- 1 Department of Physiology, Keio University School of Medicine, Shinjuku, Tokyo, Japan
- 2 Department of Cell and Developmental Biology, University of Pennsylvania School of Medicine, Philadelphia, PA, USA
MicroRNA (miRNA) is a type of short-length (~22 nt) non-coding RNA. Most miRNAs are transcribed by RNA polymerase II and processed by Drosha-DGCR8 and Dicer complexes in the cropping and dicing steps, respectively. miRNAs are exported by exportin-5 from the nucleus to the cytoplasm after cropping. Trimmed mature miRNA is loaded and targets mRNA at the 3′ or 5′ untranslated region (UTR) by recognition of base-pairing in the miRNA-loaded RISC, where it is involved in gene silencing including translational repression and/or degradation along with deadenylation. Recent studies have shown that miRNA participates in various biological functions including cell fate decision, developmental timing regulation, apoptosis, and tumorigenesis. Analyses of miRNA expression profiles have demonstrated tissue- and stage-specific miRNAs including the let-7 family, miR-124, and miR-9, which regulate the differentiation of embryonic stem cells and/or neurogenesis. This review focuses on RNA-binding protein-mediated miRNA biogenesis during neurogenesis. These miRNA biogenesis-relating proteins have also been linked to human diseases because their mutations can cause several nervous system disorders. Moreover, defects in core proteins involved in miRNA biogenesis including Drosha, DGCR8, and Dicer promote tumorigenesis. Thus, the study of not only mature miRNA function but also miRNA biogenesis steps is likely to be important.
Introduction
MicroRNAs (miRNAs) are 22–24 nucleotide (nt) single-stranded non-coding RNAs (ncRNAs) that play an important role as genetic regulators of development, differentiation, growth, and neurogenesis. Although some miRNAs are transcribed by RNA polymerase (pol) III (Borchert et al., 2006), most miRNAs are produced as primary miRNA (pri-miRNA) including 5′ cap and 3′ poly(A)-tails by pol II (Lee et al., 2004). The pri-miRNA is produced as two different transcripts (intergenic and intragenic products; please see below Transcription of miRNA; Figure 1).
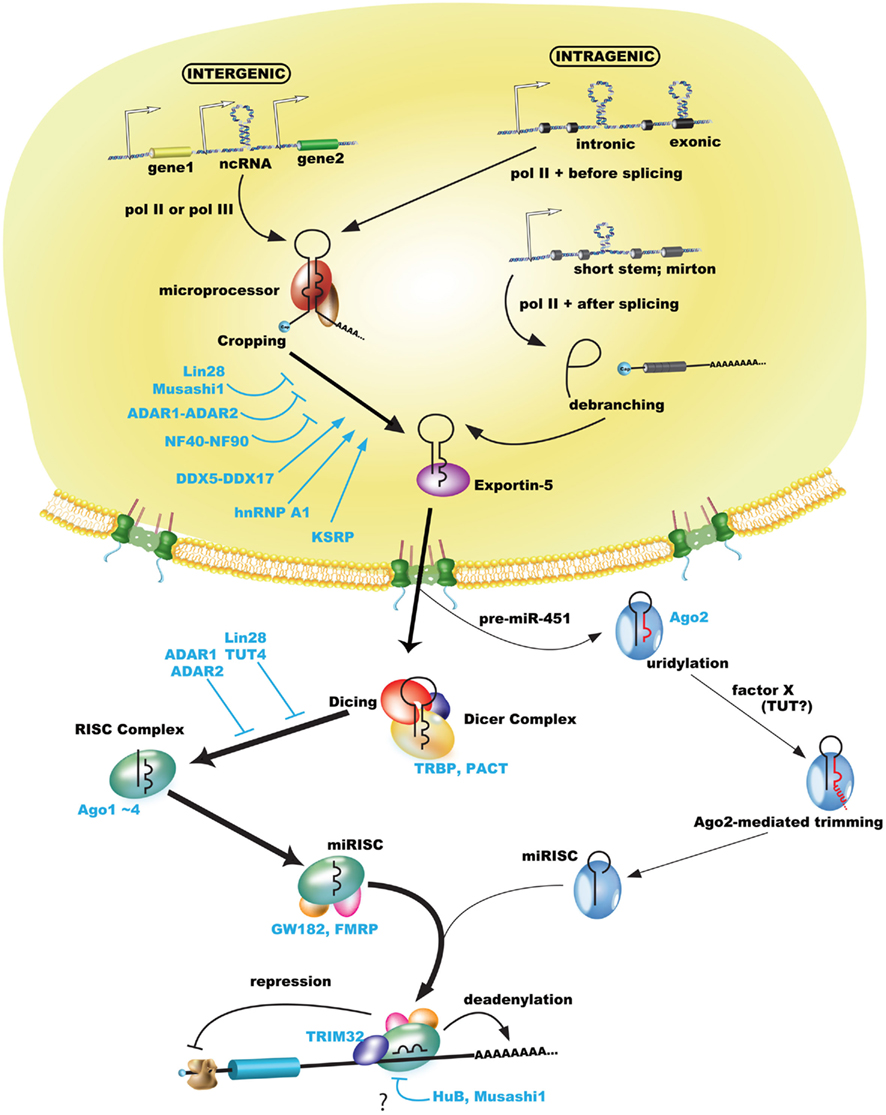
Figure 1. MicroRNA biogenesis pathway including some regulations of RNA-binding proteins. Pol II or pol III transcribe miRNAs as pri-miRNAs including a 5′ cap and poly(A)-tail, while pre-miRNAs (or mirtrons) are processed by microprocessor or canonical spliceosome (Drosha-bypassing pathway) and exported by exportin-5 from the nucleus to the cytoplasm. Exported pre-miRNAs are processed by the Dicer complex or Ago2 (Ago2-mediated cleavage in miR-451). Most trimmed miRNAs (but not miRNA* passenger strand) are sorted in the RISC complex, which includes Ago proteins, incorporated in the miRISC, and finally involved in gene silencing of their target mRNA by base-paring of the seed sequence. Gene silencing including translational repression and degradation via deadenylation of poly(A)-tail functions in the miRISC. Some RNA-binding proteins may regulate each biogenesis step (please see the text).
The processing of miRNAs includes both cropping and dicing steps. The pri-miRNA is processed as precursor-miRNA (pre-miRNA) hairpins of approximately 60–100 nt by the microprocessor in the nucleus that includes the nuclear RNase III enzyme Drosha and DGCR8 (DiGeorge syndrome critical region gene 8); this is known as the cropping step. In addition, approximately 50% of mammalian miRNAs are expressed from introns of protein-coding genes (Saini et al., 2008). In an alternate or Drosha-bypassing pathway, mirtrons, which mimic the structural features of pre-miRNAs, are generated as branched intron lariat pre-mirtrons during an ordinal splicing step (Okamura et al., 2007; Ruby et al., 2007). The pre-miRNA and mirtron hairpins are exported to the cytoplasm by exportin-5 and Ran-GTP (Figure 1; Yi et al., 2003). The pre-miRNA is then cleaved near its terminal loop by the RNase III enzyme Dicer to form mature miRNA of approximately 22-nt miRNA duplexes in the cytoplasm; this is known as the dicing step. The cleaved miRNA duplex contains mature miRNA (the guide) and miRNA* (the passenger) strands, whose strands demonstrate a 2-nt 3′ overhang due to RNase III enzyme-mediated cleavage. The mature miRNA is incorporated into the RNA-induced silencing complex (RISC), referred to as the miRNA-loaded RISC (miRISC), while the non-functional passenger strand is generally degraded. Finally, the miRISC is capable of conducting RNA-based gene silencing through base-pair matching between the 3′ (or 5′) untranslated region (UTR) of target RNA and positions 2–8 from the 5′ end of the miRNA, known as the “seed” sequence (Bartel, 2009).
Several recent studies have discussed the biological mechanisms of miRNA regulation in the nervous system. In this review, we highlight recent contributions to our understanding of the molecular mechanism of neural stem cell (NSC) maintenance and neurogenesis by mature miRNA, and how their expression is regulated during miRNA biogenesis by RNA-binding proteins.
Transcription of miRNA
In mammals, miRNAs are categorized as either intergenic or intragenic miRNAs in the genomic position: the former can be transcribed by pol II or pol III, while the latter can be done by pol II including intronic or exonic miRNAs. Intergenic miRNAs are synthesized from their own promoters, which are generally more evolutionarily conserved than intronic miRNAs, while intragenic miRNAs are synthesized with their host genes from the host transcriptional start sites (Kim et al., 2009b). Some intronic miRNAs, which have shorter precursor stem’s lengths than canonical miRNAs, are processed as spliced-out introns (so-called mirtrons; Ozsolak et al., 2008). Pol III is involved in transcribing miRNAs that have upstream Alu-, tRNA-, and mammalian-wide interspersed repeat (MWIR) promoter units. The human chromosome 19 miRNA cluster (C19MC), which contains 46 intronic miRNAs, 37 repeated exons, and many Alu sequences, is transcribed by pol III (Borchert et al., 2006; Bortolin-Cavaille et al., 2009). Interestingly, C19MC is a novel biomarker of the central nervous system primitive neuroectodermal tumor (CNS-PNET). Screening of about 40 CNS-PNET patients using microarray (SNP and dChip) analysis indicated a frequent high-level amplicon at chr19q13.41, whose locus contains C19MC. Either miR-520g or 517c overexpression enhanced cell growth and transformation in xenografted mice and inhibited differentiation of human NSCs via Wnt signaling. Remarkably, the results of Kaplan–Meier analysis (n = 25) revealed that overall survival was significantly reduced in C19MC-amplified patients, and tissue microarrays only detected the C19MC amplicon in CNS-PNET but not in 263 other brain tumors (Li et al., 2009).
Cropping of miRNA
In the cropping step associated with transcription of intergenic miRNA (pri-miRNA, average length 100–1000 nt), miRNA is recognized at a bulge site between upper stem and lower stem regions (Han et al., 2006) and cleaved by the microprocessor (Drosha and DGCR8). Then pri-miRNA is converted into trimmed pre-miRNA of approximately 60–100 nt with a 2-nt overhang at the 3′ end (Lee et al., 2003). Human Drosha-associated proteins have been biochemically identified as two discrete small and large molecular weight complexes using FLAG-tagged immunoprecipitation in FLAG-Drosha-expressing HEK293T cells (Gregory et al., 2004; Han et al., 2004). The larger Drosha complex contains multiple classes of RNA-binding proteins [hnRNPs (heterogeneous nuclear ribonucleoproteins), DDXs (DEAD-box), and RRM-type proteins], and the smaller complex contains DGCR8. However, the larger complex has an insignificant effect on the cleavage activity of pri-miRNAs; thus, the smaller complex Drosha-DGCR8 is considered the microprocessor (Gregory et al., 2004).
