- 1 Section of Integrative Biology, University of Texas, Austin, TX, USA
- 2 Department of Biology, University of Portland, Portland, OR, USA
- 3 Department of Biology, North Carolina State University, Raleigh, NC, USA
Sensory and social inputs interact with underlying gene suites to coordinate social behavior. Here we use a naturally complex system in sexual selection studies, the swordtail, to explore how genes associated with mate preference, receptivity, and social affiliation interact in the female brain under specific social conditions. We focused on 11 genes associated with mate preference in this species (neuroserpin, neuroligin-3, NMDA receptor, tPA, stathmin-2, β-1 adrenergic receptor) or with female sociosexual behaviors in other taxa (vasotocin, isotocin, brain aromatase, α-1 adrenergic receptor, tyrosine hydroxylase). We exposed females to four social conditions, including pairings of differing mate choice complexity (large males, large/small males, small males), and a social control (two females). Female mate preference differed significantly by context. Multiple discriminant analysis (MDA) of behaviors revealed a primary axis (explaining 50.2% between-group variance) highlighting differences between groups eliciting high preference behaviors (LL, LS) vs. other contexts, and a secondary axis capturing general measures distinguishing a non-favored group (SS) from other groups. Gene expression MDA revealed a major axis (68.4% between-group variance) that distinguished amongst differential male pairings and was driven by suites of “preference and receptivity genes”; whereas a second axis, distinguishing high affiliation groups (large males, females) from low (small males), was characterized by traditional affiliative-associated genes (isotocin, vasotocin). We found context-specific correlations between behavior and gene MDA, suggesting gene suites covary with behaviors in a socially relevant context. Distinct associations between “affiliative” and “preference” axes suggest mate preference may be mediated by distinct clusters from those of social affiliation. Our results highlight the need to incorporate natural complexity of mating systems into behavioral genomics.
Introduction
Social behavior is a broad descriptive that can encompass behavioral interactions among individuals in multiple contexts, be they shoaling/flocking, aggression, maternal care, mate preference, or sexual behaviors. Often certain behaviors may be common across different social interaction categories, but can serve a very different function depending on the context. Further, the interplay between the expression of social behaviors and underlying brain gene expression patterns can also vary by context. The mechanisms underlying social behaviors are complex, and studies often sample genes from multiple pathways in order to get a more complete picture of the molecular correlates of social behavior (Toth et al., 2007; Filby et al., 2010). Recent technological advances such as microarrays and other comparative genomic approaches have provided tremendous breakthroughs in understanding the genomics underlying social behavior (Robinson et al., 2008; Aubin-Horth and Renn, 2009; Filby et al., 2010; Wong and Hofmann, 2010; O’Connell and Hofmann, 2011). For example, exposure to social information causes changes in gene expression and behavioral modulation in several systems (Hunt et al., 2007; Renn et al., 2008; Ellis and Carney, 2009; Mukai et al., 2009), including mate preference behavior in swordtail fishes (Cummings et al., 2008).
The northern swordtail (Xiphophorus nigrensis) is a classic taxa for sexual selection studies, and female swordtails exhibit strong, predictable mating preferences toward larger, ornamented, courting males over smaller size class males with force copulation mating strategies (Ryan and Causey, 1989; Ryan and Rosenthal, 2001). In an earlier microarray experiment, female swordtails were exposed to different social stimuli (paired males, females) and then sacrificed for a whole brain neurogenomics study that identified suites of genes associated with female mate choice (Cummings et al., 2008). Further analyses with a subset of these genes identified significant relationships between whole brain (Cummings et al., 2008; Lynch et al., 2012) and localized (Wong et al., in review) expression patterns with individual variation in female preference behaviors.
