A bottom-up approach to data annotation in neurophysiology
- 1 Department Biology II, Ludwig-Maximilians Universität München, Martinsried, Germany
- 2 Bernstein Center for Computational Neuroscience Munich, Munich, Germany
- 3 German Neuroinformatics Node, Ludwig-Maximilians Universität München, Martinsried, Germany
Metadata providing information about the stimulus, data acquisition, and experimental conditions are indispensable for the analysis and management of experimental data within a lab. However, only rarely are metadata available in a structured, comprehensive, and machine-readable form. This poses a severe problem for finding and retrieving data, both in the laboratory and on the various emerging public data bases. Here, we propose a simple format, the “open metaData Markup Language” (odML), for collecting and exchanging metadata in an automated, computer-based fashion. In odML arbitrary metadata information is stored as extended key–value pairs in a hierarchical structure. Central to odML is a clear separation of format and content, i.e., neither keys nor values are defined by the format. This makes odML flexible enough for storing all available metadata instantly without the necessity to submit new keys to an ontology or controlled terminology. Common standard keys can be defined in odML-terminologies for guaranteeing interoperability. We started to define such terminologies for neurophysiological data, but aim at a community driven extension and refinement of the proposed definitions. By customized terminologies that map to these standard terminologies, metadata can be named and organized as required or preferred without softening the standard. Together with the respective libraries provided for common programming languages, the odML format can be integrated into the laboratory workflow, facilitating automated collection of metadata information where it becomes available. The flexibility of odML also encourages a community driven collection and definition of terms used for annotating data in the neurosciences.
1 Introduction
Data sharing in the neurosciences is the basis of every successful collaboration. However, working with another scientist’s data is usually quite cumbersome because of a high diversity of data formats and insufficient annotations. This diversity is a problem also faced by the various platforms for data sharing and publication of raw data that are being established1 (e.g., CARMEN, Fletcher et al., 2008; CRCNS, Teeters et al., 2008; G-Node, Gardner et al., 2001a; Herz et al., 2008). These initiatives are confronted with: (i) the vast variety of data formats used in the neurosciences, (ii) the lack of common, standardized ways in which data annotations are handled, and (iii) the scientific individualism together with reluctance to subdue to defined “standards.” The first point is not the scope of this paper and has been addressed by other initiatives like Neuroshare2 or SignalML3 (Durka and Ircha, 2004). In this paper we deal with the last two issues. We propose a format for metadata transfer that, on the one hand, is free of a specific, complex metadata model but, on the other hand, can be used in a standardized way to ensure interoperability. Application of this format is by no means restricted to data sharing. Rather, data annotation and metadata handling is an inevitable part of everyday scientific work in the lab (Figure 1). For example, almost every scientist knows the difficulties that can be involved in the reconstruction of the conditions or parameters under which a certain experiment or analysis in the past has been performed. Often, this information is stored in a non-standardized way in laboratory journals, metadata files, or is hidden in the source code of software tools. Evidently, providing a dataset with a complete set of metadata is a tedious business and hardly possible. Nevertheless, the more information can be retrieved easily, the more valuable the data are, and the more easily and more often the data can be used. The attempt to start recording metadata in a standardized way throughout the experiment and analysis process seems a promising approach to ensure future re-usability and availability of experimental data.
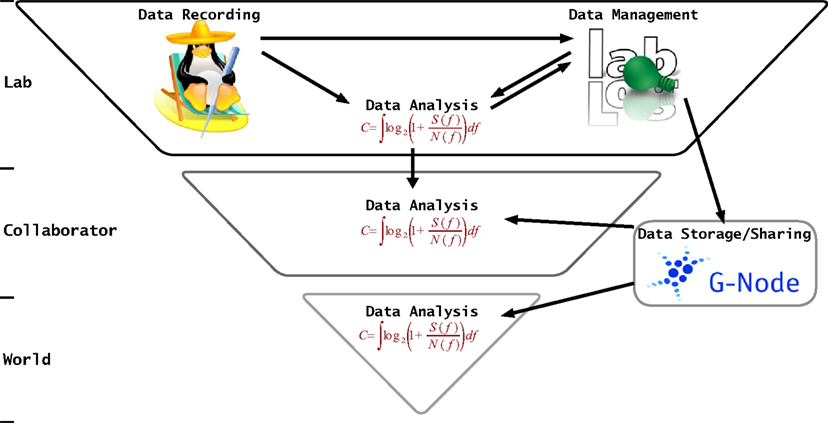
Figure 1. The flow of data and metadata in sciences. The basis of this “food chain,” on top, is the laboratory in which the data is originally recorded, stored, managed and analyzed. Here metadata are important in many respects. Data management uses them to categorize and organize the data, during data analysis stimulus information is required and further, derived, data characteristics are added which again may be useful for querying data, etc. Data may further be shared with collaborators for discussion and re-evaluation. Eventually, data may be made available via public databases like the G-Node (Herz et al., 2008). On all levels data exchange between people as well as computer programs requires a detailed annotation of the raw data with metadata.
The problems mentioned above are neither new nor specific to the neurosciences (Hey and Trefethen, 2003). Especially in the fields of Genomics and Proteomics many efforts, and much progress, have been made to organize the sharing of data (Gelbart et al., 1997; Stoesser et al., 1997). The general approach is to define ontologies, or controlled vocabularies, that specify the names of entities and their relations. The open biomedical ontologies Foundry (OBO)4 collects and coordinates ontologies in the biological domain. A downside of such a “top-down” approach in which an authority defines a “standard,” i.e., the terms and respective contexts in which these may be used, is not flexible and open enough and will inevitably lag behind the ever-changing requirements due to the progress in science. Further, this “standard” would need to be accepted by the community. We instead propose a “bottom-up” approach aiming primarily at the scientific work in the laboratories. Our goal is the convenient, semi, or fully automated handling of metadata that can be embedded into the laboratory workflow and is thus of direct use for the scientist. At the same time we want to foster interoperability by providing a possibility to apply “standards” enabling data exchange between tools or via data sharing platforms. Thus, we aim at (i) a format in which arbitrary information can be stored, and (ii) mechanisms to apply conventions regarding the content. The scope of these conventions may vary from local, laboratory needs, to a global community scope. Accordingly, our approach has two parts. The first is the rather simple open metadata Markup Language (odML) format or (meta)data model: In odML so called Properties are grouped in Sections resulting in a highly flexible hierarchical tree structure. This structure is to some extent similar to the way data annotations can be handled in the HDF55 data format. HDF5 also contains a hierarchical structure of nodes which can contain data or attributes for data annotation. Nodes and attributes are similar to our Sections and Properties, but there are distinct differences between these formats which are discussed below (see Section 5). The second component of our approach is to provide terminologies for definitions of Properties and Sections. The terminologies can be used to guarantee interoperability between tools. At the same time it is possible to immediately provide additional definitions for new Properties and Sections. Thus, odML offers the required flexibility and freedom to store any information that is necessary to describe a given dataset while supporting interoperability by using it in combination with specific terminologies. Given its acceptance, this “bottom-up” approach can lead to a community driven definition of global terminologies and general models of metadata in Neurosciences.
1.1 What are Metadata?
In the context of this paper we understand metadata as that information that describes the conditions under which a certain dataset has been recorded. This includes descriptions of environmental parameters like temperature, humidity, date, and time, etc., descriptions of the stimulus and recording protocols, settings of the used hardware and software, information about the experimental subject, and much more. Storing metadata in as much detail as possible allows replication of an experiment or the reconstruction of an analysis and thus enables reproducing results. It eases the re-use of once-acquired data and thus can increase the outcome of scientific efforts. Our goal is to provide the means to conveniently and automatically capture as many as possible of what we call the hard metadata, i.e., those parameters that can be directly measured (temperature, recording date, and time, etc.) or are known in advance (e.g., stimulus parameters). The more descriptive, soft metadata (e.g., the experimental rationale, context information, etc.) provide important background information but are much harder to capture automatically and, even if present, can hardly replace a discussion with the experimenter in person. In the interest of data sharing and reproducibility, datasets should be annotated with as much of the hard metadata as possible.
