- Department of Molecular Visual Plasticity, Netherlands Institute for Neuroscience, An Institute of the Royal Netherlands Academy of Arts and Sciences, Amsterdam, Netherlands
Many studies have been devoted to the identification of genes involved in experience-dependent plasticity in the visual cortex. To discover new candidate genes, we have reexamined data from one such study on ocular dominance (OD) plasticity in recombinant inbred BXD mouse strains. We have correlated the level of plasticity with the gene expression data in the neocortex that have become available for these same strains. We propose that genes with a high correlation are likely to play a role in OD plasticity. We have tested this hypothesis for genes whose inactivation is known to affect OD plasticity. The expression levels of these genes indeed correlated with OD plasticity if their levels showed strong differences between the BXD strains. To narrow down our candidate list of correlated genes, we have selected only those genes that were previously found to be regulated by visual experience and associated with pathways implicated in OD plasticity. This resulted in a list of 32 candidate genes. The list contained unproven, but not unexpected candidates such as the genes for IGF-1, NCAM1, NOGO-A, the gamma2 subunit of the GABA(A) receptor, acetylcholine esterase, and the catalytic subunit of cAMP-dependent protein kinase A. This demonstrates the viability of our approach. More interestingly, the following novel candidate genes were identified: Akap7, Akt1, Camk2d, Cckbr, Cd44, Crim1, Ctdsp2, Dnajc5, Gnai1, Itpka, Mapk8, Nbea, Nfatc3, Nlk, Npy5r, Phf21a, Phip, Ppm1l, Ppp1r1b, Rbbp4, Slc1a3, Slit2, Socs2, Spock3, St8sia1, Zfp207. Whether all these novel candidates indeed function in OD plasticity remains to be established, but possible roles of some of them are discussed in the article.
Introduction
During brain development there are periods in which specific regions are highly plastic and learning occurs more readily and more permanently than during adulthood. These sensitive periods allow us to effectively acquire the skills and knowledge that we build on for the rest of our lives. Unfortunately, it also means that during such sensitive periods, permanent damage to the circuits of the brain can arise if plasticity does not occur correctly. This can be caused by genetic defects, as is often the case in neurodevelopmental disorders, or due to inappropriate inputs as is the case with the development of amblyopia (lazy eye). Understanding the molecular and genetic mechanisms underlying sensitive period plasticity could lead to clinical therapies for such disorders through reopening sensitive periods and allowing plasticity to reoccur.
The most extensively used paradigm to study sensitive periods of cortical plasticity is ocular dominance (OD) plasticity, which also underlies the development of amblyopia. When one eye is closed (monocular deprivation, MD) for several days during the sensitive period, the primary visual cortex (V1) will become less responsive to this eye, while non-deprived eye responses increase (Wiesel and Hubel, 1963; Gordon and Stryker, 1996; Hensch, 2004). MD after the sensitive period results in much more limited plasticity or no plasticity at all, depending on the species and age of the animals.
Much work has been devoted to unravel the mechanisms behind sensitive period plasticity and identifying the genes and proteins involved. Pharmacology and knock-out models have been employed most frequently to test whether proteins and genes implied in specific forms of plasticity, such as LTD, LTP, or homeostasis in vitro, also affected OD plasticity (e.g., Beaver et al., 2001; Taha et al., 2002; Kaneko et al., 2008). A second approach has been to study the correlation of gene expression and periods of enhanced plasticity in order to identify candidate genes (Majdan and Shatz, 2006; Lyckman et al., 2008; Dahlhaus et al., 2011). This has, for instance, led to the recognition of the role of the IGF-1 signaling pathway in OD plasticity (Tropea et al., 2006). A third method is the forward genetics screen, where OD plasticity is measured in animals in which random genes have been inactivated. A variation on this approach is the screening of OD plasticity in a panel of recombinant inbred strains. In this method, plasticity is measured in several strains. The correlation of plasticity level with allele genotype in these strains indicates which genes might play a role in OD plasticity (Heimel et al., 2008). One important benefit of measuring in a genetic reference panel of animals is that measurements acquired by different labs can be combined and compared. For example, in BXD mouse strains there is extensive data available on gene expression levels in several brain areas. The correlation of expression levels of a certain gene with the level of plasticity across the different inbred strains could also point to a causal relationship. Such a gene could regulate or modulate sensitive period plasticity or could be partaking in the plasticity itself.
Genetic screens, however, are commonly hampered by the problem of multiple testing, because of the vast number of different genes. Genes which truly play a role in a process can have similar p-values as randomly correlating genes. If one does not correct for multiple testing when selecting candidate genes based on p-value, there will be many spurious genes contaminating the candidate list. If one does correct, however, it is very likely that some causative genes will be thrown out as insignificant. Often this problem is tackled by making an ad hoc selection of the most significant genes by using additional knowledge about their function. In this paper, we formalize this approach using an unbiased combination of several publicly available datasets of genetic information of OD plasticity. This leads to the identification of 32 genes with a high likelihood of being regulators of plasticity in the visual cortex.
Materials and Methods
We composed three lists of genes from different sources of publicly available data, which we call the Correlated, Implicated, and Regulated gene lists. The genes appearing on all three lists were considered candidate plasticity genes. For these genes, the mouse Allen Brain Atlas1 (Lein et al., 2007) was consulted in January 2011 to check whether they are indeed expressed in the visual cortex. A schematic representation of the selection procedure is shown in Figure 1.
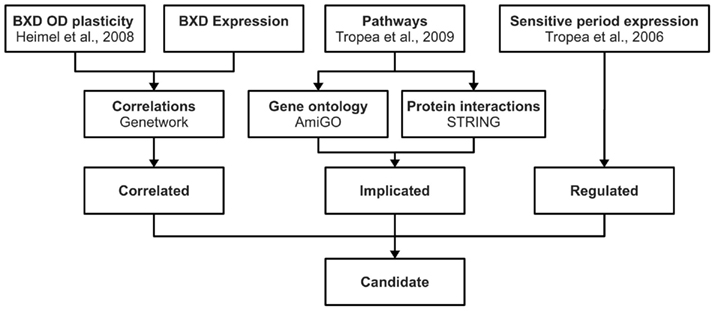
Figure 1. Candidate gene selection procedure. This figure schematically shows the approach used for identifying candidate genes for OD plasticity. Using publicly available information we composed three gene lists (correlated, implicated, regulated). Genes that were present in all three lists were considered candidate plasticity genes.
Correlated Gene List
The first list was computed by correlating functional data on OD plasticity with gene expression levels in the neocortex of BXD mice. The BXD set is a genetic reference panel of 80 recombinant inbred strains derived from C57BL/6J and DBA/2J parent strains. A wealth of data about these mice, including the data used for this paper, is publicly available from Genenetwork2 (Chesler et al., 2004).
