Practical management of heterogeneous neuroimaging metadata by global neuroimaging data repositories
- Laboratory of Neuro Imaging, Department of Neurology, David Geffen School of Medicine, University of California Los Angeles, Los Angeles, CA, USA
Rapidly evolving neuroimaging techniques are producing unprecedented quantities of digital data at the same time that many research studies are evolving into global, multi-disciplinary collaborations between geographically distributed scientists. While networked computers have made it almost trivial to transmit data across long distances, collecting and analyzing this data requires extensive metadata if the data is to be maximally shared. Though it is typically straightforward to encode text and numerical values into files and send content between different locations, it is often difficult to attach context and implicit assumptions to the content. As the number of and geographic separation between data contributors grows to national and global scales, the heterogeneity of the collected metadata increases and conformance to a single standardization becomes implausible. Neuroimaging data repositories must then not only accumulate data but must also consolidate disparate metadata into an integrated view. In this article, using specific examples from our experiences, we demonstrate how standardization alone cannot achieve full integration of neuroimaging data from multiple heterogeneous sources and why a fundamental change in the architecture of neuroimaging data repositories is needed instead.
Introduction
The role of neuroimaging in biomedical research is increasingly important as new modalities targeting different aspects of disease progression are developed and the utility of imaging as a biomarker is more widely recognized (Ryan and Fox, 2009; Brooks and Pavase, 2011). The number of magnetic resonance imaging (MRI) scanners installed in the United States increased more than 230% between 1995 and 2004 with similar increases in other countries (National Center for Health Statistics, 2007) contributing to an increase in biomedical research studies that are incorporating neuroimaging. This trend, combined with advances in networking and computing technologies, has converged toward an environment in which global, multi-disciplinary collaborations between geographically distributed scientists are not only possible, but are becoming common (Mazziotta et al., 2001; Toga, 2002; Butcher, 2007; Toga and Crawford, 2010). As multi-site, collaborative efforts proliferate, neuroimaging repositories offer a way to pool data from multiple institutions to provide larger sample sizes and shared resources. In an ideal environment, this data would be uniform, consistent, and easy to use. However, neuroimaging data often come in many flavors and in spite of harmonization efforts of controlled studies, are acquired from different scanners in different modalities, and are heterogeneous in data format representations and metadata content (Wong and Huang, 1996). The separation between collecting and analyzing data require good data management practices and extensive and standardized metadata (Atkins et al., 2003; Gray et al., 2005), otherwise data may be misinterpreted, difficult to use, or completely unusable.
Shared data repositories offer many benefits, often beyond those envisioned by those who collected or created the data (National Science Board, 2005). However, the inherent complexity of biological data—diversity of data sources, data types, and data processing (Goble and Stevens, 2008) and difficulties in communicating relevant metadata (Teeters et al., 2008)—remains a challenge. The source, content, structure, and context of the collected data must be recorded and are essential to both computerized and human utilization. Without metadata, data is useless (Gray et al., 2005). However, metadata that are inconsistent, poorly defined or ambiguous do not support data reusability. Metadata standardization in the form of shared controlled vocabularies (taxonomies, ontologies) is key to supporting common semantics and data sharing. Significant progress in developing shared controlled vocabularies has been made, however, standards develop slowly, are often perceived to be at odds with investigation and require full adoption in order to be practical (Goble and Stevens, 2008).
Neuroimaging data commonly originates in an isolated clinical setting, and is therefore subject to local constraints and institutional practices (Wiederhold, 2003; Fletcher and Wyss, 2009). For example, limitations of medical image scanners can result in metadata with sparse descriptions, which is particularly problematic with experimental acquisitions and new research protocols. The majority of the metadata is entirely determined by the manufacturers of the scanners, who choose the conventions to conform to and the terminology to use. Since the central function of neuroimaging data repositories is to aggregate data from multiple institutions and provide a schema to its users for posing queries, the data must be combined into one integrated view (Halevy et al., 2006). In effect, neuroimaging data repositories are mediators between multiple heterogeneous data contributors and those who search its contents and extract information (Wiederhold, 1997; Halevy et al., 2006). Ideally, the metadata from each new data source could be automatically mapped into the integrated schema of the data repository, but this “data mapping problem” has long been recognized as a complex and unreachable goal with current technology (Fletcher and Wyss, 2009). As such, there are at present difficulties in not only collecting and integrating neuroimaging data from multiple contributors, but also in converting between the different formats of the image files exchanged between clinicians and research scientists.
Viewed in this context, it is interesting that most recent efforts to resolve these issues have focused upon the creation and adoption of standards (Langlotz, 2006; Bug et al., 2008; Poldrack et al., 2011; Gadde et al., 2012; Turner and Laird, 2012) without a corresponding focus on the development of data mapping tools. This is likely because the focus has been on what information is being stored, as opposed to how it is being stored and exchanged. But as research studies expand their scope to include more varieties of heterogeneous data, the problems associated with data integration are becoming more obtrusive than problems stemming from lack of a lexicon. An inspection of a main resource of neuroimaging tools (http://www.nitrc.org) suggests that the neuroimaging community has not yet appreciated this trend; of the nearly 500 registered tools and resources available we were only able to find one for mapping metadata (Neu et al., 2005).