Annotating data may seem a costly process that requires the scientist to manually record a large number of values. However, most hard metadata are directly available and could thus be automatically recorded during data acquisition, with minimal manual intervention. Further information is typically derived during subsequent processing steps, for example analyses, etc. Ideally, all components of the data analysis tool-chain, from data acquisition, data analysis, and data management to data sharing, should be able to work hand in hand and exchange data and metadata in an automated fashion. The goal of odML is to provide the basic components for this automation.
2 Data Model
2.1 Two Use Cases
The basic idea of the odML approach is to combine a rather general data model with domain specific terminologies. Independence of format and content offers a maximum of flexibility. The terminologies introduce the basis for standardization that, however, can be ignored or extended when necessary. For example, if the terminology does not define a term that is needed for annotation, it should be possible to instantaneously add new terms with their respective definitions. The odML model is designed for both use cases: (i) exchanging metadata and (ii) definition of terminologies. Hence, the format contains more elements than are needed for either case alone. Using the same format for both the actual content and definitions (the terminologies) may seem confusing at first, but in our view is the key for granting immediate extensibility required to satisfy the ever-changing scientific needs.
2.2 Model Description
The elements of the odML metadata model are derived from the requirements: We need a metadata model that offers a flexible structure, can take various kinds of metadata, offers the means to ensure interoperability, and can be used for carrying metadata and for defining terminologies, while respecting conventions of the various scientific communities through customization. Figure 2 shows the data model as an entity-relation diagram. A tabular description can be found in the Appendix. We will start the description of the model with a coarse description of the structure and then go into more detail of the defined elements.
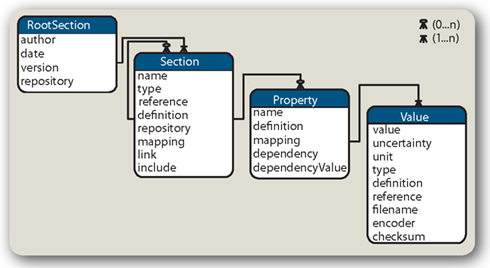
Figure 2. Open metaData Markup Language Entity-Relation diagram. The odML model is a tree structure of Sections and Properties. Connecting lines and “crow’s feet” indicate the relationship between the entities. For example: a Section can contain 0 to many (n) Properties which in turn must have at least 1 Value. The recursive connection of the Section indicates that there can be 0 to many subsections building the tree. All is embraced by a RootSection that contains some document-related elements. All elements listed in the different entities may at maximum occur once.
2.2.1 Entities and their relations
The Property is the core entity of the odML data model Figure 2. Roughly speaking, it is an extended key–value pair, like “stimulusRepetitions = 10.” A Property contains one or multiple Values. To meaningfully organize growing amounts of metadata and to allow the same Property name to be used several times, Properties are logically grouped in Sections. Besides Properties, a Section can also have subsections. Thus, a tree-like structure can be built up by nesting Sections. The RootSection, finally, constitutes the root of the tree that embraces a set of Sections. This kind of tree-like data model is rather generic and thus offers the required flexibility but also has some disadvantages (see Section 5).
2.2.2 Elements
In the following we will describe the various elements with respect to their purposes.
(1) Storing different types of metadata. Many metadata are numbers and can be specified by simple key–value pairs like “stimulusRepetitions = 10.” Others contain a unit (e.g., “temperature = 26°C”) or are measured values that comprise some uncertainty (e.g., “resting-potential = −58.0 ± 1.5 mV”). Other metadata may be purely textual or be even binary content (e.g., a microscopic picture of the recorded cell). Accordingly, the Property corresponds to a key–value pair with a name (the key) and the Value with according value-related information as the unit, uncertainty, and data-type. There is no strict restriction on the types, but we suggest the usage of the standard types as listed in Table A5 in appendix. The type allows tools to adapt their interfaces and allows performing consistency checks (see below and Table A5 in Appendix for more information). The Section groups all these metadata items (i.e., Properties). It is addressed by its name and, more importantly, defines a type element which indicates what kind of information can be found in it. For example, a Section describing a stimulus would be of the “stimulus” type (see “odML-terminologies” for more information).
(2) Interoperability and automation. These requirements demand more information about the content and a standardization we want to achieve by the terminologies (see below). For such purposes it is necessary to provide a definition for Sections and Properties. These can be omitted if the according definition of a Section or Property is provided by a terminology. To state where the underlying terminology can be found, the Section has the repository element. Sometimes Values have definitions, for example when describing the type of a recorded cell and linking to a definition in an ontology.
(3) Customization. Different labs or communities in the neurosciences use different names to address the same entities. Interoperability between these can only be achieved if an agreement on the terms to use could be reached or if the standard terms can be addressed with synonyms. Synonyms are introduced to odML by defining a mapping from, e.g., a custom Property to definitions made in a common terminology. Mappings can be provided for Sections and Properties. The odML-libraries can apply the mapping and convert the custom metadata to the standard terminologies so that tools which can handle the terms defined in the standard terminologies can work on the metadata.
(4) Miscellaneous. Finally, there are a number of elements that have been introduced to help working with the metadata. In the context of data management it is required to uniquely identify entities that are described by a Section’s or Property’s Value. For example a “subject” Section may refer to an entry in the (laboratory) database which has a certain id, primary key, etc. This information should be provided with the reference element of Values and Sections. Besides numerical or textual metadata, including binary content like the picture of a recorded cell could be necessary. Binary data can be included in two different ways: (i) in form of an url of the file location or, (ii) directly by including the binary content. In the latter case one can use the Value’s filename element to note the file name to be used when writing the binary content to disk. When binary content is included use the Value’s encoder element to state the encoder used. The checksum element takes the checksum information to verify a file’s integrity. A checksum should be given in the format: “algorithm$checksum” (e.g., “crc32$b84892a2” for a checksum calculated with a “crc” algorithm applying a 32-bit polynom). The Property further defines the dependency and dependencyValue elements to allow content validation or the adaptation of tools. With the dependency it can be stated that this Property is only meaningful if also the dependency is present. dependencyValue further restricts this in a way that the required Property should assume a certain value.
The Section defines two further elements, link and include. These are related and are introduced to allow inheritance and to distribute information across multiple files, respectively (see “relations outside the hierarchy” for more information).
The encapsulating RootSection contains elements to provide information about the whole document: these are author, date, version, and repository. The repository given in the RootSection defines a default repository valid for the whole document unless locally redefined.
3 odML-Terminologies
The odML metadata model is so general that arbitrary content can be exchanged or stored. In this form it could be used locally in a single lab or by an individual user. However, to achieve interoperability and allow sharing of metadata, standards defining property-names and section types are needed. The odML-terminologies are meant as a starting point for such standards.
An odML-terminology is specified by an odML file that provides definitions of Sections and Properties. Usually, there are separate terminologies for each defined Section type and provide the names, definitions, units, and data types of Properties and Values. All of this information can be overridden by the user, if necessary. The existence of a possibly large number of terminologies and contained terms does not imply that all these terms must be specified in an actual metadata file. The odML approach never requires any information to be provided by the user. Thus, all terms are suggestions that should be used when appropriate, but are not mandatory (see “Using odML” below).
3.1 Terminologies for Neurophysiology
Ideally, a metadata terminology should, as much as possible, be in agreement with existing terminologies and ontologies (Gardner et al., 2001b; Bug et al., 2008; Gibson et al., 2008a; Taylor et al., 2008; Frishkoff et al., 2009). However, these standardized sets will often be insufficient to fully describe the metadata for a given experiment. In particular, terminologies for the field of neurophysiology are currently still at a developmental stage (Gardner et al., 2008; Gibson et al., 2008a). To encourage usage of standardized terminologies from the very start of data and metadata collection, we started to set up several terminologies that are needed to annotate data from electrophysiological recordings6. These cover those parts of the Dublin Core7 that apply for annotation of neurophysiological data and also includes the definitions suggested by the CARMEN consortium (Gibson et al., 2008b). Compatibility with terminologies established by existing data sharing initiatives (Bowden and Dubach, 2003; Gardner et al., 2008; Gibson et al., 2008a) gives the possibility to retrieve the respective metadata items from an odML file when data are contributed. In addition, odML files can contain much more metadata that will not be lost if the odML file is made available with the data.
