysiological mechanisms implicated in MS and prior to that validating the expression stability of commonly used housekeeping genes for their use in future gene expression experiments.
We selected some frequently used housekeeping genes from literature to determine their expression stability in our experimental setting. MS is a major cause of non-traumatic neurological disability deemed to affect more than 2 million people worldwide (Blight, 2011). It manifests as a chronic inflammation in central nervous system (CNS) that leads to demyelination and neurodegeneration. The disease typically manifests at 20–40 years of age when people are in their full employment and sometimes develops into an aggressive stage that alters the lives of patients and their families. Unfortunately the current treatments are only effective in preventing relapses and slowing down progression but not completely ceasing it. Although the pathogenesis of MS is not well understood, accumulating evidence suggests a complex interplay of both genetic and environmental factors (Al-Bader and Al-Sarraf, 2005; Compston and Coles, 2008; Oksenberg et al., 2008). A plethora of gene expression studies have been undertaken in peripheral mononuclear white blood cells (Der et al., 1998; Ramanathan et al., 2001; Wandinger et al., 2001; Bomprezzi et al., 2003; Koike et al., 2003; Sturzebecher et al., 2003; Hong et al., 2004; Iglesias et al., 2004; Satoh et al., 2006), in MS brain tissues (Becker et al., 1997; Whitney et al., 1999; Chabas et al., 2001; Whitney et al., 2001; Lock et al., 2002; Mycko et al., 2003; Tajouri et al., 2003; Lindberg et al., 2004; Mycko et al., 2004) and in CSF (Brynedal et al., 2010). Proteomic approaches have also been used to identify differentially expressed proteins in the CSF of MS patients (Dumont et al., 2004; Hammack et al., 2004; Noben et al., 2006). However, the proteomics analysis of CSF obtained from MS patient is relatively challenging. Since proteins are highly abundant, diversified, and soluble, only some protein subgroups may be detected and others important proteins may fail to be identified by proteomics approach. Thus, it would be prudent to use proteomic analysis along with other approaches such as gene expression profiling using microarray. Another similar but totally distinct neurological disease known as NMO shares many pathological similarities with MS and therefore it was previously considered as its variant. For this reason clinicians often used to encounter difficulty in distinguishing MS from NMO and hence similar treatment was provided to both the category of patients. However, recent research shows that there are some NMO specific IgG antibodies present in the sera of NMO patients, which differentiate both the diseases (Lennon et al., 2004).
In MS, axonal damage is widely accepted as the major cause of persistent functional disability, although its origin is unknown. During the relapsing-remitting disease course the patient’s brain itself is capable of repairing the damage, remyelinating the axon and recovering the neurological function. CSF is in contact with brain parenchyma (Rossi et al., 2012, 2014) and a site of deposition of cellular damaged products, which can influence the cellular physiology of brain cells. It is a promising biofluid in the search for biomarkers and disease associated proteins in MS, both with respect to inflammatory and neurodegenerative processes. Exposure of CGNs with CSF from diseased states can allow us to understand the pathophysiology of MS but prior to that evaluation of housekeeping genes to accurately normalize target genes is a crucial step. Selected housekeeping genes were quantified using real time PCR to accurately normalize target genes in our experimental setting. The expression stability of reference genes was further assessed by GeNorm and NormFinder algorithms. GeNorm program defines the gene stability as the average pairwise variation of a particular gene with all other control genes and ranks the genes according to their average expression stability denoted by M (Vandesompele et al., 2002). The gene with minimum M value is considered to be highly stable whereas the gene with highest M value is least stable and can be excluded. An alternative program, NormFinder, ranks the candidate reference genes based on the combined estimates of both intra- and intergroup variations (Andersen et al., 2004).
Materials and Methods
All procedures were approved by the Committee of Animal Care of Prince Felipe Research Center (CIPF), Valencia, in accordance with the regulations of the European Union and Spanish legislation. Informed consent was obtained from all the patients and controls for this study and authorized by the Ethical Committee of the Institute.
Patient Cohort
Patient Population
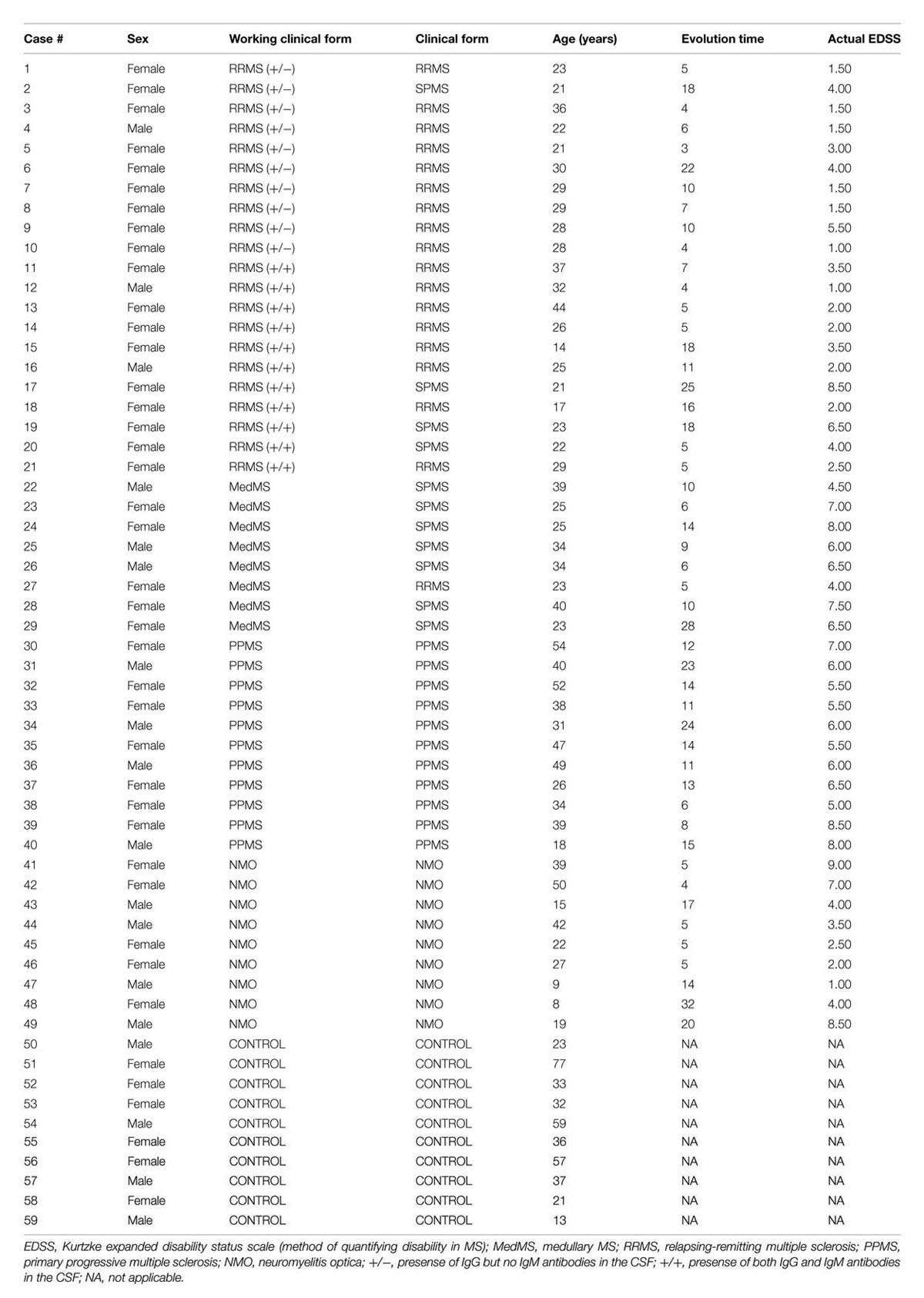
A total of 59 patients were recruited and CSF samples were obtained from the Department of Neurology, Hospital La Fe and Hospital Clinico, University of Valencia. Out of 59 patients, 21 had inflammatory MS (11 IgM+/+ and 10 IgM +/-), 8 had medullary subtype, 11 had PPMS, 9 had NMO, and 10 were non-inflammatory neurological controls (NIND patients). In CSF, apart from factors related to MS or NMO, there are factors from other diseases that produce their action. This must be considered as “background noise” as average population. Mixing of total CSF samples in all clinical forms may potentiate the factors related to MS. Therefore, we mixed CSF samples in all clinical forms.
Multiple sclerosis patients were defined and grouped in different clinical courses, according to the current criteria (Lublin and Reingold, 1996) and diagnosed according to McDonald criteria. They all met the following characteristics: oligoclonal IgG bands (OCGB) present, not in a phase of relapse, and have spent more than a month after the last dose of steroids. Wingerchuk criteria were used to diagnose patients with NMO disease (Wingerchuk et al., 2006). Patients suffered relapses of optic neuritis and myelitis, and two of the three criteria, normal MRI or that did not accomplish the Patty criteria for MRI diagnosis of MS. Table 1 illustrates the clinical characteristics of the patients.
Patient Characteristics
Inflammatory MS (RRMS and SPMS forms)
MS is categorized into: (1) Relapsing remitting MS (RRMS) that later develops into secondary progressive stage (SPMS); and (2) primary progressive MS (PPMS). Over 95% of patients with MS show oligoclonal bands (OCBs) of IgG in CSF (G+) (Kostulas et al., 1987) and 40% show IgM OCBs in CSF (M+) related to a more aggressive course of disease (Sharief and Thompson, 1991). In our project we also classified and named inflammatory MS into “IgM+/-” and “IgM+/+ subtype” (see below) on the basis of aggressivity and prognosis that is more complete than just RRMS or PPMS. In addition we have studied separately a set of patients with MS but with a predominant affectation of the spinal cord, because these patients have some peculiarities, and we wanted to explore if they have some differences in light of our experiments. The most aggressive cases termed as “medullary” have more spinal injuries.
