Control of Sleep by Dopaminergic Inputs to the Drosophila Mushroom Body
- 1Department of Cellular and Molecular Physiology, Yale University School of Medicine, New Haven, CT, USA
- 2Janelia Research Campus, Howard Hughes Medical Institute, Ashburn, VA, USA
- 3Department of Genetics, Yale University School of Medicine, New Haven, CT, USA
- 4Program in Cellular Neuroscience, Neurodegeneration and Repair, Yale University School of Medicine, New Haven, CT, USA
The Drosophila mushroom body (MB) is an associative learning network that is important for the control of sleep. We have recently identified particular intrinsic MB Kenyon cell (KC) classes that regulate sleep through synaptic activation of particular MB output neurons (MBONs) whose axons convey sleep control signals out of the MB to downstream target regions. Specifically, we found that sleep-promoting KCs increase sleep by preferentially activating cholinergic sleep-promoting MBONs, while wake-promoting KCs decrease sleep by preferentially activating glutamatergic wake-promoting MBONs. Here we use a combination of genetic and physiological approaches to identify wake-promoting dopaminergic neurons (DANs) that innervate the MB, and show that they activate wake-promoting MBONs. These studies reveal a dopaminergic sleep control mechanism that likely operates by modulation of KC-MBON microcircuits.
Introduction
Parkinson's disease in human patients is strongly associated with disruption of sleep (Schrempf et al., 2014). Furthermore, the ascending dopamine system that projects from the brain stem to the forebrain and, specifically, to cerebral cortex is known to be important for regulating sleep and arousal, and is considered a wake-promoting pathway (Dzirasa et al., 2006; España and Scammell, 2011; Dauvilliers et al., 2015). However, despite this general knowledge concerning dopaminergic regulation of mammalian sleep through projections to cerebral cortex, underlying cellular, and molecular mechanisms remain poorly understood. In part, this is because of a lack of effective tools for highly restricted cell-specific experimental control of ascending dopaminergic systems in mammalian model systems such as rodents. We thus employed Drosophila melanogaster as a convenient experimentally tractable model system for studying cellular and molecular mechanisms of dopaminergic regulation of sleep.
The Drosophila mushroom body (MB) is an associative memory network of evolutionary origin likely conserved with mammalian cerebral cortex that has also been implicated in the control of sleep (Heisenberg et al., 1985; Zars et al., 2000; Joiner et al., 2006; Pitman et al., 2006; Yuan et al., 2006; Tomer et al., 2010; Guo et al., 2011; Seugnet et al., 2011; Yi et al., 2013; Ueno and Kume, 2014; Wu et al., 2014; Haynes et al., 2015). Fly sleep exhibits all of the key features of vertebrate sleep, including circadian regulation and homestatic rebound, and can be operationally identified as periods of locomotor quiescence lasting 5 min or longer (Hendricks et al., 2000; Shaw et al., 2000). The MB contains ~2000 KCs divided into 7 anatomical classes based on the specific projections of their axons into the α/β, α′/β′, or γ lobes (Tanaka et al., 2008). KCs receive synaptic inputs from sensory systems, such as olfactory projection neurons. KCs synapse convergently onto 34 MBONs of 21 cell types, whose dendrites contiguously tile the MB lobes, defining 15 non-overlapping compartments (Tanaka et al., 2008; Aso et al., 2014a). MBON cell types are uniquely named by their stereotyped arborizations in these compartments, numbered consecutively within each lobe starting from the intersection of the horizontal and vertical lobes (Aso et al., 2014a).
These 15 compartments are also each innervated by MB dopaminergic neurons (DANs). The axon terminals of ~130 DANs divided into 20 cell types tile the lobes isomorphically to the MBON dendrites (Aso et al., 2014a). While previous studies have implicated dopamine signaling (Kume et al., 2005) and several classes of DANs in the control of fly sleep (Shang et al., 2011; Liu et al., 2012b; Ueno et al., 2012), none of them innervate the MB, although it has recently been shown that manipulation of dopamine transporter expression using broadly expressed MB drivers influences sleep (Ueno and Kume, 2014).
To comprehensively identify the KCs, MBONs, and MB DANs that control sleep, we performed a thermogenetic neuronal activation screen using a new library of intersectional “split-GAL4” fly lines, each of which drives expression of an effector transgene under the control of the upstream activating sequence (UAS) to defined MB cell classes (Luan et al., 2006; Pfeiffer et al., 2010; Aso et al., 2014a,b; Sitaraman et al., 2015). We identified several classes of sleep-controlling MBONs with dendrites in distinct lobe compartments: cholinergic sleep-promoting MBON-γ2α′1, and glutamatergic wake-promoting MBON-γ5β′2a/β′2mp/β′2mp_bilateral and MBON-γ4 > γ1γ2 (Aso et al., 2014a,b). We also determined that α′/β′ and γm KCs are wake-promoting and γd KCs are sleep-promoting, and that α′/β′ and γm KCs promote wake by activating MBON-γ5β′2a/β′2mp/β′2mp_bilateral and γd KCs promote sleep by activating MBON-γ2α′1 (Sitaraman et al., 2015). Finally, we demonstrated that sleep deprivation activates γd KCs and MBON-γ2α′1 and activation of this synaptic microcircuit is required for homeostatic rebound sleep to occur after sleep deprivation (Sitaraman et al., 2015). Here we report the results of our screen of the MB DAN cell types, revealing that DANs projecting to lobe compartments containing the dendrites of wake-promoting MBONs are wake-promoting. We also demonstrate that activation of DANs projecting to the γ5 and β′2 compartments induces greater activation of MBON-γ5β′2a/β′2mp/β′2mp_bilateral than MBON-γ2α′1. These studies reveal a previously unknown dopaminergic sleep control mechanism that functions by modulation of the mushroom bodies, and provides for the first time a cellular and molecular substrate for control of sleep by dopaminergic modulation of an associative network.