MicroRNA biogenesis is often regulated at the post-transcriptional level (Newman and Hammond, 2010; Siomi and Siomi, 2010). Some pri-miRNAs are not cropped and remain present at high levels, and pre-miRNAs are not necessarily processed into mature miRNAs at the dicing step, leading to the induction of mature miRNA expression during developmental stages (Thomson et al., 2006). Various studies suggest that the cropping step is an important step in miRNA biogenesis, whose mechanism is regulated by many RNA-binding proteins, such as DDX5, DDX17, KSRP (K-homology splicing regulator protein), hnRNP A1, hnRNP L, NF complex (NF90 and NF45), Musashi1, and Lin28 (Yamagata et al., 2009; Michlewski and Caceres, 2010; Newman and Hammond, 2010; Siomi and Siomi, 2010; Kawahara et al., 2011).
In ATP-dependent DEAD-box RNA helicase DDX5/p68- and DDX17/p78-null mice, a global survey of miRNA microarray expression indicated decreased expression of many miRNAs except for miR-19a, and DDX5–DDX17 was found to interact with mouse Drosha in vivo in co-immunoprecipitation experiments. Although other Drosha-associated proteins demonstrate consistent results between human and mouse Drosha complex (Gregory et al., 2004; Fukuda et al., 2007), the mouse Drosha complex is purified as a single large complex. The production of pre-miRNA was significantly decreased when pri-miRNA processing was performed under conditions of DDX5 or DDX17 knockout or in the presence of an ATP analog. By contrast, ATP stimulated production of pre-miRNA in a concentration-dependent manner (Fukuda et al., 2007). These results suggest that DDX5–DDX17 is required for recognition of a subset of pri-miRNAs in Drosha-mediated processing (Figure 1). How DDX15–DDX17 specifically recognizes the miRNAs is unclear.
Lin28 has been identified as a key gene in the control of the succession of patterns of division and differentiation during C. elegans larval stages, which has been to depend on repression of lin28 mRNA by lin-4 miRNA (Moss et al., 1997). The human homolog of Lin28 used as a reprogramming factor to establish iPS cells (Yu et al., 2007). The highest expression of Lin28 peak occurs at the early embryogenesis stage [e.g., embryonic stem (ES) cells] and expression gradually decreases during cell differentiation (Polesskaya et al., 2007; Viswanathan et al., 2008). Lin28 localizes mainly to the cytoplasm and cytoplasmic bodies, including processing bodies and stress granules, but also partially to the nucleus (Balzer and Moss, 2007). Particular ES cells have been shown by two groups to post-transcriptionally inhibit let-7 family miRNA biogenesis via Lin28-mediated selective inhibition of let-7 at the cropping step (Newman et al., 2008; Viswanathan et al., 2008). The mechanism by which Lin28 blocks let-7 cropping is suggested to involve competition between Lin28 and the microprocessor for the pri-let-7 interaction. Lin28 selectively binds the conserved terminal loop region of precursor let-7 family miRNAs, and a comparison of Kd values indicates that the binding activity between Lin28 and precursor let-7 is stronger than that between DGCR8 and pri-miRNA (Piskounova et al., 2008; Newman and Hammond, 2010).
Musashi1, which is highly expressed in neural stem/precursor cells (NS/PCs) and other somatic stem cells (Sakakibara et al., 1996, 2002; Okano et al., 2002, 2005), competes with eIF4G for PABP, resulting in the repression of formation of the 80S ribosome complex in target mRNAs of Musashi1, and supports the maintenance of NS/PCs in the undifferentiated state (Imai et al., 2001; Sakakibara et al., 2002; Kawahara et al., 2008). Comparison of the expression of Lin28, Musashi1, and miR-98, a let-7 family member, indicates that Lin28 and Musashi1 temporal expression patterns may finely regulate the timing of ES cell neural differentiation. Although Lin28 is abundantly expressed in ES cells during the initial retinoic acid-induction period, its expression is gradually downregulated during ES cell differentiation. During this period (EB cells on day 5), Lin28 repression of miR-98 cropping may attenuate. By contrast, Musashi1 expression gradually increases, which compensates for the lower activity of Lin28 and represses miR-98 cropping (Figure 2). Finally, around late neurogenesis, Musashi1 expression also decreases, allowing miR-98 cropping to proceed and resulting in the formation of mature miR-98 and the induction of neural marker expression. Musashi1 also enhanced the inhibitory effects of Lin28 on miR-98 biogenesis at the cropping step in a dose-dependent manner by controlling the subcellular localization of Lin28.
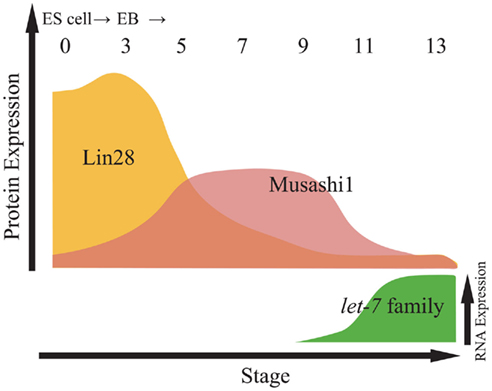
Figure 2. Relative expression of proteins and let-7 family miRNAs during embryonic stem cell (ESC) differentiation. Top panel indicates relative expression of proteins including Lin28 and Musashi1 in from embryoid body (EB) at day 0 to EB at day 13. Bottom panel indicates the expression of let-7 family miRNAs by northern blotting (Kawahara et al., 2011).
These results suggest that Lin28 and Musashi1 sequentially and synergistically repress miR-98 processing at the cropping step and determine the timing of neural differentiation (Figures 1 and 5; Kawahara et al., 2011). However, the detailed mechanism by which Musashi1 is involved in Lin28-mediated inhibition of let-7 in the cropping step has remained undetermined. The regulation of miRNA cropping in other systems outside of neurogenesis is reviewed and discussed in a recent report (Davis and Hata, 2010).
Dicing of miRNA
The RNAs processed by Drosha and DGCR8, which are referred to as pre-miRNAs, are exported from the nucleus to the cytoplasm accompanied by a nuclear export cargo receptor, exportin-5 (Yi et al., 2003; Bohnsack, 2004; Lund et al., 2004). The maturation of RNAs then proceeds to the second processing step, known as the “dicing” step. This step requires the enzymatic activity of Dicer, an RNase III-type endonuclease. Pre-miRNAs have hairpin-like structures comprised of a double-strand component and a loop component. Dicer enzymatic activity cleaves pre-miRNA to remove the loop structure. Through dicing, pre-miRNAs are processed into ~22-nt double-strand RNAs, which are often referred to as miRNA duplexes (Bernstein et al., 2001; Ketting et al., 2001).
In the dicing step, Dicer cooperates with HIV-1 TAR RNA-binding protein (TRBP; Chendrimada et al., 2005; Haase et al., 2005) and PKR activator (PACT; Lee et al., 2006). Both TRBP and PACT contain double-strand RNA-binding domains (dsRBD) and their domain structures are highly similar. Interestingly, TRBP and PACT interact with not only Dicer but also Argonaute2 (Ago2), one of the core components of RISC (Figure 1; Chendrimada et al., 2005). It is possible that the two dsRBD proteins are involved not alone in the dicing step, but also in the loading of mature miRNAs into the RISC (Chendrimada et al., 2005; Lee et al., 2006).
Recent reports using Ago2 catalytically inactive mutant knock-in mice and zebrafish have demonstrated a Dicer-independent pathway for miRNA dicing that depends on Ago2 catalytic activity (Cheloufi et al., 2010; Cifuentes et al., 2010). miR-451 has an evolutionarily conserved secondary hairpin structure that is contained in mature miR-451, and its length is insufficient for efficient cleavage by Dicer. Pre-miR-451 is processed by Ago2, resulting in mature miR-451 after further uridylation and trimming (Figure 1; Cifuentes et al., 2010). However, the details of how uridylation and trimming contribute to miR-451 maturation remain to be elucidated.
As is well known in stem cells, including ES cells and NS/PCs, many molecular mechanisms, including cytokine signal transduction, transcription regulation, epigenetic modification, and differentiation-related miRNA regulation, work to inhibit further cell differentiation. In the central nervous system (CNS), primary transcript expression of let-7, miR-124, miR-125, and miR-128 is induced during neural induction (Rybak et al., 2008), and the activities of these transcripts lead NS/PCs to the neuronal differentiation (Enciu et al., 2011). However, their activities are lower in NS/PCs than in mature cells (Rybak et al., 2008; Kawahara et al., 2011). Why does miRNA activity fail to emerge although precursor transcripts of the miRNAs exist? The inhibition of miRNA activity would appear to depend on the regulation of numerous overlapping processes, such as miRNA primary transcription, multiple miRNA processing, RISC loading, and translational repression, as well as possibly other currently unknown processes. Here, we introduce the examples that regulation of the “dicing” miRNA maturation step is involved in the acquisition of “stemness.”
Regulation of the dicing step in immature progenitor cells has been well characterized especially with respect to pre-let-7. In stem cells, “stemness,” such as the maintenance of immature status and self-renewing proliferation activity, is likely to require the downregulation of let-7 family activity (Wulczyn et al., 2007; Nishino et al., 2008; Schwamborn et al., 2009; Zhao et al., 2010). Both the cropping and dicing steps of let-7 maturation are selectively inhibited by Lin28 (Rybak et al., 2008). Lin28 directly binds to the loop region of pre-let-7a through its specific RNA-binding activity (Newman et al., 2008; Piskounova et al., 2008).