The rather unconstrained approach proposed here could be valuable in refining and adapting these standards through automatized matching with the terminologies used in the community. All terminologies described here are available on the odML homepage (see text footnote 6). The terminologies will never be complete and many more Properties and Sections can and should be specified as needed. Extension of the terminologies is necessary and has to be driven by the scientific community. The version of a terminology is part of the URL and is provided in the terminology RootSection. Within one version Properties and Sections may be added, but definitions will not be altered.
In the following paragraphs selected terminologies will be introduced to point out some conceptual aspects. A list with all currently defined section types can be found in the appendix (Table A6 in Appendix). Generally, electrophysiological data are recorded from a certain preparation of a subject by an experimenter in a recording session, using an experimental setup consisting of various hardware components with specific settings, and presenting defined stimuli. Accordingly, respective terminologies to describe these experimental conditions are needed. The description provided by these terminologies is much more detailed than in most cases needed for data sharing. However, odML is intended to be also of use for metadata management in the laboratory, where all this information should be kept available. The following descriptions start with the “Dataset” all other metadata are referring to. It is then briefly illustrated how the used hardware items and their respective settings as well as stimuli can be described using odML.
3.1.1 Dataset terminology
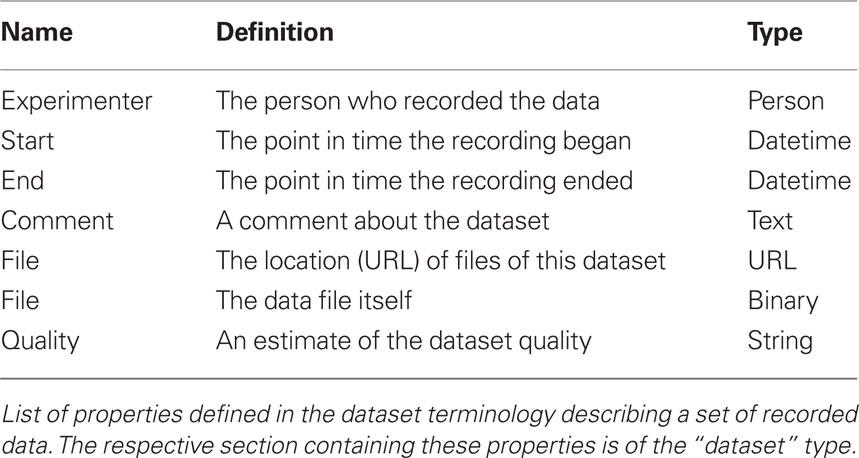
The Dataset terminology is a single Section of the “dataset”-type. It defines properties that can be used to provide general information about a recorded dataset (Table 1). The name of the Section identifies this particular dataset. Here a dataset is understood as a set of files that belong together, i.e., have been recorded in the same recording session. The “File” Property is of special interest. It occurs twice: (i) with the data type “URL” it can provide the location of a file associated with this dataset and (ii) with the data type “binary” the file content itself can be included in the metadata. Even though odML is meant to carry data about data, it is nevertheless possible to include binary content directly into an odML file. In case binary content is included, the filename element can provide a file name that should be used when extracting the data from the odML file. Generally, we recommend not to include the content of a file but to use the corresponding URL property instead. There can be several files related to the same dataset. In this case the file Property would simply have several values. The Value definition can then be used to specify what the files contain. Again, we do not aim at providing a description of the format the data is saved in. This is a challenge on its own (see, e.g., Durka and Ircha, 2004) and should rather be part of the data file itself.
3.1.2 Hardware terminologies
The hardware terminologies provide Section and Property definitions for describing hardware that was part of the experimental setup. There is a range of hardware related terminologies to describe specific hardware items. All defined hardware terminologies are specialized versions of the “hardware” type. The type of a Section to describe an amplifier is then “hardware/amplifier.” With this construct a reading tool that may have no concept of an amplifier can infer from the type that the described entity is a hardware item. This does not imply that specialized section types inherit all Properties of the parent type. The Section name could be any identifying name that is used to uniquely refer to the specific item.
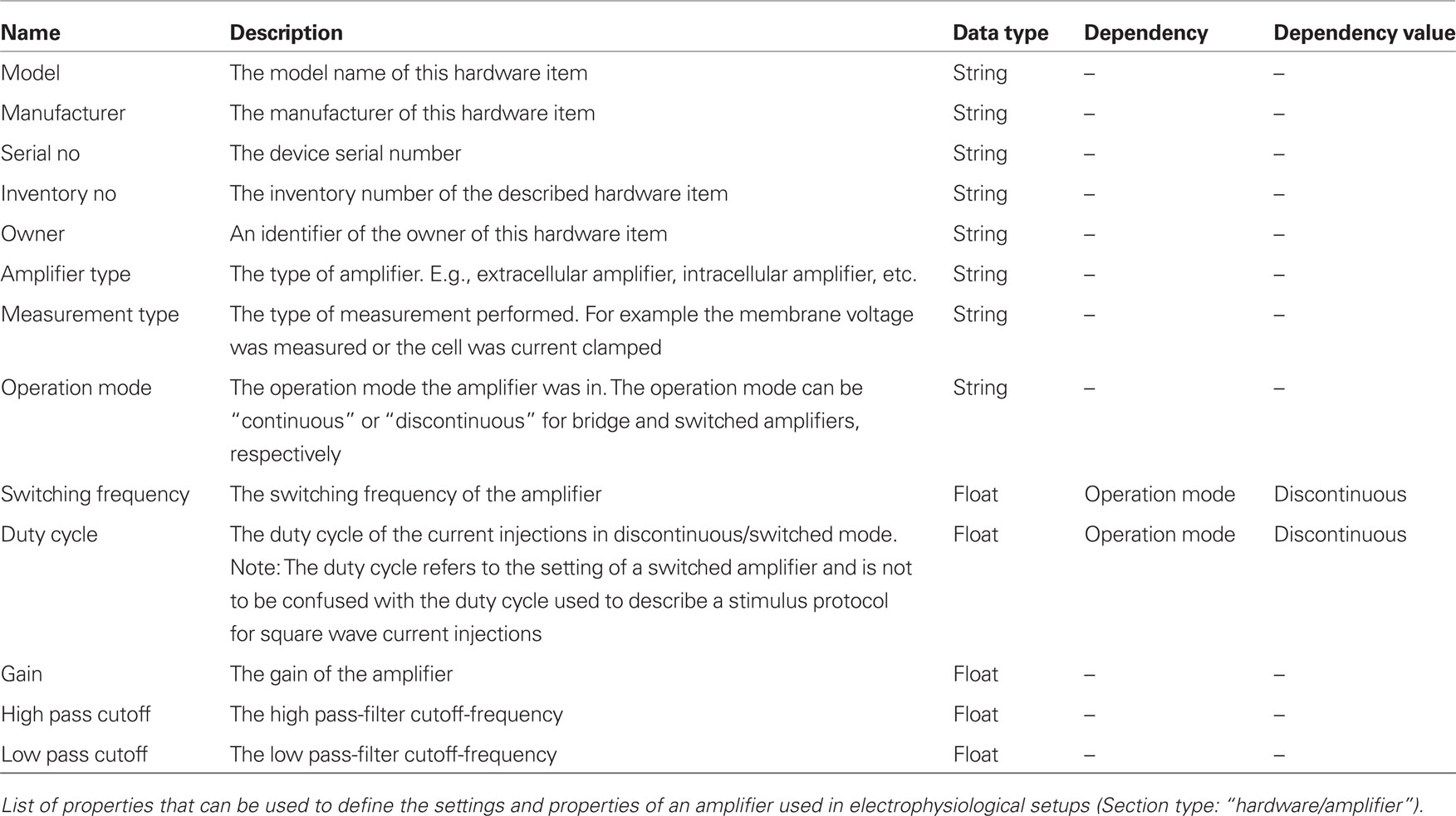
The terminology shown in Table 2 lists Properties that can be used to describe an amplifier. Most of the defined Properties are self-explanatory. A special feature is seen with the “SwitchingFrequency” and the “DutyCycle” properties. They define a “dependency” and a “dependencyValue,” indicating that they are only meaningful if the specified dependency property (here: “OperationMode”) assumes a specific value (here: “Discontinuous”). Tools using the metadata can make use of the dependency information for consistency checks or to adapt a GUI to offer appropriate values and properties.