Experimental Procedures
Molecular and Genetic Methods
Genomic enhancers used in split-GAL4 and LexA transgenic lines were selected based on expression patterns of GAL4 lines using those same enhancers (Jenett et al., 2012) and constructed as previously described (Pfeiffer et al., 2010).
Fly Stocks
Flies were maintained on conventional cornmeal-agar-dextrose medium in 12 h light: 12 h dark conditions at 21°C (for dTRPA1 experiments) or 25°C (for immunohistochemistry), with ambient humidity of 60–70%. Flies were collected and tested for sleep 3–7 days after eclosion. R58E02-LexA is described in Jenett et al. (2012) and Liu et al. (2012a). Detailed genotype information of split-GAL4 strains—including p65ADZp-ZpGAL4DBD combinations—is as described (Aso et al., 2014a). p65ADZp fragments were inserted at attP40 or VK00027 while ZpGAL4DBD fragments were inserted at attP2. The studies reported here are exempt from ethical review, as they involve only Drosophila flies, and involve no vertebrate animals or human subjects. All MB split-GAL4 fly stocks can be obtained from Janelia Research Campus using the contact information at http://splitgal4.janelia.org/cgi-bin/splitgal4.cgi.
Sleep Assays
Split-GAL4 flies were crossed to flies bearing 10X UAS-dTRPA1 (in attP16) (Hamada et al., 2008) and maintained at 21–22°C. Genomic sites were chosen to avoid transvection, as described (Mellert and Truman, 2012). Male progeny, 3–7 days post-eclosion, were placed in 65 mm × 5 mm transparent plastic tubes with standard cornmeal dextrose agar media, placed in a Drosophila Activity Monitoring system (Trikinetics), and locomotor activity data were collected in 1 min bins. Activity monitors were maintained in a 12 h:12 h light-dark cycle at 65% relative humidity. Total 24-h sleep quantity (daytime plus nighttime sleep) was extracted from locomotor activity data as described (Donelson et al., 2012): sleep is defined as a contiguous period of inactivity lasting 5 min or more (Hendricks et al., 2000; Shaw et al., 2000). Sleep profiles were generated depicting average sleep (minutes per 30 min) for day 1 (baseline), days 2 and 3 (activation), and day 4 (recovery). In addition to permissive temperature controls other genotypic controls were used for hit detection as indicated. For all screen hits, waking activity was calculated as the number of beam crossings/min when the fly was awake. Temperature changes to activate and silence neurons are as indicated in Figures 2, 3. Statistical comparisons between experimental and control genotypes were performed using Prism (Graphpad Inc, CA) by Kruskal-Wallis One-Way ANOVA followed by Dunn's post-hoc test or One-Way ANOVA followed by Dunnett's pairwise comparison test.
Stimulation of PAM Neurons by ATP/P2X2 and Simultaneous GCamp6m Imaging of MBONs
LexAop2-dsRed in attp18 (X); LexAop2-P2X2 in su(Hw)attp5(II); UAS-GCaMP6m in VK0005(III) flies were generated using standard molecular and genetic methods, with the original transgenes as described (Pfeiffer et al., 2008, 2010; Yao et al., 2012; Chen et al., 2013). These flies were crossed to flies bearing appropriate LexA and split-GAL4 driver transgenes. To gain access to the PAM neurons for ATP application whole brain explants were placed on 8 mm diameter coverslips and placed in a recording chamber containing external solution (103 mM NaCl, 3 mM KCl, 5 mM N-tris (hydroxymethyl) methyl-2-aminoethane-sulfonic acid, 8 mM trehalose, 10 mM glucose, 26 mM NaHCO3, 1 mM NaH2PO4, 2 mM CaCl2, and 4 mM MgCl2, pH 7.4). For D1 dopamine receptor antagonist experiments, 100 μM SCH23390 was added to the external solution. The ATP ejection electrode was filled with freshly prepared 10 mM ATP solution and placed close to the PAM neurons using a micromanipulator. The PAM neurons were visually identified by their dsRed fluorescence. ATP was ejected by applying a 25 ms pressure pulse at 20 psi using a picospritzer (Parker Hannifin, Precision Fluidics Division, NH). The picospritizer was triggered by Zeiss image acquisition and processing software Zen pro 2012. Calcium imaging was performed with a Zeiss Axio Examiner Z1 upright microscope with W Plan Apochromat 40× water immersion objective. GCaMP6m was excited with a 470 nm LED light source (Colibiri, Zeiss) and images were acquired using ORCA-R2 C10600-10B digital CCD camera (Hamamastu, Japan) at 3–10 Hz. For simultaneous imaging of spatially identical frames of dsRed (KCs) and GCaMP6m (MBONs) fluorescence we used a DV2 emission splitting system (Photometrics Inc). As a negative control, we imaged MBONs in brains of flies that lacked P2X2 expression in the PAM neurons and confirmed absence of Ca2+ increases in MBONs following ATP application (data not shown). The average fluorescence of all pixels for each time point in a defined ROI was subtracted from the average background fluorescence of an identically sized ROI elsewhere within the brain. The resulting pixel fluorescence value for each time point was defined as Ft. Changes in fluorescence were computed as %ΔF/Fo = ((Ft-Fo)/Fo) × 100, where Fo is defined as the average background-subtracted baseline fluorescence for the 10 frames preceding ATP application. All images were processed and quantified using Zen and Fiji (Image J).
Immunohistochemistry
Dissection and immunohistochemistry of fly brains were performed as previously described with minor modifications (Jenett et al., 2012). Brains of 3–10 day old female flies were dissected in Schneider's Insect medium and fixed in 2% paraformaldehyde in Schneider's medium for 50 min at room temperature (RT). After washing in PBT (0.5% Triton X-100 in PBS), brains were blocked in 3% normal goat serum (or normal donkey serum, depending on the secondary antibody) for 90 min. Brains were then incubated in primary antibodies diluted in PBT for 2–4 days on a nutator at 4°C, washed three times in PBT for 30 min or longer, then incubated in secondary antibodies diluted in PBT for 2–4 days on a nutator at 4°C. Brains were washed thoroughly in PBT four times for 30 min or longer, and mounted in Vectashield (Vector laboratories, CA) for imaging. The following antibodies were used: anti-GFP (1:1000; Invitrogen; #AB124), mouse anti-nc82 (1:50; Developmental Studies Hybridoma Bank, Univ. Iowa), and cross-adsorbed secondary antibodies to IgG (H+L): goat Alexa Fluor 488 anti-rabbit (1:800; Invitrogen A11034) and goat Alexa Fluor 568 (1:400; Invitrogen A11031).