In this article, we demonstrate why standardization alone cannot solve data integration problems using our experiences in the Laboratory of Neuro Imaging Image and Data Archive (LONI IDA). Our goal is to show why standardization is insufficient through the use of detailed examples that use the information currently being mediated by neuroimaging data repositories. We also hope to give readers who are not familiar with the technical aspects of neuroimaging metadata management a better understanding of why these problems exist. We start by reviewing the composition of neuroimaging files, what metadata is, and why it is needed. Then we highlight some of the obscure complexities of neuroimaging file formats in order to show how the abundance of standards has made file format conversion and the removal of patient-identifying information a formidable effort. Next we recount a situation where standardization could not keep pace with an experimental research protocol, resulting in metadata values that were either missing or incorrect. Finally, we point out where the current dominant standard, DICOM, is not addressing basic and essential grouping and labeling needs of neuroimaging data repositories and conclude that data mapping is essential when integrating neuroimaging data from multiple heterogeneous sources on a global scale.
Review of Image Metadata and Neuroimaging File Formats
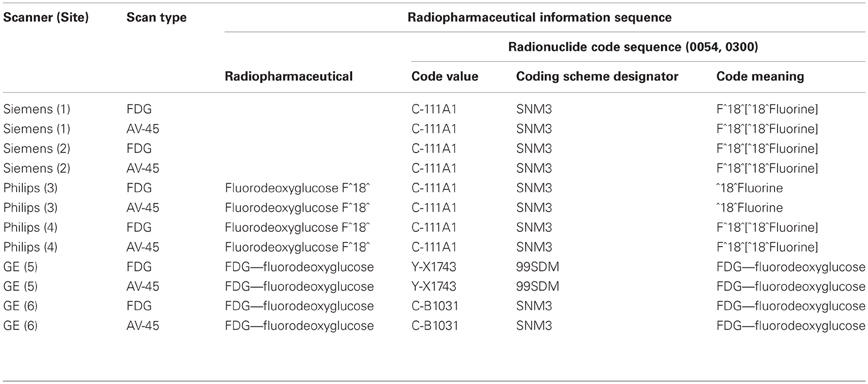
Neuroimaging data have particular requirements and constraints that are necessary to retain usability and interoperability. Digital image data without descriptive metadata is meaningless. This becomes readily apparent when considering how images are written to files on disk. Since every file consists of a line of consecutive bytes, a two-dimensional image must first be transformed into a linear array of image pixels before it can be stored. The method by which this occurs can be understood by visualizing the pixels as beads on a strand of thread. The thread starts at the first pixel in the upper-left hand corner of the image and passes through the pixels on the top image row. It loops back to the first pixel of the next row repeatedly until it reaches the last pixel of the last row. Tightly pulling the thread moves all the pixels into one straight line and the pixel values are written to the file in the thread order (Figure 1). Because the width and height of the image are lost in this process, additional information must be added to the image file in order to reconstruct and display the image. This additional information can also include other image properties such as the size of each pixel and the number of color components. If more than one image is written, the total number of images must be present. This additional information is called image metadata.
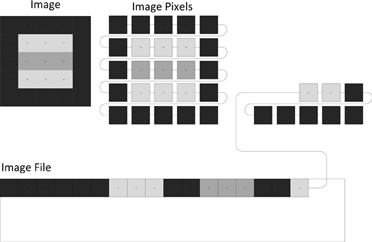
Figure 1. Unraveling the pixels of an image. A two-dimensional image consists of pixels that must be rearranged into a one-dimensional line before the pixel values can be stored in a file.
In addition, there is another type of image metadata separate from the properties of the images. This metadata describes the subject who is scanned and the acquisition of the images; for example, the name of the subject, the manufacturer of the imaging device, the date and time the images were acquired, the substance injected into the subject. Having this information stored with the images is crucial for a wide range of automated processes as well as for allowing humans to understand the context and origin of the images. Digital images are not all produced in the same way, however, they do all contain both image (pixel) data and descriptive metadata.
At present, there are many ways to store neuroimaging images and image metadata in files. A file format defines how the image metadata and pixels are stored in a file. The DICOM file format is the most dominant of these because it is the one used by most medical image scanners. Digital Imaging and Communications in Medicine (DICOM) represents a cooperative effort conceived in 1983 by a joint committee of the American College of Radiology and the National Electrical Manufacturers Association (NEMA) and has as a central goal the development of a standard that would make digital medical imaging independent of the manufacturer and thus facilitate interoperability between information systems. Since its first publication in 1985, the DICOM standard has continually evolved in parallel with developments in new scanner technologies, and though most scanner manufacturers have adopted the DICOM standard [Digital Imaging and Communications in Medicine (DICOM), 2008], differences in how scanner manufacturers apply the standard exist. However, many nuclear medicine scanners still produce image files in other file formats such as Interfile, developed by Keston, Kingston Upon Hull, UK and ECAT developed by CTI, Knoxville, TN. In the IDA, we support the archiving of ANALYZE 7.51, DICOM, ECAT2, GE3, HRRT Interfile (Cradduck et al., 1989), MGH4, MINC5, NIFTI6, and NRRD7 files.