When describing hardware items we distinguish the properties of the item from its actual settings. Properties describe the fixed characteristics of a hardware component (e.g., the “Model,” “Manufacturer,” or the maximum possible sampling rate of an analog input “AIMaxSampleRate,” etc.), whereas settings are the actual settings of adjustable features of the device (e.g., the currently used sampling rate “AISampleRate”). Accordingly there are two terminologies containing HardwareProperties or HardwareSettings typed sections. These do not define any Properties of their own but are meant as containers for respective hardware Sections. This separation allows the same name (e.g., “LowpassCutoff”) to be used as a hardware property for an amplifier with a fixed filter as well as a setting for one with an adjustable filter. Using the same Section names in both contexts (properties and settings) relates them to the same entity (Figure 3). The separation is not required but we consider it helpful when describing setups and hardware.
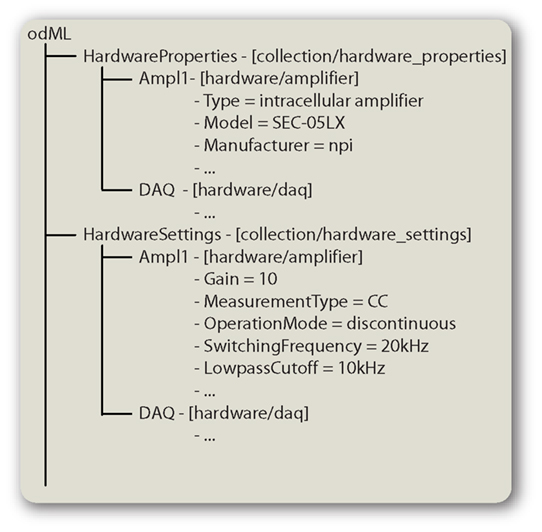
Figure 3. Hardware descriptions in odML. Hardware descriptions can be split up into the HardwareProperties and HardwareSettings. These container sections then group subsections for the individual hardware items used in the setup. Sections are shown in the form “name – [type].”
3.1.3 Stimulus terminologies
The stimulus that was used to evoke the recorded neuronal response is a highly important piece of information for which, so far, there is no standard way of description. This paragraph briefly illustrates how the often quite complex information about the stimulus can be stored in an odML metadata file. Stimuli are described by a “stimulus”-type Section and its subsections. Figure 4 shows how such a description might look like. This visual stimulus (trace on top of Figure 4) is a combination of three components. A DC shift, a sine wave, and a white noise component are combined and delivered using the same “OutputChannel” (“LED1”). The timing of the stimulus components is defined by the “TemporalOffset” and “Duration” properties. A “TemporalOffset” of zero represents the beginning of the stimulus and all other times are relative to this. If the different components were multimodal (e.g., a visual stimulus combined with a current injection) the respective components must define their own “Modality” and “OutputChannel” properties.
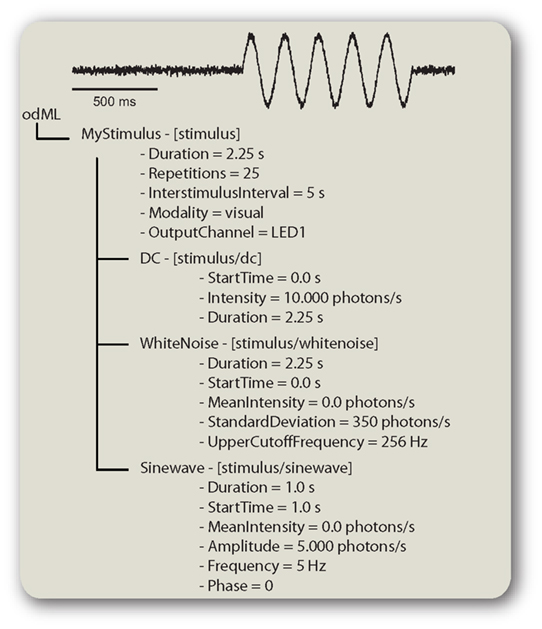
Figure 4. Describing a stimulus in odML. odML description of a visual stimulus which is an additive combination of three components. The trace on top shows how the actual stimulus might have looked like. Sections are shown in the form “name – [type].”
4 Using odML
As indicated above, the odML model contains more elements than are necessary during everyday use for data storage and analysis. Using odML should leave all options to the user. The only real restrictions imposed are almost trivial: any section must be of a specified type and should be named. Any property must have a name and a value. Optional fields offer further information: for numerical values a unit might be necessary and, if appropriate, a value definition that points to a resource that defines the value (for an example see Table A4 in Appendix). For neurophysiological datasets we strongly encourage using the terms as defined in the terminologies (see text footnote 6). However, the approach should be pragmatic:
(1) If you find an appropriate property in the respective terminologies, use it.
(2) Ignore properties that are not needed for describing your dataset.
(3) If you do not find appropriate properties in the terminologies, add new properties as needed to describe your data. For each new property provide a definition (recommended), and if you consider it of wider relevance suggest it for inclusion in the G-Node standard.
(4) If you strongly disagree with a Property’s name and prefer your own term, just provide a mapping to the respective terminology term. This ensures that the metadata can be understood by someone using the standard term.
(5) If a Property’s value is defined in some standard, e.g., an ontology like NEUROLEX8, use the Value’s definition field to provide a link to the respective definition.
4.1 Organization of the Metadata
In odML the hierarchical organization of the metadata tree plays a central role in defining which metadata belong together, e.g., which Sections are related to the same dataset. Regarding the organization of the metadata some simple rules should be obeyed:
(1) The hierarchical organization reflects the relations between sections. For example, if there is a “Cell” section that is child of a “Subject” section, then this cell is from that subject.
(2) The tree structure defines three basic relations between nodes (i.e., the Sections in odML trees) are defined: (i) parent, (ii) child, and (iii) sibling relationships. In odML these relations have different precedence. The child relation (subsections, and their subsections) is strongest. The strength of the relation between nodes descends through siblings (sections that have the same parent) to parents and their siblings. More distant relations are not considered. For example, if a dataset contains data recorded from a single cell this dataset is related that cell. In the odML tree this relation can be expressed by having the cell section being child of the dataset section. If there several datasets originating from the same cell, the dataset sections could be children of the same cell section. Siblings are treated equally and without order.
(3) Two instances of the same section type that logically belong to the same super-section (e.g., two recorded cells that contributed to one dataset) have to be siblings on the same branch of the tree.
(4) If some metadatum is described by a combination of subsections these compositions must be “pure” in the sense that all subsections must be of, or derived from, the same type. For example a stimulus description can be a composition of stimulus-derived subsections (see Figure 4).
(5) The hierarchy should be kept as flat as possible.
(6) There are some restrictions on the style in which Property and Section names as well as Section types are given. All must begin with a letter and are then treated case insensitive. In case of Section names and types, slashes (“/”) are reserved to separate paths in the tree, respectively type components (e.g., “hardware/amplifier”). By convention the Properties and Sections are given in “upper CamelCase” while Section types are all lower case.
The following paragraphs exemplify how to use odML and how to organize the metadata according to these rules when describing electrophysiological data.
4.2 Example: Information about Recorded Datasets
Figure 5 illustrates how to describe quite common situations in electrophysiological experiments. (i) a simple experiment in which several datasets were recorded from a single cell (Figure 5A). (ii) several datasets have been recorded from different cells of the same subject (Figure 5B). (iii) several datasets have been recorded in a simultaneous recording of two cells (Figure 5C). These situations are briefly described in the following paragraphs. As metadata is data about data, the linchpin of odML files discussed here are the datasets, that is the dataset-type sections.