IgM+/- clinical form of MS
Patients named as “IgM+/-subtype” had IgG antibodies (+) but no IgM (-) oligoclonal antibodies detected in the CSF of brain.
IgM+/+ clinical form of MS
Patients named as “IgM+/+ subtype” had both IgG antibodies (+) and IgM (+) oligoclonal antibodies detected in the CSF of brain.
Medullary clinical form of MS
All these patients were positive for OCGBs and negative for oligoclonal IgM bands (OCMBs) in CSF of spinal region. The patients accomplished the Swanton criteria for dissemination in time.
Primary progressive MS
These patients are characterized by progressive decline in neurological disability.
Neuromyelitis optica patients
Individuals diagnosed with NMO met at least two of the following three features. (1) Long extensive transverse myelitis (>3 vestibule bodies); (2) Antibodies against aquaporin-4; (3) Normal brain at the first event.
Controls [Non-Inflammatory Neurological Diseases (NIND)]
Individuals who were suspected to have MS but were not diagnosed with MS were classified as controls.
Cerebrospinal Fluid Samples of Patients
Cerebrospinal fluid samples were obtained by lumbar puncture at the time of diagnosis. Samples were centrifuged for 10 min at 700 × g and aliquots were frozen at -80°C until use. No patient had received treatment with immunosuppressive drugs, immunomodulators or corticosteroids for at least 1 month prior to the extraction of CSF.
Cerebrospinal Fluid Studies
All the studies were performed by immunologists who were blind to the clinical and MRI data.
Oligoclonal band studies
Paired CSF and serum samples were analyzed to detect OCBs (OCGB and OCMB) by isoelectric focusing (IEF) and immunodetection. We used a commercial kit to determine OCGB (Helena BioScience IgG-IEF Kit) and the technique described by Villar et al. (2001) to detect OCMB. Serum samples were diluted in saline before the IEF in order to reach the same concentration range as that of CSF samples. All samples were incubated with 50 mmol/L dithiothreitol at pH 9.5 to reduce IgM. Focusing was performed on a Multiphor II Electrophoresis System (GE Healthcare) at pH 5–8. Proteins were then transferred to a PVDF membrane and analyzed by Western blot. Finally, immunodetection was performed by biotin-conjugate-goat anti-human IgM and streptavidin-alkaline phosphatase (Sigma–Aldrich).
Serum studies
Anti-AQP4 antibody in NMO has a high specificity so as to contribute to early diagnosis and optimized treatment of Devic disease. Serum sample diluted 1:10 in PBS-Tween was used to detect the presence of NMO specific IgG antibodies. Indirect immunofluorescence (IFI) was performed to diagnose NMO (Figure 1B). Antibodies against aquaporin 4 were detected using a cell line, which was molecular biologically modified to produce large quantities of aquaporin 4. In this method (EuroImmun IIFT) recombinantly transfected cells act as an antigen substrate to be incubated with diluted serum samples for half an hour.
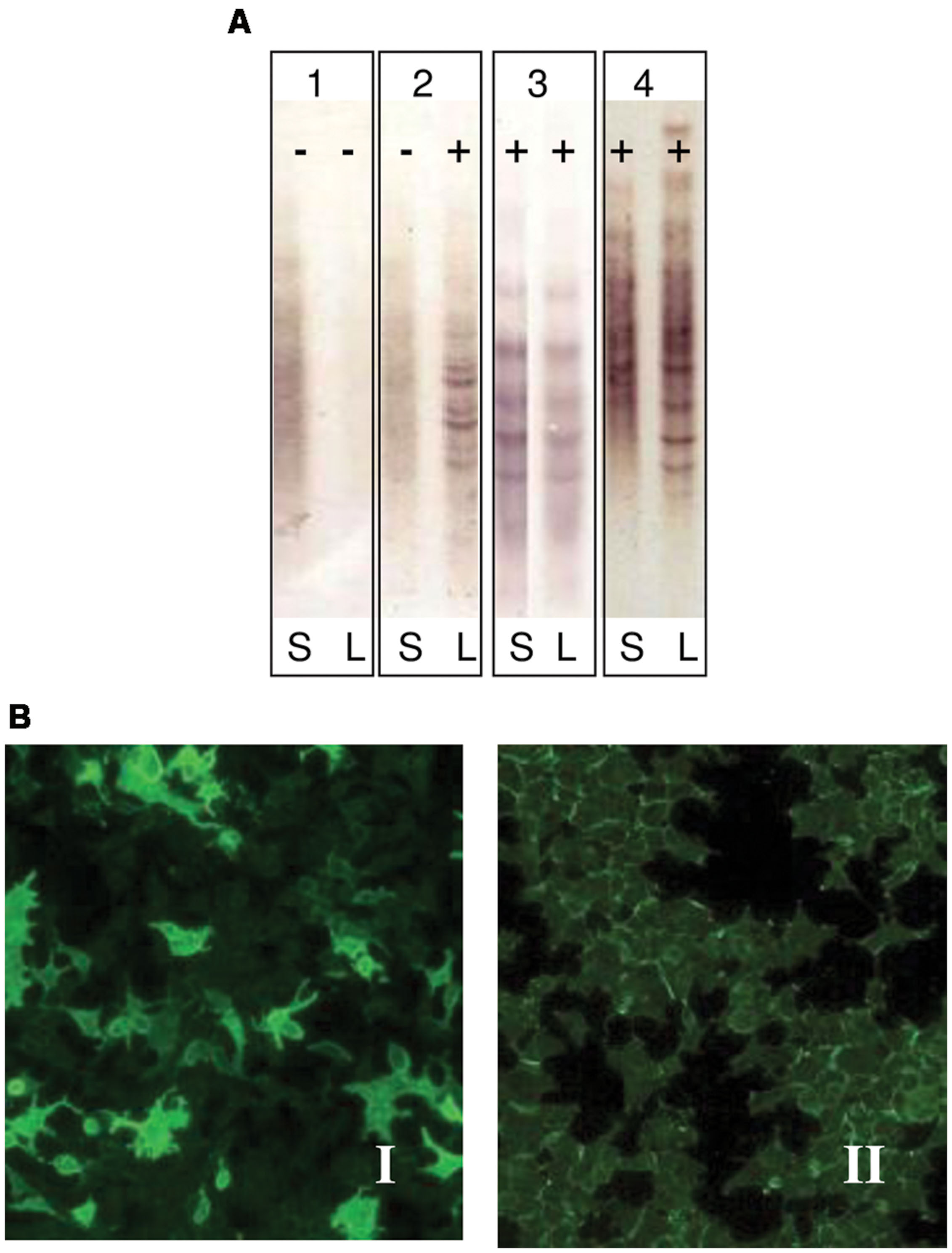
FIGURE 1. (A) Immunodetection of oligoclonal bands (OCBs) in serum (S) and cerebrospinal fluid (CSF) (L). Pattern 1: No OCBs seen (negative, polyclonal): No OCBs in CSF or serum. No intrathecal Ig synthesis; Pattern 2: OCBs in CSF only (positive): OCBs present in CSF only. Intrathecal IgG synthesis as seen in MS; Pattern 3: Identical OCBs in both (mirror images): Bands in serum mirror those in CSF. This suggests systemic Ig synthesis; Pattern 4: Identical OCBs in both with extra bands in CSF: Identical bands in both serum and CSF with extra bands in CSF. Image demonstrates both intrathecal and systemic Ig synthesis. This is identical as it is seen in MS. (B) Indirect immunofluorescence in cells transfected by aquaporin 4 (EUROIMMUN Aquaporin-4 IIFT). Panel I: Anti-AQP4 antibodies observed in the serum of NMO patients (positive sample); Panel II: Absence of anti-AQP4 antibodies in serum sample (negative sample).
Animals
Wistar rats (Harlan Iberica) with weight between 200 and 250 g were used. All animals were raised under controlled conditions with cycles of light/dark (12/12 h), temperature of 23°C and humidity of 60%. Access to water and food (standard rodent feed supplied by Harlan, Teklad 2014 Global 14% Protein Rodent Maintenance Diet) was provided. To obtain offspring, pregnant females were separated and kept in isolated cages during gestation. The maintenance of the animals was performed in the animal facilities unit of Prince Felipe Research Center, Valencia, Spain.
Primary Culture of Cerebellar Granule Neurons
All operations were performed under sterile conditions in vertical laminar flow chamber (Telstar AV-100 and Bio-II-A). The cells were kept in an incubator at 37°C in a humidified atmosphere composed of 95% air and 5% CO2 (CO2 incubator Thermo Form, model 371). Primary cultures of CGNs were obtained according to previously described modified protocol (Minana et al., 1998). Forebrains were collected from 8 days old Wistar rats, mechanically dissociated and cerebellum was dissected. Isolated cerebella were stripped of meninges, minced by mild trituration with a Pasteur pipette and treated with 3 mg/ml dispase (grade II) for 30 min at 37°C in a 5% CO2 humidified atmosphere. After half an hour, dispase was inactivated with 1mM EDTA. Granule cells were then resuspended in basal Eagleś medium (BME, Gibco, ref. 41010) with 40 μg/ml of DNaseI. The cell suspension was filtered through a mesh with a pore size of 90 μm and centrifuged at 1500 rpm for 5 min and thereafter, cell suspension was washed three times with BME. Finally, the cells were resuspended in complete BME medium with Earleś salts containing 10% heat inactivated FBS (fetal bovine serum, Gibco), 2 mM glutamine, 0.1 mg/ml gentamycin and 25 mM KCl. The neuronal cells were counted and plated onto poly-L-lysine coated 6-well (35-mm) culture dishes (Fisher) at a density of 3 × 105 cells/well and incubated at 37°C in a 5% CO2/95% humidity atmosphere. After 20 min at 37°C, the medium was removed and fresh complete medium was added. Since the purpose of our study was to obtain pure cultures of CGNs, it was necessary to add a chemical that can prevent the growth of non-neuronal cells. Twenty micro liter of cytosine arabinoside (1 mM) was added to each culture plate after 18–24 h to inhibit replication of non-neuronal cells. The cells were kept in an incubator at 37°C in a humidified atmosphere composed of 95% air and 5% CO2 (CO2 incubator Thermo Form, model 371). Cells were fed every 3–4 days in culture with 5.6 mM glucose.