Most medical imaging file formats put the image metadata at the beginning of the file (sometimes called the “image file header”), although there are some file formats (ANALYZE 7.5, Interfile, NIFTI, NRRD) that support writing the image metadata to one file and the images to another file. In the latter case, the two file names must share the same prefix and use the suffixes defined by the file format (e.g., “abc.hdr” and “abc.img”). Some file formats store the image metadata as human-readable text (HRRT Interfile, NRRD) while the others use a binary format. The dictionary of image metadata terms defined by DICOM contains several thousand terms and definitions while other file formats contain a few hundred elements (ECAT) or even less than a hundred (ANALYZE 7.5, NIFTI).
Medical imaging file formats can generally be classified into two groups. Rigid file formats (ANALYZE 7.5, ECAT, GE, MGH, NIFTI) define an unchangeable list of image metadata values (Figure 2) that must be present in the file. The location of each value defines its meaning. Even if the value for an image metadata element is not available, a value must be chosen. This situation often results in the production of files with empty text and/or zero values, which create problems for automated processing routines. Computer programmers tend to prefer rigid file formats because they are easy to implement without the use of supplemental libraries. They evolved primarily as a simple way to store images between image processing steps. Their main limitation is that they cannot be extended to add image metadata fields that are not defined by the file format. When such a need occurs, unused or vaguely defined fields are often repurposed to hold the addition information. These cases require special modifications and exceptions when archiving and processing the files.
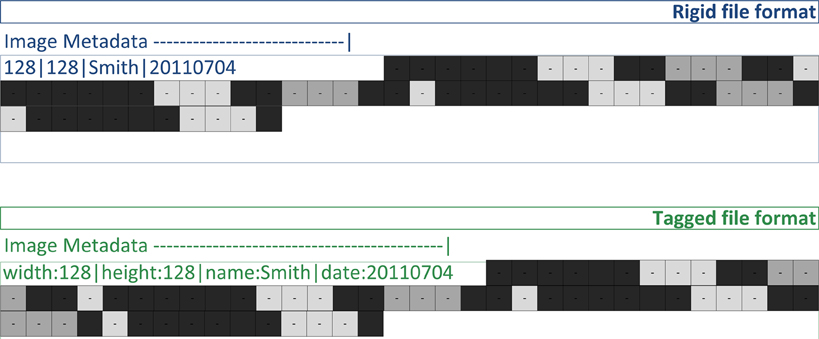
Figure 2. Image files contain both image metadata and pixel values (shown as gray squares). A rigid file format defines locations in the file for the image metadata values (128, 128, “John”). A tagged file format stores image metadata as name/value pairs (e.g., “name” associated with “John”).
Tagged file formats (DICOM, Interfile, MINC, NRRD) define a flexible set of image metadata tags (names or codes) mapped to their values (Figure 2). Unlike rigid file formats, it is optional whether or not a tag is present in a file. So only relevant tags are stored with the images in the files. DICOM has a comprehensive dictionary of tags referenced by a group and element number; for example, the DICOM tags (0010,0020) and (0010,1010) are the Patient ID and Patient's Age elements of the patient identification group (0010), respectively. When the group number is even (e.g., 0010), the tag is a public tag and has a formal definition in the DICOM data dictionary. When the group number is odd, the tag is a private tag. Scanner manufacturers use private tags when they choose to define image metadata outside the DICOM data dictionary. The definitions of private tags are obtained either directly from the scanner manufacturer or by guessing their purpose through examining file samples.
Too Many Standards
Just as there are many different neuroimaging file formats, there are also many conventions that differ between them. Information about how the image data is stored and how the subject was positioned relative to the scanner is defined in various ways, which makes converting between file formats difficult. Additional input is needed to convert from a sparse rigid file format (e.g., NIFTI) to a tagged file format (e.g., DICOM) because the latter file format can hold 2–3 times as much image metadata as the former file format. The names and the locations of the image metadata elements are different in each file format, which makes removal of patient-identifying information tedious and ungeneralizable. Errors in understanding the different standards used by neuroimaging file formats can lead to the creation of improper image files, mislabeled anatomical regions, and loss of patient confidentiality.
Inherent to most image file formats are complex and highly combinatorial schemes for storing multiple images (together comprising an image volume, see Figure 3) in a file. As previously mentioned, the pixels of all the images are written to the file in a single line; that is, all three dimensions of the image volume are reduced to one dimension. But this can be accomplished in many ways. Any order of the dimensions can be used (e.g., height/width/length or length/width/height) and each dimension can be written either forwards or backwards. Given there are three dimensions and two directions of traversal, there are 48 different ways to write the images to a file and all combinations are supported by neuroimaging file formats.
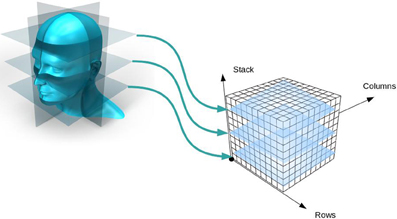
Figure 3. Creation of an image volume. As two-dimensional image slices are acquired by a scanner, they are stacked together to form an image volume.