BXD OD plasticity
Ocular dominance plasticity was previously measured in 13 BXD strains by comparing the visual responses in the left primary visual cortex at postnatal day 35 (P35) in normally treated animals to the responses in animals where the contralateral (right) eye had been closed from P28 (Heimel et al., 2008). From these published data, we used three traits for our analysis: (1) the difference in response to visual stimulation of the sutured, reopened, contralateral eye (Genenetwork RecordID/11285), (2) the difference in response to the unsutured ipsilateral eye (RecordID/11286), (3) the difference in the OD index, ODI, defined as (contralateral response − ipsilateral response)/(contralateral response + ipsilateral response)·(RecordID/11284).
BXD gene expression
Gene expression data was taken from the HQF BXD Neocortex ILM6v1.1 (Feb08) RankInv dataset (Gaglani et al., 2009) which analyzed mRNA levels in the neocortex of adult mice raised in a standard laboratory environment using the Illumina Mouse 6.1 bead micro-array. All genes from this set for which the expression level correlated (positively or negatively) at the 5% significance level or below with at least one of the OD plasticity traits, together made up the “correlated” gene list. The significance of the Pearson correlations was computed by comparing the real correlation to that of a thousand permutations of the trait values.
Validation of the “correlated” gene list
To verify that correlations between expression and plasticity can point to genes which are involved in OD plasticity, we cross-checked the correlated gene list with a list of all genes with a proven role in this process. These genes were found by a PubMed search for “OD plasticity” (on October 29, 2010) and selecting for mouse knock-out models with altered OD plasticity. If, however, there is no variation in the expression of a particular gene within the BXD strains, then the gene expression, of course, cannot correlate with plasticity. To control for this, we considered the expression levels for all the probes on the Illumina mRNA micro-array of the proven genes across the 13 strains for which OD shifts were measured (C57BL/6J, DBA/2J, and BXD strains 1, 2, 6, 14, 21, 28, 31, 33, 34, 39, 40). From these, we computed the relative expression range by taking the difference between the highest and lowest expression level and dividing this by the SEM value within a single strain, averaged across the measured strains.
Implicated Gene List
For creation of the “implicated” gene list, we used a recent review by Tropea et al. (2009). This review contains an exhaustive list of pathways and molecules with an established role in OD plasticity or its regulation. These pathways were entered (on January 7, 2011) in the gene ontology database AmiGO3 (Ashburner et al., 2000) to retrieve all genes involved in these pathways. The specific protein names listed in this review were also entered in the public protein interaction database STRING4 (Szklarczyk et al., 2011) to find their direct interaction partners (consulted on January 8, 2011). Together, these genes made up the implicated gene list. For each of the genes in this extensive list, we checked the literature to classify it as having a known, likely or uncertain role in OD plasticity.
Regulated Gene List
The third list came from an mRNA micro-array study where the objective was to search for genes regulated by visual experience and thus possibly involved in regulation of OD plasticity. Tropea et al. (2006) extracted RNA from V1 of 129/SvEv mice at P27, and compared expression levels of normally reared mice, to that of (1) mice born and reared in darkness, (2) mice in which the contralateral eye was sutured at P11–12, before eye opening, (3) mice in which the contralateral eye was sutured at P23. In this way, genes were identified that were up or down regulated after dark rearing, by long-term or short-term MD. Genes regulated with p < 0.01 were included in our regulated gene list.
Results
To identify new candidate genes playing a role in OD plasticity, we wanted to exploit the underused potential of a number of large public datasets. In particular, the data on plasticity in BXD recombinant inbred strains could possibly be mined in a novel way to obtain candidates. For a number of strains of this genetic reference panel, the changes in visual response in primary visual cortex induced by 7 days MD during the sensitive period for OD plasticity had already been measured (Heimel et al., 2008). For this study, we used the reported shifts in response to stimulation of the deprived contralateral and the undeprived ipsilateral eyes, and the shift in the OD index of the balance of the two eyes’ responses. These, as well as many other physiological and behavioral traits and, importantly, gene expression levels have been made available to study at the online Genenetwork database. Our hypothesis was that genes for which the expression in the neocortex correlates with one of these aspects of OD plasticity in different BXD strains, are likely to be involved in this type of experience-dependent plasticity.
Validation of Correlation Approach
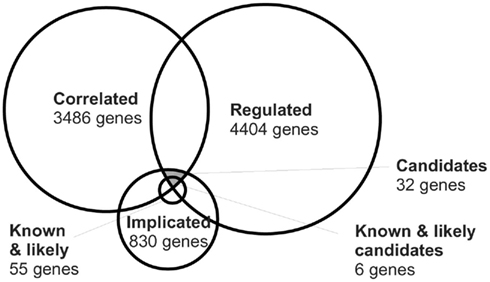
Although it is not easy to prove the hypothesis above, we could test the validity of the reverse hypothesis, i.e., do expression levels correlate with plasticity for genes which play a role in OD plasticity. To this end, we created a list of all genes which have been proven to play a role in OD plasticity by means of a knock-out mouse model. A study of the literature produced 14 such genes, shown in Table 1. For 3 (21%) of these, the expression in the adult neocortex correlated significantly at the 5% level with either the change in response to one of the eyes, or the change in the ODI, induced by the MD. This limited number of genes is comparable to that expected by chance. However, it should be taken into account that if across the BXD strains the range of expression levels of assessed genes is too small compared to the variation in the expression level measurement, it is not possible to detect a correlation between phenotype and expression. We therefore computed the relative expression level range of the 14 genes by dividing the maximal difference in expression level for the BXD strains by the average variation in the expression level per strain. If our hypothesis is correct, we expect a significant correlation between expression and plasticity for genes with a high ratio. Table 1 is sorted in descending order for this relative range. It shows that both genes with a relative range above 1, have a significant correlation, as do three from the seven genes with a relative range larger than 0.5.
Interestingly, while H2-D1 and H2-K1 both show considerable expression level variation across BXD strains, only H2-D1 strongly correlates to OD plasticity (Table 1). When both of these two major histocompatibility complex class I (MHCI) genes are knocked out, there is increased OD plasticity (Datwani et al., 2009). In particular, the area of activity-regulated Arc expression in response to stimulation of the non-deprived or non-enucleated ipsilateral eye is expanded. This fits well with the negative correlation of the expression of H2-D1 in the cortex and the increase in response to the non-deprived ipsilateral eye (r = −0.62, p = 0.032). Results of single knock-outs of H2-D1 or H2-K1 are not published. Our analysis suggests that the H2-D1 deficiency alone is causing the reported increased plasticity in the double knock-out mouse.
We also made an exhaustive list of genes which do not cause an alteration in OD plasticity when knocked out (bottom tier of Table 1). Although the list was short, this provided some justification of the original hypothesis that genes that play a role in OD plasticity are more likely to have expression levels correlating with plasticity, than genes that do not play a role. The expression level of none of these genes correlated with any of the plasticity traits. Overall, these results suggests that genes which are involved in OD plasticity and for which expression levels vary across the BXD strains, are indeed more likely than other genes to have expression levels correlating with plasticity.