Cerebellar granule neurons were stained with Texas Red and FITC dyes. The nuclei of neurofilaments were stained with DAPI. Figure 2A shows pure cultures of granule neurons isolated from cerebellum with stained neurofilaments.
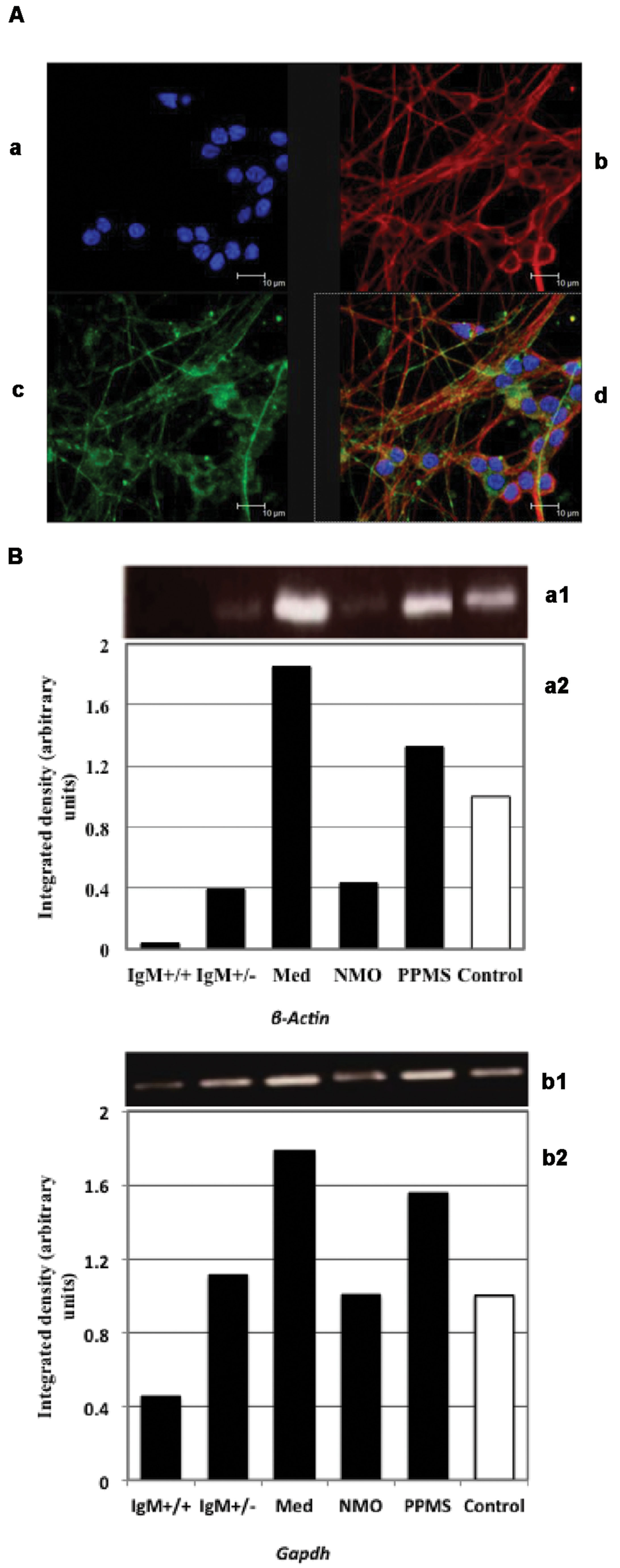
FIGURE 2. (A) Immunofluorescence of primary cultures of CGNs. (a) DAPI stained nuclei. Neurofilaments stained with (b) Texas red and (c) FITC dyes. (d) Merged images. (B) Analysis of β-actin (a) and Gapdh (b) expression by PCR: (1) Electrophoretic bands; (2) Integrated density obtained by Image J software quantification. The values correspond with the fold change vs. control values of the MS clinical forms, NMO, and control patients.
Confocal Microscopy
The living cells were always kept at 37°C and 5% CO2. Cells were analyzed on a Leica TCS SP2 confocal microscope AOBS (Leica Microsystems) inverted laser scanning confocal microscope using a 63 × Plan-Apochromat-Lambda Blue 1.4 N.A. oil objective lens. All confocal images were obtained under identical scan settings. Images of 1,024° × 1,024 pixels, 8-bits were collected for each preparation. Best focus was based on highest pixel intensity. Imaging conditions were identical for all the images, and no images were saturated. Metamorph 7.0 (Molecular Devices, Downingtown, PA, USA) was used for image analysis on the images collected.
Selection of Housekeeping Genes
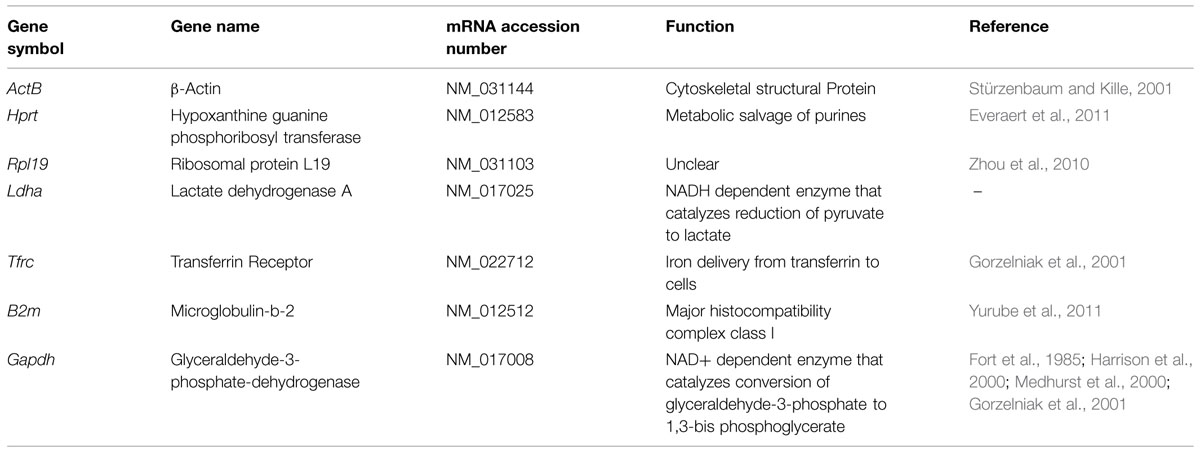
Candidate housekeeping genes were selected from those most commonly used in literature including β-actin (ActB), hypoxanthine guanine phosphoribosyl-transferase (Hprt), ribosomal protein L19 (Rpl19), lactate dehydrogenaseA (Ldha), transferrin receptor (Tfrc), microglobulin beta-2 (B2m), and glyceraldehyde-3-phosphate-dehydrogenase (Gapdh). The function and references of the genes are listed in Table 2. The primers for the selected reference genes from 5′- to 3′- end were as follows: Actb forward ATTGAACACGGCATTGTCAC, reverse ACCCTCATAGATGGGCACAG; Hprt forward CCTCTCGAAGTGTTGGATACAG, reverse TCAAATCCCTGAAGTGCTCAT; Rpl19 forward ACCTGGATGCGAAGGATGAG, reverse CCATGAGAATCCGCTTGTTT; Ldha forward AGGAGCAGTGGAAGGATGTG, reverse AGGATACATGGGACGCTGAG; Tfrc forward GTTGTTGAGGCAGACCTTCA, reverse ATGACTGAGATGGCGGAAAC; B2m forward GTCGTGCTTGCCATTCAGA, reverse ATTTGAGGTGGGTGGAACTG; Gapdh forward GGAAACCCATCACCATCTTC, reverse GTGGTTCACACCCATCACAA.
Treatment, Total RNA Isolation and cDNA Synthesis
Neuronal cell cultures were incubated with 10% v/v CSF from MS (IgM+/-, IgM+/+, medullary, PPMS) patients, NMO patients and controls for 24 h. This step was performed to identify transcriptional changes in future transcriptomic experiments. Total RNA was isolated from cell cultures exposed to the CSF of different experimental conditions (IgM+/-, IgM+/+, medullary, NMO, PPMS, Control) using Quick RNA MicroPrep Kit (Zymo Research Corp.). The RNA concentration was determined spectrophotometrically at 260 nm using the Nanodrop 1000 spectrophotometer (V3.7 software) and RNA purity was checked by means of the absorbance ratio at 260/280 nm. Isolated RNA was stored at -80° and later reverse transcribed to cDNA. The cDNA was synthesized and stored at -20°C. Primers for selected genes were designed using Primer 3 software. PCR was performed in a thermocycler (BioRad) with cycling conditions (94° for 30 s, 40 cycles at 59° for 30 s and 72° for 30 s). Each 25 μl reaction contained 12.5 μl Master Mix (Applied Biosystems), 1 μl gene-specific forward and reverse primers (0.5 μM), 1 μl undiluted cDNA and 10.5 μl DEPC (nuclease free) treated water. Negative controls with no template contained nuclease-free water instead.