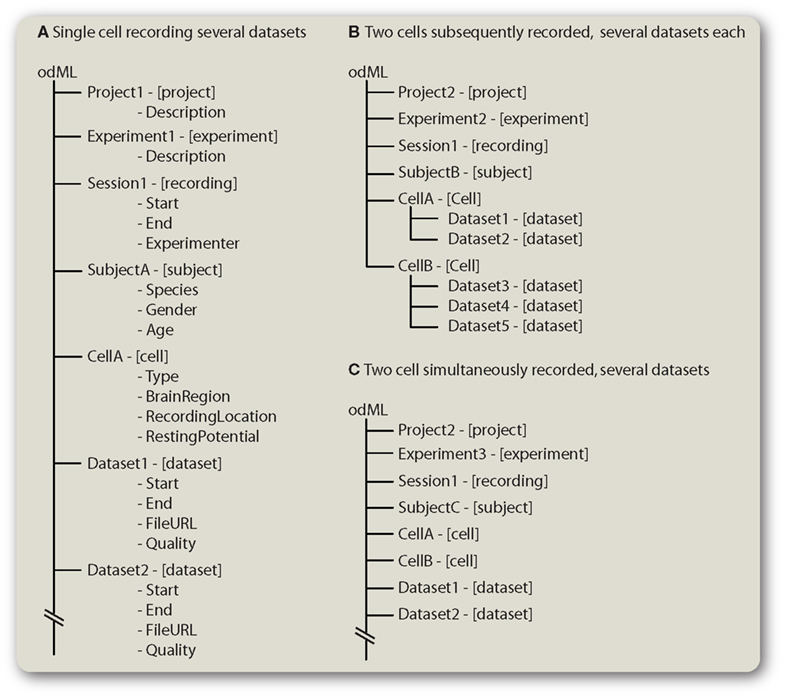
Figure 5. Transporting dataset information in odML. (A) Parts of the description of a simple electrophysiological experiment in which a single cell was recorded and several datasets were saved to disk. (B) Experiments in which several datasets have been recorded in a number of cells from the same subject. (C) Description of simultaneous recordings of two cells. Note: For clarity Properties are omitted in (B,C). Sections are shown in the form “name – [type].”
The first example (Figure 5A) illustrates a very simple case in which the hierarchy can be kept flat, since all datasets (two are shown in this example) depend on all other sections (rule 2) and there is no need for more complex structures in the actual metadata file (rules 1 and 5). Other structures to arrange these items are possible but not encouraged.
The second example (Figure 5B) shows a case where datasets originating from two different cells recorded in the same subject have to be described. Again, each of the dataset sections shown are related to the same project, experiment, recording, and subject information, according to rule 2. Their parent “cell” sections are different, reflecting the two different cells from which the datasets were obtained.
The dual recording in Figure 5C again yields a simple flat tree structure. Regarding rules 1 and 2 the two cells could have been children of the subject section but this would “violate” the rule 5 of a flat tree structure.
To document several datasets recorded in different recording sessions, it would be possible to have a number of recording sections in the same metadata file. In this case all dataset sections would need to be children of the respective recording sections (rule 1).
4.3 Relations Outside the Hierarchy
There are situations in which a strict application of the rules about the hierarchical organization (see above) would lead to complicated and redundant structures. To avoid this odML Sections define link, include, and reference elements which can be used to introduce relations that exist outside the hierarchical organization.
A link is used to refer to sections within the document and contains a path in the tree. Paths defining the position in the tree are given by Section names, separated by slashes (“/”). Paths are absolute, i.e., they begin at the root of the tree. To illustrate the use of links, we consider a case in which a number of datasets have been recorded using a rather complex stimulus. The stimulus is repeated for each dataset, but each time a single stimulus parameter, e.g., the intensity, has been changed. It would be valid to provide the full stimulus description for each dataset individually but, obviously, this would be cumbersome and inefficient. Instead, one defines the stimulus once within the document and then links to it for each dataset. The referring stimulus sections contain only the changes (i.e., the “intensity” properties). Links can only exist between sections of the same type and include all subsections and properties. Local information, given in the linking section, overrides items inherited from the linked Section.
The include element can be used to establish relations to sections (same type) that are located in an external file. Include entries can be either an URL or a path in the file system (relative or absolute). The URL is followed by a hash symbol (#) and the absolute path of the target Section. For example, a stimulus Section could contain an include element like “stimulus-metadata.xml#myStimulus.” This indicates that the stimulus information is provided in a stimulus-type section of the name “myStimulus” located in the “stimulus-metadata.xml” file in the same folder in the file system. Of course care has to be taken if the data is shared when local files are referenced. As for the link element the locally provided information overrides the one given in the included section.
Finally, Sections and Values can be descriptions of entities that are entries in a data management system. These entries usually have an id, or primary key, for unique identification. The reference element of Section and Value is meant to keep this reference information. For example, a dataset that is already stored in a database is exported and used for further analysis. If it is intended to import the respective results back into the data management system a correct linking can be easily done if the primary key is included in the reference element.
4.4 Synonyms and Mappings
Different communities, laboratories, or individuals in the neurosciences do not always agree on how to refer to the same entities. The odML approach respects this by allowing to avoid the standard without breaking it.
Furthermore, the standard odML-terminologies logically group properties into sections. This structure may in some cases not match the structure a scientist or laboratory uses to describe their metadata. In these cases, the possibility to define mappings between own Properties and Sections to those in the terminologies provides interoperability. With mappings it is possible to define synonyms for single Properties or also to define custom compact and non-hierarchical terminologies for everyday use, which defines mappings to the standard terminologies. The mapping information can be given for Sections and Properties and is provided in form of an URL. For Sections this URL points to the terminology of the target Section, whereas the reference part of the URL contains the path of the target Section (see Figure 6 mapping of “StudySubject”). For Properties the appropriate section reference is followed by a colon and the destination property (see Figure 6). The odML-libraries (see below) can apply this mapping information and convert a metadata file that was written according to the local terminology to a file that is compatible to the layout suggested by the odML-terminologies. For example, the CARMEN consortium works with a specific set of metadata (Gibson et al., 2008a). If one is used to work with the metadata terms defined there, one can use the carmen_mini terminology http://portal.g-node.org/odml/terminologies/v1.0/carmenMini/carmen_mini.xml which defines the mapping between the MINI and the standard odML-terminologies. Thus, transferring data between the CARMEN database and an odML compliant tool is directly possible. Figure 6 shows how the mapping works. The odML tree on the left side is created with the terms defined in the carmen_mini terminology. The Sections and Properties carry mapping information (the URLs) to respective Sections and Properties in the standard odML-terminologies. The odML-libraries (see below) can use these mappings to convert the tree to one that complies the odML-terminologies (right part of Figure 6). In cases in which a Property does not provide mapping information (“SpeciesIdentifier” Property in the “Subject” Section) it kept as is and is mapped into the its parent’s counterpart. The same principle applies for Sections that do not provide mapping information. Generally mapping information is provided in the terminologies. Thus, it is not necessary to provide them with the actual metadata files. In case of a conflict between the provided mapping and the one given in the terminology, the one in the actual metadata file overrides the terminology mapping.