Correlated Gene List
The correlation of expression level and plasticity for known OD plasticity genes suggested that correlation of expression and plasticity could be a fruitful starting point for identifying new candidate genes. We thus compiled a list of all genes for which the expression level in the neocortex in the BXD strains correlated positively or negatively with one or more aspects of the OD shifts measured in the same strains. This list of all significantly correlated probes with an uncorrected p-value below 0.05 consisted of 3486 unique genes (correlated gene list, Table S1 in Supplementary Material). This large list is expected to contain genes that truly influence OD plasticity, but also many that correlate by chance. A first step to reduce the latter group of genes is by performing a correction for multiple testing. However, this reduced the set to only one gene, Loc381054. Of this gene, coding a protein with the prosaic description “similar to Putative protein C21orf45,” very little is known. When applying a correction for multiple testing, many true correlations may be wrongfully discarded, because the significance of the measurements is limited by the number of measured strains and the differences in expression level. Therefore, to try to separate the true candidate genes from the randomly correlating genes, we used an alternative selection procedure that was based on intersection with two additional lists of candidate genes.
Implicated Gene List
The second list represented genes associated with signaling pathways previously implicated in OD plasticity. We began with a recent and exhaustive review of the literature by Tropea et al. (2009). It lists the following classes of molecules: acetylcholine, Arc, calcineurin, CamKII, cannabinoid receptors, CREB, CSPGs, ERK, GABA receptors, HDAC, IEGs, IGF-1, IGFBPs, MGluR, myelin-related receptors, neurotrophins, NMDAR, noradrenaline, PirB, PKA, polysialic acid (PSA), serotonin, TNFα, TPA, troponin C. For these classes, we used the online gene ontology tool AmiGO to select all related gene ontology terms to retrieve all associated genes. In addition, the protein interaction database STRING was used to identify the interaction partners for the individual proteins mentioned in the review. This implicated gene list contained 830 genes (Table S2 in Supplementary Material). Of these, 12 were known to cause a change in OD plasticity when knocked out. Moreover, 43 were genes that, based on previous studies, were highly likely to play a role in OD plasticity. In total, 55 of the 830 genes were known or likely plasticity genes. The vast majority (93%) of the genes in this list could thus represent possible new candidates.
Regulated Gene List
The third list of genes was taken from an mRNA micro-array based search for genes regulated by visual experience (Tropea et al., 2006). This study identified genes expressed in different levels after dark rearing, short MD (4 days), or long MD (16 days), compared to control conditions. It has already been shown that this list contains valuable information on the genes involved in OD plasticity. Based on this study, the IGF-1 pathway was identified and proven to play a role (Tropea et al., 2006). For our new analysis of the data, only the dark rearing and short MD lists were used, as they were most likely to contain genes which are involved in the plasticity induced by the 7-day MD that was used to generate the correlated gene list. This regulated gene list contained 4404 unique genes (Table S3 in Supplementary Material).
Intersection
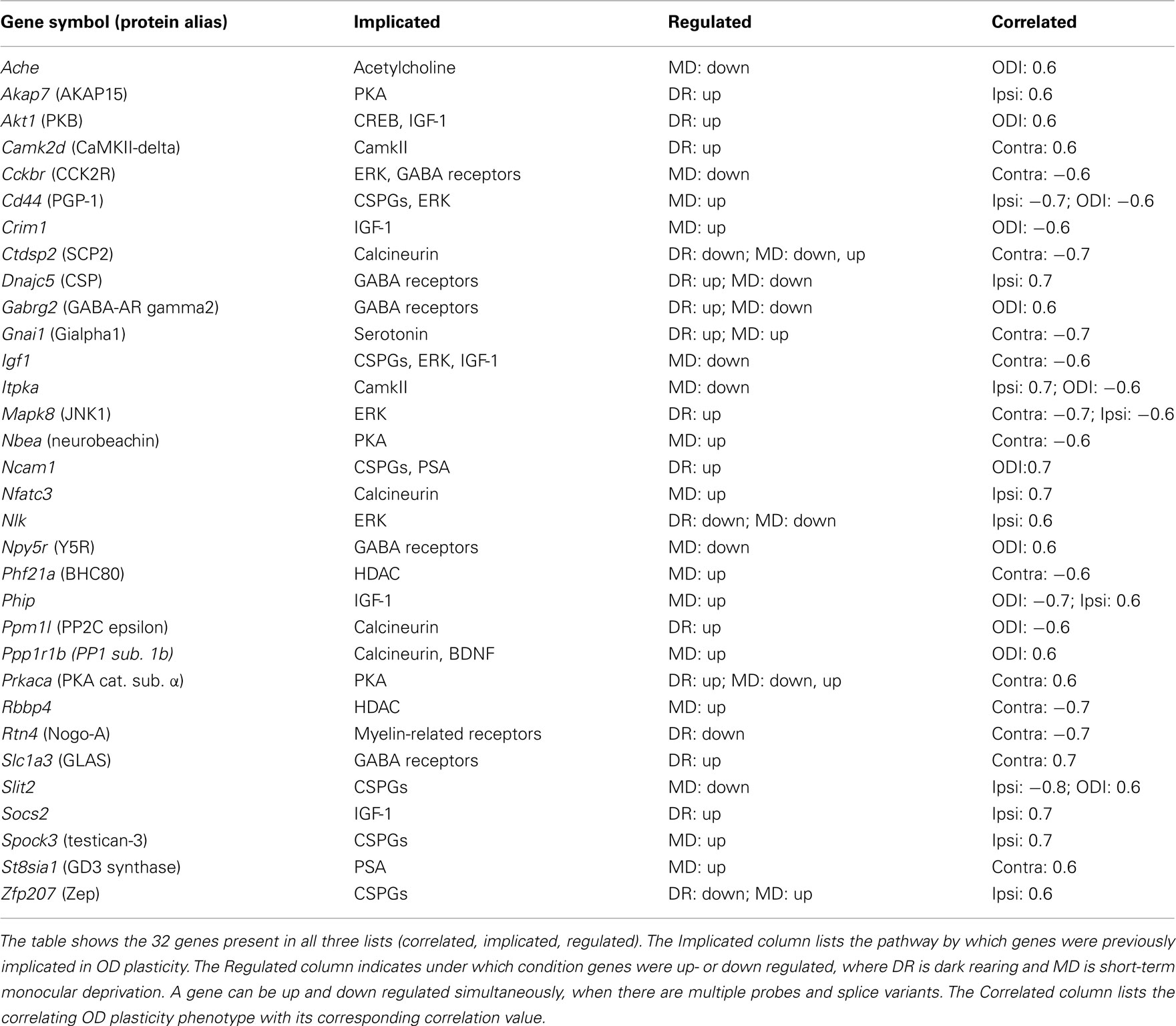
Each of the individual lists contained hundreds of genes. Many of these are involved in OD plasticity, but many are likely not to have such role. If we would make the conditions for selection of each of the individual lists more stringent, then we would remove many true candidates together with the spurious genes. Our approach was therefore to take the intersection of the lists, see Figure 2. Only the 32 genes which were correlated, implicated and regulated were considered candidates, listed in Table 2. Visual cortical expression for all these genes was evident from their inclusion in the regulated gene list (which came from samples of V1 tissue) and was confirmed additionally by consulting the mouse Allen Brain Atlas. Among the 32 candidates, there were six genes (19%), Igf1, Ncam1, Rtn4, Prkaca, Gabrg2, and Ache, which had been assigned as known or likely when compiling the list of implicated genes. This is much more than what would be expected in a random sample of the implicated genes (chi-square test, p = 0.0087), and suggests that our approach is successfully identifying good candidates. Half of this enrichment for likely and known plasticity genes is due to the intersection of the implicated list with the regulated list and half of it is due to its intersection with the correlated gene list (Table 3).