Results
Screen for MB DANs Regulating Sleep
The MB DANs are coarsely divided into PAM and PPL1 clusters based on the positions of their cell bodies (Figure 1A, Figure S1) (Budnik and White, 1988; Nassel and Elekes, 1992; Tanaka et al., 2008). Transient thermogenetic activation using the dTRPA1 temperature-gated depolarizing cation channel (Hamada et al., 2008) of some classes of PAM and PPL1 DANs suppresses sleep, in some cases very strongly (Figure 1B), and this effect is not attributable to non-specific effects on locomotion (Figure 1C). Sleep plots for MB DANs that innervate the γ5 compartment—which is also innervated by the dendrites of wake-promoting MBON-γ5β′2a/β′2mp/β′2mp_bilateral—are shown and quantified in Figures 2A,B. Lobe compartment innervation patterns of the DANs targeted by each split-GAL4 line are as previously described (Aso et al., 2014a; Yamagata et al., 2015), with images of selected lines shown in Figure 2F. Wake-promoting DANs project to various compartments in the γ and α′/β′ lobes, including γ5 and β′2, which contain dendrites of wake-promoting MBON-γ5β′2a/β′2mp/β′2mp_bilateral. None of the MB DANs increase sleep when activated, consistent with previous studies suggesting that dopamine is exclusively wake-promoting in flies (Liu et al., 2012b; Ueno et al., 2012; Ueno and Kume, 2014).
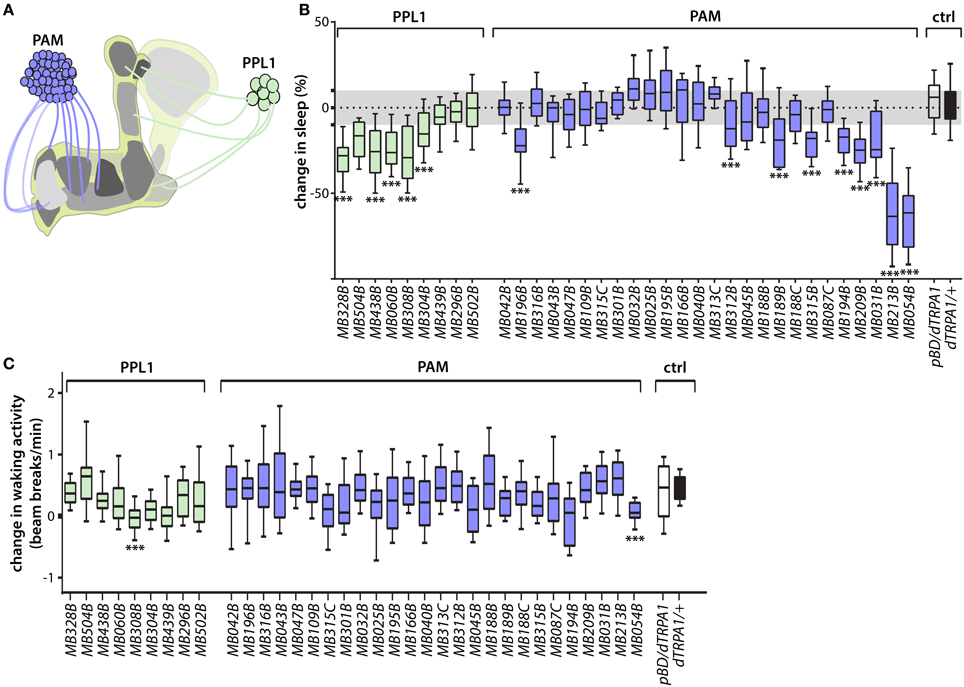
Figure 1. Identification of wake-promoting MB dopamine neurons. (A) Schematic representation of MB showing lobe-specific axon projections of PPL1 and PAM cluster dopamine neurons. (B) Change in sleep of male flies induced by activation of MB dopamine neuron subsets targeted by the indicated split-GAL4 driver lines to express dTRPA1 temperature-gated depolarizing cation channel. Percent change in sleep is defined as (sleep on day 1—sleep on day 2)/sleep on day 1, where the ambient temperature is increased on day 2 from 21.5°C to 28.5°C to activate the neurons expressing dTRPA1. Activation of various PAM and PPL1 dopamine neurons projecting to α′/β′ or γ lobes increases wake. Midline, box boundaries, and whiskers represent median, quartiles, and 10th and 90th percentiles, respectively. split-GAL4 lines are grouped by dopamine neuron cell cluster. Each split-GAL4 line was compared to an enhancerless GAL4 (pBDPGAL4U, indicated in the figures as pBDG4U or pBD) by Kruskal-Wallis non-parametric One-Way ANOVA and Dunn's post-hoc correction for multiple comparisons (***p < 0.001; n = 24–68 flies per genotype). (C) Change in waking locomotor activity of male flies induced by dTRPA1-mediated activation of DAN classes targeted by the indicated split-GAL4 driver lines. Change in activity is defined as the difference between average beam breaks/min during periods of wakefulness on day 2 (28.5°C) and day 1 (21.5°C) of the experiments. Strongly wake-promoting lines (MB308B, MB054B) exhibit significant decreases in waking locomotor activity. Midline, box boundaries, and whiskers represent median, quartiles, and 10th and 90th percentiles, respectively. Statistical comparison of each split-GAL4 line to an enhancerless GAL4 (pBDG4U) is by Kruskal-Wallis One-Way ANOVA and Dunnett's post-hoc correction for multiple comparisons (n = 24–68 flies for each genotype).