Agarose Gel Electrophoresis and Real-time Polymerase Chain Reaction of Selected Housekeeping Genes
The electrophoresis was performed in 1.5% agarose gels, and they were run at 50 V, stained with ethidium bromide, photographed and evaluated with ImageJ software. A DNA ladder control (100 bp, Invitrogen) was also used in the electrophoresis to evaluate DNA fragment size. Real time PCR was performed in a 96-well plate (Roche) incubated in thermocycler (LC480, Roche) with cycling conditions (94° for 15 s, 45 cycles at 60° for 30 s and 72° for 30 s). Each 10ml reaction contained 5 ml SYBR Green Master Mix (Applied Biosystems), 1 μl gene-specific forward and reverse primers (0.5 μM), 1 μl undiluted cDNA and 3 ml DEPC (nuclease free) treated water. Negative controls with no template contained nuclease-free water instead. All samples were run in duplicate and average values were calculated. Data was analyzed using 7300 Sequence Detection Software (SDS) Version 1.3 (Software Roche). Following qRT-PCR, a dissociation curve was run to check the PCR product specificity.
Determination of Reference Gene Expression Stability
To determine the stability of these genes on the basis of their Cp values, we employed comparative ΔCT method. Data are plotted as fold change values which were calculated by 2Δ(Ctexp -Ctcontrol). Cp value is defined as the PCR cycle at which the fluorescent signal of the reporter dye crosses an arbitrarily placed threshold. Invariable genes were later assessed by publicly available software tools named GeNorm and NormFinder.
Results
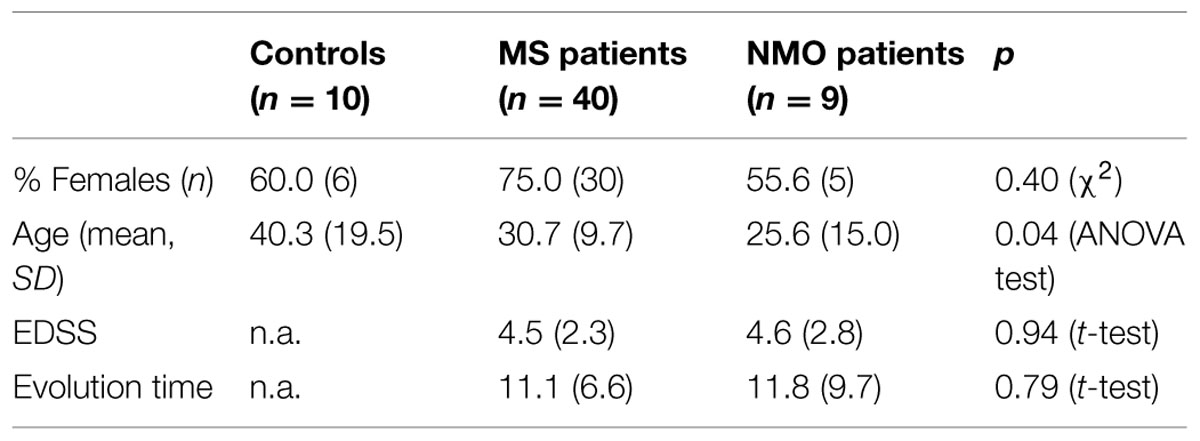
Demographic and Clinical Profiles of MS, NMO and NIND Groups
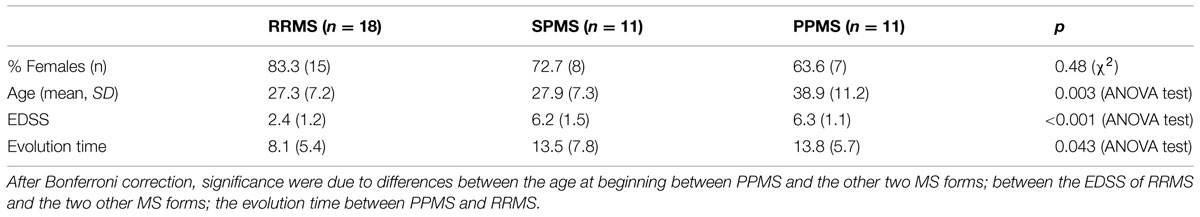
Patients were classified according to detection of OCBs (Figure 1A) and of aquaporin antibodies (Figure 1B). Baseline characteristics of the study population are described in Table 3. Prevalence of MS was found more in women (75%) than in men. Mean age of MS patients was 30.7 ± 9.7 whereas 25.6 ± 15 for NMO patients. According to the clinical classification, the general characteristics of MS patients are described in Table 4. There were significant differences observed between the age at beginning of PPMS and the other two MS forms (p < 0.003); between the EDSS of RRMS and the two other MS forms (<0.001); the evolution time between PPMS and RRMS (p = 0.043) after Bonferroni correction. Table 5 shows the characteristics of MS patients according to new proposal and working classification. After Bonferroni correction, significance was due to differences between the age at beginning and the EDSS between medullary MS and PPMS with the inflammatory MS.
We found significant differences in the age at beginning between PPMS and the other two MS forms (RRMS and SPMS) after Bonferroni correction (p < 0.003) (Table 6). People with PPMS are usually older at the time of diagnosis with an average age of 40. Furthermore, different subtypes of MS help predict disease severity and response to treatment hence their categorization is important. In our study, we found significant differences between the “Expanded Disability Status Scale” (EDSS) of RRMS and the two other MS forms (SPMS and PPMS) (p < 0.001) (Table 5). Although nerve injury always occurs, the pattern is specific for each individual with MS. Disease severity and disability increases from relapsing-remitting to secondary progressive course and in PPMS subtype, symptoms continually worsen from the time of diagnosis rather than having well-defined attacks and recovery. PPMS usually results in disability earlier than relapsing-remitting MS. Significant differences were found in the evolution time from the first to the second episode between RRMS and PPMS (p = 0.043). In patients experiencing a progressive course, evolution time was similar in secondary progressive cases and in cases that were progressive from onset (13.5 versus 13.8) (Table 5).
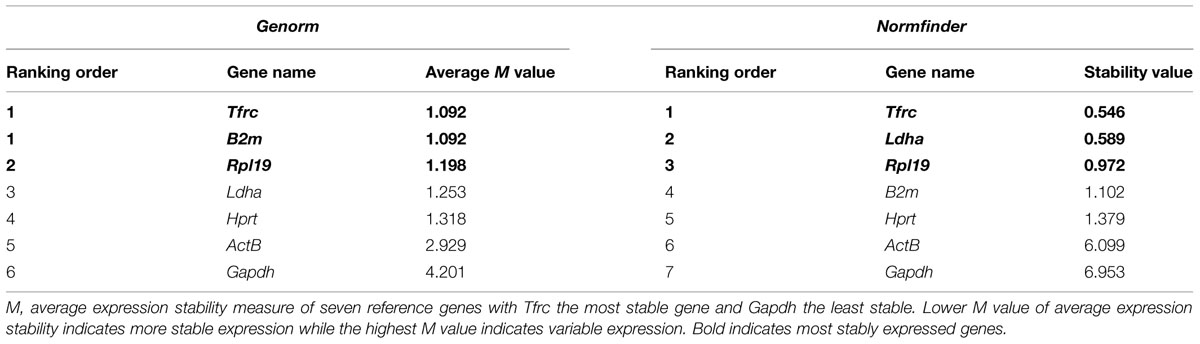
TABLE 6. Candidate housekeeping genes ranked in cerebellar granule neurons treated with CSF of MS/NMO patients according to their expression stability by Genorm and Normfinder methods.
According to the new proposal and working classification, inflammatory MS subtypes shared similar age at disease onset (mean = 26.7 versus 26.3 years; p = 0.005). Significant differences were found between the age at disease onset in medullary MS and PPMS with the inflammatory MS (p < 0.005). The degree of disability as measured by EDSS was similar in medullary MS and PPMS (6.2 versus 6.3) whereas significant differences were found between disability extent in medullary MS and PPMS with the inflammatory MS (p < 0.001). IgM+/- represents the less aggressive inflammatory subtype with OCGB in CSF with poor prognosis whereas IgM+/+ signifies a more aggressive category with OCGB and OCMB in CSF with worse prognosis. On the contrary, medullary MS represents the most aggressive subtype of MS with increased neurological disability and dysfunction as compared to inflammatory subtypes. Disability in patients experiencing PPMS worsens over time with no relapses and remission.
Identification of Stably Expressed Housekeeping Genes
PCR of Gapdh and β-actin
We first quantified ActB and Gapdh genes using conventional PCR in treated neuronal samples and ran agarose gel electrophoresis. We found that both β-Actin and Gapdh genes, which are presumed to express at constant levels showed varying band intensity in CGNs when treated with the CSF from MS and NMO patients (Figure 2B). From this data we conclude that both ActB and Gapdh genes are not suitable to normalize gene transcripts in our experimental conditions.
Quantitative PCR of Housekeeping Genes in our Experimental Conditions
Quantitative real time PCR was performed for a group of frequently used reference genes. GeNorm and NormFinder algorithms were used to assess the most stably expressed genes. Our data suggests Tfrc, B2m as the most stable genes followed by Rpl19 using GeNorm software (Average expression stability value denoted by M: 1.09 for Tfrc and B2m; M: 1.19 for Rpl19). Similarly, Tfrc showed most stable expression as assessed by NormFinder algorithm followed by Ldha and Rpl19 (M: 0.54 for Tfrc; M: 0.58 for Ldha and 0.97 for Rpl19) (Table 6). On the other hand, β-Actin and Gapdh showed highest fluctuation in our experimental conditions with 2.9 and 4.2 as the average expression stability value by GeNorm. Therefore their use is strictly discouraged while normalizing gene expression data in studies related to the current one.