The candidate genes were distributed over half of the 26 implicated molecular classes, with three or more genes associated with Calcineurin, CSPGs, ERK, GABA receptors, IGF-1, or PKA. CSPGs alone, however, already accounted for 155 of the 830 implicated genes, and even by chance a high number of genes associated with CSPGs would have been expected. The only significant enrichment was for IGF-1-associated genes (p = 0.049, chi-square test), which was already observed in the Regulated gene list (Tropea et al., 2006). None of these six pathways associated with three or more candidates was exclusively regulated by dark rearing or MD. Also, none were correlated with the change in one eye specifically. This demonstrates that these pathways are involved both in the loss of response to the deprived eye, and in the gain of the open eye response after MD. Previously, it was found that the loss of deprived eye responses, and the gain of unsutured eye responses do not correlate (Heimel et al., 2008). In our selection of candidate genes, most genes indeed correlate with the changes of one eye alone, but five genes correlate with a change in the eye balance (ODI), or with changes in both eyes. This suggests that the expression level variation of even individual genes can simultaneously affect the deprived eye loss and the open eye gain.
Discussion
Experience-dependent plasticity during sensitive periods of development is more effective and results in more persistent changes than plasticity in the adult brain. The aim of this study was to produce a selection of novel genes with a very high likelihood of regulating or participating in sensitive period plasticity in the visual cortex. This selection of genes may be used in future research to serve as a handle through which plasticity can be activated using pharmacological means or gene therapy. We combined previous literature with the results of a micro-array study on visually regulated genes and the results of a forward genetics approach to identify candidate genes potentially involved in regulating OD plasticity in the visual cortex.
The forward genetics study was a screen of C57BL/6J × DBA recombinant inbred strains for the efficiency of three components of OD plasticity, loss of responsiveness to the deprived eye, increased responsiveness of the non-deprived eye and the OD index, and correlating them with the expression levels of genes expressed in the adult neocortex. We first determined the limitations of this forward genetics approach by testing whether genes that were found to be essential for OD plasticity by reverse genetics would be identified. We found that we could indeed enrich for these genes, but not surprisingly, only if expression levels of these genes showed sufficient variability between the different BXD strains. We did not, however, pick up all of the genes with a known influence on OD plasticity. This could be due to various causes. First, genes that regulate OD plasticity exert their influence in visual cortex and during the sensitive period, while the available expression data is limited to that of the full neocortex of adult or very young mice. Expression across areas and ages will often correlate, but this certainly does not always have to be the case, in particular for genes involved in closure of the sensitive period. This is an important drawback of our approach. Any future study which would measure the expression level of genes in the visual cortex in BXD strains during the fifth postnatal week (matching the functional plasticity data) would remove this problem. A second reason for underreporting OD plasticity genes, however, is that some genes may encode proteins that are necessary for plasticity, but are not rate limiting. A knock-out of such a gene would produce a phenotype, but expression level differences would not correlate with the strength of plasticity. In the specific case of MHCI gene H2-K1 where we did not find a correlation, it may actually be because H2-K1 is not involved in OD plasticity. The evidence for its involvement comes from a study where both H2-D1 and H2-K1 were simultaneously knocked out (Datwani et al., 2009). We find a significant negative correlation of H2-D1 expression and the amount of increase in non-deprived ipsilateral response strength, in line with the increased response for the ipsilateral non-deprived eye in the double knock-out.
Candidate Selection
By correlating expression with plasticity, we created a list of 3486 genes. To narrow down this list to the most likely candidate plasticity genes we assumed that such genes would be associated with biological events or signaling pathways that were at least loosely implicated in cortical plasticity, and that the genes would be regulated by visual experience. In practical terms this means that we created two additional lists of genes that adhered to these criteria and selected the genes that were in the intersection of the three lists. The first list (implicated gene list) was based on the literature. Signaling pathways and cellular processes known to play a role in OD plasticity were selected and used as terms in gene ontology searches. This resulted in a set of 830 genes potentially related to OD plasticity. While this list limits the possibility to discover candidate genes in entirely new signaling pathways it certainly does not exclude this, as the genes on the list are obviously not restricted to the implicated pathways. The second list contained genes from a micro-array study identifying genes regulated upon MD or rearing mice in the dark. This regulated gene list contained 4404 unique genes, 404 of these also were present on the correlated gene list. In total 32 genes were present in all three gene lists and thus regulated by visual experience, correlated with OD plasticity and associated with cellular events implicated in plasticity in the visual cortex.
Known and Likely Candidates
For six of the candidate genes, Igf1, Ncam1, Rtn4, Prkaca, Gabrg2, Ache, there is good evidence for a role in OD plasticity. Evidence that the cell adhesion molecule NCAM1 is involved in OD plasticity comes from a study by Di Cristo et al. (2007). In this study, the removal of PSA polymers attached to NCAM1 resulted in enhanced inhibitory synaptic transmission and an earlier onset of OD plasticity. The second such gene is Igf1. The pathway around insulin-like growth factor-1 (IGF-1) was identified by Tropea et al. (2006) as regulated after MD. Furthermore, they showed that exogenous application of IGF-1 prevents the physiological effects of MD on OD plasticity. Ciucci et al. (2007) blocked IGF-1 in the visual cortex of rats housed in an enriched environment and showed that IGF-1 affected perisomatic inhibition and the condensation of chondroitin sulfate proteoglycans in perineuronal nets. There is also direct evidence for the involvement in OD plasticity of the third gene in the list, Rtn4, encoding Nogo-A. McGee et al. (2005) showed that mice in which NGR, the Nogo-receptor, was knocked out, showed continued OD plasticity long after the normal end of the sensitive period. The catalytic subunit of cAMP-dependent protein kinase A (PKA), coded by Prkaca, is another usual suspect. Pharmacologically inhibiting PKA in cat blocked an OD shift after MD in the sensitive period (Beaver et al., 2001) and mice lacking the RII alpha subunit of PKA have a reduced OD shift (Rao et al., 2004). The fifth gene very likely to be essential for OD plasticity is Gabrg2, coding the GABA(A) receptor subunit gamma2, which is obligatory for GABA(A) receptor expression on the cell surface (Schweizer et al., 2003). Much previous work has implicated the GABAergic system in sensitive period plasticity (e.g., Huang et al., 1999; Fagiolini and Hensch, 2000; Fagiolini et al., 2004; Harauzov et al., 2010; Heimel et al., 2011). And last, the gene encoding acetylcholinesterase (Ache), the enzyme responsible for inactivating the neurotransmitter acetylcholine, is likely to regulate OD plasticity as previous studies provide clear evidence for the role of cholinergic signaling (Bear and Singer, 1986; Gu and Singer, 1993). In particular, injection of an acetylcholinesterase inhibitor restored normal vision in amblyopic mice (Morishita et al., 2010).