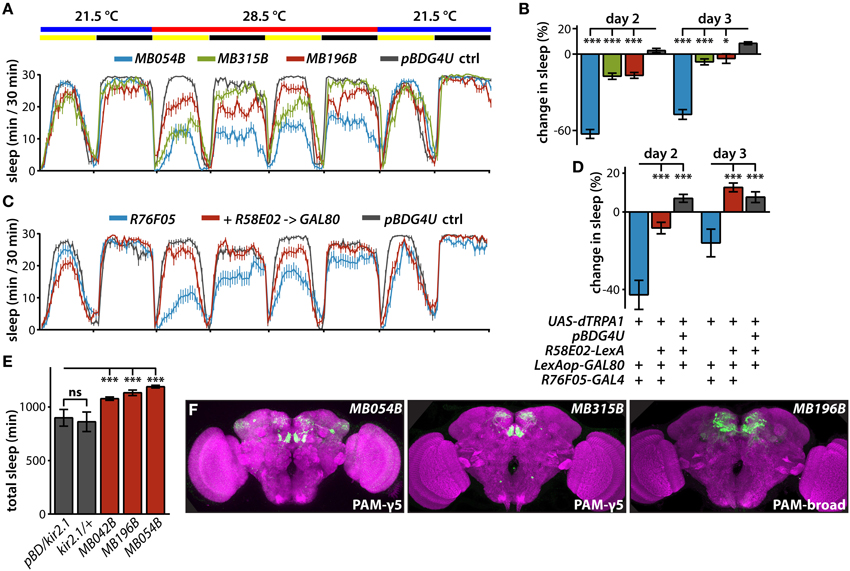
Figure 2. Sleep profiles of flies following thermogenetic activation of MB dopamine neurons. (A) Sleep profiles of male flies expressing dTRPA1 in PAM neurons driven by the indicated split-GAL4 lines (mean ± sem). Flies were maintained in 12 h:12 h light:dark (LD) conditions (indicated by the yellow and black bars, respectively), and the temperature was increased for days 2 and 3 of the 4 day experiment. DANs targeted by MB054B innervate the γ3 and γ5 compartments, MB315B the γ5 compartment, and MB196B multiple compartments. (B) Quantification of change in sleep of male flies expressing dTRPA1 in PAM neurons. Genotypes are color coded as in (A). Statistical analysis was by One-Way ANOVA and Dunnett's paired-comparison test. (C) Sleep profiles of male flies expressing dTRPA1 in PAM neurons driven by R76F05-GAL4 driver, with and without co-expression in PAM neurons of the GAL80 inhibitor of GAL4-mediated transcription driven by the R58E02-LexA driver, treated as in (A). (D) Co-expression of GAL80 in PAM neurons strongly suppresses wake induced by GAL4-mediated expression of dTRPA1. Genotypes are color coded as in (C); quantification and statistical analysis as in (B); n = 26–29 flies per genotype. (E) Expression of the hyperpolarizing kir2.1 K+ channel driven in PAM neurons by the indicated split-GAL4 lines increases total 1-day sleep. Quantification and statistical analysis as in (B); n = 30–32 flies per genotype. (F) Whole brains of flies expressing GFP under the control of indicated splitGAL4 drivers immunostained for GFP (green) and the synaptic neuropil marker BRUCHPILOT (purple). *p < 0.05; ***p < 0.001.
Expressing the GAL4 inhibitor, GAL80, with the R58E02-LexA driver suppresses the wake-promoting effect of dTRPA1-mediated activation using the R76F05-GAL4 driver (Figures 2C,D), and these drivers overlap only in MB PAM neurons (Jenett et al., 2012). Furthermore, silencing neuronal activity of various PAM DANs by expression of the hyperpolarizing kir2.1 K+ channel (Baines et al., 2001; Nitabach et al., 2002) increases sleep (Figure 2E). Interestingly, while thermogenetic activation using the MB042B driver does not affect sleep, inactivation by kir2.1 expression using this driver increases sleep. While the reason for this is unclear, it could reflect different relative levels of GAL4-mediated transcriptional activation in the various PAM subsets targeted by this broad PAM driver. Regardless, these experiments further support a role for MB PAM neurons in the control of sleep.
In addition to wake-promoting MBONs whose dendrites innervate γ5 and β′2 compartments, MBON-γ4>γ1γ2 is also wake-promoting (Aso et al., 2014b). Interestingly, transient activation of PAM neurons that project solely to the γ4 compartment containing dendrites of MBON-γ4 > γ1γ2 using the MB312B split-GAL4 suppresses sleep (Figures 3A–C). In some cases, thermogenetic activation using split-GAL4 lines targeting the same classes of MB DANs do not induce the same sleep phenotypes. For example, while MB315B, MB315C, and MB313C split-GAL4 lines each target PAM DANs projecting to the γ5 compartment, only MB315B suppresses sleep when used for thermogenetic activation (Figure 1B). Also, while MB439B and MB296B each target PPL1 DANs projecting to the γ2 and α′1 compartments innervated by sleep-promoting MBON-γ2α′1, thermogenetic activation using these split-GAL4 lines does not affect sleep (Figure 1B). These differences could be due to different levels of GAL4 expression in different split-GAL4 lines, and consequent differences in dTRPA1 expression, leading to differences in the degree of neural activation. Furthermore, there are some PPL1 lines that suppress sleep when used for thermogenetic activation (Figure 1B), while activation of the MBONs innervating the same compartments does not (Aso et al., 2014b). This suggests that strong thermogenetic activation could induce spillover of dopamine across compartments (which are not isolated from one another by glia), while endogenous dopamine secretion could be weaker and thus more specific. Future studies following up on additional specific sleep-regulating MB DAN classes is likely to resolve this question raised by the comprehensive screen we report here.