The Seoul National University Kim group found that Lin28 overexpression induced uridylation (average 14 nt) at the 3′ terminal of pre-let-7 and that Dicer could not cleave pre-let-7 bearing a uridylated tail (Heo et al., 2008). The activity of 3′-terminal uridylyl transferase 4 (TUTase 4/TUT4), also known as zinc finger CCHC domain containing 11 (Zcchc11), was also reported to accompany Lin28-mediated dicing inhibition. Elongation of the pre-miRNA 3′-terminal by TUT4-mediated uridylation decreased the accessibility of pre-miRNA to Dicer (Hagan et al., 2009) and increased pre-miRNA sensitivity to destabilizing exonuclease activity (Heo et al., 2008). By inhibiting the dicing maturation step of pre-let-7, the expression of let-7 target genes, including c-Myc, Hmga2, Ras, and TLX (Bussing et al., 2008; Zhao et al., 2010) is promoted in NS/PCs (Figure 5). Thus, regulation of the pre-let-7 dicing step is thought to be deeply associated with NS/PC proliferation and differentiation. Interestingly, tissue-specific cases, such as in the case of miR-138, whose mature form is restricted to the mouse developing CNS despite the ubiquitous expression of it precursor RNA (Obernosterer et al., 2006), suggest the existence of other, currently unknown, regulatory mechanisms of miRNA dicing.
The conditional knock out studies using Cre-recombination directed by Emx1 and Nestin promoters/enhancers revealed that Dicer is critical for neurogenesis and gliogenesis in the CNS (De Pietri Tonelli et al., 2008; Kawase-Koga et al., 2009). However, these assays showed the delayed Dicer deletion and a low level of Dicer proteins persisted in processing miRNAs. Thus, Livesey group established Dicer-null NSC lines from the embryonic mouse cerebral cortex (Andersson et al., 2010). The observations using these cell lines exhibited Dicer could be involved in protecting NS/PCs from the EGF-mediated apoptosis, and that the NS/PCs’ self-renewal ability was maintained because of increased Hmga2 expression. Hmga2 is one of the let-7 target mRNAs and its expression is promoted in NSCs. Interestingly, although these cell lines were incompetent for neurogenesis and gliogenesis, the retroviral reintroduction of Dicer allowed Dicer-null NSCs to recover from incapability of either neurogenesis or gliogenesis, suggesting that Dicer-null NSC lines have reversibly altered their neurogenic and gliogenic potential. However, recent reports showed the Dicer-bypass pathway in vivo (Figure 1; Cheloufi et al., 2010; Cifuentes et al., 2010). Furthermore, miRNA expression array used by Livesey group did not cover many kinds of miRNA (just 171 miRNAs were tested; Andersson et al., 2010). Thus, it remains still unclear whether Ago2-mediated pre-miRNA processing can be involved in self-renewal of NS/PCs and neurogenesis.
Loading and Targeting of miRNA
Processed 22-nt mature miRNAs are transferred from the Dicer processing complex to the RISC. This step is called miRNA “loading.” RISC is assembled with many proteins including Ago, GW182, MOV10, and others (Peters and Meister, 2007; Kawamata and Tomari, 2010). The main component of the RISC is Ago, which is a member of a conserved protein family. In mammals, the Ago family consists of four (human) or five (mouse) members (Ago1, Ago2, Ago3, Ago4, Ago5) that are expressed in somatic cells and four germ line-specific members (PIWIL1, PIWIL2, PIWIL3, PIWIL4; Peters and Meister, 2007; Czech and Hannon, 2011). Among Ago proteins, the major player in somatic cells is Ago2, which is ubiquitously expressed and predominantly located in the cytoplasm (Peters and Meister, 2007).
In the miRNA transition from the Dicer processing complex to the Ago2-containing RISC, the above-mentioned TRBP is required for the recruitment of Ago2 to miRNA in the Dicer complex (Chendrimada et al., 2005). After loading, only one of the miRNA duplex strands with the lower thermodynamic stability profile is usually selected and captured by the RISC (Khvorova et al., 2003; Schwarz et al., 2003). The selected strand is called the “guide strand,” while the other strand, called the “passenger strand” (passenger*), is discarded from the RISC. Ago2 protein directly binds miRNAs through their PAZ domain (binding to the 3′ miRNA terminal) and MID domain (binding to the 5′ miRNA terminal; Song et al., 2003; Peters and Meister, 2007; Wang et al., 2008a,b; Czech and Hannon, 2011). Once loaded onto Ago2, the miRNAs act as guides to target mRNAs. Then, miRNAs utilize the guide strand seed region to access and engage in complementary hybridization with the 3′ UTR of the target mRNA.
The seed region at the 5′ end (nt 2–8) of miRNA is necessary for the association between miRNA and its target mRNA. Complementary hybridization by the seed sequence is important for target gene silencing, but a perfect match between miRNA and mRNA is not necessary for target gene downregulation (Pillai, 2005; Tomari and Zamore, 2005). The miRISC-bound target mRNAs are inhibited upon translation or degraded through mRNA decay, which is accompanied by decapping and deadenylation (Bhattacharyya et al., 2006; Giraldez et al., 2006; Mishima et al., 2006; Wu et al., 2006). The miRISC is enriched in cytoplasmic foci referred to as processing bodies (P-bodies), which contain Dcp1–Dcp2 decapping enzyme and Ccr4–Caf1 deadenylation complexes but lack translation initiation factors and ribosomes (Liu et al., 2005; Chen et al., 2009). The P-body is the site of translational inhibition and mRNA decay by miRISC.
One regulatory mechanism of RISC-loaded miRNA involves an RNA-binding protein to counteract miRNA-mediated repression by its specific binding to mRNA. Dead end 1 (Dnd1), which is required for germ cell survival and migration, directly binds to the U-rich region near the miR-221 complementary site of p27 mRNA, which encodes a cell cycle inhibitor, and inhibits accessibility of p27 mRNA to miR-221-loaded RISC. In another case, the RNA-binding protein HuR relieved miR-122 microRNA-mediated translational repression of CAT-1 mRNA in human hepatoma under stress conditions (Kedde et al., 2007). By contrast, a recent report demonstrated that HuR recruited let-7-loaded RISC onto c-Myc mRNA and that HuR and let-7 cooperatively inhibited c-Myc expression (Kim et al., 2009a). The current understanding of the regulation of miRNA loading and mRNA targeting may be incomplete because not all of the mechanisms involving RNA-binding proteins or other types of molecules have been discovered.
Modification of miRNA Precursors by Editing
Adenosine deaminases acting on RNA (ADARs) convert adenosines to inosines (A → I editing) in double-stranded RNA (dsRNA) substrates (Nishikura, 2010). The human and mouse miR-376 cluster are transcribed into a long primary transcript, and the seed sequences of some miR-370 members are edited at a highly edited site by ADAR1 and ADAR2 in cerebral cortex and medulla but not in other tissues (Blow et al., 2006; Kawahara et al., 2007b). ADAR2 also edits other sites. For instance, edited miR-376 represses a different set of targets including phosphoribosyl pyrophosphate synthetase 1 (PRPS1). ADAR1−/− is embryonic lethal in mice, but analyses using ADAR2−/− mice demonstrate no editing of miR-376 and no repression of PRPS1 mRNA expression in mouse cerebral cortex (Kawahara et al., 2007b). Additionally, mutation of PRPS1 leads to hereditary peripheral neuropathy (Kim et al., 2007). The regulation of miRNA precursor editing suppresses miRNA biogenesis at both the cropping and dicing steps.
Pri-miR-151 is highly edited in many tissues (Blow et al., 2006); the editing frequency of pre-miR-151 or pri-miR-151 within mature miRNA at the +3 site is 100 and 38%, respectively, in human amygdala. The editing of pre-miR-151 at the dicing step inhibited pre-miR-151 processing by the Dicer complex (Kawahara et al., 2007a). Analysis using pri-miR-142 demonstrated that pri-miR-142 cropping is inhibited by editing in a dose-dependent manner, and the edited pri-miR-142 is further targeted and degraded by Tudor-SN, which contains five staphylococcal/micrococcal nuclease domains and is an evolutionarily conserved component of RISC in many animal species (Caudy et al., 2003). miRNA-142 expression increases in spleen and thymus but not in liver in ADAR2−/− mice and ADAR1−/− conditional knockout mice (Yang et al., 2006). These reports indicate that miRNA editing, particularly by ADARs, plays an important role in tissue-specific gene expression via regulation of miRNA biogenesis.
Regulation by Let-7 Family miRNAs
TRIM32 contains four domains: RING, BBOX, a coiled-coil, and NHL (NCL1, HT2A, and Lin41) domains. The RING-domain drives ubiquitin ligase activity, and the NHL domain regulates miRNA loading by associating with Ago proteins. Mutations in the NHL domain of TRIM32 cause limb-girdle muscular dystrophy type 2H (LGMD2H; Frosk et al., 2002), and a missense mutation in the BBOX domain causes Bardet–Biedl syndrome type 11 (BBS11; Chiang et al., 2006). In the mouse telencephalon, TRIM32 is strongly expressed in differentiating neurons in the ventricular zone (VZ) at E12.5 and focally in cortical layers at E18.5. TRIM32 overexpression using in utero electroporation resulted in a 6.3-fold decrease of Nestin-positive neural progenitors cells (NPCs) and in a 12.8-fold increase of Tuj1-positive early differentiating neurons when compared with the control, and TRIM32-overexpressing cells migrated to the cortical plate at E15.5. By contrast, knockdown of TRIM32 by electroporation resulted in a 2.3-fold increase of Nestin-positive NPCs and in a 3.6-fold decrease of Tuj1-positive early differentiating neurons, and TRIM32-knockdown cells remained localized to the VZ and subventricular zone (SVZ) at E18.5. Additionally, doublet analysis of paired cells demonstrated higher TRIM32 expression in the daughter cell that underwent neuronal differentiation (Schwamborn et al., 2009). These results indicate that TRIM32 represses proliferation and induces mouse NS/PC differentiation.
The RING-domain of TRIM32 ubiquitinates c-Myc protein but not Ago1 protein, and a RING-domain mutation or coexpression of c-Myc and TRIM32 significantly decreased Tuj1-positive differentiating neurons in vivo (Schwamborn et al., 2009). Let-7 family miRNAs are enriched in NS/PCs (Wulczyn et al., 2007; Marson et al., 2008) and inhibit c-Myc protein expression (Melton et al., 2010). Let-7a is present in the TRIM32-Ago1 complex via its association with the NHL domain of TRIM32, and let-7a induces increase of Tuj1-positive neurons (Schwamborn et al., 2009). These reports suggest that TRIM32 regulates NSC differentiation through a pathway involving let-7 and c-Myc, and that differentiation is accompanied by TRIM32-mediated degradation (Figure 5). Interestingly, the Drosophila ortholog Mei-P26, also acts by inhibiting the many miRNAs pathway, such as dr-let-7, and control ovarian stem cell proliferation (Neumuller et al., 2008).