Kinase Candidates
The possible role of each of the other 26 candidate genes is not as immediately obvious, despite the fact that they are by definition associated with signaling pathways or cellular events known to be linked to OD plasticity. On the list are five kinases besides PKA. Protein kinase B (AKT1) is a serine/threonine kinase involved in many pathways and was present in the implicated gene list as member of the IGF-1 signaling cascade (Cheng et al., 2000). Phosphorylated AKT1 was significantly reduced by MD and restored by addition of IGF-1 (Tropea et al., 2006), probably by activation of PI3K. Interestingly, one role of AKT1 in the brain is the phosphorylation of GABA-A receptors which leads to enhanced GABAergic synaptic transmission (Wang et al., 2003). This could be the pathway through which AKT1 exerts its possible role in OD plasticity. A second kinase on the list is the enzyme inositol trisphosphate 3-kinase A (ITPKA). This enzyme phosphorylates IP3, a second messenger molecule which is also downstream of PI3K and thus possibly affected by IGF-1. ITPKA was already suggested to play a role in structural plasticity (Kim et al., 2004). It is highly enriched in dendritic spines and also regulates the F-actin structure independently of its kinase activity (Johnson and Schell, 2009). It is unclear which of these functions, if any, would be its role in OD plasticity, but Itpka knock-out mice have reduced synaptic plasticity in the hippocampus (Kim et al., 2009). A third kinase, c-Jun N-terminal kinase 1 (JNK1, or MAPK8) is one of the three JNKs, and a member of the MAPK family. JNKs have been reported to regulate short-term memory (Bevilaqua et al., 2003). MAPK8 mutant mice show progressive learning impairment (Chang et al., 2003). MAPK8 contributes to mGluR-dependent LTD in the hippocampus (Li et al., 2007) and could play a similar role in the cortex. OD plasticity can be prevented by inhibiting MAPK (ERK) signaling by MEK inhibitors (Di Cristo et al., 2001), but whether MAPK8 is also involved in OD plasticity remains to be tested. For the other kinases on the list, there is little in the literature to suggest how they could act on OD plasticity. This is the case in particular for nemo like kinase (NLK), and Ca2+/Calmodulin dependent kinase 2 delta (CaMKII-delta), although the latter has already been reported to be highly upregulated during the critical period in the rat (Ossipow et al., 2004). The candidate list also contains two anchoring proteins that help cAMP-dependent protein kinase (PKA) to be in proximity to its targets (Edwards and Scott, 2000). A-kinase anchoring protein 15 (AKAP15), also known as AKAP7, localizes PKA to sodium channels where it may directly affect plasticity levels. The PKA anchoring protein Neurobeachin (NBEA) is involved in neuronal membrane protein trafficking (Wang et al., 2000) and is required for the formation and functioning of central synapses (Medrihan et al., 2009). Mutations in Nbea are thought to cause autism (Castermans et al., 2003; Medrihan et al., 2009), but a role in plasticity has not yet been shown.
Other Candidates
Among the other candidates is the gene Ppp1r1b, which codes for the regulatory (inhibitor) subunit 1B of protein phosphatase 1 (PP1), better known as DARPP-32. PP1 is a serine/threonine phosphatase which can suppress learning and memory (Genoux et al., 2002), possibly through dephosphorylating CREB (Hagiwara et al., 1992). PP1 has a critical role in NMDA-dependent LTD (Morishita et al., 2001). This makes a role for PP1 in OD plasticity likely (Heynen et al., 2003), but still unproven. The gene for ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 (St8sia1) entered our implicated gene list only through its ontological link with St8sia2 (STX) and St8sia4 (PST), the enzymes responsible for the polysialylation of NCAM1. It is better known under the name GD3 synthase, and is one of the enzymes responsible for producing Sialic acid-containing glycosphingolipids (gangliosides). Through double knock-out experiments of St8sia1 and GM2/GD2 synthase (B4galnt1), it was previously established that the expression of complex gangliosides is essential for the integrity of the nervous system (Tajima et al., 2009). Moreover, Inokuchi et al. (1998) showed that gangliosides play a role in synaptic plasticity by manipulating their biosynthesis in cultured cortical neurons, which resulted in upregulated neurite outgrowth and functional synapse formation. Interestingly, elimination of GD3 synthase also improves memory and reduces amyloid-beta plaque-load in a mouse model for Alzheimer’s disease (Bernardo et al., 2009). Another interesting candidate gene is Cd44, coding a transmembrane hyaluronan-binding glycoprotein involved in axon routing (Sretavan et al., 1994). This glycoprotein is regulated after nerve dissection and implicated in axon regeneration (Jones et al., 2000). It is thus possible that regulation of CD44 may affect structural plasticity underlying OD plasticity. The final candidates for which we can easily envision a role in OD plasticity are receptors for peptides expressed in cortical interneurons, neuropeptide (Npy)5-receptor, and cholecystokinin (CCK)2-receptor. The NPY5 receptor is highly expressed in the entire rodent neocortex (Wolak et al., 2003). It was already suggested that this receptor modulates postsynaptic activity and that the neuropeptide Y/Y5 system has an effect on the modulation of certain GABAergic neurons in the cortex (Grove et al., 2000), through which it may modulate OD plasticity. CCK is a gut-brain peptide expressed in a subset of interneurons. The CCK2-receptor has been implicated in anxiety, learning, and memory, mediation of pain and regulation of feeding (Noble and Roques, 1999). It has been found to have effects on dopaminergic (Altar and Boyar, 1989) and GABAergic transmission (Miller et al., 1997; Foldy et al., 2007) through which it may affect plasticity in the visual cortex.
In conclusion, by taking a subsection of genes correlated with the amount of OD plasticity, regulated in conjunction with the critical period, and implicated in pathways associated with ODP, we identified 32 candidate genes that are possibly involved in OD plasticity. For six of the candidates, it would be surprising if they do not play a role. The other genes are novel candidates for regulating cortical plasticity and it will be of great interest to confirm their involvement in plasticity in vivo and to dissect the underlying mechanisms.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Glenn Rosen for the neocortex expression data, Koen Bossers, and Sarah Janssen for help with the gene ontology analyses, Matt Self for critical reading of the manuscript, and we especially thank Rob Williams for developing Genenetwork and stimulating research on recombinant inbred strains. J. Alexander Heimel, J.-P. Sommeijer, and the Neuro-Bsik Mouse Phenomics consortium were supported by grant BSIK 03053 from SenterNovem (The Netherlands). The Neuro-Bsik Mouse Phenomics consortium is composed of the laboratories of A. B. Brussaard, J. G. G. Borst, Y. Elgersma, N. Galjart, G. T. van der Horst, C. N. Levelt, C. M. Pennartz, A. B. Smit, B. M. Spruijt, M. Verhage, and C. I. de Zeeuw, and the companies Noldus Information Technology and Synaptologics.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/neurogenomics/10.3389/fnins.2012.00011/abstract
Footnotes
References
Altar, C. A., and Boyar, W. C. (1989). Brain CCK-B receptors mediate the suppression of dopamine release by cholecystokinin. Brain Res. 483, 321–326.