Table 6 illustrates candidate housekeeping genes ranked in CGNs treated with CSF from MS/NMO patients according to their expression stability by GeNorm and NormFinder methods. The Ct values of all the experimental conditions obtained from qPCR experiment were normalized to control. Then we plotted the fold change values for each reference gene tested in distinct disease courses of MS and NMO patients (Figure 3). Fold change was calculated by 2Δ(Ctexp -Ctcontrol).
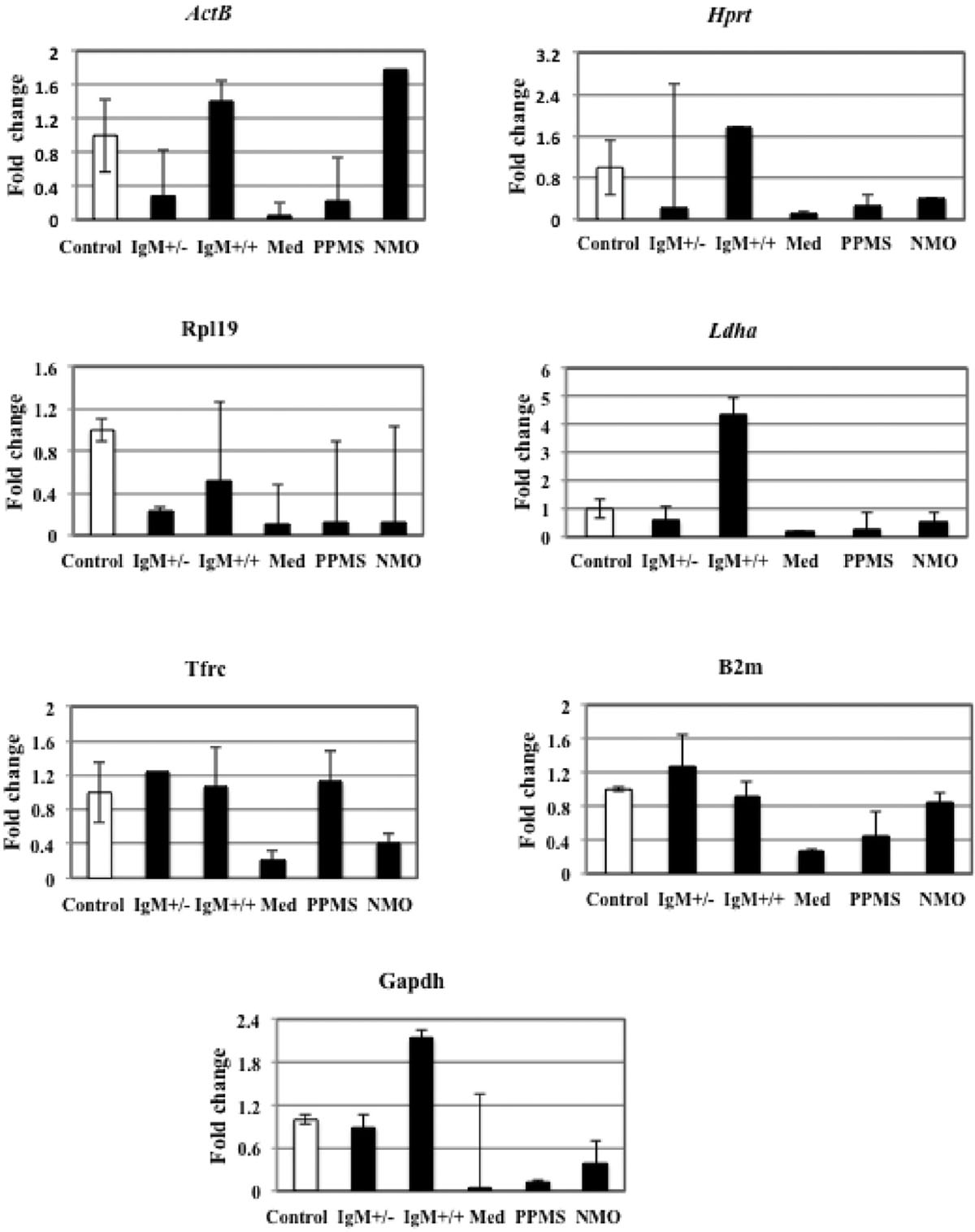
FIGURE 3. Fold change for each reference gene tested in distinct disease courses of multiple sclerosis (MS). IgM+/+ and IgM+/-: Inflammatory forms of relapsing remitting MS; Med: medullary form; NMO: neuromyelitis optica; Control: other non-inflammatory neurological diseases (NIND).
ActB
We found that the expression of ActB gene dropped to 0.2 folds in neurons treated with IgM+/- MS patients and increased again to 1.4 folds in IgM+/+ treated neurons, as compared to control. In neurons treated with medullary CSF the gene expression dropped to 0.04 folds and 0.2 folds in PPMS and increased to 1.78 folds in NMO patients compared to control. Although the variation in the expression level of this gene in all the different experimental conditions is not large as seen by qPCR data, we employed GeNorm software to compare the expression stability of all the reference genes with each other and identify the best reference gene out of a group of commonly used reference genes to avoid getting biased results. The software GeNorm ranked ActB gene as second last unstable gene as compared to the expression levels of other selected reference genes (M value: 2.92 using GeNorm). We conclude that this gene varies in our experimental conditions with respect to other selected reference genes. Hence it should not be used to normalize gene expression data in our experimental conditions.
Hprt
The data indicates that Hprt gene was 0.23 folds downregulated in neurons treated with IgM+/- CSF of MS patients as compared to control. The expression was up regulated 1.7 times in IgM+/+ treated neurons and again down regulated by 0.1 folds, 0.27 folds and 0.4 folds in neurons treated with the CSF of medullary, PPMS and NMO patients as compared to control. According to GeNorm program, Hprt was ranked third last unstable reference genes with respect to other reference genes (Average expression stability value: 1.3).
Rpl19
Rpl19 gene expression was down regulated by 0.2 and 0.5 folds in IgM+/- and IgM+/+ treated neurons as compared to control. There was only 0.1 folds decrease in Rpl19 gene expression when neurons were treated with medullary, PPMS and NMO patients as compared to control. Hence, there is a negligible variation in all the experimental conditions as indicated by qPCR data. In agreement with this data GeNorm identifies this gene as the third most stable gene (M value: 1.19).
Ldha
There was 0.5 folds downregulation of Ldha gene in neurons treated with the CSF of IgM+/- patients with respect to control. The expression level increased up to fourfolds in neurons treated with IgM+/+ treated neurons. In medullary, there was a 0.17 folds decrease in gene expression and we found 0.28 folds and 0.54 folds decrease gene expression in PPMS and NMO patients. The data signifies that the expression of this gene is not constant in all the experimental conditions. The average expression stability (M) value of this gene was 1.25 and was ranked as the fourth stable gene according to GeNorm software.
Tfrc
Tfrc gene was up regulated by 1.2 folds in neurons treated with IgM+/- treated neurons as compared to control. In IgM+/+ treated neurons the expression level almost remained the same as compared to control. In medullary patients the expression was reduced by 0.2 folds and in PPMS treated neurons the level was increased by only 1.1 folds which was almost similar as compared to control. There was a downregulation of this gene by 0.4 folds in neurons treated with the CSF of MS patients. Overall, there was a negligible variation in the gene expression in different experimental conditions. According to the GeNorm algorithm the average expression stability value was 1.092 and it was ranked the best reference gene with respect to others.
B2m
The data indicates that there was 1.2 folds up regulation of B2m gene in IgM+/- treated neurons as compared to control. The expression was down regulated by 0.9 folds in IgM+/+ treated neurons which was not a large variation as compared to control. It dropped to 0.2 folds in medullary treated neurons and 0.4 folds in PPMS treated neurons. The expression decreased by 0.8 folds in NMO treated neurons as compared to control. According to the GeNorm algorithm the average expression stability value was similar to Tfrc average expression stability (M: 1.092) and it was also ranked the best reference gene with respect to others. We conclude that both Tfrc and B2m with similar average expression stability values should be used to normalize gene expression data in our experimental conditions.
Gapdh
The expression level of Gapdh gene was 0.89 folds lower in IgM+/- treated neurons as compared to control. The expression level increased by twofolds in IgM+/+ treated neuons as compared to control. In neurons treated with the CSF of medullary MS patients the gene downregulated by 0.02 folds and by 0.1 fold in neurons treated with the CSF of PPMS patients. Similarly the expression level declined by 0.38 folds in NMO treated patients. According to qPCR data that there is a huge fluctuation in Gapdh gene expression in our experimental conditions. Normally, Gapdh is used as a housekeeping gene but we find that it is not a housekeeping gene in our experimental conditions. GeNorm ranked this gene as the least stable gene with 4.2 as the average expression stability value.