Pluripotency factors Oct4/Sox2/Nanog/Tcf3, which control the identity of ES cells and are involved in the induction of iPS cells, promote the expression of miRNAs in ES cells enriched in these miRNAs (ES cell-miRNAs). The connection between these factors and ES cell-miRNAs (mouse miR-290 to 295) maintains pluripotency by repressing the pathway of Lin28-let-7 family miRNAs (Marson et al., 2008). DGCR8-deficient ES cells continue ES cell self-renewal despite induction of differentiation by retinoic acid, with no reduction in pluripotency factor expression (Wang et al., 2007). Additionally, rescue screening with a combination of let-7 and ES cell-miRNA families indicated that they had opposing roles in regulating the self-renewal of ES cells. Pluripotency factor expression decreased with let-7 family treatment, but both let-7 and ES cell-miRNA treatment abolished let-7 family function. ChIP target overlap analysis comparing previously generated ChIP-on-chip data indicated that the let-7 family repressed downstream targets of the pluripotency factors Lin28 and Myc, whereas miR-294, known as an ES cell-miRNA, had no significant effects on either pluripotency factors or downstream targets (Melton et al., 2010). Let-7 and Lin28 are involved in a negative autoregulatory circuit during neurogenesis (Rybak et al., 2008). Thus, these reports suggest that repression of the let-7 family by Lin28 promotes reprogramming to induce pluripotency (Figure 5).
Regulation by miR-124
Polypyrimidine tract-binding protein (PTBP) is an RNA-binding protein with multiple functions, including transcriptional regulation, mRNA localization, mRNA stability, translational regulation, polyadenylation, and splicing regulation (Dreyfuss et al., 2002). Three isoforms of mammalian PTBP have been identified as PTBP1, PTBP2/nPTBP/brPTBP, and PTBP3/ROD1. Four splice variants (PTB1 to PTB4) and one pseudogene (ψPTBP1) are expressed in PTBP1, four splice variants (nPTB5 to nPTB8) are expressed in PTBP2, and three splice variants (ROD1-1 to ROD1-3) are expressed in PTBP3 (Romanelli et al., 2000). PTBP1 expression is downregulated during neurogenesis, while PTBP2 expression is upregulated during neurogenesis and involved in neural-specific splicing (Chou et al., 2000; Makeyev et al., 2007).
Microarray expression analysis identified PTBP2 as an enrichment target in miR-124-mediated neural differentiation of CAD cells. Although miR-124 led to neuronal differentiation of mouse neuroblastoma cell lines CAD and Neuro2a (Makeyev et al., 2007), it did not affect neuronal fate in chick neural tube (Cao et al., 2007). In various mouse neuroblastoma cell lines and primary cortical cultured cells, PTBP1 strongly represses the production of neural-specific splice variants of its target pre-mRNAs including Mtap4, Rufy3, and Nogo, but induces the production of their global splice variants. Likewise, PTBP2 also moderately repressed the production of neural-specific splice variants. Moreover, PTBP1-mediated exon skipping induced nonsense-mediated decay of PTBP2.
Chick and mouse miRNA-124 is expressed in the neural tube mantle zone (Cao et al., 2007; Makeyev et al., 2007). miR-124 and neural-specific splice variants of PTBP-targeted mRNA colocalized to the neural tube, while miR-124 and PTBP1 mRNA were expressed almost mutually exclusively. Additionally, PTBP1 expression is upregulated and neural-specific splice variants of its targets are reduced in Dicer−/− mice that do not produce all miRNAs (Makeyev et al., 2007). These results suggest that miR-124 promotes neuronal differentiation by suppression of PTBP1, which represses neural-specific alternative splicing of its targets and induces PTBP1-mediated nonsense-mediated decay of PTBP2, and then PTBP2 can fine-tune neurogenesis (Figure 5). However, discrepancies in miR-124-mediated neurogenesis between mouse and chick still remain.
Rncr3 is an approximately 4-kb ncRNA that contains pre-miR-124a in its exon (Blackshaw et al., 2004) and is expressed in retina and CNS, including cerebrum and cerebellum. Although Rncr3−/− mice maintained 20–60% of miR-124a expression in some tissues, considerable loss of miR-124a expression was observed in the dentate gyrus (DG). Cell death and normal cell proliferation in the DG and cortex led to a small brain size and dyskinesia of the limb-clasping response. Expression of Lhx2, a target of miR-124a, was slightly upregulated (28%) in Rncr3−/− cultured hippocampal cells by transfection with luciferase construct with Lhx2 of 3′ UTR, but endogenous LHX2 expression was significantly increased in the Rncr3−/− DG. Although the brain size, clasping, and apoptosis in the cortex were not recovered in Rncr3−/− mice when miR-124a-2 was transgenically expressed, the mossy fiber elongation in the Rncr3−/− DG was rescued by knockdown of Lhx2 (Sanuki et al., 2011). These results suggest that miR-124a-mediated LHX2 regulation is involved in DG maturation and survival.
Not only miR-124a but miR-132 is also participated in neuronal morphogenesis in developing neurons of DG including SGZ (subgranular zone) where adult neurogenesis occurs. The CREB-signaling regulates the maturation and survival of newborn neurons in DG and miR-132 transcription is regulated by CREB at the consensus CRE sequences (Vo et al., 2005). Since miR-132 and miR-212 locus are proximal, floxed miR-132/212 conditional knockout mice was generated. In the mice, the number of newborn hippocampal neurons had dramatically decreased dendritic arbors and spine density (Magill et al., 2010). Additionally, miR-132 was the predominantly active product in hippocampal neurons (Vo et al., 2005; Magill et al., 2010). By using a bidirectional ratiometric sensors capable of distinguishing miR-132, Magill et al. elucidated that miR-132 but not miR-132*, -212, or -212* was active in Doublecortin-positive immature hippocampal granule neurons. The result was coincident with other papers (Edbauer et al., 2010; Luikart et al., 2011). Thus, miR-132 plays an important role in DG including SGZ.
Microglia, tissue-specific resident macrophages in the CNS, pass the blood–brain barrier to serve as immunocompetent cells in the CNS, where they are maintained in a quiescent state. Upon nerve injury, microglia are activated by CCAAT/enhancer binding protein-α (C/EBP-α), a master transcription factor involved in myeloid cell differentiation. Microglia then migrate to the lesioned area and are involved in cell clearance via cytophagocytosis. miR-124 is expressed in inactivated microglia but not in activated microglia or bone marrow-derived macrophages (BMDMs). In autoimmune encephalomyelitis, miR-124 overexpression inhibits microglial activation and macrophage stimulation with accompanying suppression of C/EBP and leads to amelioration of spinal cord inflammation. By contrast, miR-124 knockdown causes activation of microglia and macrophages in co-cultures of BMDMs with either an astroglial or neuronal cell line. Furthermore, miR-124 was found to inhibit expression of C/EBP-α (Ponomarev et al., 2011) in the NIE115 mouse neuroblastoma cell line transfected with a construct containing the full-length C/EBP-α 3′ UTR sequence downstream of firefly luciferase. These results suggest that miR-124 is a key regulator of microglial cell quiescence in the CNS that acts via direct repression of C/EBP-α mRNA at the 3′ UTR.
Strong expression of miR-124 has been observed in early born neurons and CNS during embryonic neurogenesis stage (Makeyev et al., 2007; Rybak et al., 2008), and miR-124 expression was also observed in the SVZ during the conversion of GFAP-positive astrocyte stem cells into neuroblasts and newly born neurons in adult neurogenesis. Gene ontology analysis of computationally predicted targets of miR-124 detected Sox9 as a novel molecule of unknown function, and miR-124-mediated direct repression of Sox9 was verified by luciferase assays in HEK293T cells. Sox9 expression was observed in approximately 30% of transit amplifying cells but not in neuroblasts. Sox9 knockdown by retroviral infection led to a moderate increase of Tuj1-positive neurons, and miR-124 overexpression led to increased neurogenesis. Also, neurogenesis induced by miR-124 overexpression was silenced in Sox9-overexpressing cells (Cheng et al., 2009). These results suggest that miR-124 regulates adult neurogenesis in the SVZ stem cell population as well as during embryogenesis (Figure 5).
Regulation by miR-9
The neural-specific miRNA miR-9 is evolutionarily conserved in NSC and neurogenesis function. Drosophila miR-9 is expressed in epithelial cells in the peripheral nervous system, including in cells adjacent to sensory organ precursor (SOP) cells, and is involved in neuron production from SOPs via repression of the senseless 3′ UTR, a proneural gene and transcriptional activator (Li et al., 2006). Zebrafish miR-9 expressed in regions adjacent to the midbrain–hindbrain boundary (MHB) inhibits FGF signals including FGF8 and FGFR1. Additionally, z-miR-9 regulates neuron production via the repression of the mammalian Hes ortholog, bHLH hairy/E(spl) transcription factors Her5 and 9 (Leucht et al., 2008). These two independent pathways play a role in the delimitation and maintenance of MHB. Xenopus miR-9 also regulates neurogenesis at the forebrain–hindbrain boundary by controlling the expression of hairy1 mRNA. Like zebrafish, x-miR-9 is expressed in regions adjacent to the MHB and in the neural tube including forebrain, midbrain, and hindbrain along the anterior–posterior (A–P) axis, but not in spinal cord. Z-miR-9 knockdown using morpholino oligonucleotide injection did not affect expression levels of progenitor markers Sox3 and pH3 (phospho-Histone H3) in the forebrain, while z-miR-9 knockdown led to an increase in the levels of these makers in the hindbrain and TUNEL-positive cells in the forebrain. Expression of hairy1 and miR-9 is mutually exclusive in the neural tube, and z-miR-9 knockdown induced hairy1 expression. Thus, z-miR-9 is involved in neural progenitor survival in the forebrain and in the transition of progenitors to neurons across the A–P axis in the hindbrain via repression of hairy1 (Bonev et al., 2011).