In order to preserve orientation, file formats define a coordinate system (x, y, z) with respect to the scanner (see Figure 4) and then define how the images and patient are oriented with respect to the coordinate system. DICOM uses direction cosines (cosines of the angles between each image vector and the coordinate axes). NIFTI offers three choices: an implicit fixed orientation, quaternions, and the option of using quaternions and direction cosines in combination. MINC assigns the labels “xspace,” “yspace,” and “zspace” to each dimension of the image volume and maps a set of direction cosines to each label (be warned that if the step size in a direction is negative then the direction is flipped). Table 1 lists how nine common neuroimaging file formats define the image orientation and also how each file format assigns the subject's orientation to the coordinate system. There are two conventions used by most file formats, one from radiology and one from neurology. Radiologists perform examinations while facing the patient and so point the x axis from the subject's right to left, the y axis from the subject's front to back, and the z axis from the subject's feet to head (Figure 5). Neurologists prefer to orient themselves with their subjects and so point the x axis from the subject's left to right, the y axis from the subject's back to front, and the z axis from the subject's feet to head (Figure 5). DICOM uses the radiological convention whereas NIFTI uses the neurological convention.
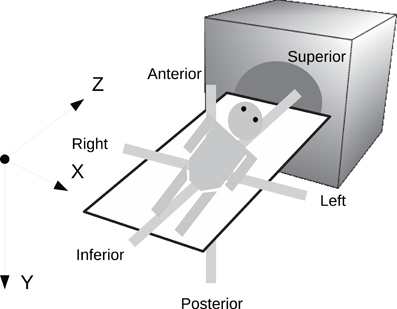
Figure 4. Subject orientation. A coordinate system (X, Y, Z) is defined when a subject is scanned. The subject's orientation is recorded relative to the coordinate system (e.g., +X = left, −X = right, +Y = Posterior, etc.).
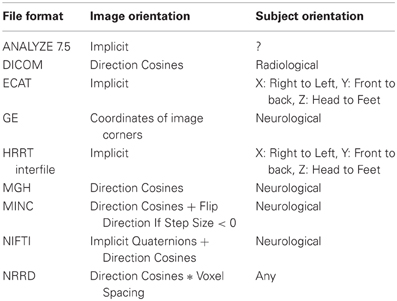
Table 1. Comparison of how image and subject orientation information is captured in different neuroimaging file formats.
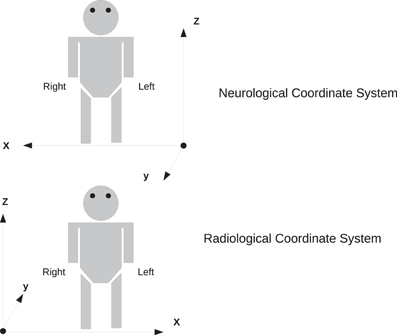
Figure 5. Two coordinate systems for determining subject orientation. The neurological coordinate system defines the positive X, Y, and Z axes along the subject's right, anterior, and superior, respectively. The radiological coordinate system defines the positive X, Y, and Z axes along the subject's left, posterior, and superior, respectively.
Unfortunately, ANALYZE 7.5 does not store information about the subject's orientation. This leads to confusion when one uses multiple image viewers due to the high symmetry (left versus right) of the human head. Figure 6 shows how three different image viewers (BrainSuite, FSLView, 3D Slicer) display the same ANALYZE 7.5 file. Although each viewer is designed to show orientation markers (R, L, A, P, etc.) on top of the images, none are displayed because that information is unavailable. As the images are inconsistently displayed between viewers, only through fiducial comparisons can one determine exactly how each viewer is orienting the subject.
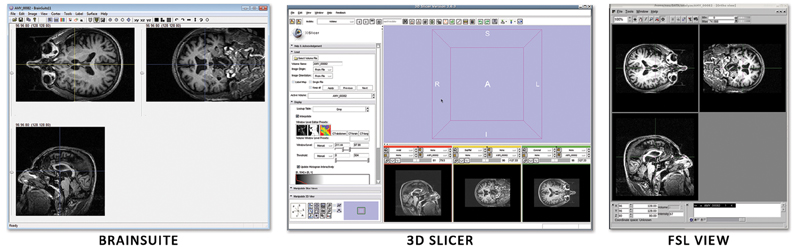
Figure 6. Three image viewers display the same ANALYZE 7.5 file: (A) BrainSuite, (B) FSLView, (C) 3D Slicer. Without image markers it is difficult to deduce the subject's right from the subject's left and easy to get the two reversed when moving between viewers.
The diversity of neuroimaging file formats and differences in how image metadata (including subject identifiers) are stored by each file format require that any approach to remove patient-identifying information (de-identification) involve a thorough understanding of file formats and their image metadata. There is no single standard for ensuring data are de-identified in a manner that meets regulations (Kulynych, 2002), however, software tools for manipulating image metadata exist with varying degrees of flexibility (Neu et al., 2005). Replacing the subject name and ID in files is relatively straightforward because most file formats define a specific location for those elements. However, other metadata elements can contain free-form text that is not suitable for sharing, and rules for removing or replacing these need to be defined. The DICOM standard defines a number of different value representations (VR) that specify the type of metadata that may be stored in each tag. These are codes that restrict the allowed values; for example, codes that specify integers, dates, and people's names. We have found that many of the string value tags are subject to interpretation and it cannot be guaranteed that names or other subject-identifying information won't be present. In our experience, a good general policy is to remove or replace all but a few specified string tags while preserving all the numeric and code tags in order to safeguard subject privacy and allow the image files to be shared to the widest extent possible.