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T., Harris, M. A., Hill, D. P., Issel-Tarver, L., Kasarskis, A., Lewis, S., Matese, J. C., Richardson, J. E., Ringwald, M., Rubin, G. M., and Sherlock, G. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29.
Bear, M. F., and Singer, W. (1986). Modulation of visual cortical plasticity by acetylcholine and noradrenaline. Nature 320, 172–176.
Beaver, C. J., Ji, Q., Fischer, Q. S., and Daw, N. W. (2001). Cyclic AMP-dependent protein kinase mediates ocular dominance shifts in cat visual cortex. Nat. Neurosci. 4, 159–163.
Bernardo, A., Harrison, F. E., McCord, M., Zhao, J., Bruchey, A., Davies, S. S., Jackson Roberts, L. II, Mathews, P. M., Matsuoka, Y., Ariga, T., Yu, R. K., Thompson, R., and McDonald, M. P. (2009). Elimination of GD3 synthase improves memory and reduces amyloid-beta plaque load in transgenic mice. Neurobiol. Aging 30, 1777–1791.
Bevilaqua, L. R., Kerr, D. S., Medina, J. H., Izquierdo, I., and Cammarota, M. (2003). Inhibition of hippocampal Jun N-terminal kinase enhances short-term memory but blocks long-term memory formation and retrieval of an inhibitory avoidance task. Eur. J. Neurosci. 17, 897–902.
Carulli, D., Pizzorusso, T., Kwok, J. C., Putignano, E., Poli, A., Forostyak, S., Andrews, M. R., Deepa, S. S., Glant, T. T., and Fawcett, J. W. (2010). Animals lacking link protein have attenuated perineuronal nets and persistent plasticity. Brain 133, 2331–2347.
Castermans, D., Wilquet, V., Parthoens, E., Huysmans, C., Steyaert, J., Swinnen, L., Fryns, J. P., Van de Ven, W., and Devriendt, K. (2003). The neurobeachin gene is disrupted by a translocation in a patient with idiopathic autism. J. Med. Genet. 40, 352–356.
Chang, L., Jones, Y., Ellisman, M. H., Goldstein, L. S., and Karin, M. (2003). JNK1 is required for maintenance of neuronal microtubules and controls phosphorylation of microtubule-associated proteins. Dev. Cell 4, 521–533.
Cheng, C. M., Reinhardt, R. R., Lee, W. H., Joncas, G., Patel, S. C., and Bondy, C. A. (2000). Insulin-like growth factor 1 regulates developing brain glucose metabolism. Proc. Natl. Acad. Sci. U.S.A. 97, 10236–10241.
Chesler, E. J., Lu, L., Wang, J., Williams, R. W., and Manly, K. F. (2004). WebQTL: rapid exploratory analysis of gene expression and genetic networks for brain and behavior. Nat. Neurosci. 7, 485–486.
Ciucci, F., Putignano, E., Baroncelli, L., Landi, S., Berardi, N., and Maffei, L. (2007). Insulin-like growth factor 1 (IGF-1) mediates the effects of enriched environment (EE) on visual cortical development. PLoS ONE 2, e475. doi:10.1371/journal.pone.0000475
Dahlhaus, M., Wan Li, K., van der Schors, R. C., Saiepour, M. H., van Nierop, P., Heimel, J. A., Hermans, J. M., Loos, M., Smit, A. B., and Levelt, C. N. (2011). The synaptic proteome during development and plasticity of the mouse visual cortex. Mol. Cell. Proteomics 10, M110.005413.
Datwani, A., McConnell, M. J., Kanold, P. O., Micheva, K. D., Busse, B., Shamloo, M., Smith, S. J., and Shatz, C. J. (2009). Classical MHCI molecules regulate retinogeniculate refinement and limit ocular dominance plasticity. Neuron 64, 463–470.
Di Cristo, G., Berardi, N., Cancedda, L., Pizzorusso, T., Putignano, E., Ratto, G. M., and Maffei, L. (2001). Requirement of ERK activation for visual cortical plasticity. Science 292, 2337–2340.
Di Cristo, G., Chattopadhyaya, B., Kuhlman, S. J., Fu, Y., Bélanger, M. C., Wu, C. Z., Rutishauser, U., Maffei, L., and Huang, Z. J. (2007). Activity-dependent PSA expression regulates inhibitory maturation and onset of critical period plasticity. Nat. Neurosci. 10, 1569–1577.
Edwards, A. S., and Scott, J. D. (2000). A-kinase anchoring proteins: protein kinase A and beyond. Curr. Opin. Cell Biol. 12, 217–221.
Fagiolini, M., Fritschy, J. M., Löw, K., Möhler, H., Rudolph, U., and Hensch, T. K. (2004). Specific GABAA circuits for visual cortical plasticity. Science 303, 1681–1683.
Fagiolini, M., and Hensch, T. K. (2000). Inhibitory threshold for critical-period activation in primary visual cortex. Nature 9;404, 183–186.
Fagiolini, M., Katagiri, H., Miyamoto, H., Mori, H., Grant, S. G., Mishina, M., and Hensch, T. K. (2003). Separable features of visual cortical plasticity revealed by N-methyl-D-aspartate receptor 2A signaling. Proc. Natl. Acad. Sci. U.S.A. 100, 2854–2859.
Fischer, Q. S., Beaver, C. J., Yang, Y., Rao, Y., Jakobsdottir, K. B., Storm, D. R., McKnight, G. S., and Daw, N. W. (2004). Requirement for the RII beta isoform of PKA, but not calcium-stimulated adenylyl cyclase, in visual cortical plasticity. J. Neurosci. 24, 9049–9058.
Foldy, C., Lee, S. Y., Szabadics, J., Neu, A., and Soltesz, I. (2007). Cell type-specific gating of perisomatic inhibition by cholecystokinin. Nat. Neurosci. 10, 1128–1130.
Gaglani, S. M., Lu, L., Williams, R. W., and Rosen, G. D. (2009). The genetic control of neocortex volume and covariation with neocortical gene expression in mice. BMC Neurosci. 10, 44. doi:10.1186/1471-2202-10-44
Genoux, D., Haditsch, U., Knobloch, M., Michalon, A., Storm, D., and Mansuy, I. M. (2002). Protein phosphatase 1 is a molecular constraint on learning and memory. Nature 418, 970–975.
Gordon, J. A., and Stryker, M. P. (1996). Experience-dependent plasticity of binocular responses in the primary visual cortex of the mouse. J. Neurosci. 16, 3274–3286.
Gordon, J. A., Cioffi, D., Silva, A. J., and Stryker, M. P. (1996). Deficient plasticity in the primary visual cortex of alpha-calcium/calmodulin-dependent protein kinase II mutant mice. Neuron 17, 491–499.
Grove, K. L., Campbell, R. E., Ffrench-Mullen, J. M., Cowley, M. A., and Smith, M. S. (2000). Neuropeptide Y Y5 receptor protein in the cortical/limbic system and brainstem of the rat: expression on gamma-aminobutyric acid and corticotropin-releasing hormone neurons. Neuroscience 100, 731–740.