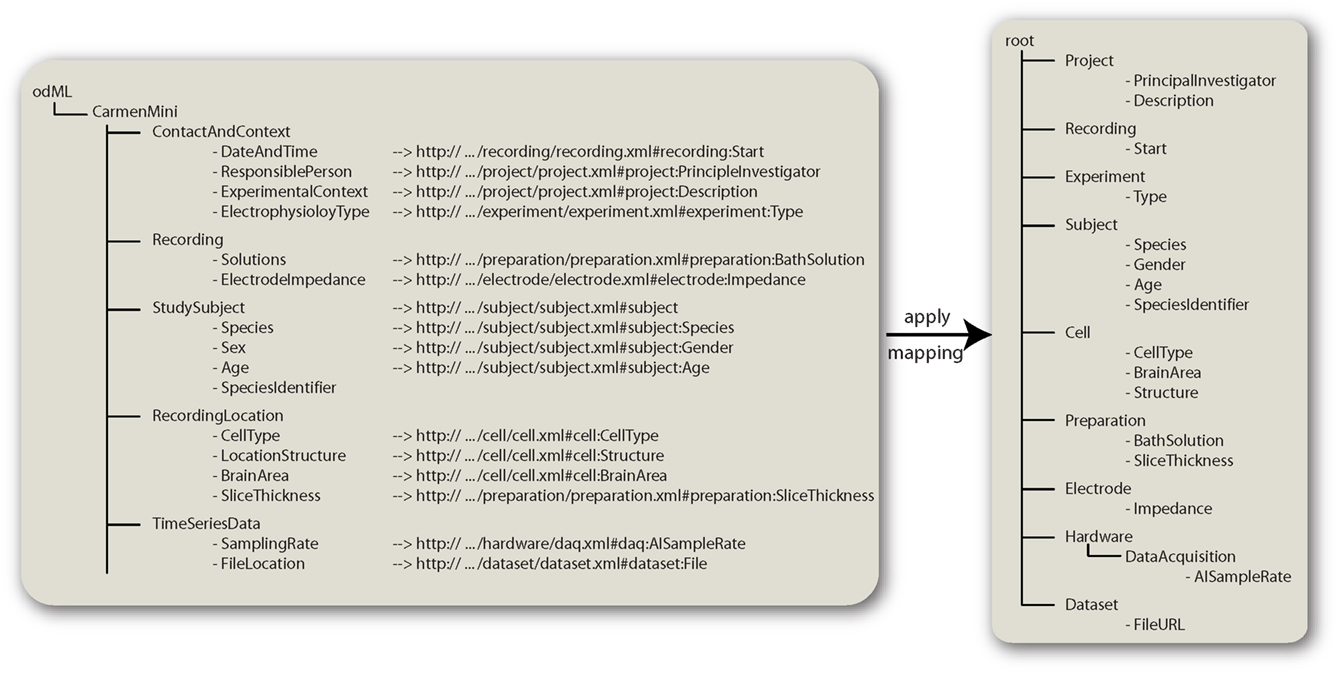
Figure 6. Using mappings. This figure shows how mappings can be applied to convert a metadata tree from one layout to another. The left panel shows metadata that are organized as suggested by the CARMEN “Mini” metadata standard. The metadata file is in the odML format and refers to the CarmenMini terminology which defines mappings for properties and sections. These are URLs to the respective properties in the odML-terminologies. Applying this mapping information converts the tree to the layout suggested by the odML-terminologies (right panel).
4.5 Supporting Tools
On the odML website at www.g-node.org/odml we offer libraries and tools to manipulate odML metadata. So far, there is a Java implementation which can be easily used with Matlab. We currently work on implementations of these odML-libraries in C/C++ and Python. By means of these libraries custom software can be easily extended to automatically write or read odML files. We also offer an editor to create, view, and manipulate odML files. All this software is freely available and open-source.
In our laboratory, odML is used to transfer metadata from the recording program that automatically writes metadata to disk (RELACS by Jan Benda)9 and our data management tool (The LabLog by Jan Grewe)10. The G-Node data management system11 supports odML as a format for metadata import and export. Furthermore, it is planned to integrate odML support into the Vision Egg (Straw, 2008), a free tool to generate and present visual stimuli.
4.6 Example Scenario
This paragraph briefly describes an example scenario when using odML in the lab. For this example it is assumed that a set of datasets has been recorded and that the recording tool has written the metadata into an odML file. The following listings describe how one could use the metadata during data analysis in Matlab. The underlying odML library is written in Java and can be easily used in Matlab.
Line 2: Call of a Matlab function that imports the odML library and the used logger to the Matlab classpath. Line 5: the reader variable is an instance of the Reader class which handles reading of odML files. Line 6: load-function reads the metadata and, as is defined by the option flag, includes external files, resolves links, and converts the tree, if mapping information is provided. Mappings are either specified in the metadata itself or in the used terminologies. Line 9 and following: the dataset sections defined in the metadata are found. These are retrieved by looking for the “dataset” section type. One could also look for them by name, if known. The returned set (a Java Vector) contains references to the respective sections in the metadata tree which can be passed to the analysis function. Such a function is sketched in Listing 2. In this example loading data is handled by a further matlab function (that is not part of the odML library line 13). The analysis function returns, besides the results, the analysis metadata (line 14) which are appended to the dataset metadata (line 18). Finally, the new metadata is written to a file using an instance of the Writer class (lines 22, 23).
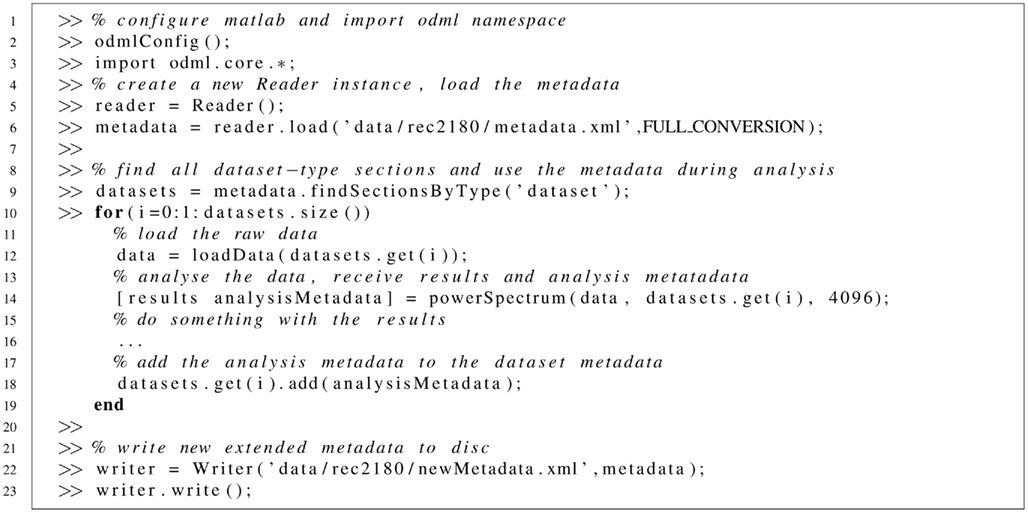
Listing 1. Using odML in Matlab. Example code shows how odML could be used during everyday work in the lab. The listing shows Matlab command line calls.
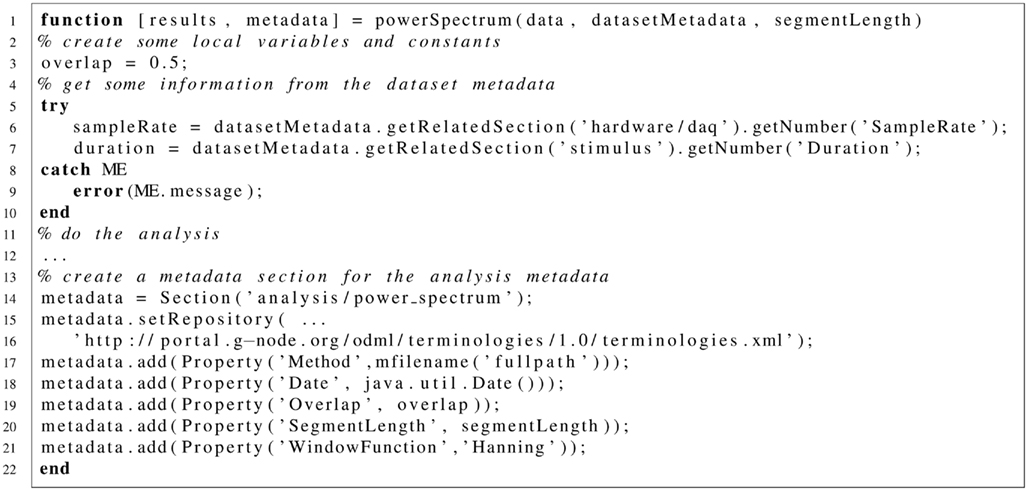
Listing 2. Dummy Matlab function “powerSpectrum.m” to illustrate how metadata can be retrieved and used during data analysis.
