Engineered neuronal circuits: a new platform for studying the role of modular topology
- 1 School of Electrical Engineering, Tel-Aviv University, Tel-Aviv, Israel
- 2 School of Physics and Astronomy, Tel-Aviv University, Tel-Aviv, Israel
Neuron–glia cultures serve as a valuable model system for exploring the bio-molecular activity of single cells. Since neurons in culture can be conveniently recorded with great fidelity from many sites simultaneously, it has long been suggested that uniform cultured neurons may also be used to investigate network-level mechanisms pertinent to information processing, activity propagation, memory, and learning. But how much of the functionality of neural circuits can be retained in vitro remains an open question. Recent studies utilizing patterned networks suggest that they provide a most useful platform to address fundamental questions in neuroscience. Here we review recent efforts in the realm of patterned networks’ activity investigations. We give a brief overview of the patterning methods and experimental approaches commonly employed in the field, and summarize the main results reported in the literature. The general picture that emerges from these reports indicates that patterned networks with uniform connectivity do not exhibit unique activity patterns. Rather, their activity is very similar to that of unpatterned uniform networks. However, by breaking the connectivity homogeneity, using a modular architecture, it is possible to introduce pronounced topology-related gating and delay effects. These findings suggest that patterned cultured networks may serve as a new platform for studying the role of modularity in neuronal circuits.
Structure and Function of Neuronal Circuits in vivo
Understanding the interplay between the morphological and functional features of neuronal circuits is a major challenge in contemporary neuroscience. While the network’s morphological constraints clearly shape its activity patterns, these activity patterns also reciprocally affect the architecture of the network. Investigating morphology–activity relationship is therefore an important yet challenging task, in particular in light of the modular and hierarchical organization of the brain (Mountcastle, 1997; Bullmore and Sporns, 2009; Meunier et al., 2011). Such investigations are particularly cumbersome in intact neural tissue owing to their overwhelming complex structure. While it is easier to examine isolated circuits with controlled inputs and outputs, in vivo circuits are extremely difficult to isolate due to the multiple activation pathways impinging on any recorded region. Despite recent advances in electrophysiology, imaging, and drug delivery, the accessibility of specific in vivo circuits to visualization, recording, and manipulation is still limited and the connectivity maps of neurons in vivo are highly untraceable in a three dimensional architecture. Finally, since natural circuits are not prone to design, different wiring schemes can not be systematically studied. Owing to these limitations, alternative methods are needed to allow methodical investigation of morphology–activity relationships in neuronal networks. One such approach is the study of cultures which allows network engineering, mapping, and simultaneous multi-site electrical recordings.
Engineering Neuronal Circuits in vitro
Beginning with the pioneering work of Kleinfeld et al. (1988), much effort has been dedicated to constructing engineered neuronal circuits with defined connectivity patterns (see Wheeler and Brewer, 2010 for review). Although many techniques can be used to achieve patterning, two main requirements must be satisfied to guarantee their practicability for long-term investigations: stability and yield. Cell migration and extension redirection, resulting from the inherent motility of cells and tension forces exerted by neurite bundles (Sorkin et al., 2006, 2009; Jun et al., 2007b; Anava et al., 2009), often compromise patterning quality (Ruardij et al., 2002; Wheeler and Brewer, 2010). Accordingly, physically caging neurons by introducing barriers (Maher et al., 1999; Zeck and Fromherz, 2001), combining cell repelling surfaces with strongly linked adhesive chemistries (Nam et al., 2006) or patterning rough surface coatings (Greenbaum et al., 2009) have been used to create stable patterns. In general, the finer the patterns the less stable they are (Khatami, 2008; Wheeler and Brewer, 2010). While patterns with a feature size of several microns begin to breakdown as early as 1 week in culture, patterns with feature sizes of tens of micros can be stable for several months in culture (Khatami, 2008).
All the above approaches aim at keeping cells at specific locations by restricting their natural motility. An alternative approach is to harness this innate motility mechanism to promote patterning. If the constraints to keep cells at specific locations are relaxed, neurons and glia in culture tend to self-organize into clusters (Segev et al., 2003; Rutten et al., 2006; Sorkin et al., 2006; Soussou et al., 2007; Shein et al., 2009; Shein Idelson et al., 2010). Using simple protein or carbon-nanotube patterning, it is possible to direct cluster locations and form highly stable and well organized networks (Gabay et al., 2005; Sorkin et al., 2006; Macis et al., 2007; Shein et al., 2009) which can be faithfully recorded for over 1 month in culture (Macis et al., 2007; Shein et al., 2009). It should be noted that clustering is such a strong propensity of neuronal networks, that some degree of clustering is almost always observed for most patterning techniques after some time in culture (Ruardij et al., 2002; Jun et al., 2007a). Clustering may also be enhanced by biological substrates with ingredients similar to those found in the brain extracellular matrix (Soussou et al., 2007).
Cell patterning methods are especially attractive when coupled with recording elements such as transistor or multi-electrode arrays (MEAs; Chang et al., 2006; Jun et al., 2007a; Jungblut et al., 2009; Shein Idelson et al., 2010). Such devices provide activity recording of many neurons simultaneously, non-invasively, and for very long times (Potter, 2001). Patterning also allows direct coupling of neurons to recording sites, thus increasing the fraction of recorded neurons relative to the total number of neurons in the network (Jungblut et al., 2009). Alternatively, optical imaging can be used, allowing high spatial resolution at the price of low temporal resolution (Jimbo et al., 1993; Feinerman et al., 2007; Zeringue et al., 2011).