Some mammalian miR-9s (miR-9-1 to miR-9-3) are separately transcribed from different promoters. Drosophila contains miR-9a to miR-9c, and zebrafish contains miR-9-1 to miR-9-7. Various miR-9 targets are known, including TLX/Nre1, Foxg1, REST/NRSF (repression element silencing transcriptional factor/neural restrictive silencer factor), CoREST, Meis2, Gsh2, Islet1, and Id4 (Shibata et al., 2008, 2011). TLX/Nre1 represses the transcription of genes including p21, Pten, and Gfap via recruitment of HDAC (histone deacetylase) to each of the promoters of these genes. TLX also represses pri-miR-9-1 expression, and the TLX–HDAC complex promotes proliferation of NSCs (Qu and Shi, 2009). Therefore, TLX forms a negative regulatory loop with miR-9. Additionally, TLX is expressed in type B cells in the SVZ and stimulates proliferation and self-renewal of adult NSCs (Qu et al., 2010). TLX is also involved in glioma stem cell genesis (Liu et al., 2010; Park et al., 2010).
Staining using various neuronal makers in miR-9-2 and miR-9-3 double-mutant mice revealed almost normal generation of cerebral layer VI, with decreased layers I to IV and subplate, poorly formed axons of commissural neurons, and thin corticofugal axons (CFAs) in the VZ and SVZ of ganglionic eminences. Although miR-9-1 overexpression by electroporation led to expression of Reelin, p21, p73, and βIII-tubulin, targets of Foxg1 (Shibata et al., 2008), double-mutant mice did not demonstrate increased Foxg1 during neurogenesis at E16.5. Additionally, a luciferase assay of P19 cells indicated that neural RNA-binding proteins including Musashi1 and Hu proteins competed with miR-9 for interaction with the 3′ UTR of target mRNA of miR-9 (Figure 1). Hu proteins that bind the AU-rich element of the 3′ UTR of this target mRNAs are positively involved in neurogenesis via mRNA stability and translational activation. However, detailed characterization of the spatio-temporal expression patterns of all Hu proteins (HuA/HuR, HuB, Huc, HuD) is lacking, and the relationship between Hu binding proteins and miR-9 is not clear. For instance, there is a discrepancy in HuB expression between a previous report and this analysis: the Darnell group showed that HuB expression is gradually decreased from E10 to E16 (Okano and Darnell, 1997; Shibata et al., 2011). How does HuB function as a neural repressor protein? Additionally, hnRNP K, a HuB-binding protein, participates in stemness, but hnRNP K-mediated function is suspended by HuB (Figure 3; Yano et al., 2005). The relationship between miR-9 and hnRNP K remains to be determined. Moreover, miR-9 does not control the expression of Foxg1 in Xenopus (Bonev et al., 2011).
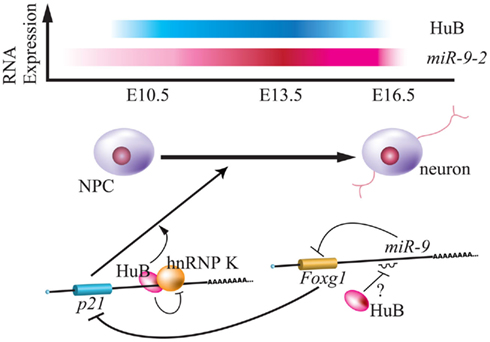
Figure 3. Relationship between Hu protein and miR-9-2 during neurogenesis. Top panel indicates the relative expression of mouse HuB and miR-9-2 throughout the embryonic stage (Okano and Darnell, 1997; Shibata et al., 2011). Bottom panel indicates the model of translational regulation by HuB-hnRNP K and miR-9 via p21 and Foxg1 mRNA during neurogenesis (Yano et al., 2005; Shibata et al., 2011). HuB induces the expression of p21CIP1, is a positive regulator of neuronal differentiation, by inhibiting function of hnRNP K in the 3′ UTR. HuB can also attenuate the miR-9 suppression of Foxg1, blocks expression of p21CIP1 (Seoane et al., 2004).
As mentioned above, the studies in several model animals showed miR-9 possesses the neuronal differentiation and migration promoting activity (Li et al., 2006; Leucht et al., 2008; Shibata et al., 2008; Qu and Shi, 2009). Human miR-9 also functions proliferation and maturation of early NPCs, and moreover regulates migration of NPCs in vitro and in vivo (Delaloy et al., 2010). Stathmin, whose expression was inversely correlated with h-miR-9 in hNPCs culture (between rosette formation and neuronal formation stage), was identified as one of the direct target of h-miR-9 (Delaloy et al., 2010). Loss of h-miR-9 function promoted hNPCs migration and proliferation in a two-dimensional migration assay, and moreover both h-miR-9 and stathmin knockdown rescued the effect of loss of h-miR-9 activity. Thus, stathmin is an essential target of h-miR-9 in modulating hNPCs migration and proliferation (Delaloy et al., 2010).
Combination of miR-9* and miR-124
The Crabtree group reported that miR-124 and miR-9 co-act to regulate neurogenesis via chromatin-remodeling of REST and CoREST. The ATP-dependent chromatin-remodeling enzyme, Brahma-associated factor (BAF) complex, represses transcription by associating with the nucleosome. Stage-specific BAF complex is formed by switching of some stage-specific subunits: at the ES cell stage, esBAF; at the neural progenitor stage, npBAF; and at the post-mitotic neuronal stage, nBAF (Figure 4; Ho and Crabtree, 2010). For instance, although the BAF53a subunit is expressed in NPCs residing at the neuroepithelial layer at E11.5, the BAF53b subunit, miR-124, and miR-9* are colocalized to the spinal cord mantle layer. A luciferase assay of CHO cells indicated that miR-124 and miR-9* repressed BAF53a translation via direct binding of its 3′ UTR. Transgenic embryos containing a BAF53a 3′-UTR mutant with miRNA-binding defect exhibit significantly decreased endogenous BAF53b expression. Moreover, REST overexpression induces endogenous BAF53a in the neuroepithelial layer, while co-expression of REST, miR-124, and miR-9* has no significant effect on BAF53a expression. Additionally, overexpression of BAF53a and its 3′ UTR induced an extension of dendritic outgrowth in transfected neurons upon KCl stimulation, while by contrast, the overexpression of BAF53a and its mutated 3′ UTR had no effect (Yoo et al., 2009). These results suggest that REST represses miR-124 and miR-9*. Thus, BAF53a can maintain NS/PCs via inhibition of BAF53b expression, while miR-124 and miR-9* represses BAF53a during neurogenesis, allowing BAF53b to induce dendritic outgrowth (Figure 4).
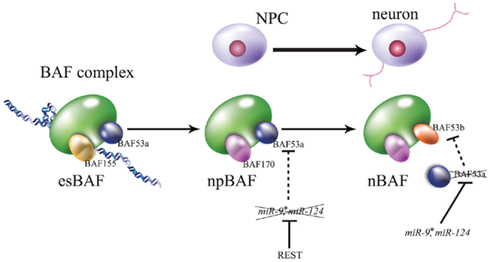
Figure 4. Brahma-associated factor (BAF) complexes during embryogenesis. Convertible subunits are focused on each stage. The esBAF complex contains BAF155 and BAF53a in embryonic stem cells, npBAF contains BAF170 (substituent of BAF155) and BAF53a in neural progenitor cells (NPCs), and nBAF contains BAF170 and BAF53b (substituent of BAF55a) in neurons. REST/NRSF represses the expression of miR-124 and miR-9*, these neural miRNAs repress BAF53a via its translation, and BAF54a represses the transcription of BAF53b. For greater detail please see this text and the review of Ho and Crabtree (2010).
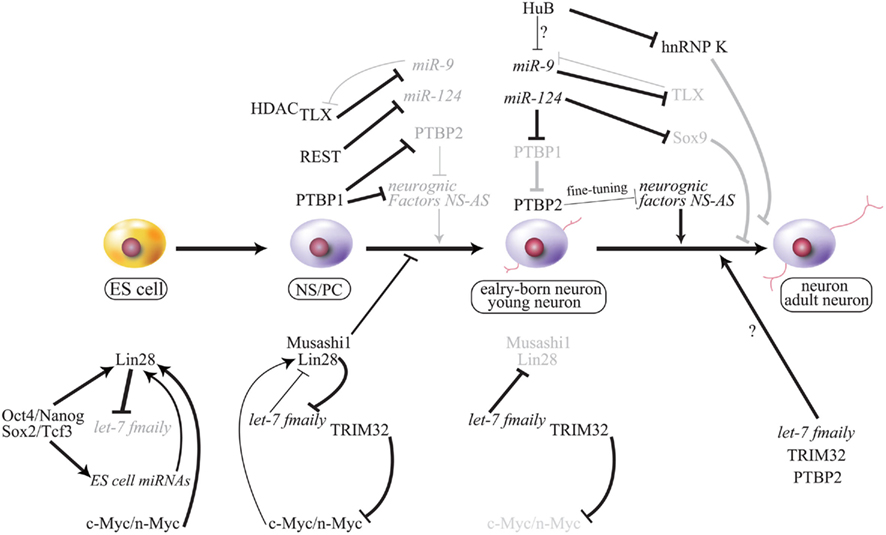
Figure 5. Working model of cell fate decision by miRNA and the gene regulatory network. Important phenomena during neurogenesis are summarized, and regulation of each is described in this text. NS-AS indicates that neuron-specific alternative splicing is repressed by PTBP1 (strongly) or PTBP2 (moderately) in some neurogenic factors that are involved in neurogenesis. PTBP2 may regulate neurogenesis via spatial–temporal fine-tuning during neurogenesis. Gray font and line indicate attenuation. Bold lines indicate strong activation or repression, and thin lines indicate mild or weak regulation. ES, embryonic stem cells; NS/PC, neural stem/progenitor cell.
Although overexpression of miR-124 and miR-9/9* by lentivirus infection converted low levels of human neonatal foreskin fibroblasts to MAP2-positive neurons, the co-overexpression of miR-124, miR-9/9*, and NeuroD2 increased the conversion frequency to 50%. These miR-124 and miR-9/9*-induced neurons expressed voltage-gated sodium channels and potassium channels (Yoo et al., 2011). DAM neurogenic transcription factors (ASCL1 and MYT1L) functioned significantly in transdifferentiation of mouse embryonic fibroblasts (Vierbuchen et al., 2010), and co-expression systems including miR-9/9*, miR-124, NeuroD2, Ascl1, and DAM exhibited 80% conversion frequency. This conversion method can also be adapted to adult fibroblasts, but converted adult neurons are expressed in almost all cortical layers and synthesize voltage-gated channels markers. Importantly, conversion methods were ineffective without miR-124 and miR-9/9* (Yoo et al., 2011). Thus, not only are these neural-specific miRNAs involved in neural fate determination, but various combinations of these miRNAs and different sets of transcription factors may induce various types of neurons.