Gu, Q., and Singer, W. (1993). Effects of intracortical infusion of anticholinergic drugs on neuronal plasticity in kitten striate cortex. Eur. J. Neurosci. 5, 475–485.
Hagiwara, M., Alberts, A., Brindle, P., Meinkoth, J., Feramisco, J., Deng, T., Karin, M., Shenolikar, S., and Montminy, M. (1992). Transcriptional attenuation following cAMP induction requires PP-1-mediated dephosphorylation of CREB. Cell 70, 105–113.
Harauzov, A., Spolidoro, M., DiCristo, G., De Pasquale, R., Cancedda, L., Pizzorusso, T., Viegi, A., Berardi, N., and Maffei, L. (2010). Reducing intracortical inhibition in the adult visual cortex promotes ocular dominance plasticity. J. Neurosci. 30, 361–371.
Heimel, J. A., Hermans, J. M., Sommeijer, J.-P., Neuro-Bsik Mouse Phenomics Consortium, and Levelt, C. N. (2008). Genetic control of experience-dependent plasticity in the visual cortex. Genes Brain Behav. 7, 915–923.
Heimel, J. A., van Versendaal, D., and Levelt, C. N. (2011). The role of GABAergic inhibition in ocular dominance plasticity. Neural Plast. 2011, 391763.
Hensch, T. K., Fagiolini, M., Mataga, N., Stryker, M. P., Baekkeskov, S., and Kash, S. F. (1998a). Local GABA circuit control of experience-dependent plasticity in developing visual cortex. Science 282, 1504–1508.
Hensch, T. K., Gordon, J. A., Brandon, E. P., McKnight, G. S., Idzerda, R. L., and Stryker, M. P. (1998b). Comparison of plasticity in vivo and in vitro in the developing visual cortex of normal and protein kinase A RIbeta-deficient mice. J. Neurosci. 18, 2108–2117.
Heynen, A. J., Yoon, B. J., Liu, C. H., Chung, H. J., Huganir, R. L., and Bear, M. F. (2003). Molecular mechanism for loss of visual cortical responsiveness following brief monocular deprivation. Nat. Neurosci. 6, 854–862.
Huang, Z. J., Kirkwood, A., Pizzorusso, T., Porciatti, V., Morales, B., Bear, M. F., Maffei, L., and Tonegawa, S. (1999). BDNF regulates the maturation of inhibition and the critical period of plasticity in mouse visual cortex. Cell 98, 739–755.
Inokuchi, J., Kuroda, Y., Kosaka, S., and Fujiwara, M. (1998). L-threo-1-phenyl-2-decanoylamino-3-morpholino-1-propanol stimulates ganglioside biosynthesis, neurite outgrowth and synapse formation in cultured cortical neurons, and ameliorates memory deficits in ischemic rats. Acta Biochim. Pol. 45, 479–492.
Johnson, H. W., and Schell, M. J. (2009). Neuronal IP3 3-kinase is an F-actin-bundling protein: role in dendritic targeting and regulation of spine morphology. Mol. Biol. Cell 20, 5166–5180.
Jones, L. L., Liu Shen, J., Werner, A., Kreutzberg, G. W., and Raivich, G. (2000). Regulation of the cell adhesion molecule CD44 after nerve transection and direct trauma to the mouse brain. J. Comp. Neurol. 426, 468–492.
Kaneko, M., Stellwagen, D., Malenka, R. C., and Stryker, M. P. (2008). Tumor necrosis factor-alpha mediates one component of competitive, experience-dependent plasticity in developing visual cortex. Neuron 58, 673–680.
Kim, I. H., Park, S. K., Hong, S. T., Jo, Y. S., Kim, E. J., Park, E. H., Han, S. B., Shin, H. S., Sun, W., Kim, H. T., Soderling, S. H., and Kim, H. (2009). Inositol 1,4,5-trisphosphate 3-kinase a functions as a scaffold for synaptic Rac signaling. J. Neurosci. 29, 14039–14049.
Kim, I. H., Park, S. K., Sun, W., Kang, Y., Kim, H. T., and Kim, H. (2004). Spatial learning enhances the expression of inositol 1,4,5-trisphosphate 3-kinase A in the hippocampal formation of rat. Brain Res. Mol. Brain Res. 124, 12–19.
Lein, E. S., Hawrylycz, M. J., Ao, N., Ayres, M., Bensinger, A., Bernard, A., Boe, A. F., Boguski, M. S., Brockway, K. S., Byrnes, E. J., Chen, L., Chen, L., Chen, T. M., Chin, M. C., Chong, J., Crook, B. E., Czaplinska, A., Dang, C. N., Datta, S., Dee, N. R., Desaki, A. L., Desta, T., Diep, E., Dolbeare, T. A., Donelan, M. J., Dong, H. W., Dougherty, J. G., Duncan, B. J., Ebbert, A. J., Eichele, G., Estin, L. K., Faber, C., Facer, B. A., Fields, R., Fischer, S. R., Fliss, T. P., Frensley, C., Gates, S. N., Glattfelder, K. J., Halverson, K. R., Hart, M. R., Hohmann, J. G., Howell, M. P., Jeung, D. P., Johnson, R. A., Karr, P. T., Kawal, R., Kidney, J. M., Knapik, R. H., Kuan, C. L., Lake, J. H., Laramee, A. R., Larsen, K. D., Lau, C., Lemon, T. A., Liang, A. J., Liu, Y., Luong, L. T., Michaels, J., Morgan, J. J., Morgan, R. J., Mortrud, M. T., Mosqueda, N. F., Ng, L. L., Ng, R., Orta, G. J., Overly, C. C., Pak, T. H., Parry, S. E., Pathak, S. D., Pearson, O. C., Puchalski, R. B., Riley, Z. L., Rockett, H. R., Rowland, S. A., Royall, J. J., Ruiz, M. J., Sarno, N. R., Schaffnit, K., Shapovalova, N. V., Sivisay, T., Slaughterbeck, C. R., Smith, S. C., Smith, K. A., Smith, B. I., Sodt, A. J., Stewart, N. N., Stumpf, K. R., Sunkin, S. M., Sutram, M., Tam, A., Teemer, C. D., Thaller, C., Thompson, C. L., Varnam, L. R., Visel, A., Whitlock, R. M., Wohnoutka, P. E., Wolkey, C. K., Wong, V. Y., Wood, M., Yaylaoglu, M. B., Young, R. C., Youngstrom, B. L., Yuan, X. F., Zhang, B., Zwingman, T. A., and Jones, A. R. (2007). Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176.
Li, X. M., Li, C. C., Yu, S. S., Chen, J. T., Sabapathy, K., and Ruan, D. Y. (2007). JNK1 contributes to metabotropic glutamate receptor-dependent long-term depression and short-term synaptic plasticity in the mice area hippocampal CA1. Eur. J. Neurosci. 25, 391–396.