Approaches for Studying Electrical Activity in Patterned Networks
Despite their attractiveness, until recently patterned networks were rarely used to address basic questions in neuroscience. Most studies were limited to demonstrating recording and stimulation capacities with only basic activity analysis. However, several recent studies which have explored the electrical activity of patterned networks have been able to demonstrate the great potential of these systems. Three main approaches are commonly used (Figure 1). In the first (Figure 1A), networks of different sizes are used to study scaling properties (Segev et al., 2002; Rutten et al., 2006; Wilson et al., 2007; Shein Idelson et al., 2010). In these networks, cells are uniformly distributed on the surface (no preferential adhesion to any specific location), and neurons at different locations have the same probability to connect to other neurons in the network (no preferential directionality or connectivity), similarly to the widely used uniform networks.
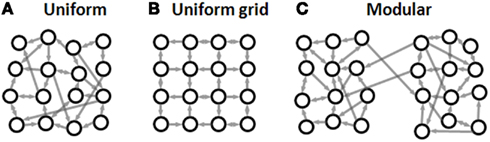
Figure 1. Common experimental approaches in neuronal network patterning. (A) Finite uniform network. Cells are uniformly distributed without preferential adhesion to any specific location and without preferential directionality or connectivity. (B) Uniform grid. Cells are patterned at the nodes of a grid with connections restricted to the grid lines. (C) Modular network. Two or more uniform sub-networks with high intra-sub-network connectivity and low inter-sub-network connectivity.
In the second approach (Figure 1B), single neurons are patterned at the nodes of a grid with connections restricted to grow along grid lines (Vogt et al., 2005; Nam et al., 2006; Jun et al., 2007a). This setup allows convenient mapping of the connections between the neurons and recording from a large fraction of the neurons in the network. Such an experimental setup is ideal for studying signal propagation between single cells and for studying plasticity within a small cell assembly (Vogt et al., 2005; Claverol-Tinture et al., 2007; Suzuki and Yasuda, 2007). However, due to low cell density, the spontaneous activity levels of such patterned systems are usually very low (Jun et al., 2007a; Cohen et al., 2008), making detailed, long-term statistical activity analysis difficult.
The third approach (Figure 1C), concerns neither complete mapping nor scaling properties of a single neuronal assembly but rather aims to examine how connected sub-networks interact. This is achieved by patterning modular networks with hierarchical connectivity patterns. Such modular networks are implemented using two or more uniform networks coupled by a neurite bridge (Yvon et al., 2005; Berdondini et al., 2006; Baruchi et al., 2008; Feinerman et al., 2008) or using clusters connected by neurite bundles (Soussou et al., 2007; Tsai et al., 2008; Shein Idelson et al., 2010).
Network Bursts in Patterned Circuits
To better understand how the activity of patterned networks differs from that of uniform networks, we begin by briefly summarizing the main activity features of uniform networks. A more detailed review is presented in the “Discussion” section. Large uniform neuronal networks in culture are typified by network bursts (NBs, Figures 2A,B). These short (several hundreds of milliseconds) spontaneous epochs of network-wide synchronized activity, are considered as the hallmark of developing brain circuits (Ben-Ari, 2001) and have been observed both in vitro (Maeda et al., 1995; Kamioka et al., 1996; Corner, 2008) and in vivo (Leinekugel et al., 2002; Buzsaki, 2004; Ikegaya et al., 2004; Luczak et al., 2007). Figure 2B shows typical NB activity of a uniform cortical culture. The extent of synchronization is most apparent in Figures 2C,D. It has been found that NBs are mostly characterized by a fast rise time (tens of milliseconds) and a more gradual decay (several hundreds of milliseconds, Figures 2C,D) and are important for the morphological maturation of the network (Katz and Shatz, 1996; Hu et al., 2004; Spitzer, 2006). An interesting question addressed by several groups is whether NBs retain their characteristics for different network sizes, cell arrangement, connectivity, and topology.
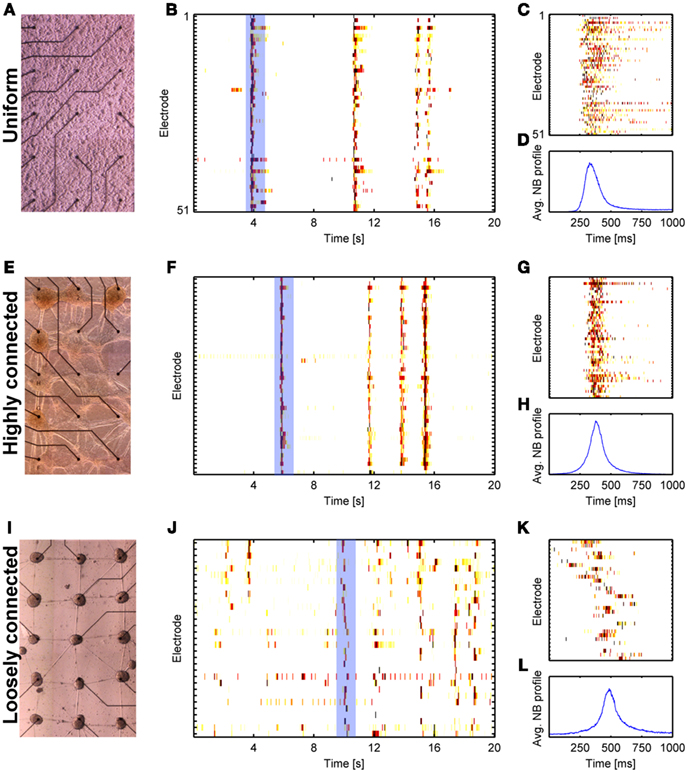
Figure 2. Spontaneous bursting in large uniform and clustered networks. Results from three network morphologies are shown: Uniform (A–D), highly connected clustered network (E–H), and loosely connected clustered network (I–L). (A,E,I) Bright field images (distance between electrodes is 500 μm). (B,F,J) Raster plots of recorded activity from the networks in (A,E,I), respectively. (C,G,K) Zoom into one NB from the raster plots shown in (B,F,J), respectively (marked by the light blue rectangle). (D,H,L) Average network activity for 500 consecutive NBs (see Shein Idelson et al., 2010 for methods).