Discussion
Quantitative RT-PCR has recently become the most widely accepted method of quantification for its sensitive, accurate and reliable determination of gene expression levels in cells and tissues. To avoid sample-to-sample variation, normalization of gene transcripts is required. The conventional way to perform normalization is to select a housekeeping gene whose expression is believed to remain stable in all cell types/tissues, during cellular development and under various experimental conditions then relate the expression of gene of interest to that of a housekeeping gene. For many years it has been assumed that the genes such as β-Actin and Gapdh express constitutively in all cells and tissues. β-Actin (ActB) is a cytoskeletal protein that maintains the structure and integrity of cells. GADPH, on the other hand, is a key glycolytic enzyme involved mainly in the production of energy. Since both ActB and Gapdh are involved in maintaining the basic metabolic functions of a cell, they are presumed to express at stable levels. Therefore, they are employed as common internal controls in most of the laboratories. However, several lines of evidence show that their rate of transcription is affected by a variety of factors such as epidermal growth factor, transforming growth factor-β and platelet-derived growth factor while constitutively expressed (Elder et al., 1984; Leof et al., 1986; Keski-Oja et al., 1988). Therefore, their expression may not necessarily be constant in all conditions. Furthermore, GADPH is implicated in non-metabolic processes independent of its metabolic function, such as transcription activation, vesicle transport from endoplasmic reticulum to Golgi apparatus and polymerization of tubulin into microtubules (Kumagai and Sakai, 1983; Durrieu et al., 1987; Muronetz et al., 1994; Zheng et al., 2003; Tisdale and Artalejo, 2007). Previous literature reveals that neuronal apoptosis is associated with suppressed glycolytic activity of GADPH (Burke, 1983; Dastoor and Dreyer, 2001; Makhina et al., 2009). It has been observed that GADPH interacts with other proteins which results in reduced glycolytic activity (Hara et al., 2005). This process may lead to neuroaxonal damage in neurodegenerative diseases such as Huntington’s, Parkinson’s, and Alzheimer’s disease (Vécsei and Pál, 1993; Mazzola and Sirover, 2003; Senatorov et al., 2003; Li et al., 2004; Tsuchiya et al., 2005; Kolln et al., 2010). The realization that these reference genes may fluctuate in different experiments has led to their pre-validation for their expression stability.
This is the first study to the best of our knowledge that reports the most stable HK genes in CGNs treated with CSF from MS/NMO patients. Seven commonly used housekeeping genes were chosen from the available literature. Expression levels of HK genes in different MS clinical forms were quantified by qRT-PCR. Our results reveal that Gapdh expression levels changed in all forms (RRMS, PPMS, NMO) as compared to controls. This gene was not among the best reference genes therefore it is strongly advised not to employ it as a control in studies related to current one. Moreover, β-Actin, that is often used as a loading control also showed unstable expression in all conditions, though to a lesser extent than Gapdh. Transferrin receptor (Tfrc) gene was up regulated by 1.2 folds in neurons treated with IgM+/- treated neurons as compared to control. In IgM+/+ treated neurons the expression level almost remained the same as compared to control. In medullary patients the expression was reduced by 0.2 folds and in PPMS treated neurons the level was increased by only 1.1 folds which was almost similar as compared to control. There was a downregulation of this gene by 0.4 folds in neurons treated with the CSF of MS patients. Overall, we see from the data that there was a negligible variation in the Tfrc gene expression in different experimental conditions. Tfrc is required for iron delivery from transferrin to cells. Microglobulin beta-2 (B2m), a component of MHC class I molecule, showed higher expression in IgM+/- treated neurons as compared to control. The expression was down regulated by 0.9 folds in IgM+/+ treated neurons which was not a large variation as compared to control. It dropped to 0.2 folds in medullary treated neurons and 0.4 folds in PPMS treated neurons. The expression decreased by 0.8 folds in NMO treated neurons as compared to control. According to the GeNorm algorithm the average expression stability value was similar to Tfrc average expression stability (M: 1.092) and it was also ranked the best reference gene with respect to others. We conclude that both Tfrc and B2m with similar average expression stability values should be used to normalize gene expression data in our experimental conditions. Hypoxanthine guanine phosphoribosyl-transferase (Hprt) gene showed fluctuated expression level in different experimental conditions. It plays an important role in purine salvage pathway. According to GeNorm program, Hprt was ranked third last unstable reference genes with respect to other reference genes (Average expression stability value: 1.3). Ribosomal protein L19 (Rpl19) showed negligible down regulation in all the different experimental conditions. GeNorm identifies this gene as the third most stable gene (M value: 1.19). On the contrary, Ldha gene was upregulated in IgM+/+ but down regulated in IgM+/- and medullar clinical form of RRMS. Its expression further lowered in PPMS and NMO. The data signifies that the expression of this gene was not constant in all the experimental conditions and not suitable for normalization of gene transcripts in studies related to the current one.
Overall, geNorm and NormFinder algorithms identified Tfrc and B2m the best housekeeping genes and Gapdh and ActB the most unsuitable genes in our experimental model of MS and therefore the current study demonstrates the necessity for pre-validation of HK genes for any experimental system. Since both the algorithms are based on different mathematical approaches, the order of genes was not exactly similar. However, both geNorm and NormFinder rank the traditional reference genes GAPDH and β-actin as most unstable genes. Therefore, we strongly advise to check the expression stability of these genes before using them for normalization purposes.
We conclude from data provided in this study that transferrin receptor (Tfrc) and microglobulin beta-2 (B2m) as the most stably expressed housekeeping genes in CGNs treated with CSF of MS patients. On the other hand, Gapdh and β-actin showed highly fluctuated expression indicating their unsuitability for such studies. This study demonstrates the usefulness of pre-validating the expression stability of housekeeping genes for normalization of target gene transcripts in gene expression studies. Our data suggest that it is required to determine the suitability of any common HK genes to be used for normalization in “Omic” studies, and even such pre-selection should be a routine step for any experimental system in a laboratory.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Grant sponser: Health Research Fund of the Institute of Health Carlos III. R + D + I 2009–2012; PS09/00976. The authors thank Maria Jose Agullo for technical assistance with primary neuronal cultures, Alberto Hernandez and Eva Lafuente for confocal microscopy. The authors thank Dr. Sanjib Kumar Agarwalla and Institute of Physics, India for providing required software for data analysis.
References
Al-Bader, M. D., and Al-Sarraf, H. A. (2005). Housekeeping gene expression during fetal brain development in the rat—validation by semi-quantitative RT-PCR. Dev. Brain Res. 156, 38–45. doi: 10.1016/j.devbrainres.2005.01.010
Alcazar, A., Regidor, I., Masjuan, J., Salinas, M., and Alvarez-Cermeno, J. C. (2000). Axonal damage induced by cerebrospinal fluid from patients with relapsing-remitting multiple sclerosis. J. Neuroimmunol. 104, 58–67. doi: 10.1016/S0165-5728(99)00225-8
Andersen, C. L., Jensen, J. L., and Orntoft, T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. doi: 10.1158/0008-5472.CAN-04-0496
Becker, K. G., Mattson, D. H., Powers, J. M., Gado, A. M., and Biddison, W. E. (1997). Analysis of a sequenced cDNA library from multiple sclerosis lesions. J. Neuroimmunol. 77, 27–38. doi: 10.1016/S0165-5728(97)00045-3
Biedler, J. L., Helson, L., and Spengler, B. A. (1973). Morphology and growth, tumorigenicity, and cytogenetics of human neuroblastoma cells in continuous culture. Cancer Res. 33, 2643–2652.
Blight, A. R. (2011). Treatment of walking impairment in multiple sclerosis with dalfampridine. Ther. Adv. Neurol. Disord. 4, 99–109. doi: 10.1177/1756285611403960
Bomprezzi, R., Ringner, M., Kim, S., Bittner, M. L., Khan, J., Chen, Y., et al. (2003). Gene expression profile in multiple sclerosis patients and healthy controls: identifying pathways relevant to disease. Hum. Mol. Genet. 12, 2191–2199. doi: 10.1093/hmg/ddg221
Brynedal, B., Khademi, M., Wallström, E., Hillert, J., Olsson, T., and Duvefelt, K. (2010). Gene expression profiling in multiple sclerosis: a disease of the central nervous system, but with relapses triggered in the periphery? Neurobiol. Dis. 37, 613–621. doi: 10.1016/j.nbd.2009.11.014
Burke, D. (1983). Demyelination in optic neuritis and its effects on the visual evoked potential. Aust. J. Ophthalmol. 11, 341–345. doi: 10.1111/j.1442-9071.1983.tb01104.x
Chabas, D., Baranzini, S. E., Mitchell, D., Bernard, C. C., Rittling, S. R., Denhardt, D. T., et al. (2001). The influence of the proinflammatory cytokine, osteopontin, on autoimmune demyelinating disease. Science 294, 1731–1735. doi: 10.1126/science.1062960
Compston, A., and Coles, A. (2008). Multiple sclerosis. Lancet 372, 1502–1517. doi: 10.1016/S0140-6736(08)61620-7
Dastoor, Z., and Dreyer, J. L. (2001). Potential role of nuclear translocation of glyceraldehyde-3-phosphate dehydrogenase in apoptosis and oxidative stress. J. Cell Sci. 114, 1643–1653.
Deindl, E., Boengler, K., van Royen, N., and Schaper, W. (2002). Differential expression of GAPDH and beta3-actin in growing collateral arteries. Mol. Cell. Biochem. 236, 139–146.