However, female mate preference is a subset of the more general category of sociality, and it is unclear how precopulatory preference-associated genes might relate to other emerging pathways in behavioral genomics. Mate choice consists of being around other individuals (sociality/affiliation), displaying a preference for a particular individual(s), and exhibiting sexual behavior toward a preferred individual. In the current study, we sample behaviors and genes associated with three realms of social behavior (Figure 1) to investigate how female mate preference is related to more general female sexual and social spheres, and test whether these genes–behavior relationships are general or context-specific. Swordtail females are a social species with strong female mate preference, yet female preference is not dependent on reproductive status (Ramsey et al., 2011) and females do not exhibit post-copulatory pair bonding. Therefore we predict (1) distinct behavioral and genetic patterns under mate preference conditions than under general social context, and (2) distinct female preference-associated gene patterns from those traditionally associated with social affiliation and (post-copulatory) partner preference (e.g., vasotocin, oxytocin).
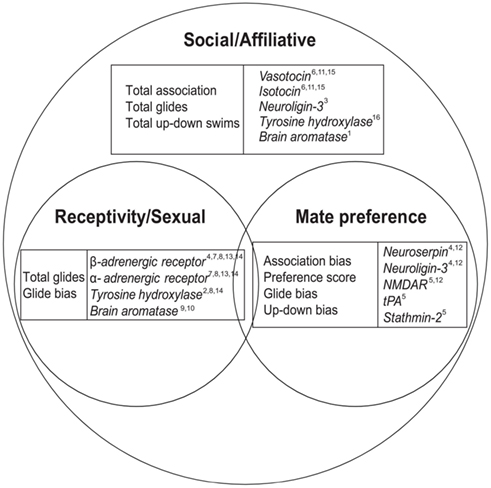
Figure 1. Female social behaviors, brain gene expression, and the subcategories of mate preference and reproduction/sexual behaviors. Venn diagram depiction of the relationships between general female sociality and the subcategories of mate preference and reproduction. We categorize the primary sphere for each of our genes, but some genes (i.e., tyrosine hydroxylase, brain aromatase) may be listed more than once if they are clearly linked to multiple behavioral categories. Representative citations for category assignment (broad social/affiliative vs. reproduction/sexual behavior or mate preference) are included for reference: 1 Adkins-Regan (2009), 2 Becker et al. (2001), 3 Biswas et al. (2010), 4 Cummings et al. (2008), 5Cummings, unpublished data, 6 Donaldson and Young (2008), 7 Etgen et al. (2001), 8 Everitt et al. (1975), 9 Forlano et al. (2006), 10 Forlano and Bass (2011), 11 Goodson and Bass (2000), 12 Lynch et al. (2012), 13 Meston et al. (1996), 14 Riters et al. (2007), 15 Thompson and Walton (2004), 16 Young and Wang (2004). See Table A1 in Appendix for gene nomenclature.
Social behavior is multi-faceted and rarely the product of a single behavioral expression. Rather, social behavior is usually expressed as a suite of displays with components that may vary not only by context but also in their independent expression. For instance, mate choice assays often use association bias as a behavioral measure of preference (e.g., Ryan and Wagner, 1987; Basolo, 2004; Morris et al., 2006). However, an association bias observed in one context (e.g., with stimulus males) may represent a different response than an association bias observed in another social context (e.g., with stimulus females), and examining other behavioral displays during these association times can help distinguish between social contexts at a behavioral level. Therefore, in order to characterize social behaviors more accurately, we need to quantify the suite of behavioral responses present in different contexts so that we can accurately phenotype social behaviors and examine how they vary by social contexts. To this end, we exposed females to four distinct social pairings that represent varying levels of mate choice complexity [two large males (LL), a large and small male (LS), two small males (SS)] or a social control [two females (FF)] and measured eight behaviors. Of these behaviors, four were preference metrics that measured biased behaviors toward a favored stimulus, three were putative affiliative metrics measuring social/sexual interest regardless of target, and one assessed overall activity.