A first interesting observation, reported by Ben-Jacob and co-workers is the robustness of NB patterns in networks of different sizes. It was shown that networks ranging from 50 to 106 cells can sustain network bursting with statistical features which are almost size independent (Segev et al., 2002). A similar result was reported for isolated clusters in which NB statistics did not dramatically change for networks larger than 100 cells (Shein Idelson et al., 2010; Figures 3A,B). This suggests the existence of regulatory mechanisms that modify the network morphology to sustain specific activity patterns. Indeed, such mechanisms were identified for the excitability of single cells and for the connectivity between them (Turrigiano, 1999; Shein et al., 2008; Ivenshitz and Segal, 2010). Using small rectangular networks of different sizes, it was shown that although the number of synapses per neuron increased with network size, their strength decreased, assumingly to compensate for the potential increase in excitability (Wilson et al., 2007).
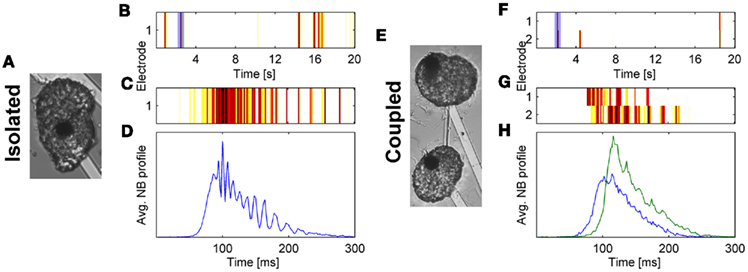
Figure 3. Oscillations and master–slave relations in small isolated (A–D) and coupled (E–H) clusters. (A,E) Bright field images (Electrode diameter, 30 μm). (B,F) Raster plots of recorded activity from the networks in (A,E), respectively. (C,G) Zoom into one NB from the raster plots shown in (B,F), respectively (marked by the light blue rectangle). (D,H) Average NB activity for 500 consecutive NBs (see Shein Idelson et al., 2010 for methods).
A second important question is whether a minimum cell number is required to maintain such a compensatory mechanism. Several studies have indicated that such a limit exists (Rutten et al., 2006; Kang et al., 2009; Shein Idelson et al., 2010). We have recently shown that very small neuro-glia networks (<20 cells) do not show synchronized activity but mostly tonic spiking (Shein Idelson et al., 2010). The exact cell limit appears to vary for different preparations and different compositions of culturing medium (Lau and Bi, 2005; Kang et al., 2009). We have additionally demonstrated that small networks of few tens of cells are not only able to generate NBs but also support oscillatory behavior during bursting (Shein Idelson et al., 2010; Figures 3C,D). Other studies have shown that such networks sustain long lasting reverberatory activity (Lau and Bi, 2005; Zeringue et al., 2011).
Patterning single neurons on a grid (Figure 1B) allows to examine activity under constraints of ordered cell arrangement and connectivity and to compare it to that of randomly arranged uniform networks. In these ordered networks, a critical cell density of ∼200 cells/mm2 is required for the network to be spontaneously active (Jun et al., 2007a; Jungblut et al., 2009; Wheeler and Brewer, 2010). Below that limit either no activity or only single asynchronous spikes are observed (Nam et al., 2006; Jun et al., 2007a; Jungblut et al., 2009). This density limit appears to correspond to the cell number limit discussed above for small uniform networks and neuronal clusters. In both cases it appears that the underlying activity limit is related to insufficient connectivity. This notion is supported by the fact that no network activity is observed if the network is patterned with thin grid lines which limit the connectivity (Jungblut et al., 2009; Wheeler and Brewer, 2010). Interestingly, for wide enough grid lines and high enough cell density, the activity patterns are characterized by NBs similar to those of uniform networks or isolated clusters (Jun et al., 2007a; Jungblut et al., 2009). This similarity is not surprising since the connectivity of these ordered networks is quite uniform (the average connectivity level for all the neurons is similar). It becomes therefore apparent that in order to fundamentally change the highly synchronized activity of neuronal network, patterning approaches that radically break the network uniformity are required.
A practical approach to breaking the network uniformity is to pattern it as two or more coupled uniform networks. Such networks are characterized by high connectivity within each sub-network and weaker connectivity between sub-networks (Figure 1C). At the limit of strong coupling, the two sub-networks are simply one big network for which network-wide synchrony is observed. For weaker coupling, the global synchrony is partially broken and individual features of the sub-networks begin to emerge (Figures 2I–L). Each sub-network continues to exhibit NBs, however, these NBs do not necessary propagate to other sub-networks (Yvon et al., 2005; Berdondini et al., 2006; Baruchi et al., 2008; Feinerman et al., 2008). The percentage of these propagation failures is modulated by past activity levels (Yvon et al., 2005). Following a treatment of prolonged activity inhibition, the number of failures is decreased due to underdevelopment of inhibitory connections between clusters (Yvon et al., 2005). When NBs do propagate, the propagation delays between the sub-networks are much longer than those expected from axonal conductance delays or propagation delays within uniform networks (Yvon et al., 2005; Berdondini et al., 2006). The same features are seen in networks of two connected neuro-glia clusters (Shein Idelson et al., 2010; Figures 3E–H).