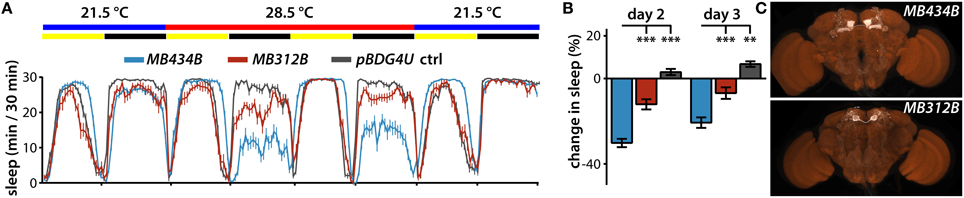
Figure 3. Regulation of sleep by MBONs and DANs projecting to the γ4 compartment. (A) Sleep profiles of male flies expressing dTRPA1 in MBON-γ4 > γ1γ2 using MB434B split-GAL4 driver (n = 32 flies) or in PAM DANs projecting to the γ4 compartment using MB312B (n = 31 flies). (B) Quantification of change in sleep of male flies expressing dTRPA1 in MBON-γ4 > γ1γ2 using MB434B split-GAL4 driver (n = 32 flies) or in PAM DANs projecting to the γ4 compartment using MB312B (n = 31 flies). Genotypes are color coded as in (A). Statistical analysis was by One-Way ANOVA and Dunnett's paired-comparison test. (C) Whole brains of flies expressing GFP under the control of indicated splitGAL4 drivers immunostained for GFP (white) and the synaptic neuropil marker BRUCHPILOT (brown). **p < 0.01; ***p < 0.001.
MB DANs Activate Wake-promoting MBONs
To test the hypothesis that MB DANs promote wake by increasing activation of the wake-promoting γ5β′2 microcircuit, we activated P2X2-expressing MB DANs and measured Ca2+ responses in MBON-γ5β′2a/β′2mp/β′2mp_bilateral (Figure 4A). Because of the lack of available LexA drivers specific for DANs projecting to γ5 or β′2, we drove P2X2 expression using the relatively broad R58E02-LexA PAM driver that projects to the γ5 and β′2 compartments, among others, but is not active in non-PAM neurons (Liu et al., 2012a). Consistent with our hypothesis, transient activation of PAM DANs induces robust transient Ca2+ increases in the dendrites of MBON-γ5β′2a/β′2mp/β′2mp_bilateral in both the γ5 and β′2 compartments (Figures 4B,C). Complete block by bath application of the competitive D1-specific dopamine receptor antagonist SCH23390 at 100 μM (Boto et al., 2014) establishes the underlying molecular basis for this response, and confirms that it is mediated by dopamine acting on D1 receptors (Figure 4B).
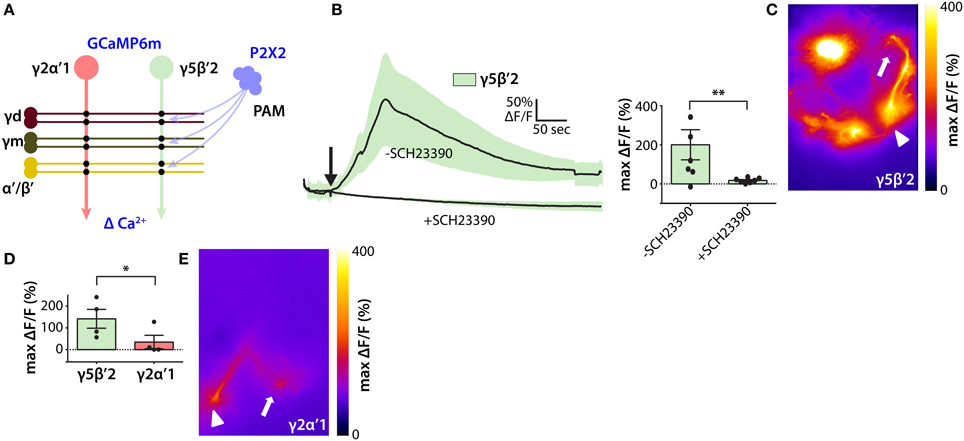
Figure 4. Functional inputs of PAM dopamine neurons to the wake-promoting MB microcircuit. (A) Schematic of experimental design to probe functional connections between wake-promoting PAM neurons and sleep-regulating MBONs. P2X2 ATP-gated depolarizing cation channel was expressed in PAM dopamine neurons, and GCaMP6m fluorescent Ca2+ indicator in MBONs. (B) Ca2+ responses of GCaMP6m-expressing γ5β′2-MBONs following ATP-mediated activation of P2X2 expressed in PAM neurons using R58E02-LexA. Line graph depicts ΔF/F time course in the presence and absence of dopamine receptor D1 subtype blocker SCH23390 (mean ± sem); bar graph depicts peak ΔF/F (mean ± sem). Statistical comparison of peak responses was by unpaired t-test (n = 6 brains per condition). (C) Heat map showing peak Ca2+ response of representative GCaMP6m-expressing γ5β′2-MBONs following ATP-mediated PAM activation. Arrowhead indicates MBON dendrites and arrow indicates axon terminals; panel width is 50 μm. (D) Peak Ca2+ responses of GCaMP6m-expressing γ2α′1 and γ5β′2-MBONs after ATP-mediated activation of P2X2-expressing PAM neurons. γ5β′2-MBONs respond more strongly than γ2α′1 to PAM activation. Quantification and statistical analysis as in (B) (n = 4 brains per genotype). (E) Heat map showing peak Ca2+ response of GCaMP6m-expressing MBON-γ2α′1 following ATP-mediated PAM activation. Symbols as in (C); panel width is 50 μm. *p < 0.05; **p < 0.01.