We then sampled 11 genes from these three aspects of female social behavior: preference, sociality/affiliation, and receptivity/sexual behavior (Figure 1) and compared brain expression patterns in relation to differential social exposure. We selected these particular genes because they fit one of two criteria: (1) previous association with preference behavior in X. nigrensis females [neuroserpin (serpin1), neuroligin-3 (nlgn3), NMDAR (N-methyl d-aspartate receptor; grin1), tPA (plat), stathmin-2 (stmn2, scg10), β-1 adrenergic receptor (Adrb1)], or (2) functional association with social or sexual behavior in other taxa [vasotocin (AVT), isotocin (IT), brain aromatase (Cyp191b), α-1 adrenergic receptor (Adra1b), tyrosine hydroxylase (TH)]. Our different social treatments allowed us to contrast the gene expression profiles of non-sexual affiliation (female-only environments) with precopulatory mate preference (large male environments), as well as determine which gene suites covary with our different behavioral measures (e.g., general association behaviors vs. mate preference behaviors).
Materials and Methods
Behavior
Xiphophorus nigrensis females were either wild-caught at the Naciemento de Rio Choy in the state of San Luis Potosi, Mexico or obtained from semi-wild populations held at the Brackenridge Field Laboratories (University of Texas). Once brought into the lab, females were kept isolated from males for at least 2 weeks before behavior trial to ensure motivation.
Behavior trials were conducted as described in Cummings et al. (2008) with slight modifications. Briefly, behavior trials consisted of a non-contact dichotomous choice trial wherein females were isolated in a center tank region with visual exposure to stimuli on either end of the tank (see Cummings et al., 2008; Ramsey et al., 2011). Immediately before behavioral testing, we measured circulating estradiol levels via a waterborne assay previously validated for this species (Ramsey et al., 2011; see below). Females were exposed to one of five testing conditions (total n = 37): LS (large male–small male; n = 8), SS (small male–small male; n = 7), LL (large male–large male; n = 7), FF (female–female; n = 10), and home tank (HT control; n = 5). In the LL group, one male was placed behind a UV pass barrier and the other behind a UV block barrier. The contrasting UV pass and UV block barriers were intended to provide a potential visual preference between the two large-class male stimuli (Cummings et al., 2003). Large male sizes ranged from 32.6 to 37.1 mm SL (standard length), and small male sizes ranged from 20.7 to 25.7 mm SL. For size-matched stimuli (SS, LL, FF), paired individuals differed no more than 1.2 mm in SL, and for the size-contrast group (LS), the average size difference between the large and small stimulus males was 14.8 mm. Females were allowed to acclimate to the testing tank for 5 min while shielded from visual contact with the stimuli by a PVC cylinder. Females were then released from the cylinder and allowed to swim freely in the center tank region for one more minute before the trial began. Trials lasted 30 min, and midway through the trial the stimuli were switched to avoid side bias (for the LL group, the UV pass/UV block barriers were switched rather than the males). Females were sacrificed immediately after the trial for total RNA extraction from the whole brain. HT females were placed back into their housing environment for 30 min prior to sacrifice. All experimental procedures were approved by IACUC (#07110101) at the University of Texas at Austin.
We measured eight behaviors during each trial. These included four preference measures (association bias, preference score, glide bias, and up-down bias), three social/sexual affiliation measures including an overall measure of sociality (total association = proportion of total time spent in either association zone/total time of the trial), an activity-based measure of motivation (total up-down swims), and a receptivity proxy (total glides performed toward both stimuli = total glides). Finally, we also included a measure of total activity (total transits = total swims out of either association zone into the middle of the tank). In X. nigrensis, glides are defined as a social display behavior where the female orients toward the male, then swims away from him, turns, and then returns to the barrier, and have been shown to predict copulation events (Cummings and Mollaghan, 2006). Association bias is defined as the proportion of time spent in the association zone with stimulus a/(time with stimulus a + stimulus b), where time with stimulus a > time with stimulus b. Preference score is a composite time + behavior score, where preference score = association bias + log [(1 + # glides to stimulus a)/total transits]. Glide bias is defined as the number of glides performed toward stimulus a/(# glides with stimulus a plus stimulus b). Up-down swims are defined as a behavior where the female orients to the barrier and makes a vertical swim either up or down at least one body length in distance, but always returning to the same relative location oriented toward the barrier. While this behavior is an artifact of experimental conditions (not observed in the wild), it has been used as a proxy for motivation of a fish to interact with the fish on the other side of the barrier in dichotomous choice condition (Cummings et al., 2006). Up-down bias is a proportion measure defined as the number of up-down swims toward stimulus a/total number of up-down swims to both stimuli. We include four different preference measures to quantify the biased performance of all commonly displayed female behaviors (time in proximity to a stimulus, glides, up-down swims) in our analysis. Each trial was scored live by an observer and taped for later review.