Der, S. D., Zhou, A., Williams, B. R. G., and Silverman, R. H. (1998). Identification of genes differentially regulated by interferon α, β, or γ using the oligonucleotide arrays. PNAS 95, 15623–15628. doi: 10.1073/pnas.95.26.15623
Dumont, D., Noben, J. P., Raus, J., Stinissen, P., and Robben, J. (2004). Proteomic analysis of cerebrospinal fluid from multiple sclerosis patients. Proteomics 4, 2117–2124. doi: 10.1002/pmic.200300715
Durrieu, C., Bernier-Valentin, F., and Rousset, B. (1987). Microtubules bind glyceraldehyde 3-phosphate dehydrogenase and modulate its enzyme activity and quaternary structure. Arch. Biochem. Biophys. 252, 32–40. doi: 10.1016/0003-9861(87)90005-1
Elder, P. K., Schmidt, L. J., Ono, T., and Getz, M. J. (1984). Specific stimulation of actin gene transcription by epidermal growth factor and cycloheximide. Proc. Natl. Acad. Sci. U.S.A. 81, 7476–7480. doi: 10.1073/pnas.81.23.7476
Everaert, B. R., Boulet, G. A., Timmermans, J. P., and Vrints, C. J. (2011). Importance of suitable reference gene selection for quantitative real-time PCR: special reference to mouse myocardial infarction studies. PLoS ONE 6:e23793. doi: 10.1371/journal.pone.0023793
Fort, P., Marty, L., Piechaczyk, M., el Sabrouty, S., Dani, C., Jeanteur, P., et al. (1985). Various rat adult tissues express only one major mRNA species from the glyceraldehyde-3-phosphate-dehydrogenase multigenic family. Nucleic Acids Res. 13, 1431–1442. doi: 10.1093/nar/13.5.1431
Glare, E. M., Divjak, M., and Bailey, M. J. (2002). Walters EH. beta-Actin and GAPDH housekeeping gene expression in asthmatic airways is variable and not suitable for normalising mRNA levels. Thorax 57, 765–770. doi: 10.1136/thorax.57.9.765
Gorzelniak, K., Janke, J., Engeli, S., and Sharma, A. M. (2001). Validation of endogenous controls for gene expression studies in human adipocytes and preadipocytes. Horm. Metab. Res. 33, 625–627. doi: 10.1055/s-2001-17911
Gubern, C., Hurtado, O., Rodríguez, R., Morales, J. R., Romera, V. G., Moro, M. A., et al. (2009). Validation of housekeeping genes for quantitative real-time PCR in in-vivo and in-vitro models of cerebral ischaemia. BMC Mol. Biol. 10:57. doi: 10.1186/1471-2199-10-57
Hamalainen, H. K., Tubman, J. C., Vikman, S., Kyrola, T., Ylikoski, E., Warrington, J. A., et al. (2001). Identification and validation of endogenous reference genes for expression profiling of T helper cell differentiation by quantitative real-time RT-PCR. Anal. Biochem. 299, 63–70. doi: 10.1006/abio.2001.5369
Hammack, B. N., Fung, K. Y., Hunsucker, S. W., Duncan, M. W., Burgoon, M. P., Owens, G. P., et al. (2004). Proteomic analysis of multiple sclerosis cerebrospinal fluid. Mult. Scler. 10, 245–260. doi: 10.1191/1352458504ms1023oa
Hara, M. R., Agrawal, N., Kim, S. F., Cascio, M. B., Fujimuro, M., Ozeki, Y., et al. (2005). S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell Biol. 7, 665–674.
Harrison, D. C., Medhurst, A. D., Bond, B. C., Campbell, C. A., Davis, R. P., and Philpott, K. L. (2000). The use of quantitative RT-PCR to measure mRNA expression in a rat model of focal ischemia – caspase-3 as a case study. Brain Res. Mol. Brain Res. 75, 143–149. doi: 10.1016/S0169-328X(99)00305-8
Hong, J., Zang, Y. C., Hutton, G., Rivera, V. M., and Zhang, J. Z. (2004). Gene expression profiling of relevant biomarkers for treatment evaluation in multiple sclerosis. J. Neuroimmunol. 152, 126–139. doi: 10.1016/j.jneuroim.2004.03.004
Iglesias, A. H., Camelo, S., Hwang, D., Villanueva, R., Stephanopoulos, G., and Dangond, F. (2004). Microarray detection of E2F pathway activation and other targets in multiple sclerosis peripheral blood mononuclear cells. J. Neuroimmunol. 150, 163–177. doi: 10.1016/j.jneuroim.2004.01.017
Keski-Oja, J., Raghow, R., Sawdey, M., Loskutoff, D. J., Postlethwaite, A. E., Kang, A. H., et al. (1988). Regulation of mRNAs for type-1 plasminogen activator inhibitor, fibronectin, and type I procollagen by transforming growth factor-beta. Divergent responses in lung fibroblasts and carcinoma cells. J. Biol. Chem. 263, 3111–3115.
Koike, F., Satoh, J., Miyake, S., Yamamoto, T., Kawai, M., Kikuchi, S., et al. (2003). Microarray analysis identifies interferon beta-regulated genes in multiple sclerosis. J. Neuroimmunol. 139, 109–118. doi: 10.1016/S0165-5728(03)00155-3
Kolln, J., Zhang, Y., Thai, G., Demetriou, M., Hermanowicz, N., Duquette, P., et al. (2010). Inhibition of glyceraldehyde-3-phosphate dehydrogenase activity by antibodies present in the cerebrospinal fluid of patients with multiple sclerosis. J. Immunol. 185, 1968–1975. doi: 10.4049/jimmunol.0904083
Kostulas, V. K., Link, H., and Lefvert, A. K. (1987). Oligoclonal IgG bands in cerebrospinal fluid. Principles for demonstration and interpretation based on findings in 1114 neurological patients. Arch. Neurol. 44, 1041–1044.
Kumagai, H., and Sakai, H. (1983). A porcine brain protein (35 K protein) which bundles microtubules and its identification as glyceraldehyde 3-phosphate dehydrogenase. J. Biochem. 93, 1259–1269.
Lennon, V. A., Wingerchuk, D. M., Kryzer, T. J., Pittock, S. J., Lucchinetti, C. F., Fujihara, K., et al. (2004). A serum autoantibody marker of neuromyelitis optica: distinction from multiple sclerosis. Lancet 364, 2106–2112. doi: 10.1016/S0140-6736(04)17551-X
Leof, E. B., Proper, J. A., Getz, M. J., and Moses, H. L. (1986). Transforming growth factor type beta regulation of actin mRNA. J. Cell. Physiol. 127, 83–88. doi: 10.1002/jcp.1041270111
Li, Y., Nowotny, P., Holmans, P., Smemo, S., Kauwe, J. S., Hinrichs, A. L., et al. (2004). Association of late onset Alzheimer’s disease with genetic variation in multiple members of the GAPD gene family. Proc. Natl. Acad. Sci. U.S.A. 101, 15688–15693. doi: 10.1073/pnas.0403535101
Lindberg, R. L., De Groot, C. J., Certa, U., Ravid, R., Hoffmann, F., Kappos, L., et al. (2004). Multiple sclerosis as a generalized CNS disease-comparative microarray analysis of normal appearing white matter and lesions in secondary progressive MS. J. Neuroimmunol. 152, 154–167. doi: 10.1016/j.jneuroim.2004.03.011
Lock, C., Hermans, G., Pedotti, R., Brendolan, A., Schadt, E., Garren, H., et al. (2002). Gene-microarray analysis of multiple sclerosis lesions yields new targets validated in autoimmune encephalomyelitis. Nat. Med. 8, 500–508. doi: 10.1038/nm0502-500
Lublin, F. D., and Reingold, S. C. (1996). Defining the clinical course of multiple sclerosis: results of an international survey. National Multiple Sclerosis Society (USA) advisory committee on clinical trials of new agents in multiple sclerosis. Neurology 46, 907–911. doi: 10.1212/WNL.46.4.907
Makhina, T., Loers, G., Schulze, C., Ueberle, B., Schachner, M., and Kleene, R. (2009). Extracellular GAPDH binds to L1 and enhances neurite outgrowth. Mol. Cell. Neurosci. 41, 206–218. doi: 10.1016/j.mcn.2009.02.010
Mazzola, J. L., and Sirover, M. A. (2003). Subcellular alteration of glyceraldehyde-3-phosphate dehydrogenase in Alzheimer’s disease fibroblasts. J. Neurosci. Res. 71, 279–285. doi: 10.1002/jnr.10484
Medhurst, A. D., Harrison, D. C., Read, S. J., Campbell, C. A., Robbins, M. J., and Pangalos, M. N. (2000). The use of TaqMan RT-PCR assays for semiquantitative analysis of gene expression in CNS tissues and disease models. J. Neurosci. Methods 98, 9–20. doi: 10.1016/S0165-0270(00)00178-3
Minana, R., Sancho-Tello, M., Climent, E., Segui, J. M., Renau-Piqueras, J., and Guerri, C. (1998). Intracellular location, temporal expression, and polysialylation of neural cell adhesion molecule in astrocytes in primary culture. Glia 24, 415–427. doi: 10.1002/(SICI)1098-1136(199812)24:4<415::AID-GLIA7>3.3.CO;2-1
Muronetz, V. I., Wang, Z. X., Keith, T. J., Knull, H. R., and Srivastava, D. K. (1994). Binding constants and stoichiometries of glyceraldehyde 3-phosphate dehydrogenase- tubulin complexes. Arch. Biochem. Biophys. 313, 253–260. doi: 10.1006/abbi.1994.1385
Mycko, M. P., Cwiklinska, H., Szymanski, J., Szymanska, B., Kudla, G., Kilianek, L., et al. (2004). Inducible heat shock protein 70 promotes myelin autoantigen presentation by the HLA class II. J. Immunol. 172, 202–213. doi: 10.4049/jimmunol.172.1.202
Mycko, M. P., Papoian, R., Boschert, U., Raine, C. S., and Selmaj, K. W. (2003). cDNA microarray analysis in multiple sclerosis lesions: detection of genes associated with disease activity. Brain 126(Pt 5), 1048–1057. doi: 10.1093/brain/awg107
Noben, J. P., Dumont, D., Kwasnikowska, N., Verhaert, P., Somers, V., Hupperts, R., et al. (2006). Lumbar cerebrospinal fluid proteome in multiple sclerosis: characterization by ultrafiltration, liquid chromatography, and mass spectrometry. J. Proteome Res. 5, 1647–1657. doi: 10.1021/pr0504788
Ohl, F., Jung, M., Xu, C., Stephan, C., Rabien, A., Burkhardt, M., et al. (2005). Gene expression studies in prostate cancer tissue: which reference gene should be selected for normalization? J. Mol. Med. 83, 1014–1024. doi: 10.1007/s00109-005-0703-z
Oksenberg, J. R., Baranzini, S. E., Sawcer, S., and Hauser, S. L. (2008). The genetics of multiple sclerosis: SNPs to pathways to pathogenesis Nat. Rev. Genet. 9, 516–526. doi: 10.1038/nrg2395
Radonic, A., Thulke, S., Mackay, I. M., Landt, O., Siegert, W., and Nitsche, A. (2004). Guideline to reference gene selection for quantitative realtime PCR. Biochem. Biophys. Res. Commun. 313, 856–862. doi: 10.1016/j.bbrc.2003.11.177
Ramanathan, M., Weinstock-Guttman, B., Nguyen, L. T., Badgett, D., Miller, C., Patrick, K., et al. (2001). In vivo gene expression revealed by cDNA arrays: the pattern in relapsing-remitting multiple sclerosis patients compared with normal subjects. J. Neuroimmunol. 116, 213–219. doi: 10.1016/S0165-5728(01)00308-3
Rossi, S., Furlan, R., De Chiara, V., Motta, C., Studer, V., Mori, F., et al. (2012). Interleukin-1beta causes synaptic hyperexcitability in multiple sclerosis. Ann. Neurol. 71, 76–83. doi: 10.1002/ana.22512
Rossi, S., Motta, C., Studer, V., Barbieri, F., Buttari, F., Bergami, A., et al. (2014). Tumor necrosis factor is elevated in progressive multiple sclerosis and causes excitotoxic neurodegeneration. Mult. Scler. 20, 304–312. doi: 10.1177/1352458513498128
Satoh, J., Nakanishi, M., Koike, F., Onoue, H., Aranami, T., Yamamoto, T., et al. (2006). T cell gene expression profiling identifies distinct subgroups of Japanese multiple sclerosis patients. J. Neuroimmunol. 174, 108–118. doi: 10.1016/j.jneuroim.2006.02.004
Senatorov, V. V., Charles, V., Reddy, P. H., Tagle, D. A., and Chuang, D. M. (2003). Overexpression and nuclear accumulation of glyceraldehyde-3-phosphate dehydrogenase in a transgenic mouse model of Huntington’s disease. Mol. Cell. Neurosci. 22, 285–297. doi: 10.1016/S1044-7431(02)00013-1
Sharief, M. K., and Thompson, E. J. (1991). Intrathecal immunoglobulin M synthesis in multiple sclerosis. Brain 114, 181–195.