Since PAM DANs do not project to the γ2 or α′1 compartments (Aso et al., 2014a) we assessed the compartmental specificity of dopamine secretion into the MB by comparing Ca2+ responses of MBON-γ5β′2a/β′2mp/β′2mp_bilateral and MBON-γ2α′1 to PAM DAN activation. Consistent with compartmental specificity, transient activation of PAM DANs induces significantly greater Ca2+ increases in MBON-γ5β′2a/β′2mp/β′2mp_bilateral than in MBON-γ2α′1 (Figures 4D,E). Taken together, our results support the hypothesis that MB DANs promote wake by activating γ5β′2 and γ4 > γ1γ2 wake-promoting MB microcircuits. Note, however, that our results do not exclude an indirect pathway whereby dopamine secreted by sleep-regulating MB DANs activates sleep-regulating MBONs by activating D1 receptors in other neurons presynaptic to MBONs, specifically KCs. Future studies, such as the suppression of D1 receptor expression specifically in MBONs using UAS-RNAi hairpin transgenes, are likely to resolve this question.
Discussion
We have used a combination of sophisticated cell-specific genetic manipulations with behavioral sleep analysis and optical electrophysiology to identify compartment-specific wake-promoting MB DANs that activate wake-promoting microcircuits. Previous studies have implicated DANs innervating the central complex (CX)—a brain region involved in locomotor control (Strauss and Heisenberg, 1993)—in regulating sleep (Liu et al., 2012b; Ueno et al., 2012), and other non-dopamingeric CX neurons have been implicated in homeostatic control of sleep (Donlea et al., 2011, 2014). In addition, it has recently been shown that manipulations of dopamine signaling in the MB alter sleep, although the precise DANs involved remains unclear (Ueno and Kume, 2014). We have now identified specific wake-promoting MB DANs and shown that they innervate lobe compartments also innervated by wake-promoting KCs and MBONs (Figures 1–3). Importantly, we have also shown that dopamine secretion by DANs innervating a particular MB lobe compartment acts through D1 subtype receptors to activate the wake-promoting microcircuit specific to that compartment to a much greater extent than it activates the sleep-promoting microcircuit residing in different compartments (Figure 4). This provides direct physiological evidence for compartment-specific dopamine signaling in the regulation of sleep by the MB, and is consistent with a previous study in the context of learning and memory (Boto et al., 2014). Future studies are required to determine additional cellular and molecular details of how dopamine signals modulate sleep-regulating microcircuits.
On the basis of recently published studies of MB control of sleep and the results presented here, we propose a unified mechanistic model for homeostatic control of sleep by excitatory microcircuits in the Drosophila MB. Wake-promoting MBON-γ5β′2a/β′2mp/β′2mp_bilateral and sleep-promoting MBON-γ2α′1 each receive anatomical inputs from both wake-promoting γm and α′/β′ KCs and sleep-promoting γd KCs (Aso et al., 2014a). However, segregation of sleep control information into separate microcircuits is enforced by greater synaptic weights between γm and α′/β′ KCs and MBON-γ5β′2a/β′2mp/β′2mp_bilateral, and between γd KCs and MBON-γ2α′1 (Sitaraman et al., 2015). We thus hypothesize that compartment-specific dopamine signals from MB DANs could potentially determine these differences in synaptic weight. Future studies will test this hypothesis.
Interestingly, other fly behaviors have recently been found to be regulated by sleep-controlling compartment-specific MB microcircuits. For example, the integration of food odor to suppress innate avoidance of CO2 is mediated by MBON-γ5β′2a/β′2mp/β′2mp_bilateral and PAM DANs that innervate the β′2 compartment (Lewis et al., 2015). Optogenetic activation experiments reveal that wake-promoting MBON-γ5β′2a/β′2mp/β′2mp_bilateral mediates innate avoidance, while MBON-γ2α′1 mediates attraction (Aso et al., 2014b). However, thermogenetic inactivation studies reveal that both MBON-γ5β′2a/β′2mp/β′2mp_bilateral and MBON-γ2α′1 are important for various forms of associative memory formation (Aso et al., 2014b). These diverse waking behaviors that involve the activity of sleep-regulating neurons raises the interesting question whether such roles are independent, or causally linked, which future studies can address.
Importantly, we have provided for the first time a cellular and molecular mechanism for for dopaminergic control of sleep through modulation of an associative network. While dopaminergic projections to cerebral cortex are known to be important for regulating sleep and arousal in mammals, underlying cellular and molecular mechanisms remain poorly understood (España and Scammell, 2011; Dauvilliers et al., 2015), although D2 subtype dopamine receptors have been implicated in the control of REM sleep (Dzirasa et al., 2006). Because of the possible evolutionary relationship between the MB and vertebrate forebrain associative networks (such as mammalian cerebral cortex), these studies thus provide a framework for the design of analogous experiments in genetically tractable vertebrate model systems such as zebrafish and mice.
Author Contributions
Design of experiments: DS, YA, GR, MN. Performing experiments: DS, YA. Analyzing data: DS, YA. Drafting manuscript: DS, YA, GR, MN.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Heather Dionne, Ming Wu, Tomoko Ohyama, and Romain Franconville for many helpful discussions, as well as fly stocks. We thank Leslie Vosshall, Nelson Spruston, and Marina Picciotto for comments on the manuscript. The Janelia Fly facility (Karen Hibbard, Todd Laverty, and other members of the fly core) helped in fly husbandry, and the FlyLight Project Team performed brain dissections, histological preparations, and confocal imaging. We thank members of the Nitabach lab for helpful comments on experimental approaches and conceptual frameworks, specifically Davide Raccuglia, Christina Paquin, Karl Barber, and Michael Kunst. Work in the laboratory of MN at Yale University was supported in part by the National Institute of Neurological Disorders and Stroke, National Institutes of Health (NIH) (R01NS055035, R01NS056443, R01NS091070), the National Institute of General Medical Sciences, NIH (R01GM098931), and the Kavli Institute for Neuroscience. DS and MN performed portions of this work as participants in the Janelia Visiting Scientist Program.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fncir.2015.00073
Figure S1. Lobe compartment innervation pattern of all screened MB DAN split-GAL4 lines.