Hormone Assays
Estradiol (E2) levels were measured using a non-invasive water assay previously validated for this species (Ramsey et al., 2011). Briefly, females were placed into a 250 ml glass beaker containing 150 ml of reservoir water (aerated, dechlorinated tapwater that is the source for all home and experimental tank waters) for 1 h. Females were then removed from the beaker and placed immediately into the behavior tank for testing. Water samples were then filtered with Whatman P5 filter paper and stored at −20°C in polypropylene bottles. Steroid hormones were extracted from water samples using C18 Solid Phase Extraction columns (Sep-Pak® Plus C18 cartridge 55–105 μm; Waters Corporation, Milford, MA, USA), as described in Ramsey et al. (2011). The E2 concentration of the resulting material was measured using the Correlate-EIA 17β-estradiol Enzyme Immunoassay Kit (Assay Designs) according to the manufacturer’s protocol (see Ramsey et al., 2011 for detailed protocol). Duplicate well values were then averaged for the final E2 concentration/sample, which was then normalized by deriving the residuals from a linear regression of the measured E2 concentration by fish weight. Hormone samples were run on a 96-well EIA assay plate, and the intra-assay CV was 3.6%. There were no significant differences in female pre-testing E2 levels (p = 0.8612).
Gene Cloning and qPCR Analysis of Whole Brain Gene Expression
Our initial microarray experiment was conducted on whole brain tissue because our fish are quite small and therefore their brains are small – even using whole brain tissue individuals had to be pooled to obtain enough RNA for microarray hybridization. More importantly, our initial genomics