This conditional synchrony between sub-networks is important from a population-level computational perspective. It supports the possibility to gate the propagation according to activity levels (only intense enough NBs propagate between sub-networks, see Feinerman et al., 2008), to introduce variable delays (Izhikevich, 2006), and to perform logical computational operations between sub-populations (Feinerman et al., 2008). An additional important feature that emerges from partitioning to sub-networks is the break down of symmetry between the sub-populations. It has been demonstrated that even if two sub-populations are constructed in a similar manner, one sub-network will initiate more NBs than the other (Baruchi et al., 2008; Tsai et al., 2008; Shein Idelson et al., 2010). It has been suggested that such master–slave asymmetry emerges due to amplification of very small asymmetries in the network (Feinerman et al., 2007), or may be related to variations in the topology of each of the networks or their connecting bridge (Baruchi et al., 2008).
Clearly, coupled sub-networks show unique activity patterns unseen in uniform networks. How do these features scale upon a transition to large networks of many connected sub-units? Several studies have examined the activity of such networks (Segev et al., 2003; Rutten et al., 2006; Macis et al., 2007; Soussou et al., 2007; Tsai et al., 2008; Shein et al., 2009). While in all reports, these networks exhibit NBs, the degree of synchrony and time delays vary between reports. In some cases (Segev et al., 2003; Rutten et al., 2006; Soussou et al., 2007) the networks exhibited synchronized activity resembling that of uniform networks. Other studies reported an age dependent increase in synchronization and confined stimulation effects in which stimulated bursts propagated only to neighboring clusters (Macis et al., 2007). In such networks, long propagation delays between clusters were observed and were shown to increase by reducing connectivity using synaptic blockers (Tsai et al., 2008). This apparent discrepancy may be related to the degree of connectivity. By systematically studying patterned networks with variable connectivity levels, it is possible to show that networks with higher connectivity exhibit highly synchronous NBs resembling those observed in uniform networks. Such a comparison is shown in Figure 2. On the other hand, lowering the degree of connectivity results in longer delays and conditional activation of adjacent clusters (Figure 2J). Namely, different NBs are initiated at different clusters and propagate to different distances. This conditional propagation is the network-level manifestation of the gating observed between two coupled networks (Figures 3E–H). Interestingly, clustered networks exhibit repeated propagation patterns (Tsai et al., 2008). Although similar repeating patterns have also been observed in uniform networks (Beggs and Plenz, 2004; Segev et al., 2004; Madhavan et al., 2007), in clustered networks these patterns are directly related to the spatial organization of the network (Tsai et al., 2008).
Discussion
It is widely accepted that network-level approaches are required to understand information processing in neuronal circuits. In the past few decades, a multitude of studies have addressed this issue by studying multi-site recordings from developing neuronal networks in culture (Maeda et al., 1995; Kamioka et al., 1996; Gross and Kowalski, 1999; Jimbo et al., 1999; Streit et al., 2001; Segev et al., 2004; Madhavan et al., 2007; Corner, 2008; Shein et al., 2008; le Feber et al., 2010). The underlying motivation was that in vitro cultures retain many physiological features of the brain while being highly accessible to manipulation and control (Potter, 2001; Marom and Shahaf, 2002). Cultured neurons show similar activity patterns across different preparations (Ben-Ari, 2001), manifesting short events of network-wide synchronization (Figure 2B). Consequently, most studies focused on examining the spatio-temporal patterns within these synchronous events and their statistics.
These studies revealed the potential of uniform networks to regulate activity, process information, and sustain biological memory. Examination of inter-burst statistics revealed that NB time series’ exhibit long-range temporal auto-correlations and complex temporal organization (Segev et al., 2002; Ayali et al., 2004; Hulata et al., 2004). Inspired by the theory of synfire chains, the intra-burst firing patterns were investigated and were found to be far from random. Highly repetitive spatio-temporal patterns with defined propagation schemes were detected in the spontaneous activity of the networks (Streit et al., 2001; Segev et al., 2004; Baruchi et al., 2006; Feinerman et al., 2007). Furthermore, such patterns were artificially induced by targeted electrical (Madhavan et al., 2007) and chemical (Baruchi and Ben-Jacob, 2007) stimulations demonstrating a potential to imprint network-level memory.
Uniform cultures were also found to support learning. Following the discovery of plasticity rules in coupling between single neurons (Bi and Poo, 2001), a network-level analog was found in which path specific potentiation and depression were induced using electrical stimulations (Jimbo et al., 1999). Other studies reported that plastic changes can be induced by controlled removal of a driving stimulus using feedback from the network activity (Shahaf and Marom, 2001; le Feber et al., 2010). Interestingly, plastic changes were not restricted to coupling between neurons but were also applicable to time delays of axonal conductances (Bakkum et al., 2008). In addition to these plastic changes, a series of regulatory mechanisms constantly tune synapses and intrinsic neuronal excitability to achieve single cell and network-level homeostasis (Turrigiano, 1999; Shein et al., 2008). Overall, the wealth of information extracted from uniform networks supports the notion that they are a good model for studying mechanisms of memory, information processing, and learning.
Despite these advantages, uniform networks have several limitations. Foremost, the uniform organization of cells in culture is markedly different from in vivo networks. Unlike the uniform topology of most culture preparations, many of the circuits involved in information processing are characterized by modular organization. Namely, they are made of small sub-networks with high intra-network connectivity and low inter-network connectivity (Mountcastle, 1997; Meunier et al., 2011). Furthermore, cells in uniform networks are strongly anchored to the surface which substantially restricts their motility.