Perspectives
Expression of miRNA is strictly controlled in a developmental and/or tissue-specific manner, and disruption of miRNA expression can induce many human diseases including cancer. Indeed, knockout of major components of miRNA biogenesis such Dicer, Drosha, DGCR8, and regulator Lin28 can cause tumorigenesis (Kumar et al., 2007; Viswanathan et al., 2009). TDP-43, one component of the Drosha complex (Gregory et al., 2004), binds the let-7b sequence itself or to the stem-loop of miR-663, which serves to recruit/keep the pri-miRNAs in the cropping step (Buratti et al., 2010). TDP-43 is found in ubiquitin-positive inclusions in frontotemporal lobar degeneration (FTLD) and amyotrophic lateral sclerosis (ALS). Mutations in angiogenin are also related to ALS (Greenway et al., 2006). Angiogenin is a secreted ribonuclease that induces production of stress-induced small RNAs (tiRNAs) from cleaved tRNA, serves not to translational arrest under some stress conditions (Yamasaki et al., 2009; McLaughlin et al., 2010). Angiogenin and tiRNA have been suggested to contribute to the survival of motor neurons.
The fragile X mental retardation protein (FMRP), mutation of which can cause, fragile x syndrome, is a component of the RISC that interacts with Ago1 (Jin et al., 2004). Drosophila FMRP1 is involved in maintenance of the steady-state neural-specific dr-miR-124a expression in the nervous system (Xu et al., 2008). Mutation of PACT, one of the core proteins of the Dicer complex also known as DYT16, causes young-onset dystonia–parkinsonism disorder (Camargos et al., 2008). As mentioned above, mutation of TRIM32 causes nervous system disorders including BBS11. Mutation of PRPS1, which is regulated by ADARs, leads to hereditary peripheral neuropathy. These reports suggest that miRNA biogenesis is closely related to the pathogenesis of nervous system diseases. Thus, analysis of miRNA biogenesis may provide potential cures for these diseases.
Interestingly, miR-9-2 expression is relatively higher than that of miR-9-1 and miR-9-3 at all differentiated neurogenic stages (Shibata et al., 2011); by contrast, pri-miR-9-2 expression is incomplete, but other pri-miR-9 are expressed in undifferentiated and differentiated neurons (Ko et al., 2008). The expression difference between pri-miRNA and mature miRNA has not yet been addressed. Additionally, there are not enough reports focusing on the relationship between neurogenesis and miRNA biogenesis. Addressing these relationships could be important for a full understanding of the regulation of NSCs and neurogenesis.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Mary C. Mullins and Drs. Yohei Okada and Masato Yano for comments on the manuscript. This work was supported by MEXT (Ministry of Education, Culture, Sports, Science, and Technology of Japan) grants to Hideyuki Okano and Takao Imai. Hironori Kawahara is supported by the Uehara Memorial and Mitsukoshi Health and Welfare Foundations.
References
Andersson, T., Rahman, S., Sansom, S. N., Alsio, J. M., Kaneda, M., Smith, J., O’Carroll, D., Tarakhovsky, A., and Livesey, F. J. (2010). Reversible block of mouse neural stem cell differentiation in the absence of dicer and microRNAs. PLoS ONE 5, e13453. doi:10.1371/journal.pone.0013453
Balzer, E., and Moss, E. G. (2007). Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biol. 4, 16–25.
Bernstein, E., Caudy, A. A., Hammond, S. M., and Hannon, G. J. (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366.
Bhattacharyya, S. N., Habermacher, R., Martine, U., Closs, E. I., and Filipowicz, W. (2006). Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125, 1111–1124.
Blackshaw, S., Harpavat, S., Trimarchi, J., Cai, L., Huang, H., Kuo, W. P., Weber, G., Lee, K., Fraioli, R. E., Cho, S. H., Yung, R., Asch, E., Ohno-Machado, L., Wong, W. H., and Cepko, C. L. (2004). Genomic analysis of mouse retinal development. PLoS Biol. 2, e247. doi:10.1371/journal.pbio.0020247
Blow, M. J., Grocock, R. J., van Dongen, S., Enright, A. J., Dicks, E., Futreal, P. A., Wooster, R., and Stratton, M. R. (2006). RNA editing of human microRNAs. Genome Biol. 7, R27.
Bohnsack, M. T. (2004). Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10, 185–191.
Bonev, B., Pisco, A., and Papalopulu, N. (2011). MicroRNA-9 reveals regional diversity of neural progenitors along the anterior-posterior axis. Dev. Cell 20, 19–32.
Borchert, G. M., Lanier, W., and Davidson, B. L. (2006). RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 13, 1097–1101.
Bortolin-Cavaille, M. L., Dance, M., Weber, M., and Cavaille, J. (2009). C19MC microRNAs are processed from introns of large Pol-II, non-protein-coding transcripts. Nucleic Acids Res. 37, 3464–3473.
Buratti, E., De Conti, L., Stuani, C., Romano, M., Baralle, M., and Baralle, F. (2010). Nuclear factor TDP-43 can affect selected microRNA levels. FEBS J. 277, 2268–2281.
Bussing, I., Slack, F. J., and Grosshans, H. (2008). let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 14, 400–409.
Camargos, S., Scholz, S., Simon-Sanchez, J., Paisan-Ruiz, C., Lewis, P., Hernandez, D., Ding, J., Gibbs, J. R., Cookson, M. R., Bras, J., Guerreiro, R., Oliveira, C. R., Lees, A., Hardy, J., Cardoso, F., and Singleton, A. B. (2008). DYT16, a novel young-onset dystonia-parkinsonism disorder: identification of a segregating mutation in the stress-response protein PRKRA. Lancet Neurol. 7, 207–215.
Cao, X., Pfaff, S. L., and Gage, F. H. (2007). A functional study of miR-124 in the developing neural tube. Genes Dev. 21, 531–536.
Caudy, A. A., Ketting, R. F., Hammond, S. M., Denli, A. M., Bathoorn, A. M., Tops, B. B., Silva, J. M., Myers, M. M., Hannon, G. J., and Plasterk, R. H. (2003). A micrococcal nuclease homologue in RNAi effector complexes. Nature 425, 411–414.
Cheloufi, S., Dos Santos, C. O., Chong, M. M., and Hannon, G. J. (2010). A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465, 584–589.
Chen, C. Y., Zheng, D., Xia, Z., and Shyu, A. B. (2009). Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat. Struct. Mol. Biol. 16, 1160–1166.
Chendrimada, T. P., Gregory, R. I., Kumaraswamy, E., Norman, J., Cooch, N., Nishikura, K., and Shiekhattar, R. (2005). TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436, 740–744.
Cheng, L. C., Pastrana, E., Tavazoie, M., and Doetsch, F. (2009). miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 12, 399–408.
Chiang, A. P., Beck, J. S., Yen, H. J., Tayeh, M. K., Scheetz, T. E., Swiderski, R. E., Nishimura, D. Y., Braun, T. A., Kim, K. Y., Huang, J., Elbedour, K., Carmi, R., Slusarski, D. C., Casavant, T. L., Stone, E. M., and Sheffield, V. C. (2006). Homozygosity mapping with SNP arrays identifies TRIM32, an E3 ubiquitin ligase, as a Bardet-Biedl syndrome gene (BBS11). Proc. Natl. Acad. Sci. U.S.A. 103, 6287–6292.
Chou, M. Y., Underwood, J. G., Nikolic, J., Luu, M. H., and Black, D. L. (2000). Multisite RNA binding and release of polypyrimidine tract binding protein during the regulation of c-src neural-specific splicing. Mol. Cell 5, 949–957.
Cifuentes, D., Xue, H., Taylor, D. W., Patnode, H., Mishima, Y., Cheloufi, S., Ma, E., Mane, S., Hannon, G. J., Lawson, N. D., Wolfe, S. A., and Giraldez, A. J. (2010). A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 328, 1694–1698.
Czech, B., and Hannon, G. J. (2011). Small RNA sorting: matchmaking for Argonautes. Nat. Rev. Genet. 12, 19–31.
Davis, B. N., and Hata, A. (2010). MicroRNA in cancer – the involvement of aberrant microRNA biogenesis regulatory pathways. Genes Cancer 1, 1100–1114.
De Pietri Tonelli, D., Pulvers, J. N., Haffner, C., Murchison, E. P., Hannon, G. J., and Huttner, W. B. (2008). miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development 135, 3911–3921.
Delaloy, C., Liu, L., Lee, J. A., Su, H., Shen, F., Yang, G. Y., Young, W. L., Ivey, K. N., and Gao, F. B. (2010). MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell 6, 323–335.
Dreyfuss, G., Kim, V. N., and Kataoka, N. (2002). Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3, 195–205.
Edbauer, D., Neilson, J. R., Foster, K. A., Wang, C. F., Seeburg, D. P., Batterton, M. N., Tada, T., Dolan, B. M., Sharp, P. A., and Sheng, M. (2010). Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 65, 373–384.
Enciu, A. M., Popescu, B. O., and Gheorghisan-Galateanu, A. (2011). MicroRNAs in brain development and degeneration. Mol. Biol. Rep. 39, 2243–2252.
Frosk, P., Weiler, T., Nylen, E., Sudha, T., Greenberg, C. R., Morgan, K., Fujiwara, T. M., and Wrogemann, K. (2002). Limb-girdle muscular dystrophy type 2H associated with mutation in TRIM32, a putative E3-ubiquitin-ligase gene. Am. J. Hum. Genet. 70, 663–672.
Fukuda, T., Yamagata, K., Fujiyama, S., Matsumoto, T., Koshida, I., Yoshimura, K., Mihara, M., Naitou, M., Endoh, H., Nakamura, T., Akimoto, C., Yamamoto, Y., Katagiri, T., Foulds, C., Takezawa, S., Kitagawa, H., Takeyama, K., O’Malley, B. W., and Kato, S. (2007). DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat. Cell Biol. 9, 604–611.