experiment utilized whole brain tissue because the neural network underlying precopulatory mate preference in this species is not known, therefore we did not have specific regions to target. The genes in the current experiment were chosen because they had already been identified as being differentially expressed during mate choice conditions at the whole brain level, and we then chose candidate genes from other key realms of female social behavior (i.e., more general social- or reproduction-associated genes) to address the broad question of how mate preference and social context might relate to other aspects of female social behaviors.
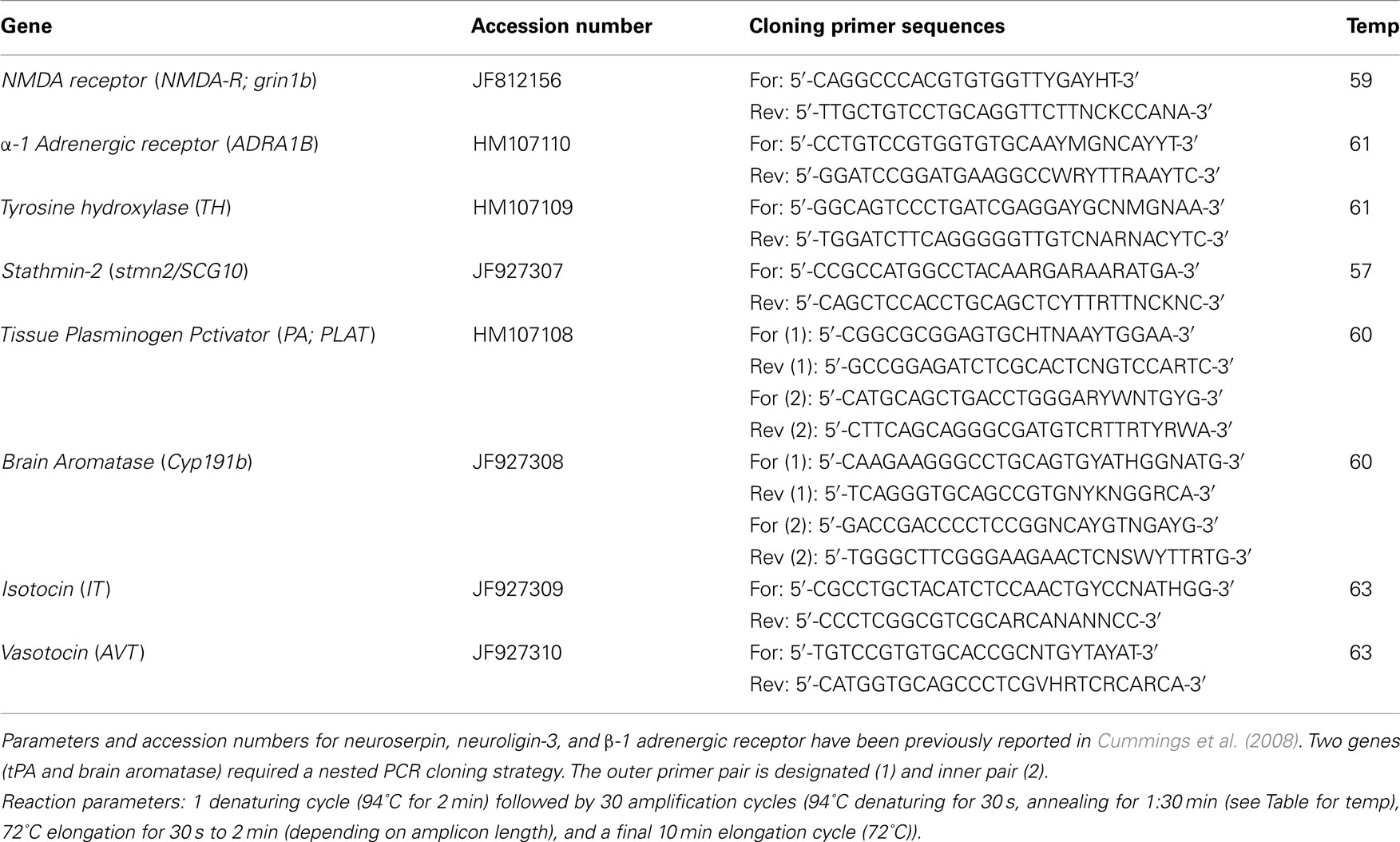
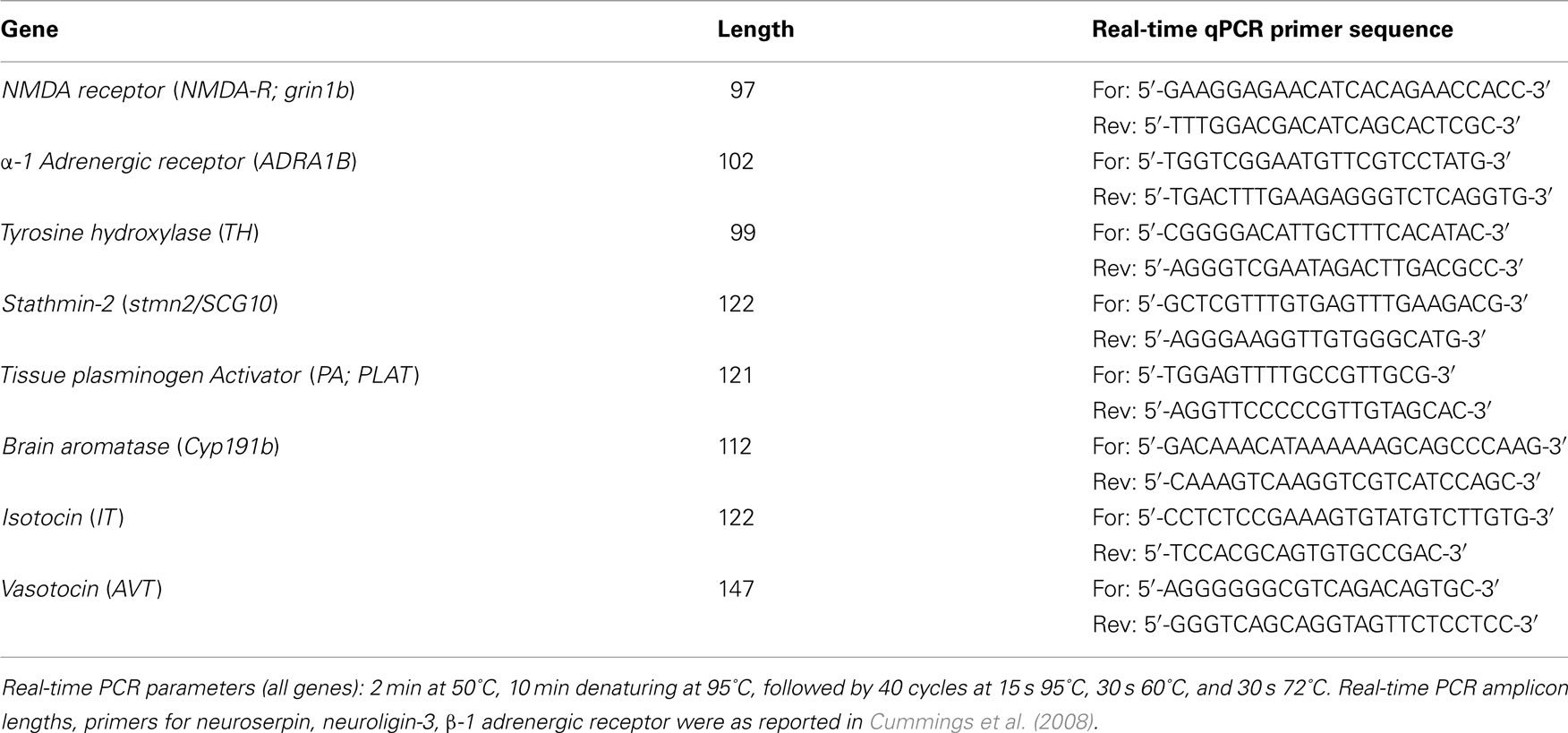
For gene cloning experiments, degenerate primers were designed using CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primer)1, with consensus sequences derived from known fish sequences (see Table A1 in Appendix for gene nomenclature, accession numbers, primer sequences, and cloning PCR reaction parameters). Primers were designed using MacVector, and the primers were purchased from Integrated DNA Technologies (IDT)2.
For the qPCR experiment, females were sacrificed and whole brain tissue dissected and placed in RNAlater (Ambion) solution overnight at 4°C. The RNAlater solution was then removed and brain tissue stored at −80°C until processed. Total RNA extraction and qPCR measures are described in Cummings et al. (2008). Briefly, total RNA was extracted using Trizol reagent (Invitrogen) according to the manufacturer’s protocol, and then DNAse-treated using turbo DNA-free kit (Ambion). Please see Table A3 in Appendix for pre-testing E2 levels. Total RNA from each individual