File format conversion involves mapping the image metadata from one file format into another, and the removal of patient-identifying information from a file is essentially a mapping that removes or replaces the image metadata in the file. The existence of standards for storing neuroimaging data is not sufficient enough to support these operations without data mapping.
Lack of Standardization
It is often the case with standardization that changes take a long time to be adopted by the community and longer still to be incorporated. When studies use a new or experimental radiopharmaceutical, the scanners may not include the compound in the list of choices available via the console or the technician may not enter the information correctly or completely. If either of these occurs, then the metadata stored in the image files will either contain incorrect or missing information describing the radiopharmaceutical agent. For example, results of the first study utilizing Pittsburg Compound-B (PIB), a PET imaging tracer used to show amyloid deposits, were published in 2004 (Klunk et al., 2004) and the compound was adopted by a number of studies including the Alzheimer's Disease Neuroimaging Initiative (ADNI) (Weiner et al., 2010) which began collecting PIB scans in 2007. Of the PIB scans archived in the IDA from 2007 to present, less that 50% of the DICOM files contain metadata identifying the PIB compound in the radiopharmaceutical tag (0018,0031).
The DICOM standard supports the use of controlled terminologies as a means of standardizing metadata. This may be done by incorporating coded values in a Code Sequence Attribute which is comprised of a triplet of items for each element: (1) the Coding Scheme Designator which identifies the coding scheme from which the term is obtained; (2) the Code Value which is the code as designated within that scheme, and; (3) the Code Meaning which is the human-understandable meaning of the code. DICOM has defined an internal Controlled Terminology in part 3.16 of the DICOM standard, and defines 36 additional coding schemes for use in DICOM. In addition, the DICOM standard allows the use of any coding scheme that has an entry in the Health Level Seven (HL7) Registry of Coding Schemes (National Electrical Manufacturer's Association, 2011). The presence of standardized terms in imaging files could add much to the richness of the metadata and support improved methods for querying, grouping, and interpreting data. In our experience, however, very few image files contain Code Sequence Attribute sequences, and we've found few software tools that are designed to make use of them. When files do contain Code Sequence Attribute sequences, we have often found them to be inconsistent. The ADNI study began using AV-45 compound (Avid Radiopharmaceuticals, Philadelphia) in early 2010. AV-45 targets the same amyloid plaques as PIB, however, the carrier is fluorine, the same carrier used in regular FDG scans. The Systematized Nomenclature of Medicine-Clinical Terms (SNOMED CT) terminology is the preferred coding scheme within DICOM for pharmaceutical/biologic products and for anatomy and clinical terms. In SNOMED-CT, Fluorine is defined as a radioactive isotope having seven subtypes. The Code Values and Code Meanings of these radioisotope subtypes, as defined within SNOMED CT, are shown in Table 2.
It was expected that all scanners utilizing the same controlled terminology would use the same codes to represent the same compounds and the defined meaning of those terms would be consistent. In our experience, the application of these terms is neither consistent nor accurate, even within scanner manufacturer. Table 3 shows actual values taken from DICOM files in which radiopharmaceutical information was encoded using the Code Sequence Attribute and referencing the SNOMED terminologies. The files were obtained from six different sites participating in the ADNI study and following the same PET scanning protocol. The protocol includes one scan of the patient injected with F^18^ Fluorodeoxyglucose (18F-FDG) and a second scan with the patient injected with F^18^ Florebetapir (18F-AV45), both isotopes of Fluorine. Each scan occurs at separate scanning sessions but on the same scanner.
In each case, the same Code Value is used for both the AV-45 and FDG scans. It should be noted that there is no entry in SNOMED CT for F^18 Florebetapir (AV-45), likely the explanation for why the same Code Value for is used for different radioisotope subtypes. However, it does not explain the use of different Code Meanings for the same Code Value and serves to emphasize the gaps that exist between developing, supporting, and adopting standardized terminologies. Although each of the DICOM files listed in Table 3 incorporates terms from standardized terminologies, the terms as used are insufficient to determine the radioisotope used. The existence of standardized terminologies alone cannot solve standardization problems. Full adoption and appropriate use are necessary.
In order to identify these scans as one of “FDG,” “PIB” or “AV45,” we constructed the data maps illustrated in Figure 7. The radiopharmaceutical agent for three different file formats is inferred from acquisition properties such as the number of frames in each scan, text searches on descriptions and codes, and range restrictions on the total length of each scan. Our general approach to solving this problem involves maintaining a cache of small image metadata samples from each set of files in the archive. The maps used to identify image types and classify image groups are constructed by inspecting and testing the data in the cache. If, after the maps have been established, files with image metadata that run contrary to the rules are received, previous assumptions must be revisited and new maps established using the new image metadata. This may involve reworking the current maps or adding new image metadata fields to the cache until new maps can be created. Because image metadata is different across different file formats, we have one cache for each of the file formats we archive. For the DICOM file format, this most often involves exploring vendor-specific private tags whose full definition is unknown but whose tag values give clues as to their meanings. Private tags tend to be more reliable than public text tags (such as study and series descriptions) because the private tag values are less variable across different institutions. However, in some instances it may be necessary to construct maps based upon both private and public tags and/or even use the location of the scanner as a last resort.