Giraldez, A. J., Mishima, Y., Rihel, J., Grocock, R. J., Van Dongen, S., Inoue, K., Enright, A. J., and Schier, A. F. (2006). Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312, 75–79.
Greenway, M. J., Andersen, P. M., Russ, C., Ennis, S., Cashman, S., Donaghy, C., Patterson, V., Swingler, R., Kieran, D., Prehn, J., Morrison, K. E., Green, A., Acharya, K. R., Brown, R. H. Jr., and Hardiman, O. (2006). ANG mutations segregate with familial and “sporadic” amyotrophic lateral sclerosis. Nat. Genet. 38, 411–413.
Gregory, R. I., Yan, K. P., Amuthan, G., Chendrimada, T., Doratotaj, B., Cooch, N., and Shiekhattar, R. (2004). The microprocessor complex mediates the genesis of microRNAs. Nature 432, 235–240.
Haase, A. D., Jaskiewicz, L., Zhang, H., Laine, S., Sack, R., Gatignol, A., and Filipowicz, W. (2005). TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 6, 961–967.
Hagan, J. P., Piskounova, E., and Gregory, R. I. (2009). Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 16, 1021–1025.
Han, J., Lee, Y., Yeom, K. H., Kim, Y. K., Jin, H., and Kim, V. N. (2004). The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 18, 3016–3027.
Han, J., Lee, Y., Yeom, K. H., Nam, J. W., Heo, I., Rhee, J. K., Sohn, S. Y., Cho, Y., Zhang, B. T., and Kim, V. N. (2006). Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125, 887–901.
Heo, I., Joo, C., Cho, J., Ha, M., Han, J., and Kim, V. N. (2008). Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol. Cell 32, 276–284.
Imai, T., Tokunaga, A., Yoshida, T., Hashimoto, M., Mikoshiba, K., Weinmaster, G., Nakafuku, M., and Okano, H. (2001). The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol. Cell. Biol. 21, 3888–3900.
Jin, P., Zarnescu, D. C., Ceman, S., Nakamoto, M., Mowrey, J., Jongens, T. A., Nelson, D. L., Moses, K., and Warren, S. T. (2004). Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat. Neurosci. 7, 113–117.
Kawahara, H., Imai, T., Imataka, H., Tsujimoto, M., Matsumoto, K., and Okano, H. (2008). Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP. J. Cell Biol. 181, 639–653.
Kawahara, H., Okada, Y., Imai, T., Iwanami, A., Mischel, P. S., and Okano, H. (2011). Musashi1 cooperates in abnormal cell lineage protein 28 (Lin28)-mediated Let-7 family microRNA biogenesis in early neural differentiation. J. Biol. Chem. 286, 16121–16130.
Kawahara, Y., Zinshteyn, B., Chendrimada, T. P., Shiekhattar, R., and Nishikura, K. (2007a). RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 8, 763–769.
Kawahara, Y., Zinshteyn, B., Sethupathy, P., Iizasa, H., Hatzigeorgiou, A. G., and Nishikura, K. (2007b). Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science 315, 1137–1140.
Kawase-Koga, Y., Otaegi, G., and Sun, T. (2009). Different timings of Dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Dev. Dyn. 238, 2800–2812.
Kedde, M., Strasser, M. J., Boldajipour, B., Oude Vrielink, J. A., Slanchev, K., le Sage, C., Nagel, R., Voorhoeve, P. M., van Duijse, J., Orom, U. A., Lund, A. H., Perrakis, A., Raz, E., and Agami, R. (2007). RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 131, 1273–1286.
Ketting, R. F., Fischer, S. E., Bernstein, E., Sijen, T., Hannon, G. J., and Plasterk, R. H. (2001). Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15, 2654–2659.
Khvorova, A., Reynolds, A., and Jayasena, S. D. (2003). Functional siRNAs and miRNAs exhibit strand bias. Cell 115, 209–216.
Kim, H. H., Kuwano, Y., Srikantan, S., Lee, E. K., Martindale, J. L., and Gorospe, M. (2009a). HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 23, 1743–1748.
Kim, V. N., Han, J., and Siomi, M. C. (2009b). Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10, 126–139.
Kim, H. J., Sohn, K. M., Shy, M. E., Krajewski, K. M., Hwang, M., Park, J. H., Jang, S. Y., Won, H. H., Choi, B. O., Hong, S. H., Kim, B. J., Suh, Y. L., Ki, C. S., Lee, S. Y., Kim, S. H., and Kim, J. W. (2007). Mutations in PRPS1, which encodes the phosphoribosyl pyrophosphate synthetase enzyme critical for nucleotide biosynthesis, cause hereditary peripheral neuropathy with hearing loss and optic neuropathy (cmtx5). Am. J. Hum. Genet. 81, 552–558.
Ko, M. H., Kim, S., Hwang do, W., Ko, H. Y., Kim, Y. H., and Lee, D. S. (2008). Bioimaging of the unbalanced expression of microRNA9 and microRNA9* during the neuronal differentiation of P19 cells. FEBS J. 275, 2605–2616.
Kumar, M. S., Lu, J., Mercer, K. L., Golub, T. R., and Jacks, T. (2007). Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 39, 673–677.
Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., Yim, J., Lee, J., Provost, P., Rådmark, O., Kim, S., and Kim, V. N. (2003). The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419.
Lee, Y., Hur, I., Park, S. Y., Kim, Y. K., Suh, M. R., and Kim, V. N. (2006). The role of PACT in the RNA silencing pathway. EMBO J. 25, 522–532.
Lee, Y., Kim, M., Han, J., Yeom, K. H., Lee, S., Baek, S. H., and Kim, V. N. (2004). MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23, 4051–4060.
Leucht, C., Stigloher, C., Wizenmann, A., Klafke, R., Folchert, A., and Bally-Cuif, L. (2008). MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat. Neurosci. 11, 641–648.
Li, M., Lee, K. F., Lu, Y., Clarke, I., Shih, D., Eberhart, C., Collins, V. P., Van Meter, T., Picard, D., Zhou, L., Boutros, P. C., Modena, P., Liang, M. L., Scherer, S. W., Bouffet, E., Rutka, J. T., Pomeroy, S. L., Lau, C. C., Taylor, M. D., Gajjar, A., Dirks, P. B., Hawkins, C. E., and Huang, A. (2009). Frequent amplification of a chr19q13.41 microRNA polycistron in aggressive primitive neuroectodermal brain tumors. Cancer Cell 16, 533–546.
Li, Y., Wang, F., Lee, J. A., and Gao, F. B. (2006). MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 20, 2793–2805.
Liu, H. K., Wang, Y., Belz, T., Bock, D., Takacs, A., Radlwimmer, B., Barbus, S., Reifenberger, G., Lichter, P., and Schutz, G. (2010). The nuclear receptor tailless induces long-term neural stem cell expansion and brain tumor initiation. Genes Dev. 24, 683–695.
Liu, J., Rivas, F. V., Wohlschlegel, J., Yates, J. R. III, Parker, R., and Hannon, G. J. (2005). A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 7, 1261–1266.
Luikart, B. W., Bensen, A. L., Washburn, E. K., Perederiy, J. V., Su, K. G., Li, Y., Kernie, S. G., Parada, L. F., and Westbrook, G. L. (2011). miR-132 mediates the integration of newborn neurons into the adult dentate gyrus. PLoS ONE 6, e19077. doi:10.1371/journal.pone.0019077
Lund, E., Guttinger, S., Calado, A., Dahlberg, J. E., and Kutay, U. (2004). Nuclear export of microRNA precursors. Science 303, 95–98.
Magill, S. T., Cambronne, X. A., Luikart, B. W., Lioy, D. T., Leighton, B. H., Westbrook, G. L., Mandel, G., and Goodman, R. H. (2010). MicroRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc. Natl. Acad. Sci. U.S.A. 107, 20382–20387.
Makeyev, E. V., Zhang, J., Carrasco, M. A., and Maniatis, T. (2007). The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell 27, 435–448.
Marson, A., Levine, S. S., Cole, M. F., Frampton, G. M., Brambrink, T., Johnstone, S., Guenther, M. G., Johnston, W. K., Wernig, M., Newman, J., Calabrese, J. M., Dennis, L. M., Volkert, T. L., Gupta, S., Love, J., Hannett, N., Sharp, P. A., Bartel, D. P., Jaenisch, R., and Young, R. A. (2008). Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134, 521–533.
McLaughlin, R. L., Phukan, J., McCormack, W., Lynch, D. S., Greenway, M., Cronin, S., Saunders, J., Slowik, A., Tomik, B., Andersen, P. M., Bradley, D. G., Jakeman, P., and Hardiman, O. (2010). Angiogenin levels and ANG genotypes: dysregulation in amyotrophic lateral sclerosis. PLoS ONE 5, e15402. doi:10.1371/journal.pone.0015402
Melton, C., Judson, R. L., and Blelloch, R. (2010). Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 463, 621–626.
Michlewski, G., and Caceres, J. F. (2010). Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat. Struct. Mol. Biol. 17, 1011–1018.
Mishima, Y., Giraldez, A. J., Takeda, Y., Fujiwara, T., Sakamoto, H., Schier, A. F., and Inoue, K. (2006). Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr. Biol. 16, 2135–2142.
Moss, E. G., Lee, R. C., and Ambros, V. (1997). The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88, 637–646.
Neumuller, R. A., Betschinger, J., Fischer, A., Bushati, N., Poernbacher, I., Mechtler, K., Cohen, S. M., and Knoblich, J. A. (2008). Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature 454, 241–245.
Newman, M. A., and Hammond, S. M. (2010). Emerging paradigms of regulated microRNA processing. Genes Dev. 24, 1086–1092.
Newman, M. A., Thomson, J. M., and Hammond, S. M. (2008). Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14, 1539–1549.
Nishikura, K. (2010). Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 79, 321–349.
Nishino, J., Kim, I., Chada, K., and Morrizson, S. J. (2008). Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf expression. Cell 135, 227–239.
Obernosterer, G., Leuschner, P. J., Alenius, M., and Martinez, J. (2006). Post-transcriptional regulation of microRNA expression. RNA 12, 1161–1167.
Okamura, K., Hagen, J. W., Duan, H., Tyler, D. M., and Lai, E. C. (2007). The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130, 89–100.