In addition to these structural limitations, functional limitations are also encountered. In vivo neuronal networks are capable of exhibiting network-wide, but also localized activation. The latter is missing from the activity repertoire of uniform cultures which is marked by network-wide recruitment (with the exception of aborted bursts, see Eytan and Marom, 2006). Such network-wide synchronization does not support parallel and hierarchical information processing (Meunier et al., 2011). It is tempting to assert that these functional limitations are strongly linked to the structural constraints discussed above. This assertion suggests that the uniform network model should be extended to incorporate structural features to make it more relevant.
Thanks to recent advances in cell patterning techniques and multi-site recordings, a large set of tools have become available for controlling neuronal network architecture in culture (Wheeler and Brewer, 2010). However, activity analysis of patterned networks has, so far, only sparsely been addressed. Nevertheless, the few existing reports demonstrate the great potential of this approach. Foremost, these reports show that NBs are highly robust (Segev et al., 2002; Shein Idelson et al., 2010) and that the network’s connectivity is self-modulated on the synaptic level to sustain them (Wilson et al., 2007). In fact, even though networks of a few tens of cells with strong enough connectivity are required to spontaneously maintain NBs, they are already sufficient to support many of the activity features of large uniform networks in addition to intra-burst oscillations (Lau and Bi, 2005; Kang et al., 2009; Shein Idelson et al., 2010; Zeringue et al., 2011). This suggests that finite uniform networks may serve as a unitary system for studying network-level activity. Interestingly, it has been proposed that such minimal assembly size may have a functional role in real brains, where it may serve as a basic processing module (Shaw et al., 1982). From this perspective, the self-regulatory nature of NBs is certainly a desirable property. However, at the same time it suggests that uniformly adding cells or changing connectivity levels, can not profoundly alter network-wide synchrony. Only by breaking the network homogeneity, other activity patterns can be expected. Such a uniformity break can be achieved by introducing an additional hierarchy to uniform networks through implementation of a modular architecture.
Theoretical investigation also helps to understand the importance of network inhomogeneity. Such studies suggested that information processing in networks benefits from a modular architecture (Volman et al., 2005; Fuchs et al., 2009; Meunier et al., 2011). Firstly, modularity can give rise to time-scale separation between fast intra-modular and slow inter-modular processing (Pan and Sinha, 2009). Secondly, modularity maintains a fine balance within the spectrum of network-wide (integrated) and module-restricted (segregated) activation (Kaiser, 2007; Shanahan, 2008). In the latter, synchronized activity is confined to isolated modules, while in the former it spreads to all other modules in the network. These two features break the network-wide synchronization in time and space and promote the network’s dynamical complexity (Shanahan, 2008). Another benefit of modular networks is the short path length between neurons in the network, resulting in low wiring cost. However, while this benefit is shared by all small world networks (Watts and Strogatz, 1998), the former two benefits are exclusive to modular networks.
Interestingly, experimental studies in modular patterned cultures confirm these predictions. Modular networks in culture support time-scale separations (Yvon et al., 2005; Berdondini et al., 2006; Shein Idelson et al., 2010). The propagation delay in the activation of connected modules is of the order of several tens of milliseconds (Figures 3G,H). This delay is similar to the intra-module recruitment time (Eytan and Marom, 2006; Shein Idelson et al., 2010), and results in temporal separation between intra and inter module activation. In fact, in large modular networks, this delay is accumulated during activity propagation in a way that removes the temporal overlap between early and late activated modules (Figure 2K). It is worth noting, that long delays can also be implemented without using loose connections to break uniformity (Feinerman et al., 2007; Raichman and Ben-Jacob, 2008). However, in these implementations the delays are due to the time it takes the activity front to propagate, and therefore requires especially large networks. In addition, this implementation does not support other features of modular networks, such as activity segregation.
While connected modules have the potential to synchronize, they also exhibit segregated activity in which NBs within a module do not propagate to other modules (Yvon et al., 2005; Berdondini et al., 2006; Feinerman et al., 2008; Shein et al., 2008; Figures 2J and 3F). This suggests that a gating mechanism controls the degree of synchronization and the spread of activity. Interestingly, NBs with higher intensity have a greater probability to propagate to connected clusters (Feinerman et al., 2008). Such intensity related gating introduces a new set of computational features. For example, a module connected to two other modules can be conditionally activated only if both connected modules are synchronously activated (Feinerman et al., 2008). An additional feature of modular networks is the emergence of master–slave relationships where one module drives the other (Baruchi et al., 2008; Shein et al., 2008). Such a feature may be utilized to control the propagation directionality in the network.
In summary, the studies we have reviewed here suggest that modular cultured networks are a promising platform for studying information processing in vitro. Unlike networks in vivo, in vitro systems can be constructed and monitored with great precision allowing systematic investigations. Patterning networks of different sizes allows studying the scaling properties of uniform networks. Such studies bring to light the robustness of network-wide synchrony in uniform cultures and the self-regulatory mechanisms that maintain it. This robustness suggests that even small uniform networks can serve as a unitary module which supports NBs. However, at the same time it suggests that a break in uniformity is required to enrich the network’s activity patterns. Indeed, coupling small highly connected uniform networks through loose connections results in modular networks with unique activity characteristics. Such networks support synchronized bursting within each module, however, they also support mechanisms of activity gating and time delays between the modules. The use of this hierarchical topology further elevates the richness and complexity of activity in uniform networks.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Shimon Marom for fruitful discussions. The investigation reported here was partially supported by an ISF grant (827/10) and the Tauber Family Foundation.