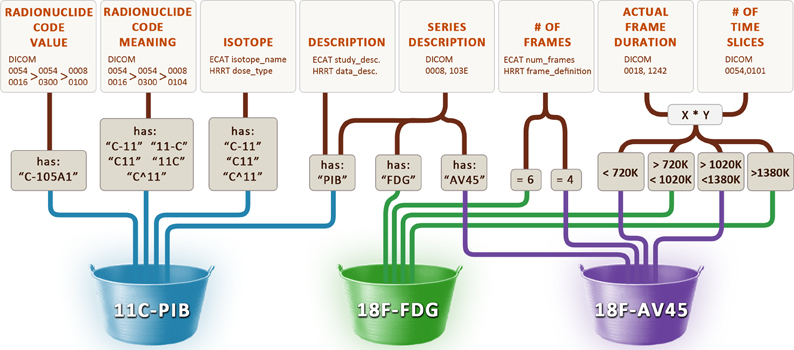
Figure 7. Rules used to determine the radiopharmaceutical agent. Various image metadata fields from the DICOM, ECAT, and HRRT Interfile file formats are used to infer the name of the radiopharmaceutical agent (11C-PIB, 18F-FDG, 18F-AV45).
The DICOM Standard
Repositories of neuroimaging data are often created for research purposes. In order to make comparisons and test hypotheses between different cohorts, it is often necessary to search through all of the collected metadata. However, the information obtained during image acquisition and the way it is organized at the image scanner is not necessarily optimal for research purposes. Search criteria that extend beyond the set of terms present in neuroimaging files are often needed for efficient and productive searching, grouping, and analysis. Currently most neuroimaging data is acquired and stored using the DICOM standard, but we have found it to be inadequate to group and label our data in a manner that facilitates searching.
Grouping images together is a non-trivial matter, and in the DICOM standard there is little information about exactly how to do this. In its “DICOM model of the real-world” (National Electrical Manufacturers Association, 2011), patients have studies performed during their visits, and each study contains one or more series. However, although each series contains one or more images, documents, or measurements, no specification is given as to how the components should be grouped together by series. At best a series is defined (National Electrical Manufacturers Association, 2011) as a set of “composite instances” that (1) must be of the same modality, (2) must be spatially or temporally related to one another, (3) must be created by the same equipment, and (4) must have the same series information. In the IDA we create a series identifier by mapping together the subject, study, series date, and series description of each image. This was motivated by how investigators expect to search and receive search results. We've also added an additional sub-series level in which DICOM images are grouped together using the echo time (structural MRI), series UID (0020,000E), coordinate axes, and image size which are all mapped into a grouping identifier. This definition divides a PD and T2 scan into separate sub-series.
Although many journals contain articles with multiple references to imaging modalities such as “fMRI,” “MRA”, and “DTI,” NEMA recognizes none of these as true modalities. In fact, in each of these cases the DICOM modality tag (0008,0060) has the value “MR”. At least one expert in the field (Clunie, 2011) acknowledges that MRA, fMRI, and DTI are best considered “sub-types” of a single MR modality. The contradiction between the labels researchers use to classify MR data and the DICOM image metadata describing the MR data has required us to construct maps to divide MR DICOM files into these sub-types. Unfortunately, these maps are non-trivial and cumbersome because of the need to heavily rely on the private tags used by each scanner manufacturer. For example, despite the existence of public DICOM tags for diffusion [e.g., diffusion b-value (0018,9087) and diffusion directionality (0018,9075)], we have found in practice these tags are most often missing from DTI image metadata. Instead, for GE DTI images, we rely upon the GE private tags (0019,10E0) for the number of diffusion directions and tag (0021,105A) for the diffusion direction. Philips DTI images were found to have the diffusion direction in their private tag (2001,1004) and Siemens DTI images required us to reverse engineer the encoded value of their private tag (0029,1020) and use the element “lDiffWeightings” to detect DTI image metadata and the element “lDiffDirections” for the number of diffusion directions.
Conclusion
Wiederhold (Wiederhold, 2003) gives many reasons why global consistency is impossible; those applicable to the establishment of a single neuroimaging standard are: (1) problems occur when interacting between domains (clinical, research) that fundamentally have different objectives, (2) since research is less regulated and more dynamic than clinical practice, terminologies are necessarily different, and (3) committees from diverse groups that establish terminological consistency tend to define compromises rather than precise definitions. These observations are consistent with what we have found in practice.
Multiple neuroimaging file formats were defined primarily because of the different needs of those using the image data. The DICOM file format originated to support a clinical environment, crafted by scanner manufacturers who wanted to provide detailed descriptions of the image acquisitions in a consistent and flexible way. The popularity among researchers of the ANALYZE 7.5 file format (and its derivative, the NIFTI file format) was due to the simplicity of the format definition (easy to implement) and the convenience of storing multiple images in one or two files. Since the methodologies and goals of radiologists and researchers are different, the subject's orientation was defined in DICOM using the radiological convention and in NIFTI using the neurological convention. The need to add more metadata to the rigid file formats and the aim to reconcile all of the various file formats into one motivated the creation of other neuroimaging file formats, but their continued coexistence with their predecessors is a testament to the impracticality of establishing a single standard.