References
Aso, Y., Hattori, D., Yu, Y., Johnston, R. M., Iyer, N. A., Ngo, T. T., et al. (2014a). The neuronal architecture of the mushroom body provides a logic for associative learning. eLife 3:e04577. doi: 10.7554/eLife.04577
Aso, Y., Sitaraman, D., Ichinose, T., Kaun, K. R., Vogt, K., Belliart-Guérin, G., et al. (2014b). Mushroom body output neurons encode valence and guide memory-based action selection in Drosophila. eLife 3:e04580. doi: 10.7554/eLife.04580
Baines, R. A., Uhler, J. P., Thompson, A., Sweeney, S. T., and Bate, M. (2001). Altered electrical properties in Drosophila neurons developing without synaptic transmission. J. Neurosci. 21, 1523–1531.
Boto, T., Louis, T., Jindachomthong, K., Jalink, K., and Tomchik, S. M. (2014). Dopaminergic modulation of cAMP drives nonlinear plasticity across the Drosophila mushroom body lobes. Curr. Biol. 24, 822–831. doi: 10.1016/j.cub.2014.03.021
Budnik, V., and White, K. (1988). Catecholamine-containing neurons in Drosophila melanogaster: distribution and development. J. Comp. Neurol. 268, 400–413. doi: 10.1002/cne.902680309
Chen, T. W., Wardill, T. J., Sun, Y., Pulver, S. R., Renninger, S. L., Baohan, A., et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. doi: 10.1038/nature12354
Dauvilliers, Y., Tafti, M., and Landolt, H. P. (2015). Catechol-O-methyltransferase, dopamine, and sleep-wake regulation. Sleep Med. Rev. 22, 47–53. doi: 10.1016/j.smrv.2014.10.006
Donelson, N. C., Kim, E. Z., Slawson, J. B., Vecsey, C. G., Huber, R., and Griffith, L. C. (2012). High-resolution positional tracking for long-term analysis of Drosophila sleep and locomotion using the “tracker” program. PLoS ONE 7:e37250. doi: 10.1371/annotation/4c62d454-931e-4c48-841a-a701cb658a1c
Donlea, J. M., Pimentel, D., and Miesenböck, G. (2014). Neuronal machinery of sleep homeostasis in Drosophila. Neuron 81, 860–872. doi: 10.1016/j.neuron.2013.12.013
Donlea, J. M., Thimgan, M. S., Suzuki, Y., Gottschalk, L., and Shaw, P. J. (2011). Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science 332, 1571–1576. doi: 10.1126/science.1202249
Dzirasa, K., Ribeiro, S., Costa, R., Santos, L. M., Lin, S. C., Grosmark, A., et al. (2006). Dopaminergic control of sleep-wake states. J. Neurosci. 26, 10577–10589. doi: 10.1523/JNEUROSCI.1767-06.2006
España, R. A., and Scammell, T. E. (2011). Sleep neurobiology from a clinical perspective. Sleep 34, 845–858. doi: 10.5665/sleep.1112
Guo, F., Yi, W., Zhou, M., and Guo, A. (2011). Go signaling in mushroom bodies regulates sleep in Drosophila. Sleep 34, 273–281.
Hamada, F. N., Rosenzweig, M., Kang, K., Pulver, S. R., Ghezzi, A., Jegla, T. J., et al. (2008). An internal thermal sensor controlling temperature preference in Drosophila. Nature 454, 217–220. doi: 10.1038/nature07001
Haynes, P. R., Christmann, B. L., and Griffith, L. C. (2015). A single pair of neurons links sleep to memory consolidation in Drosophila melanogaster. eLife 4:e03868. doi: 10.7554/eLife.03868
Heisenberg, M., Borst, A., Wagner, S., and Byers, D. (1985). Drosophila mushroom body mutants are deficient in olfactory learning. J. Neurogenet. 2, 1–30. doi: 10.3109/01677068509100140
Hendricks, J. C., Finn, S. M., Panckeri, K. A., Chavkin, J., Williams, J. A., Sehgal, A., et al. (2000). Rest in Drosophila is a sleep-like state. Neuron 25, 129–138. doi: 10.1016/S0896-6273(00)80877-6
Jenett, A., Rubin, G. M., Ngo, T. T., Shepherd, D., Murphy, C., Dionne, H., et al. (2012). A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2, 991–1001. doi: 10.1016/j.celrep.2012.09.011
Joiner, W. J., Crocker, A., White, B. H., and Sehgal, A. (2006). Sleep in Drosophila is regulated by adult mushroom bodies. Nature 441, 757–760. doi: 10.1038/nature04811
Kume, K., Kume, S., Park, S. K., Hirsh, J., and Jackson, F. R. (2005). Dopamine is a regulator of arousal in the fruit fly. J. Neurosci. 25, 7377–7384. doi: 10.1523/JNEUROSCI.2048-05.2005
Lewis, L. P., Siju, K. P., Aso, Y., Friedrich, A. B., Bulteel, A. J., Rubin, G. M., et al. (2015). A Higher brain circuit for immediate integration of conflicting sensory information in Drosophila. Curr. Biol. 25, 2203–2214. doi: 10.1016/j.cub.2015.07.015
Liu, C., Plaçais, P. Y., Yamagata, N., Pfeiffer, B. D., Aso, Y., Friedrich, A. B., et al. (2012a). A subset of dopamine neurons signals reward for odour memory in Drosophila. Nature 488, 512–516. doi: 10.1038/nature11304
Liu, Q., Liu, S., Kodama, L., Driscoll, M. R., and Wu, M. N. (2012b). Two dopaminergic neurons signal to the dorsal fan-shaped body to promote wakefulness in Drosophila. Curr. Biol. 22, 2114–2123. doi: 10.1016/j.cub.2012.09.008
Luan, H., Peabody, N. C., Vinson, C. R., and White, B. H. (2006). Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron 52, 425–436. doi: 10.1016/j.neuron.2006.08.028
Mellert, D. J., and Truman, J. W. (2012). Transvection is common throughout the Drosophila genome. Genetics 191, 1129–1141. doi: 10.1534/genetics.112.140475