Okano, H., Imai, T., and Okabe, M. (2002). Musashi: a translational regulator of cell fate. J. Cell. Sci. 115, 1355–1359.
Okano, H., Kawahara, H., Toriya, M., Nakao, K., Shibata, S., and Imai, T. (2005). Function of RNA-binding protein Musashi-1 in stem cells. Exp. Cell Res. 306, 349–356.
Okano, H. J., and Darnell, R. B. (1997). A hierarchy of Hu RNA binding proteins in developing and adult neurons. J. Neurosci. 17, 3024–3037.
Ozsolak, F., Poling, L. L., Wang, Z., Liu, H., Liu, X. S., Roeder, R. G., Zhang, X., Song, J. S., and Fisher, D. E. (2008). Chromatin structure analyses identify miRNA promoters. Genes Dev. 22, 3172–3183.
Park, H. J., Kim, J. K., Jeon, H. M., Oh, S. Y., Kim, S. H., Nam, D. H., and Kim, H. (2010). The neural stem cell fate determinant TLX promotes tumorigenesis and genesis of cells resembling glioma stem cells. Mol. Cells 30, 403–408.
Peters, L., and Meister, G. (2007). Argonaute proteins: mediators of RNA silencing. Mol. Cell 26, 611–623.
Piskounova, E., Viswanathan, S. R., Janas, M., LaPierre, R. J., Daley, G. Q., Sliz, P., and Gregory, R. I. (2008). Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J. Biol. Chem. 283, 21310–21314.
Polesskaya, A., Cuvellier, S., Naguibneva, I., Duquet, A., Moss, E. G., and Harel-Bellan, A. (2007). Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 21, 1125–1138.
Ponomarev, E. D., Veremeyko, T., Barteneva, N., Krichevsky, A. M., and Weiner, H. L. (2011). MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-alpha-PU.1 pathway. Nat. Med. 17, 64–70.
Qu, Q., and Shi, Y. (2009). Neural stem cells in the developing and adult brains. J. Cell. Physiol. 221, 5–9.
Qu, Q., Sun, G., Li, W., Yang, S., Ye, P., Zhao, C., Yu, R. T., Gage, F. H., Evans, R. M., and Shi, Y. (2010). Orphan nuclear receptor TLX activates Wnt/beta-catenin signalling to stimulate neural stem cell proliferation and self-renewal. Nat. Cell Biol. 12, 31–40; sup 31–39.
Romanelli, M. G., Lorenzi, P., and Morandi, C. (2000). Organization of the human gene encoding heterogeneous nuclear ribonucleoprotein type I (hnRNP I) and characterization of hnRNP I related pseudogene. Gene 255, 267–272.
Ruby, J. G., Jan, C. H., and Bartel, D. P. (2007). Intronic microRNA precursors that bypass Drosha processing. Nature 448, 83–86.
Rybak, A., Fuchs, H., Smirnova, L., Brandt, C., Pohl, E. E., Nitsch, R., and Wulczyn, F. G. (2008). A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 10, 987–993.
Saini, H. K., Enright, A. J., and Griffiths-Jones, S. (2008). Annotation of mammalian primary microRNAs. BMC Genomics 9, 564. doi:10.1186/1471-2164-9-564
Sakakibara, S., Imai, T., Hamaguchi, K., Okabe, M., Aruga, J., Nakajima, K., Yasutomi, D., Nagata, T., Kurihara, Y., Uesugi, S., Miyata, T., Ogawa, M., Mikoshiba, K., and Okano, H. (1996). Mouse-Musashi-1, a neural RNA-binding protein highly enriched in the mammalian CNS stem cell. Dev. Biol. 176, 230–242.
Sakakibara, S., Nakamura, Y., Yoshida, T., Shibata, S., Koike, M., Takano, H., Ueda, S., Uchiyama, Y., Noda, T., and Okano, H. (2002). RNA-binding protein Musashi family: roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proc. Natl. Acad. Sci. U.S.A. 99, 15194–15199.
Sanuki, R., Onishi, A., Koike, C., Muramatsu, R., Watanabe, S., Muranishi, Y., Irie, S., Uneo, S., Koyasu, T., Matsui, R., Chérasse, Y., Urade, Y., Watanabe, D., Kondo, M., Yamashita, T., and Furukawa, T. (2011). miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nat. Neurosci. 14, 1125–1134.
Schwamborn, J. C., Berezikov, E., and Knoblich, J. A. (2009). The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 136, 913–925.
Schwarz, D. S., Hutvagner, G., Du, T., Xu, Z., Aronin, N., and Zamore, P. D. (2003). Asymmetry in the assembly of the RNAi enzyme complex. Cell 115, 199–208.
Seoane, J., Le, H. V., Shen, L., Anderson, S. A., and Massague, J. (2004). Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 117, 211–223.
Shibata, M., Kurokawa, D., Nakao, H., Ohmura, T., and Aizawa, S. (2008). MicroRNA-9 modulates Cajal-Retzius cell differentiation by suppressing Foxg1 expression in mouse medial pallium. J. Neurosci. 28, 10415–10421.
Shibata, M., Nakao, H., Kiyonari, H., Abe, T., and Aizawa, S. (2011). MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J. Neurosci. 31, 3407–3422.
Siomi, H., and Siomi, M. C. (2010). Posttranscriptional regulation of microRNA biogenesis in animals. Mol. Cell 38, 323–332.
Song, J. J., Liu, J., Tolia, N. H., Schneiderman, J., Smith, S. K., Martienssen, R. A., Hannon, G. J., and Joshua-Tor, L. (2003). The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. 10, 1026–1032.
Thomson, J. M., Newman, M., Parker, J. S., Morin-Kensicki, E. M., Wright, T., and Hammond, S. M. (2006). Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 20, 2202–2207.
Vierbuchen, T., Ostermeier, A., Pang, Z. P., Kokubu, Y., Sudhof, T. C., and Wernig, M. (2010). Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041.
Viswanathan, S. R., Daley, G. Q., and Gregory, R. I. (2008). Selective blockade of microRNA processing by Lin28. Science 320, 97–100.
Viswanathan, S. R., Powers, J. T., Einhorn, W., Hoshida, Y., Ng, T. L., Toffanin, S., O’Sullivan, M., Lu, J., Phillips, L. A., Lockhart, V. L., Shah, S. P., Tanwar, P. S., Mermel, C. H., Beroukhim, R., Azam, M., Teixeira, J., Meyerson, M., Hughes, T. P., Llovet, J. M., Radich, J., Mullighan, C. G., Golub, T. R., Sorensen, P. H., and Daley, G. Q. (2009). Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet. 41, 843–848.
Vo, N., Klein, M. E., Varlamova, O., Keller, D. M., Yamamoto, T., Goodman, R. H., and Impey, S. (2005). A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 102, 16426–16431.
Wang, Y., Juranek, S., Li, H., Sheng, G., Tuschl, T., and Patel, D. J. (2008a). Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature 456, 921–926.
Wang, Y., Sheng, G., Juranek, S., Tuschl, T., and Patel, D. J. (2008b). Structure of the guide-strand-containing argonaute silencing complex. Nature 456, 209–213.
Wang, Y., Medvid, R., Melton, C., Jaenisch, R., and Blelloch, R. (2007). DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 39, 380–385.
Wu, L., Fan, J., and Belasco, J. G. (2006). MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. U.S.A. 103, 4034–4039.
Wulczyn, F. G., Smirnova, L., Rybak, A., Brandt, C., Kwidzinski, E., Ninnemann, O., Strehle, M., Seiler, A., Schumacher, S., and Nitsch, R. (2007). Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J. 21, 415–426.
Xu, X. L., Li, Y., Wang, F., and Gao, F. B. (2008). The steady-state level of the nervous-system-specific microRNA-124a is regulated by dFMR1 in Drosophila. J. Neurosci. 28, 11883–11889.
Yamagata, K., Fujiyama, S., Ito, S., Ueda, T., Murata, T., Naitou, M., Takeyama, K., Minami, Y., O’Malley, B. W., and Kato, S. (2009). Maturation of microRNA is hormonally regulated by a nuclear receptor. Mol. Cell 36, 340–347.
Yamasaki, S., Ivanov, P., Hu, G. F., and Anderson, P. (2009). Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 185, 35–42.
Yang, W., Chendrimada, T. P., Wang, Q., Higuchi, M., Seeburg, P. H., Shiekhattar, R., and Nishikura, K. (2006). Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 13, 13–21.
Yano, M., Okano, H. J., and Okano, H. (2005). Involvement of Hu and heterogeneous nuclear ribonucleoprotein K in neuronal differentiation through p21 mRNA post-transcriptional regulation. J. Biol. Chem. 280, 12690–12699.
Yi, R., Qin, Y., Macara, I. G., and Cullen, B. R. (2003). Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17, 3011–3016.
Yoo, A. S., Staahl, B. T., Chen, L., and Crabtree, G. R. (2009). MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 460, 642–646.
Yoo, A. S., Sun, A. X., Li, L., Shcheglovitov, A., Portmann, T., Li, Y., Lee-Messer, C., Dolmetsch, R. E., Tsien, R. W., and Crabtree, G. R. (2011). MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476, 228–231.
Yu, J., Vodyanik, M. A., Smuga-Otto, K., Antosiewicz-Bourget, J., Frane, J. L., Tian, S., Nie, J., Jonsdottir, G. A., Ruotti, V., Stewart, R., Slukvin, I. I., and Thomson, J. A. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920.
Keywords: microRNA biogenesis, neural stem cells, neural progenitor cells, neurogenesis, Musashi1, Let-7, Lin28
Citation: Kawahara H, Imai T and Okano H (2012) MicroRNAs in neural stem cells and neurogenesis. Front. Neurosci. 6:30. doi: 10.3389/fnins.2012.00030
Received: 13 October 2011; Accepted: 17 February 2012;
Published online: 12 March 2012.
Edited by:
Yanhong Shi, City of Hope, USAReviewed by:
Ashok K. Shetty, Texas A&M Health Science Center College of Medicine at Scott and White, USAFrancesca Ciccolini, University of Heidelberg, Germany
Copyright: © 2012 Kawahara, Imai and Okano. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Hideyuki Okano, Department of Physiology, Keio University School of Medicine, 35 Shinanomachi, Shinjuku, Tokyo 160-8582, Japan. e-mail: hidokano@a2.keio.jp