The dynamics of scientific research and advances in image scanner technology will continue to produce image metadata that vary over time. New variations of existing image metadata are required by experimental acquisitions and new research protocols, and in some cases new image acquisition methods (e.g., DTI) will produce entirely new sets of descriptive metadata and new nomenclature for neuroimaging file formats (e.g., new DICOM tags). So any collection of neuroimaging data that is added to over time will contain dynamic image metadata, some of which may be necessary to interpret and organize the data. When changes occur in newly collected data, the repository needs to internally reorganize in order to correctly identify and classify the new data, as well as provide the requisite new search capability. For cases where needed information is completely unavailable (i.e., not explicitly represented in the image metadata), ad hoc rules based upon what is available must be constructed and maintained accordingly over time. The dynamic nature of research makes standardization impractical.
Standards represent consensus and consensus requires negotiation over time between interested parties who voluntarily adopt them (ISO, 2011). The DICOM standard evolved as a compromise between scanner manufacturers who were using their own proprietary file formats and wanted to establish consistency amongst themselves. However, this standardization process has been slowly developing. The fact that each manufacturer still uses private DICOM tags to store essential information (and Siemens continues to use its own proprietary encoding in at least two private DICOM tags) demonstrates the degree to which complete standardization has been achieved. The difficulties encountered in determining the “sub-types” of the MR modality exemplify how slowly the standardization is proceeding. It should be noted that these problems are not unique to the DICOM standard; the NIFTI file format was defined through the consensus of the most influential neuroimaging processing software developers at the time. The motivation behind the NIFTI effort was to create a backwards-compatible file format to ANALYZE 7.5 with some desired new features (addition of a coordinate system and new image data types). But backwards-compatibility meant retaining some of the obsolete metadata elements of ANALYZE 7.5, thus limiting the pace of standardization.
Can further metadata standardization solve the class of problems we have described in this article, or will the intrinsic nature of neuroimaging research perpetually add more problems than can be resolved with standardization at any given time? These issues may not be problematic in cases where a small group of investigators are working closely together on the same study, but when sharing moves outside the lab and across the globe they become readily apparent. Where standardization fails, data mapping is required. Therefore, neuroimaging data repositories must implement adaptive metadata management tools if they are to effectively collect, manage, and distribute data on a national and global scale.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This article was supported by NIH grants (P41 RR013642, U01 AG024904, and M01 RR000865), the CHDI Foundation, Inc. and the Michael J. Fox Foundation for Parkinson's Research. The authors wish to thank our global community of collaborators for providing us with an ever-changing stream of data with which to practice our metadata wrangling. We would also like to express our thanks to our colleagues at the Laboratory of Neuro Imaging at UCLA.
Footnotes
References
Atkins, D. E., Droegemeier, K. K., Feldman, S. I., Garcia-Molina, H., Klein, M. L., Messerschmitt, D. G., Messina, P., Ostriker, J. P., and Wright, M. H. (2003). Revolutionizing Science and Engineering through Cyberinfrastructure: Report of the National Science Foundation Blue-Ribbon Advisory Panel on Cyberinfrastructure. Arlington, VA: National Science Foundation.
Brooks, D. J., and Pavase, N. (2011). Imaging biomarkers in Parkinson's disease. Prog. Neurobiol. 95, 613–628.
Bug, W. J., Ascoli, G. A., Grethe, J. S., Gupta, A., Fennema-Notestine, C., Laird, A. R., Larson, S. D., Rubin, D., Shepard, G. M., Turner, J. A., and Martone, M. E. (2008). The Nifstd and Birnlex vocabularies: building comprehensive ontologies for neuroscience. Neuroinformatics 6, 175–194.
Butcher, J. (2007). Alzheimer's researchers open the doors to data sharing. Lancet Neurol. 6, 480–481.
Clunie September 25, 2010. (5:21 a.m). “A Modality, by any other name (would smell as sweet?),” in David Clunie's BLOG, September 25, 2011. http://dclunie.blogspot.com/2011/09/modality-by-any-oth-er-name-would-smell.html
Cradduck, T. D., Bailey, D. L., Hutton, B. F., Deconinck, F., Busemann Sokole, E., Bergmann, H., and Noelpp, U. (1989). A standard protocol for the exchange of nuclear medicine image files. Nucl. Med. Commun. 10, 703–713.
Digital Imaging and Communications in Medicine (DICOM). (2008). A Brief History of DICOM. Berlin: Springer. E-Book.
Digital Imaging and Communications in Medicine (DICOM) PS 3.3 – 2011. (2011). 7 DICOM Model of the Real-World, p. 64 and A.1.2.3 Series IE, p. 119. VA, USA: National Electrical Manufacturers Association. http://medical.nema.org.dicom/2011/11_03pu.pdf
Fletcher, G. H. L., and Wyss, C. M. (2009). Towards a general framework for effective solutions to the data mapping problem. J. Data Semantics XIV, 37–73.
Gadde, S., Aucoin, N., Grethe, J. S., Keator, D. B., Marcus, D. S., and Pieper, S. (2012). Xcede: an extensible schema for biomedical data. Neuroinformatics 10, 19–32.
Goble, C., and Stevens, R. (2008). The state of the nation in data integration in bioinformatics. J. Biomed. Inform. 41, 687–693.