Sturzebecher, S., Wandinger, K. P., Rosenwald, A., Sathyamoorthy, M., Tzou, A., Mattar, P., et al. (2003). Expression profiling identifies responder and non-responder phenotypes to interferon-beta in multiple sclerosis. Brain 126(Pt 6), 1419–1429. doi: 10.1093/brain/awg147
Stürzenbaum, S. R., and Kille, P. (2001). Control genes in quantitative molecular biological techniques: the variability of invariance. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 130, 281–289. doi: 10.1016/S1096-4959(01)00440-7
Tajouri, L., Mellick, A. S., Ashton, K. J., Tannenberg, A. E., Nagra, R. M., Tourtellotte, W. W., et al. (2003). Quantitative and qualitative changes in gene expression patterns characterize the activity of plaques in multiple sclerosis. Brain Res. Mol. Brain Res. 119, 170–183. doi: 10.1016/j.molbrainres.2003.09.008
Tisdale, E. J., and Artalejo, C. R. (2007). A GAPDH mutant defective in Src-dependent tyrosine phosphorylation impedes Rab2-mediated events. Traffic 8, 733–741. doi: 10.1111/j.1600-0854.2007.00569.x
Toegel, S., Huang, W., Piana, C., Unger, F. M., Wirth, M., Goldring, M. B., et al. (2007). Selection of reliable reference genes for qPCR studies on chondroprotective action. BMC Mol. Biol. 8:13. doi: 10.1186/1471-2199-8-13
Torres, J. M., Gomez-Capilla, J. A., Ruiz, E., and Ortega, E. (2003). Semiquantitative RT-PCR method coupled to capillary electrophoresis to study 5alpha-reductase mRNA isozymes in rat ventral prostate in different androgen status. Mol. Cell. Biochem. 250, 125–130. doi: 10.1023/A:1024902419502
Tricarico, C., Pinzani, P., Bianchi, S., Paglierani, M., Distante, V., Pazzagli, M., et al. (2002). Quantitative real-time reverse transcription polymerase chain reaction: normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal. Biochem. 309, 293–300. doi: 10.1016/S0003-2697(02)00311-1
Tsuchiya, K., Tajima, H., Kuwae, T., Takeshima, T., Nakano, T., Tanaka, M., et al. (2005). Pro-apoptotic protein glyceraldehyde-3-phosphate dehydrogenase promotes the formation of Lewy body-like inclusions. Eur. J. Neurosci. 21, 317–326. doi: 10.1111/j.1460-9568.2005.03870.x
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034
Vécsei, L., and Pál, E. (1993). [Current data on the pathogenesis of neurodegenerative diseases and some muscular disorders: therapeutic prospectives]. Orv. Hetil. 134, 1683–1687.
Vidaurre, O. G., Haines, J. D., Sand, I. K., Adula, K. P., Huynh, J. L., McGraw, C. A., et al. (2014). Cerebrospinal fluid ceramides from patients with multiple sclerosis impair neuronal bioenergetics. Brain 137(Pt 8), 2271–2286. doi: 10.1093/brain/awu139
Villar, L. M., González-Porqué, P., Masjuán, J., Alvarez-Cermeño, J. C., Bootello, A., and Keir, G. (2001). A sensitive and reproducible method for the detection of oligoclonal IgM bands. J. Immunol. Methods 258, 151–155. doi: 10.1016/S0022-1759(01)00492-6
Wandinger, K. P., Sturzebecher, C. S., Bielekova, B., Detore, G., Rosenwald, A., Staudt, L. M., et al. (2001). Complex immunomodulatory effects of interferon-beta in multiple sclerosis include the upregulation of T helper 1-associated marker genes. Ann. Neurol. 50, 349–357. doi: 10.1002/ana.1096
Whitney, L. W., Becker, K. G., Tresser, N. J., Caballero-Ramos, C. I., Munson, P. J., Prabhu, V. V., et al. (1999). Analysis of gene expression in mutiple sclerosis lesions using cDNA microarrays. Ann. Neurol. 46, 425–428. doi: 10.1002/1531-8249(199909)46:3<425::AID-ANA22>3.0.CO;2-O
Whitney, L. W., Ludwin, S. K., McFarland, H. F., and Biddison, W. E. (2001). Microarray analysis of gene expression in multiple sclerosis and EAE identifies 5-lipoxygenase as a component of inflammatory lesions. J. Neuroimmunol. 121, 40–48. doi: 10.1016/S0165-5728(01)00438-6
Wingerchuk, D. M., Lennon, V. A., Pittock, S. J., Lucchinetti, C. F., and Weinshenker, B. G. (2006). Revised diagnostic criteria for neuromyelitis optica. Neurology 66, 1485–1489. doi: 10.1212/01.wnl.0000216139.44259.74
Xiao, B. G., Zhang, G. X., Ma, C. G., and Link, H. (1996). The cerebrospinal fluid from patients with multiple sclerosis promotes neuronal and oligodendrocyte damage by delayed production of nitric oxide in vitro. J. Neurol. Sci. 142, 114–120. doi: 10.1016/0022-510X(96)00164-5
Yurube, T., Takada, T., Hirata, H., Kakutani, K., Maeno, K., Zhang, Z., et al. (2011). Modified house-keeping gene expression in a rat tail compression loading-induced disc degeneration model. J. Orthop. Res. 29, 1284–1290. doi: 10.1002/jor.21406
Zheng, L., Roeder, R. G., and Luo, Y. (2003). S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell 114, 255–266. doi: 10.1016/S0092-8674(03)00552-X
Zhong, H., and Simons, J. W. (1999). Direct comparison of GAPDH, beta-actin, cyclophilin, and 28S rRNA as internal standards for quantifying RNA levels under hypoxia. Biochem. Biophys. Res. Commun. 259, 523–526. doi: 10.1006/bbrc.1999.0815
Keywords: housekeeping genes, multiple sclerosis, normalization, GeNorm, NormFinder
Citation: Mathur D, Urena-Peralta JR, Lopez-Rodas G, Casanova B, Coret-Ferrer F and Burgal-Marti M (2015) Bypassing hazard of housekeeping genes: their evaluation in rat granule neurons treated with cerebrospinal fluid of multiple sclerosis subjects. Front. Cell. Neurosci. 9:375. doi: 10.3389/fncel.2015.00375
Received: 17 July 2015; Accepted: 07 September 2015;
Published: 23 September 2015.
Edited by:
Rosanna Parlato, Ulm University, GermanyReviewed by:
Robert Weissert, University of Regensburg, GermanySilvia Zucchelli, University of Eastern Piedmont, Italy
Copyright © 2015 Mathur, Urena-Peralta, Lopez-Rodas, Casanova, Coret-Ferrer and Burgal-Marti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gerardo Lopez-Rodas, Department of Biochemistry and Molecular Biology, University of Valencia and INCLIVA Biomedical Research Institute, Valencia 46100, Spain, gerardo.lopez@uv.es
 Deepali Mathur
Deepali Mathur Juan R. Urena-Peralta
Juan R. Urena-Peralta Gerardo Lopez-Rodas
Gerardo Lopez-Rodas Bonaventura Casanova
Bonaventura Casanova Francisco Coret-Ferrer5
Francisco Coret-Ferrer5  Maria Burgal-Marti
Maria Burgal-Marti