References
Anava, S., Greenbaum, A., Ben Jacob, E., Hanein, Y., and Ayali, A. (2009). The regulative role of neurite mechanical tension in network development. Biophys. J. 96, 1661–1670.
Ayali, A., Zilberstein, Y., Robinson, A., Shefi, O., Hulata, E., Baruchi, I., and Ben jacob, E. (2004). Contextual regularity and complexity of neuronal activity: from stand-alone cultures to task-performing animals. Complexity 9, 25–32.
Bakkum, D. J., Chao, Z. C., and Potter, S. M. (2008). Long-term activity-dependent plasticity of action potential propagation delay and amplitude in cortical networks. PLoS ONE 3, e2088.
Baruchi, I., and Ben-Jacob, E. (2007). Towards neuro-memory-chip: imprinting multiple memories in cultured neural networks. Phys. Rev. E. Stat. Nonlin. Soft Matter Phys. 75(Pt 1), :050901.
Baruchi, I., Grossman, D., Volman, V., Shein, M., Hunter, J., Towle, V. L., and Ben-Jacob, E. (2006). Functional holography analysis: simplifying the complexity of dynamical networks. Chaos 16, 15112.
Baruchi, I., Volman, V., Raichman, N., Shein, M., and Ben-Jacob, E. (2008). The emergence and properties of mutual synchronization in in vitro coupled cortical networks. Eur. J. Neurosci. 28, 1825–1835.
Beggs, J. M., and Plenz, D. (2004). Neuronal avalanches are diverse and precise activity patterns that are stable for many hours in cortical slice cultures. J. Neurosci. 24, 5216–5229.
Berdondini, L., Chippalone, M., van der Wal, P. D., Imfeld, K., de Rooij, N. F., Koudelka-Hep, M., Tedesco, M., Martinoia, S., van Pelt, J., Le Masson, G., and Garenne, A. (2006). A microelectrode array (MEA) integrated with clustering structures for investigating in vitro neurodynamics in confined interconnected sub-populations of neurons. Sens. Actuators B Chem. 114, 530–541.
Bi, G., and Poo, M. (2001). Synaptic modification by correlated activity: Hebb’s postulate revisited. Annu. Rev. Neurosci. 24, 139–66.
Bullmore, E., and Sporns, O. (2009). Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 10, 186–198.
Chang, J. C., Brewer, G. J., and Wheeler, B. C. (2006). Neuronal network structuring induces greater neuronal activity through enhanced astroglial development. J. Neural Eng. 3, 217–226.
Claverol-Tinture, E., Cabestany, J., and Rosell, X. (2007). Multisite recording of extracellular potentials produced by microchannel-confined neurons in-vitro. IEEE Trans. Biomed. Eng. 54, 331–335.
Cohen, E., Ivenshitz, M., Amor-Baroukh, V., Greenberger, V., and Segal, M. (2008). Determinants of spontaneous activity in networks of cultured hippocampus. Brain Res. 1235, 21–30.
Corner, M. A. (2008). Spontaneous neuronal burst discharges as dependent and independent variables in the maturation of cerebral cortex tissue cultured in vitro: a review of activity-dependent studies in live “model” systems for the development of intrinsically generated bioelectric slow-wave sleep patterns. Brain Res. Rev. 59, 221–244.
Eytan, D., and Marom, S. (2006). Dynamics and effective topology underlying synchronization in networks of cortical neurons. J. Neurosci. 26, 8465–8476.
Feinerman, O., Rotem, A., and Moses, E. (2008). Reliable neuronal logic devices from patterned hippocampal cultures. Nat. Phys. 4, 967–973.
Feinerman, O., Segal, M., and Moses, E. (2007). Identification and dynamics of spontaneous burst initiation zones in uni-dimensional neuronal cultures. J. Neurophysiol. 97, 2937–2948.
Fuchs, E., Ayali, A., Ben-Jacob, E., and Boccaletti, S. (2009). The formation of synchronization cliques during the development of modular neural networks. Phys. Biol. 6, 036018.
Gabay, T., Jakobs, E., Ben-Jacob, E., and Hanein, Y. (2005). Engineered self-organization of neural networks using carbon nanotube clusters. Physica A 350, 611–621.
Greenbaum, A., Anava, S., Ayali, A., Shein, M., David-Pur, M., Ben-Jacob, E., and Hanein, Y. (2009). One-to-one neuron-electrode interfacing. J. Neurosci. Methods 182, 219–224.
Gross, G. W., and Kowalski, J. M. (1999). Origins of activity patterns in self-organizing neuronal networks in vitro. J. Intell. Mater. Syst. Struct. 10, 558–564.
Hu, H., Ni, Y., Montana, V., Haddon, R. C., and Parpura, V. (2004). Chemically functionalized carbon nanotubes as substrates for neuronal growth. Nano Lett. 4, 507–511.
Hulata, E., Baruchi, I., Segev, R., Shapira, Y., and Ben-Jacob, E. (2004). Self-regulated complexity in cultured neuronal networks. Phys. Rev. Lett. 92, 198105.
Ikegaya, Y., Aaron, G., Cossart, R., Aronov, D., Lampl, I., Ferster, D., and Yuste, R. (2004). Synfire chains and cortical songs: temporal modules of cortical activity. Science 304, 559–564.
Ivenshitz, M., and Segal, M. (2010). Neuronal density determines network connectivity and spontaneous activity in cultured hippocampus. J. Neurophysiol. 104, 1052–1060.
Jimbo, Y., Robinson, H. P. C., and Kawana, A. (1993). Simultaneous measurement of intracellular calcium and electrical-activity from patterned neural networks in culture. IEEE Trans. Biomed. Eng. 40, 804–810.