brain was reverse-transcribed using Superscript First-Strand Synthesis for RT-PCR (Invitrogen) in a 40 μl-size reaction according to a modified manufacturer’s protocol for transcribing cDNA to be used as qPCR template. cDNA synthesis was primed with both oligo-dT and random hexamers. Following the reverse transcription reaction, cDNA was purified using Amicon Ultracentrifugal filters (Millipore) according to the manufacturer’s protocol. Post-centrifugation, all samples were topped off to 70 μl total volume with Nuclease-Free water (Ambion). Real-time qPCR reactions were conducted with SYBR green detection chemistry using a 7900HT real-time PCR machine (ABI) with the following reaction recipe: 10 μl total volume reaction containing 1 μl cDNA template, 5 μl 2× POWER SYBR Green PCR master mix (ABI), and 5 pmol primers. See Table A2 in Appendix for real-time primer sequences, amplicon lengths, and real-time qPCR reaction parameters. Each sample was run in triplicate. Results were first analyzed using Applied Biosystems Sequence Detection Software (SDS vs. 2.3), and gene expression levels were then normalized as in Lynch et al. (2012) by deriving the residuals from a linear regression of the measured gene concentration by input cDNA concentration. Four genes (neuroserpin, β-1 adrenergic receptor, vasotocin, isotocin) had a significant interaction with treatment group in our input cDNA measures, therefore we derived residuals for these genes while including treatment as a covariate (ANCOVA). Input cDNA concentration was measured using Quanti-IT RiboGreen RNA reagent (Molecular Probes) using a modified version of the manufacturer’s protocol, as in Cummings et al. (2008) and Lynch et al. (2012).