The “powerSpectrum” function sketched in listing 2 takes the data, the respective metadata Section and a constant. For the performed tasks it needs to extract some information from the metadata. Lines 6 and 7: For this, the library is asked for a related Section of the specified type. For example, the rate with which the data is sampled should be found in a Property called “SampleRate” that is part of a hardware/daq (data acquisition) type section. In odML there are no strict rules where this section is located in the metadata tree. The library tries to return the section with the strongest relation according to the rules defined above. If this operation fails the code will raise an exception which is caught by the surrounding try–catch clause. Line 14: Here, a Section is created that will contain the analysis metadata. Line 15: Setting the repository URL indicates that the terminology defining a section of this type can be found at the specified location. Line 17: this line uses the matlab function “mfilename” to find out the actual function name which is inserted into the Method–Property. Line 18: An instance of the java.util. Date class, which contains the current date, is passed to the Date-Property.
5 Discussion
The key aspect of our approach to metadata handling is a common format for both the actual metadata and terminologies. This allows for flexible storage of any metadata, since new keys (Property-names) can be immediately added, without the need to extend a terminology or schema beforehand. Terminologies guarantee interoperability and are built-in a bottom-up way by the scientist that work with the data. With odML we provide a format and tools for automated metadata handling, so that the threshold for collecting metadata is considerably lowered. We hope that the flexibility of odML will convince scientists to embed metadata handling into their recording, data analysis, and management tools, thereby laying the foundation for large-scale collaborations, in particular in the neurosciences.
5.1 Advantages and Disadvantages of Using a Generic Metadata Model
We propose to use a generic data model for the metadata. The nested tree-like organization, in our opinion, is easily comprehensible and flexible. It further has a very limited number of structural elements. An alternative would have been a fully defined data model with clearly defined terms and constructs, as used in most annotation approaches (Gardner et al., 2001a; Gibson et al., 2008a). Such a design has clear advantages, for example, a guaranteed structure with detailed validation options. However, in our view it also has severe disadvantages: (i) it is hard to foresee what entities any one scientist might need to describe and (ii) using a completely specified model means that the user has to internalize all of its elements and dependencies and to accept the designated terms defined in the model. The odML data model itself does not impose standards. The terminologies introduce options for standardization and validation in a “soft” fashion which may be overridden by the user. This bears the risk of inconsistencies and leaves responsibility on validity to the user. However, this loose standardization is the key for flexibility and immediate extensibility, which we consider essential for practical application in the laboratory. Restricting as little as possible may be the key to convince researchers of annotating their data to allow reproducibility and support data sharing.
One further advantage of the generic model lies in the interchange of (meta)data between databases. Instead of writing and maintaining converters between all the different data models, it would suffice if each tool or platform could import and export metadata from the generic model. odML is very different form metamodel approaches like XMI12 (which are designed for data transfer between different data models). XMI, however, requires the existence of detailed data models which do not yet exist for neurophysiological metadata.
5.2 Terminologies and Ontologies
Compared to other disciplines in biology, such as Genomics and Proteomics (e.g., Gelbart et al., 1997; Stoesser et al., 1997), the neurosciences lag behind regarding the use of databases for the organization and exchange of data. Only recently attempts have been started to integrate neuroscience databases structures (Amari et al., 2002; Gupta et al., 2008), and there are now several initiatives that set out to foster the sharing of neurophysiological data (Gardner et al., 2001a, 2008; Gibson et al., 2008a; Herz et al., 2008; Teeters et al., 2008). A particular issue in the sharing of neurophysiology data is the relatively large amount of additional information necessary to describe an experiment. Moreover, this meta-information is often highly complex and can vary from laboratory to laboratory and from experiment to experiment, depending on the specific scientific context in which the experiment was conducted. This situation is complicated further by the lack of a standardized terminology in the field, where different sub disciplines may use different terms to describe the same neural structure, brain location, or neuron type (Bezgin et al., 2009). For some sub themes of the neurosciences standards and ontologies have been developed (Gardner et al., 2001b; Crook et al., 2005; Bowden et al., 2007; Bug et al., 2008; Frishkoff et al., 2009; Larson and Martone, 2009). The main difference between the mentioned ontologies and the odML-terminologies is that the ontologies define terms for what would be a Value in an odML – Property. The odML-terminologies will not be extended in this direction. Rather, it is suggested to have values referring to entities defined in an ontology (for example by providing the respective resource location in NEUROLEX (see text footnote 8) or BrainInfo13 in the definition element of the Value).
The odML-terminologies are no replacement for ontologies but can support ontology development. For example, currently there are no ontologies for setup, hardware, or stimulus descriptions. Our proposed method for specifying metadata could in turn provide an efficient method to support these developments by analyzing the metadata that scientists actually use in certain contexts.
5.3 Annotations in Other Data Formats
Many data formats like the Exif format for image metadata14 or ID3 tags for music annotation15 use pre-defined key–value pairs for data annotation. This approach is especially useful for cases in which the type of data that requires description and thus the required metadata terms are known in advance. The situation is more difficult when describing scientific data which is variable in many respects. The odML approach of storing metadata in the hierarchical way is to some extent similar to the way data annotations can be handled in other formats. HDF5 specifies the tree structure of nodes. These can contain attributes which are essentially key–value pairs. Nodes and attributes are thus similar to Sections and Properties but lack the opportunity of standardization. The “Scientific Data Set” extension of HDF516 offers pre-defined attributes, in addition to the “free” attributes. This gives control over the terms used in the attributes but requires that the user commits himself to the pre-defined terminology. In our view these solutions are not sufficient for flexible and extensible data annotations. For this we decided to keep the metadata separated from the actual data and to use a format that does not restrict the user but allows to apply the terminologies for standardization. For future development we aim at solutions (in the form of an API) that bring data and metadata again closer together while ideally being independent of the actual format in which data and metadata are stored.
5.4 Application to Data sharing in Cellular and Systems Neurophysiology
For databases and data sharing platforms in the neurosciences to be useful in the long run, machine-readable data annotation should not require additional manual effort when data are uploaded to a database. Instead, data annotation would be ideally integrated within the data acquisition and analysis workflow in the laboratory. This would have the further benefit of facilitated data management and data analysis for the individual scientist, independent of whether the data are uploaded to a data sharing platform or not. Moreover, scientists may be much more willing to contribute to data sharing initiatives if the upload is just a single command or button click because the metadata already exist in a machine-readable form and do not have to be entered manually. odML provides a format that enables this computer-based metadata management and exchange from the local laboratory to global neuroscience databases.
Currently, none of the data sharing portals for neurophysiology (Gardner et al., 2001a; Gibson et al., 2008a; Teeters et al., 2008) offers the possibility to routinely enter the metadata into a database by providing them in a machine-readable format. At the CARMEN platform, users can define metadata templates to ease the manual metadata entry for similar data sets. At the (see text footnote 1) database, data, and metadata upload is possible via an XML interface. This means, however, that only the set of pre-specified metadata elements can be provided and additional information, which may be essential to meaningfully analyze the data, cannot be entered easily.
The main design goal of odML is its immediate extensibility. Any metadata item can be included into a valid odML file, no matter whether it is already defined in a standard terminology. At the same time no information is mandatory. With odML, the only restriction is the metadata model, not its content. A big advantage of this approach may be the future possibility of machine-aided construction, extension, and refinement of ontologies. Analyzing the structure and terminology of the metadata provided by a large number of scientists from a given neuroscience subfield may enable a “bottom-up” development of ontologies for this subfield, which may be an efficient complement to the “top-down” approach of gathering experience and contributions from selected experts. In order to have the necessary metadata in machine-readable formats in the future, it is time to start collecting them in the laboratories now.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Andrey Sobolev, Christine Seitz, Christian Kellner, and Christian Tatarau for programming and discussions, Adrian Stoewer, Alvaro Tejero-Cantero, Colin Ingram, Fritz Sommer, Gwen Jacobs, Marianne Martone, Piotr Durka, and Raphael Ritz for fruitful discussions. Raphael Ritz and Zbigniew Jędrzejewsky-Szmek for comments on an earlier version of the manuscript. Supported by BMBF grants 01GQ0802 and 01GQ0801.