Jimbo, Y., Tateno, T., and Robinson, H. P. (1999). Simultaneous induction of pathway-specific potentiation and depression in networks of cortical neurons. Biophys. J. 76, 670–678.
Jun, S. B., Hynd, M. R., Dowell-Mesfin, N., Smith, K. L., Turner, J. N., Shain, W., and Kim, S. J. (2007a). Low-density neuronal networks cultured using patterned poly-l-lysine on microelectrode arrays. J. Neurosci. Methods 160, 317–326.
Jun, S. B., Hynd, M. R., Smith, K. L., Song, J. K., Turner, J. N., Shain, W., and Kim, S. J. (2007b). Electrical stimulation-induced cell clustering in cultured neural networks. Med. Biol. Eng. Comput. 45, 1015–1021.
Jungblut, M., Knoll, W., Thielemann, C., and Pottek, M. (2009). Triangular neuronal networks on microelectrode arrays: an approach to improve the properties of low-density networks for extracellular recording. Biomed. Microdevices 11, 1269–1278.
Kaiser, M. (2007). Brain architecture: a design for natural computation. Philos. Transact. A Math. Phys. Eng. Sci. 365, 3033–3045.
Kamioka, H., Maeda, E., Jimbo, Y., Robinson, H. P., and Kawana, A. (1996). Spontaneous periodic synchronized bursting during formation of mature patterns of connections in cortical cultures. Neurosci. Lett. 206, 109–112.
Kang, G., Lee, J. H., Lee, C. S., and Nam, Y. (2009). Agarose microwell based neuronal micro-circuit arrays on microelectrode arrays for high throughput drug testing. Lab. Chip 9, 3236–3242.
Katz, L. C., and Shatz, C. J. (1996). Synaptic activity and the construction of cortical circuits. Science 274, 1133–1138.
Khatami, D. B. (2008). Design, Implementation and Analysis of Functional Activity and Connectivity in Patterened Neural Networks. Ph.D. thesis, University of Illinois, Urbana-Champaign, IL.
Kleinfeld, D., Kahler, K. H., and Hockberger, P. E. (1988). Controlled outgrowth of dissociated neurons on patterned substrates. J. Neurosci. 8, 4098–4120.
Lau, P. M., and Bi, G. Q. (2005). Synaptic mechanisms of persistent reverberatory activity in neuronal networks. Proc. Natl. Acad. Sci. U.S.A. 102, 10333–10338.
le Feber, J., Stegenga, J., and Rutten, W. L. C. (2010). The effect of slow electrical stimuli to achieve learning in cultured networks of rat cortical neurons. PLoS ONE 5, e8871.
Leinekugel, X., Khazipov, R., Cannon, R., Hirase, H., Ben-Ari, Y., and Buzsaki, G. (2002). Correlated bursts of activity in the neonatal hippocampus in vivo. Science 296, 2049–2052.
Luczak, A., Bartho, P., Marguet, S. L., Buzsaki, G., and Harris, K. D. (2007). Sequential structure of neocortical spontaneous activity in vivo. Proc. Natl. Acad. Sci. U.S.A. 104, 347–352.
Macis, E., Tedesco, M., Massobrio, P., Ralteri, R., and Martinoia, S. (2007). An automated microdrop delivery system for neuronal network patterning on microelectrode arrays. J. Neurosci. Methods 161, 88–95.
Madhavan, R., Chao, Z. C., and Potter, S. M. (2007). Plasticity of recurring spatiotemporal activity patterns in cortical networks. Phys. Biol. 4, 181–193.
Maeda, E., Robinson, H. P., and Kawana, A. (1995). The mechanisms of generation and propagation of synchronized bursting in developing networks of cortical neurons. J. Neurosci. 15, 6834–6845.
Maher, M. P., Pine, J., Wright, J., and Tai, Y. C. (1999). The neurochip: a new multielectrode device for stimulating and recording from cultured neurons. J. Neurosci. Methods 87, 45–56.
Marom, S., and Shahaf, G. (2002). Development, learning and memory in large random networks of cortical neurons: lessons beyond anatomy. Q. Rev. Biophys. 35, 63–87.
Meunier, D., Lambiotte, R., and Bullmore, E. T. (2011). Modular and hierarchically modular organization of brain networks. Front. Neurosci. 4:200.
Nam, Y., Branch, D. W., and Wheeler, B. C. (2006). Epoxy-silane linking of biomolecules is simple and effective for patterning neuronal cultures. Biosens. Bioelectron. 22, 589–597.
Pan, R. K., and Sinha, S. (2009). Modularity produces small-world networks with dynamical time-scale separation. Europhys. Lett. 85, 68006.
Potter, S. M. (2001). Distributed processing in cultured neuronal networks. Prog. Brain Res. 130, 49–62.
Raichman, N., and Ben-Jacob, E. (2008). Identifying repeating motifs in the activation of synchronized bursts in cultured neuronal networks. J. Neurosci. Methods 170, 96–110.
Ruardij, T. G., van den Boogaart, M. A., and Rutten, W. L. (2002). Adhesion and growth of electrically active cortical neurons on polyethylenimine patterns microprinted onto peo-ppo-peo triblock copolymer-coated hydrophobic surfaces. IEEE Trans. Nanobioscience 1, 4–11.
Rutten, W. L. C., Ruardij, T. G., Marani, E., and Roelofsen, B. H. (2006). Cultured neural networks: optimization of patterned network adhesiveness and characterization of their neural activity. Appl. Bionics Biomech. 3, 1–7.