Statistics
Cross-group behavioral comparisons were conducted with ANOVA followed by post hoc Tukey’s tests, and included all stimulus-exposed females (n = 32). To keep all of the behavioral measures on a similar scale, we log-transformed total glides, total up-down swims and total transits. Five of eight behaviors (association bias, log total glides, glide bias, log total up-down swims, and up-down bias) were not normally distributed. Non-parametric Kruskal–Wallis tests yielded identical patterns to ANOVA tests for these behaviors (data not shown), therefore for consistency we report only ANOVA results in the text.
For gene expression analyses, we removed 2 outliers (1 LL-exposed female with gene values greater than ±2 SD from the mean in 8 of the 11 genes, and 1 FF-exposed female with gene values greater than ±2 SD from the mean in 9 of 11 genes) for a final total consisting of 8 LS-, 7 SS-, 6 LL-, 9 FF-, and 5 HT-exposed females (a no-behavior group also removed for the MDA analyses). The behavior and gene multivariate analyses were run on this reduced data set. Multivariate analyses were conducted using multiple discriminant analysis (MDA; Watson, 1982; McLachlan, 2005). Sometimes also referred to as discriminant factor analysis (DFA) or canonical discriminant analysis (CDA), MDA is a dimension reduction technique with some similarities to principal components analysis (PCA), but in MDA the axes are created to sequentially maximize the capture of variation between predefined groups rather than total variance explained across all samples. MDA is used primarily to classify patterns in multidimensional data.
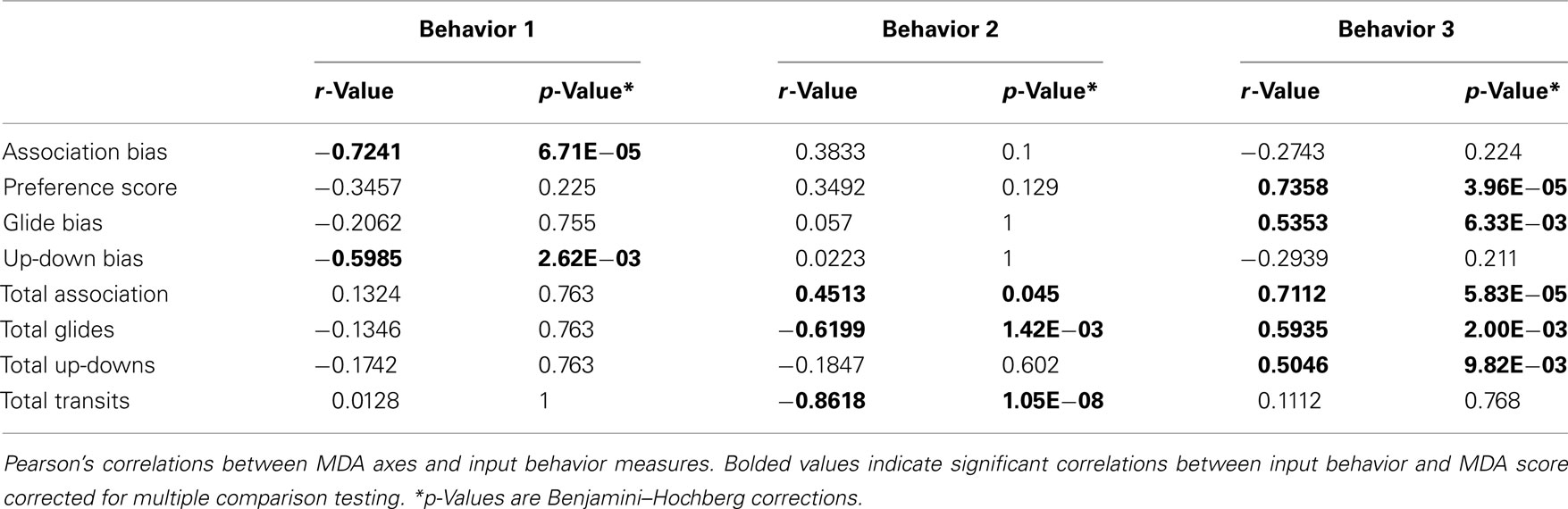
Multiple discriminant analyses were run separately for the eight behavior measures recorded during each trial (behavior MDA) and then on the normalized residual gene values (n = 11 genes; gene MDA) derived as described above, with social exposure category (LL, LS, SS, or FF) as the classification variable for both analyses. We do not include the HT group in the gene MDA so that we can more readily compare the behavior MDA and gene MDA results. However, a gene MDA with HT group included also differentiated by treatment group (Wilks’ MANOVA F = 4.25, p = 5.94E−06). All multivariate statistics were carried out using R software: MASS package (lda, lda.predict) for the MDA score generation and class prediction analyses, and the candisc package for graphical assessment of group centroid (multivariate least squares means) and gene loading patterns (i.e., variable vectors). Treatment differences for MDA scores were first assessed with a MANOVA (Wilks) on the overall MDA model scores. We then utilized ANOVA followed by post hoc pairwise Tukey’s tests to explore group differences within each MDA axis. We assessed the relative contributions of the input independent variables onto the MDA axes (variable loadings) using Pearson’s correlations. Several input behaviors in our Behavior MDA were significantly correlated with axis scores, therefore to control for false discovery rate (FDR) in our multiple comparisons, we used a Benjamini–Hochberg corrected p-value in Table 1 (Benjamini et al., 2001).