Nässel, D. R., and Elekes, K. (1992). Aminergic neurons in the brain of blowflies and Drosophila: dopamine- and tyrosine hydroxylase-immunoreactive neurons and their relationship with putative histaminergic neurons. Cell Tissue Res. 267, 147–167. doi: 10.1007/BF00318701
Nitabach, M. N., Blau, J., and Holmes, T. C. (2002). Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell 109, 485–495. doi: 10.1016/S0092-8674(02)00737-7
Pfeiffer, B. D., Jenett, A., Hammonds, A. S., Ngo, T. T., Misra, S., Murphy, C., et al. (2008). Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 105, 9715–9720. doi: 10.1073/pnas.0803697105
Pfeiffer, B. D., Ngo, T. T., Hibbard, K. L., Murphy, C., Jenett, A., Truman, J. W., et al. (2010). Refinement of tools for targeted gene expression in Drosophila. Genetics 186, 735–755. doi: 10.1534/genetics.110.119917
Pitman, J. L., McGill, J. J., Keegan, K. P., and Allada, R. (2006). A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature 441, 753–756. doi: 10.1038/nature04739
Schrempf, W., Brandt, M. D., Storch, A., and Reichmann, H. (2014). Sleep disorders in Parkinson's disease. J. Parkinsons. Dis. 4, 211–221. doi: 10.3233/JPD-130301
Seugnet, L., Suzuki, Y., Merlin, G., Gottschalk, L., Duntley, S. P., and Shaw, P. J. (2011). Notch signaling modulates sleep homeostasis and learning after sleep deprivation in Drosophila. Curr. Biol. 21, 835–840. doi: 10.1016/j.cub.2011.04.001
Shang, Y., Haynes, P., Pírez, N., Harrington, K. I., Guo, F., Pollack, J., et al. (2011). Imaging analysis of clock neurons reveals light buffers the wake-promoting effect of dopamine. Nat. Neurosci. 14, 889–895. doi: 10.1038/nn.2860
Shaw, P. J., Cirelli, C., Greenspan, R. J., and Tononi, G. (2000). Correlates of sleep and waking in Drosophila melanogaster. Science 287, 1834–1837. doi: 10.1126/science.287.5459.1834
Sitaraman, D., Aso, Y., Jin, X., Chen, N., Felix, M., Rubin, G. M., et al. (2015). Propagation of Homeostatic sleep signals by segregated synaptic microcircuits of the Drosophila mushroom body. Curr. Biol. doi: 10.1016/j.cub.2015.09.017. [Epub ahead of print].
Strauss, R., and Heisenberg, M. (1993). A higher control center of locomotor behavior in the Drosophila brain. J. Neurosci. 13, 1852–1861.
Tanaka, N. K., Tanimoto, H., and Ito, K. (2008). Neuronal assemblies of the Drosophila mushroom body. J. Comp. Neurol. 508, 711–755. doi: 10.1002/cne.21692
Tomer, R., Denes, A. S., Tessmar-Raible, K., and Arendt, D. (2010). Profiling by image registration reveals common origin of annelid mushroom bodies and vertebrate pallium. Cell 142, 800–809. doi: 10.1016/j.cell.2010.07.043
Ueno, T., and Kume, K. (2014). Functional characterization of dopamine transporter in vivo using Drosophila melanogaster behavioral assays. Front. Behav. Neurosci. 8:303. doi: 10.3389/fnbeh.2014.00303
Ueno, T., Tomita, J., Tanimoto, H., Endo, K., Ito, K., Kume, S., et al. (2012). Identification of a dopamine pathway that regulates sleep and arousal in Drosophila. Nat. Neurosci. 15, 1516–1523. doi: 10.1038/nn.3238
Wu, M., Robinson, J. E., and Joiner, W. J. (2014). SLEEPLESS is a bifunctional regulator of excitability and cholinergic synaptic transmission. Curr. Biol. 24, 621–629. doi: 10.1016/j.cub.2014.02.026
Yamagata, N., Ichinose, T., Aso, Y., Plaçais, P. Y., Friedrich, A. B., Sima, R. J., et al. (2015). Distinct dopamine neurons mediate reward signals for short- and long-term memories. Proc. Natl. Acad. Sci. U.S.A. 112, 578–583. doi: 10.1073/pnas.1421930112
Yao, Z., Macara, A. M., Lelito, K. R., Minosyan, T. Y., and Shafer, O. T. (2012). Analysis of functional neuronal connectivity in the Drosophila brain. J. Neurophysiol. 108, 684–696. doi: 10.1152/jn.00110.2012
Yi, W., Zhang, Y., Tian, Y., Guo, J., Li, Y., and Guo, A. (2013). A subset of cholinergic mushroom body neurons requires Go signaling to regulate sleep in Drosophila. Sleep 36, 1809–1821. doi: 10.5665/sleep.3206
Yuan, Q., Joiner, W. J., and Sehgal, A. (2006). A sleep-promoting role for the Drosophila serotonin receptor 1A. Curr. Biol. 16, 1051–1062. doi: 10.1016/j.cub.2006.04.032
Keywords: sleep, Drosophila melanogaster, mushroom body, dopamine, synaptic plasticity
Citation: Sitaraman D, Aso Y, Rubin GM and Nitabach MN (2015) Control of Sleep by Dopaminergic Inputs to the Drosophila Mushroom Body. Front. Neural Circuits 9:73. doi: 10.3389/fncir.2015.00073
Received: 18 September 2015; Accepted: 23 October 2015;
Published: 09 November 2015.
Edited by:
Patrick O. Kanold, University of Maryland, USAReviewed by:
Antón Barreiro-Iglesias, University of Santiago de Compostela, SpainJeffrey Donlea, University of Oxford, UK
Quentin Gaudry, Harvard Medical School, USA
Copyright © 2015 Sitaraman, Aso, Rubin and Nitabach. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael N. Nitabach, michael.nitabach@yale.edu
†Present Address: Divya Sitaraman, Department of Psychological Sciences, University of San Diego, San Diego, CA, USA
 Divya Sitaraman
Divya Sitaraman Yoshinori Aso
Yoshinori Aso Gerald M. Rubin
Gerald M. Rubin Michael N. Nitabach
Michael N. Nitabach