Segev, R., Baruchi, I., Hulata, E., and Ben-Jacob, E. (2004). Hidden neuronal correlations in cultured networks. Phys. Rev. Lett. 92, 118102.
Segev, R., Benveniste, M., Hulata, E., Cohen, N., Palevski, A., Kapon, E., Shapira, Y., and Ben-Jacob, E. (2002). Long term behavior of lithographically prepared in vitro neuronal networks. Phys. Rev. Lett. 88, 118102.
Segev, R., Benveniste, M., Shapira, Y., and Ben-Jacob, E. (2003). Formation of electrically active clusterized neural networks. Phys. Rev. Lett. 90, 168101.
Shahaf, G., and Marom, S. (2001). Learning in networks of cortical neurons. J. Neurosci. 21, 8782–8788.
Shanahan, M. (2008). Dynamical complexity in small-world networks of spiking neurons. Phys. Rev. E 78, 041924.
Shaw, G. L., Harth, E., and Scheibel, A. B. (1982). Cooperativity in brain function: assemblies of approximately 30 neurons. Exp. Neurol. 77, 324–358.
Shein, M., Greenbaum, A., Gabay, T., Sorkin, R., David-Pur, M., Ben-Jacob, E., and Hanein, Y. (2009). Engineered neuronal circuits shaped and interfaced with carbon nanotube microelectrode arrays. Biomed. Microdevices 11, 495–501.
Shein, M., Volman, V., Raichman, N., Hanein, Y., and Ben-Jacob, E. (2008). Management of synchronized network activity by highly active neurons. Phys. Biol. 5, 36008.
Shein Idelson, M., Ben-Jacob, E., and Hanein, Y. (2010). Innate synchronous oscillations in freely-organized small neuronal circuits. PLoS ONE 5, e14443.
Sorkin, R., Gabay, T., Blinder, P., Baranes, D., Ben-Jacob, E., and Hanein, Y. (2006). Compact self-wiring in cultured neural networks. J. Neural Eng. 3, 95–101.
Sorkin, R., Greenbaum, A., David-Pur, M., Anava, S., Ayali, A., Ben-Jacob, E., and Hanein, Y. (2009). Process entanglement as a neuronal anchorage mechanism to rough surfaces. Nanotechnology 20, 015101.
Soussou, W. V., Yoon, G. J., Brinton, R. D., and Berger, T. W. (2007). Neuronal network morphology and electrophysiologyof hippocampal neurons cultured on surface-treated multielectrode arrays. IEEE Trans. Biomed. Eng. 54, 1309–1320.
Streit, J., Tscherter, A., Heuschkel, M. O., and Renaud, P. (2001). The generation of rhythmic activity in dissociated cultures of rat spinal cord. Eur. J. Neurosci. 14, 191–202.
Suzuki, I., and Yasuda, K. (2007). Detection of tetanus-induced effects in linearly lined-up micropatterned neuronal networks: application of a multi-electrode array chip combined with agarose microstructures. Biochem. Biophys. Res. Commun. 356, 470–475.
Tsai, C. Y., Chang, M. C., and Lin, I. (2008). Robustness and variability of pathways in the spontaneous synchronous bursting of clusterized cortical neuronal networks in vitro. J. Physical Soc. Japan 77, 084803.
Turrigiano, G. G. (1999). Homeostatic plasticity in neuronal networks: the more things change, the more they stay the same. Trends Neurosci. 22, 221–227.
Vogt, A. K., Brewer, G. J., and Offenhausser, A. (2005). Connectivity patterns in neuronal networks of experimentally defined geometry. Tissue Eng. 11, 1757–1767.
Volman, V., Baruchi, I., and Ben-Jacob, E. (2005). Manifestation of function-follow-form in cultured neuronal networks. Phys. Biol. 2, 98–110.
Watts, D. J., and Strogatz, S. H. (1998). Collective dynamics of “small-world” networks. Nature 393, 440–442.
Wheeler, B. C., and Brewer, G. J. (2010). Designing neural networks in culture. Proc. IEEE 98, 398–406.
Wilson, N. R., Ty, M. T., Ingber, D. E., Sur, M., and Liu, G. (2007). Synaptic reorganization in scaled networks of controlled size. J. Neurosci. 27, 13581–13589.
Yvon, C., Rubli, R., and Streit, J. (2005). Patterns of spontaneous activity in unstructured and minimally structured spinal networks in culture. Exp. Brain Res. 165, 139–151.
Zeck, G., and Fromherz, P. (2001). Noninvasive neuroelectronic interfacing with synaptically connected snail neurons immobilized on a semiconductor chip. Proc. Natl. Acad. Sci. U.S.A. 98, 10457–10462.
Keywords: uniform networks, modular networks, hierarchical networks, neural engineering, carbon-nanotubes, electrical activity, clusters
Citation: Shein-Idelson M, Ben-Jacob E and Hanein Y (2011) Engineered neuronal circuits: a new platform for studying the role of modular topology. Front. Neuroeng. 4:10. doi: 10.3389/fneng.2011.00010
Received: 24 July 2011; Paper pending published: 10 August 2011;
Accepted: 23 August 2011; Published online: 27 September 2011.
Edited by:
Shimon Marom, Technion–Israel Institute of Technology, IsraelReviewed by:
Luca Berdondini, Italian Institute of Technology, ItalyBruce C. Wheeler, University of Illinois, USA
Copyright: © 2011 Shein-Idelson, Ben-Jacob and Hanein. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Mark Shein-Idelson, School of Electrical Engineering, Tel-Aviv University, Tel-Aviv 69978, Israel. e-mail: shein.mark@gmail.com