Gray, J., Liu, D. T., Nieto-Santisteban, M., Szalay, A., Heber, G., and DeWitt, D. J. (2005). Scientific data management in the coming decade. ACM SIGMOD Rec. 34, 34–41.
Halevy, A. Y., Rajaraman, A., and Ordille, J. J. (2006). Data Integration: The Teenage Years, Proceedings of the 32nd International Conference on Very Large Databases, Seoul. 9–16.
ISO. (2011). ISO/IEC Directives, Part 1. Procedures for the Technical Work. 6th Edn. Geneva: International Organization for Standardization.
Klunk, W. E., Engler, H., Nordberg, A., Wang, Y., Blomqvist, G., Holt, D. P., Bergstrom, M., Savitcheva, I., Huang, G. F., Estrada, S., Ausen, B., Debnath, M. L., Barletta, J., Price, J. C., Sandell, J., Lopresti, B. J., Wall, A., Koivisto, P., Antoni, G., Mathis, C. A., and Langstrom, B. (2004). Imaging brain amyloid in Alzheimer's disease with Pittsburgh Compound-B. Ann. Neurol. 55, 306–319.
Kulynych, J. (2002). Legal and ethical issues in neuroimaging research: human subjects protection, medical privacy, and the public communication of research results. Brain Cogn. 50, 345–357.
Langlotz, C. P. (2006). Radlex: a new method for indexing online educational materials. Radiographics 26, 1595–1597.
Mazziotta, J., Toga, A. W., Evans, A., Fox, P., Lancaster, J., Ziles, K., Woods, R. P., Paus, T., Simpson, G., Pike, B., Holmes, C., Collins, L., Thompson, P. M., MacDonald, D., Iacoboni, M., Schormann, T., Amunts, K., Palomero-Gallagher, N., Geyer, S., Parsons, L. M., Narr, K., Kabani, N., LeGoualher, G., Boomsma, D., Cannon, T., Kawashima, R., and Mazoyer, B. (2001). A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM). Philos. Trans. R. Soc. Lond. B Biol. Sci. 356, 1293–1322.
National Center for Health Statistics. (2007). Health, United States, 2007 with Chartbook on Trends in the Health of Americans. Hyattsville, MD: National Center for Health Statistics (US).
National Electrical Manufacturer's Association. (2011). Digital Imaging and Communications in Medicine (DICOM). Rosslyn, VA: NEMA, PS 3.16 – 2011.
National Science Board. (2005). Long-Lived Digital Data Collections Enabling Research and Education in the 21st Century. Technical report, National Science Foundation.
Neu, S. C., Valentino, D. J., and Toga, A. W. (2005). The LONI Debabeler: a mediator for neuroimaging software. Neuroimage 24, 1170–1179.
Poldrack, R. A., Kittur, A., Kalar, D., Miller, E., Seppa, C., Gil, Y., Parker, D. S., Sabb, F. W., and Bilder, R. M. (2011). The cognitive atlas: toward a knowledge foundation for cognitive neuroscience. Front. Neuroinform. 5:17. doi: 10.3389/fninf.2011.00017
Ryan, N. S., and Fox, N. C. (2009). Imaging biomarkers in Alzheimer's disease. Ann. N.Y. Acad. Sci. 1180, 20–27.
Teeters, J. L., Harris, K. D., Millman, K. J., Olshausen, B. A., and Sommer, F. T. (2008). Data sharing for computational neuroscience. Neuroinformatics 6, 47–55.
Toga, A. W. (2002). Neuroimage databases: the good, the bad and the ugly. Nat. Rev. Neurosci. 3, 302–309.
Toga, A. W., and Crawford, K. L. (2010). The informatics core of the Alzheimer's disease neuroimaging initiative. Alzheimers Dement. 6, 247–256.
Turner, J. A., and Laird, A. R. (2012). The cognitive paradigm ontology: design and application. Neuroinformatics 10, 57–66.
Weiner, M. W., Aisen, P. S., Jack, C. R. Jr., Jagust, W. J., Trojanowski, J. Q., Shaw, L., Saykin, A. J., Morris, J. C., Cairns, N., Beckett, L. A., Toga, A., Green, R., Walter, S., Soares, H., Snyder, P., Siemers, E., Potter, W., Cole, P. E., Schmidt, M., and Alzheimer's Disease Neuroimaging Initiative (2010). The Alzheimer's disease neuroimaging initiative: progress report and future plans. Alzheimers Dement. 6, 202–211. e7.
Keywords: image metadata, file format, data archive
Citation: Neu SC, Crawford KL and Toga AW (2012) Practical management of heterogeneous neuroimaging metadata by global neuroimaging data repositories. Front. Neuroinform. 6:8.doi: 10.3389/fninf.2012.00008
Received: 01 October 2011; Accepted: 08 March 2012;
Published online: 23 March 2012.
Edited by:
John Van Horn, University of California at Los Angeles, USAReviewed by:
Tor Wager, Columbia University, USADavid B. Keator, University of California, USA;
Angela R. Laird, The University of Texas Health Science Center, USA
Copyright: © 2012 Neu, Crawford and Toga. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Arthur W. Toga, Department of Neurology, University of California, Los Angeles, 635 Charles E. Young Drive SW, Suite 225, Los Angeles, CA 90095-7334, USA. e-mail: toga@loni.ucla.edu