Lyckman, A. W., Horng, S., Leamey, C. A., Tropea, D., Watakabe, A., Van Wart, A., McCurry, C., Yamamori, T., and Sur, M. (2008). Gene expression patterns in visual cortex during the critical period: synaptic stabilization and reversal by visual deprivation. Proc. Natl. Acad. Sci. U.S.A. 105, 9409–9414.
Majdan, M., and Shatz, C. J. (2006). Effects of visual experience on activity-dependent gene regulation in cortex. Nat. Neurosci. 9, 650–659.
Mataga, N., Fujishima, S., Condie, B. G., and Hensch, T. K. (2001). Experience-dependent plasticity of mouse visual cortex in the absence of the neuronal activity-dependent marker egr1/zif268. J. Neurosci. 21, 9724–9732.
Mataga, N., Nagai, N., and Hensch, T. K. (2002). Permissive proteolytic activity for visual cortical plasticity. Proc. Natl. Acad. Sci. U.S.A. 99, 7717–7721.
McCurry, C. L., Shepherd, J. D., Tropea, D., Wang, K. H., Bear, M. F., and Sur, M. (2010). Loss of Arc renders the visual cortex impervious to the effects of sensory experience or deprivation. Nat. Neurosci. 13, 450–457.
McGee, A. W., Yang, Y., Fischer, Q. S., Daw, N. W., and Strittmatter, S. M. (2005). Experience-driven plasticity of visual cortex limited by myelin and Nogo receptor. Science 309, 2222–2226.
Medrihan, L., Rohlmann, A., Fairless, R., Andrae, J., Döring, M., Missler, M., Zhang, W., and Kilimann, M. W. (2009). Neurobeachin, a protein implicated in membrane protein traffic and autism, is required for the formation and functioning of central synapses. J. Physiol. (Lond.) 587, 5095–5106.
Miller, K. K., Hoffer, A., Svoboda, K. R., and Lupica, C. R. (1997). Cholecystokinin increases GABA release by inhibiting a resting K+ conductance in hippocampal interneurons. J. Neurosci. 17, 4994–5003.
Morishita, H., Miwa, J. M., Heintz, N., and Hensch, T. K. (2010). Lynx1, a cholinergic brake, limits plasticity in adult visual cortex. Science 330, 1238–1240.
Morishita, W., Connor, J. H., Xia, H., Quinlan, E. M., Shenolikar, S., and Malenka, R. C. (2001). Regulation of synaptic strength by protein phosphatase1. Neuron 32, 1133–1148.
Noble, F., and Roques, B. P. (1999). CCK-B receptor: chemistry, molecular biology, biochemistry and pharmacology. Prog. Neurobiol. 58, 349–379.
Ossipow, V., Pellisier, F., Schaad, O., and Ballivet, M. (2004). Gene expression analysis of the critical period in the visual cortex. Mol. Cell. Neurosci. 27, 70–83.
Rao, Y., Fischer, Q. S., Yang, Y., McKnight, G. S., LaRue, A., and Daw, N. W. (2004). Reduced ocular dominance plasticity and long-term potentiation in the developing visual cortex of protein kinase A RII alpha mutant mice. Eur. J. Neurosci. 20, 837–842.
Renger, J. J., Hartman, K. N., Tsuchimoto, Y., Yokoi, M., Nakanishi, S., and Hensch, T. K. (2002). Experience-dependent plasticity without long-term depression by type 2 metabotropic glutamate receptors in developing visual cortex. Proc. Natl. Acad. Sci. U.S.A. 99, 1041–1046.
Schweizer, C., Balsiger, S., Bluethmann, H., Mansuy, I. M., Fritschy, J. M., Mohler, H., and Lüscher, B. (2003). The gamma 2 subunit of GABA(A) receptors is required for maintenance of receptors at mature synapses. Mol. Cell. Neurosci. 24, 442–450.
Sretavan, D. W., Feng, L., Puré, E., and Reichardt, L. F. (1994). Embryonic neurons of the developing optic chiasm express L1 and CD44, cell surface molecules with opposing effects on retinal axon growth. Neuron 12, 957–975.
Syken, J., Grandpre, T., Kanold, P. O., and Shatz, C. J. (2006). PirB restricts ocular-dominance plasticity in visual cortex. Science 313, 1795–1800.
Szklarczyk, D., Franceschini, A., Kuhn, M., Simonovic, M., Roth, A., Minguez, P., Doerks, T., Stark, M., Muller, J., Bork, P., Jensen, L. J., and von Mering, C. (2011). The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39, D561–D568.
Taha, S., Hanover, J. L., Silva, A. J., and Stryker, M. P. (2002). Autophosphorylation of alphaCaMKII is required for ocular dominance plasticity. Neuron 36, 483–491.
Tajima, O., Egashira, N., Ohmi, Y., Fukue, Y., Mishima, K., Iwasaki, K., Fujiwara, M., Inokuchi, J., Sugiura, Y., Furukawa, K., and Furukawa, K. (2009). Reduced motor and sensory functions and emotional response in GM3-only mice: emergence from early stage of life and exacerbation with aging. Behav. Brain Res. 198, 74–82.
Tropea, D., Kreiman, G., Lyckman, A., Mukherjee, S., Yu, H., Horng, S., and Sur, M. (2006). Gene expression changes and molecular pathways mediating activity-dependent plasticity in visual cortex. Nat. Neurosci. 9, 660–668.
Tropea, D., Van Wart, A., and Sur, M. (2009). Molecular mechanisms of experience-dependent plasticity in visual cortex. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 341–355.
Wang, Q., Liu, L., Pei, L., Ju, W., Ahmadian, G., Lu, J., Wang, Y., Liu, F., and Wang, Y. T. (2003). Control of synaptic strength, a novel function of Akt. Neuron 38, 915–928.
Wang, X., Herberg, F. W., Laue, M. M., Wullner, C., Hu, B., Petrasch-Parwez, E., and Kilimann, M. W. (2000). Neurobeachin: a protein kinase A-anchoring, beige/Chediak-Higashi protein homolog implicated in neuronal membrane traffic. J. Neurosci. 20, 8551–8565.
Wiesel, T. N., and Hubel, D. H. (1963). Single-cell responses in striate cortex of kittens deprived of vision in one eye. J. Neurophysiol. 26, 1003–1017.
Keywords: plasticity, ocular dominance, visual cortex, recombinant inbred
Citation: Rietman ML, Sommeijer J-P, Neuro-Bsik Mouse Phenomics consortium, Levelt CN and Heimel JA (2012) Candidate genes in ocular dominance plasticity. Front. Neurosci. 6:11. doi: 10.3389/fnins.2012.00011
Received: 15 August 2011;
Accepted: 16 January 2012;
Published online: 01 February 2012.
Edited by:
Hua Lou, Case Western Reserve University, USAReviewed by:
Sarah London, University of Illinois, USAQingzhong Kong, Case Western Reserve University, USA
Copyright: © 2012 Rietman, Sommeijer, Neuro-Bsik Mouse Phenomics Consortium, Levelt and Heimel. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: J. Alexander Heimel, Netherlands Institute for Neuroscience, Meibergdreef 47, 1105 BA Amsterdam, Netherlands. e-mail: heimel@nin.knaw.nl