Footnotes
- ^www.neurodatabase.org
- ^www.neuroshare.org
- ^www.signalml.org
- ^www.obofoundry.org
- ^www.hdfgroup.org
- ^www.g-node.org/odml
- ^www.dublincore.org
- ^www.neurolex.org
- ^www.relacs.net
- ^www.lablog.sourceforge.net
- ^www.g-node.org/data
- ^www.omg.org/spec/XMI
- ^www.braininfo.rprc.washington.edu
- ^www.exif.org
- ^www.id3.org
- ^www.hdfgroup.org/sds_api.html
References
Amari, S.-I., Beltrame, F., Bjaalie, J. G., Dalkara, T., De Schutter, E., Egan, G. F., Goddard, N. H., Gonzalez, C., Grillner, S., Herz, A., Hoffmann, K.-P., Jaaskelainen, I., Koslow, S. H., Lee, S.-Y., Matthiessen, L., Miller, P. L., Da Silva, F. M., Novak, M., Ravindranath, V., Ritz, R., Ruotsalainen, U., Sebestra, V., Subramaniam, S., Tang, Y., Toga, A. W., Usui, S., Van Pelt, J., Verschure, P., Willshaw, D., and Wrobel, A. (2002). Neuroinformatics: the integration of shared databases and tools towards integrative neuroscience. J. Integr. Neurosci. 1, 117–128.
Bezgin, G., Reid, A. T., Schubert, D., and Kötter, R. (2009). Matching spatial with ontological brain regions using java tools for visualization, database access, and integrated data analysis. Neuroinformatics 7, 7–22.
Bowden, D. M., Dubach, M., and Park, J. (2007). Creating neuroscience ontologies. Methods Mol. Biol. 401, 67–87.
Bug, W. J., Ascoli, G. A., Grethe, J. S., Gupta, A., Fennema-Notestine, C., Laird, A. R., Larson, S. D., Rubin, D., Shepherd, G. M., Turner, J. A., and Martone, M. E. (2008). The nifstd and birnlex vocabularies: building comprehensive ontologies for neuroscience. Neuroinformatics 6, 175–194.
Crook, S., Beeman, D., Gleeson, P., and Howell, F. (2005). XML for model specification in neuroscience. Brains Minds Media 1. Available at: http://www.brains-minds-media.org/archive/228
Durka, P., and Ircha, D. (2004). Signalml: metaformat for description of biomedical time series. Comput. Methods Programs Biomed. 76, 253–259.
Fletcher, M., Liang, B., Smith, L., Knowles, A., Jackson, T., Jessop, M., and Austin, J. (2008). Neural network based pattern matching and spike detection tools and services – in the carmen neuroinformatics project. Neural. Netw. 21, 1076–1084.
Frishkoff, G., LePendu, P., Frank, R., Liu, H., and Dou, D. (2009). “Development of neural electromagnetic ontologies (NEMO): ontology-based tools for representation and integration of event-related brain potentials,” in Proceedings of the International Conference on Biomedical Ontologies, Buffalo, NY.
Gardner, D., Abato, M., Knuth, K. H., DeBellis, R., and Erde, S. M. (2001a). Dynamic publication model for neurophysiology databases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356, 1229–1247.
Gardner, D., Knuth, K. H., Abato, M., Erde, S. M., White, T., DeBellis, R., and Gardner, E. P. (2001b). Common data model for neuroscience data and data model exchange. J. Am. Med. Inform. Assoc. 8, 17–33.
Gardner, D., Goldberg, D. H., Grafstein, B., Robert, A., and Gardner, E. P. (2008). Terminology for neuroscience data discovery: multi-tree syntax and investigator-derived semantics. Neuroinformatics 6, 161–174.
Gelbart, W. M., Crosby, M., Matthews, B., Rindone, W. P., Chillemi, J., Twombly, S. R., Emmert, D., Ashburner, M., Drysdale, R. A., Whitfield, E., Millburn, G. H., de Grey, A., Kaufman, T., Matthews, K., Gilbert, D., Strelets, V., and Tolstoshev, C. (1997). Flybase: a drosophila database. the flybase consortium. Nucleic Acids Res. 25, 63–66.
Gibson, F., Austin, J., Ingram, C., Fletcher, M., Jackson, T., Jessop, M., Knowles, A., Liang, B., Lord, P., Pitsilis, G., Periorellis, P., Simonotto, J., Watson, P., and Smith, L. (2008a). “The carmen virtual laboratory: web-based paradigms for collaboration in neurophysiology,” in 6th International Meeting on Substrate-Integrated Microelectrodes, 2008, Reutlingen.
Gibson, F., Overton, P., Smulders, T., Schultz, S., Eglen, S., Ingram, C., Panzeri, S., Bream, P., Whittington, M., Sernagor, E., Cunningham, M., Adams, C., Echtermeyer, C., Simonotto, J., Kaiser, M., Swan, D., Fletcher, M., and Lord, P. (2008b). Minimum information about a neuroscience investigation (MINI): electrophysiology. Nat. Precedings. Available at: http://hdl.handle.net/10101/npre.2009.1720.2
Gupta, A., Bug, W., Marenco, L., Qian, X., Condit, C., Rangarajan, A., Müller, H. M., Miller, P. L., Sanders, B., Grethe, J. S., Astakhov, V., Shepherd, G., Sternberg, P. W., and Martone, M. E. (2008). Federated access to heterogeneous information resources in the neuroscience information framework (nif). Neuroinformatics 6, 205–217.
Herz, A. V. M., Meier, R., Nawrot, M. P., Schiegel, W., and Zito, T. (2008). G-node: an integrated tool-sharing platform to support cellular and systems neurophysiology in the age of global neuroinformatics. Neural. Netw. 21, 1070–1075.
Hey, T., and Trefethen, A. (2003). The Data Deluge: An E-Science Perspective. Chichester, West Sussex: Wiley & Sons, 809–824.
Larson, S. D., and Martone, M. E. (2009). Ontologies for neuroscience: what are they and what are they good for? Front. Neurosci. 3:1. doi: 10.3389/neuro.01.007.2009
Stoesser, G., Sterk, P., Tuli, M. A., Stoehr, P. J., and Cameron, G. N. (1997). The embl nucleotide sequence database. Nucleic Acids Res. 25, 7–14.
Straw, A. D. (2008). Vision egg: an open-source library for realtime visual stimulus generation. Front. Neuroinformatics 2:4. doi: 10.3389/neuro.11.004.2008
Taylor, C. F., Field, D., Sansone, S.-A., Aerts, J., Apweiler, R., Ashburner, M., Ball, C. A., Binz, P.-A., Bogue, M., Booth, T., Brazma, A., Brinkman, R. R., Clark, A. M., Deutsch, E. W., Fiehn, O., Fostel, J., Ghazal, P., Gibson, F., Gray, T., Grimes, G., Hancock, J. M., Hardy, N. W., Hermjakob, H., Julian, R. K., Kane, M., Kettner, C., Kinsinger, C., Kolker, E., Kuiper, M., Novère, N. L., Leebens-Mack, J., Lewis, S. E., Lord, P., Mallon, A.-M., Marthandan, N., Masuya, H., McNally, R., Mehrle, A., Morrison, N., Orchard, S., Quackenbush, J., Reecy, J. M., Robertson, D. G., Rocca-Serra, P., Rodriguez, H., Rosenfelder, H., Santoyo-Lopez, J., Scheuermann, R. H., Schober, D., Smith, B., Snape, J., Stoeckert, C. J., Tipton, K., Sterk, P., Untergasser, A., Vandesompele, J., and Wiemann, S. (2008). Promoting coherent minimum reporting guidelines for biological and biomedical investigations: the mibbi project. Nat. Biotechnol. 26, 889–896.
Keywords: metadata, ontology, neuroscience, datamodel, datasharing
Citation: Grewe J, Wachtler T and Benda J (2011) A bottom-up approach to data annotation in neurophysiology. Front. Neuroinform. 5:16. doi: 10.3389/fninf.2011.00016
Received: 25 March 2010; Accepted: 12 August 2011;
Published online: 30 August 2011.
Edited by:
Ulla Ruotsalainen, Tampere University of Technology, FinlandReviewed by:
Friedrich T. Sommer, University of California at Berkeley, USAJari Peltonen, Tampere University of Technology, Finland
Copyright: © 2011 Grewe, Wachtler and Benda. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Jan Grewe, Department Biology II, Ludwig-Maximilians Universität München, Großhaderner Str. 2, 82152 Martinsried, Germany. e-mail: grewe@bio.lmu.de