To assess whether groups more similar in behavior were more similar in gene expression patterns, multivariate behavior, and gene relationships were analyzed either through direct comparison of the input behavior and gene matrices (Mantel test; ade 4 package, canonical correlation analysis; CCA package) or with treatment group as an explicit factor in the model. For the simple input matrices comparisons, we calculated Euclidean distances between the input treatment group centroids for both behavior and gene expression measures and then used a mantel test with 1000 permutations to assess whether these two matrices were significantly correlated. Second, we employed CCA to identify axes for which the input behavior and gene expression data sets were significantly correlated, and then used Bartlett’s Chi-square test to assess significance. During the CCA, we detected a statistical outlier (±2 SD from the mean for both the X and Y score generation) that was not detected during any of our other multivariate analyses. Therefore we report the CCA results with the outlier included and omitted. For the context-specific comparisons, we utilized Pearson’s correlation between the behavior and gene MDA scores.
Results
Preference Behaviors Differ Across Social Conditions
We assessed preference for one stimulus individual over another via four different preference measures: preference score, association bias, glide bias, and up-down bias. Female behaviors across the four social conditions (LS, LL, SS, FF) were significantly different for three of four preference measures (ANOVA preference score, F = 4.25, p = 0.014; Figure 2A; association bias, F = 5.76, p = 0.003; Figure 2C; up-down bias, F = 3.00, p = 0.047; Figure 2E; glide bias, F = 1.35, p = 0.277). For preference score and association bias, post hoc Tukey’s tests showed that females expressed higher preference behavior when exposed to the LS social condition than when exposed to the SS stimuli condition (preference score LS vs. SS, p = 0.013; association bias LS vs. SS, p = 0.004). Association bias and up-down bias post hoc Tukey’s tests showed that females also expressed higher preference behaviors when exposed to the LS social condition than when exposed to the conspecific FF control stimuli (association bias LS vs. FF, p = 0.017; up-down bias LS vs. FF, p = 0.036). There was a trend for group differences in the amount of total association time spent with any stimuli (ANOVA F = 2.92, p = 0.051; Figure 2D), with females spending the least amount of total association time in the SS condition. Receptivity displays (log total glides, Figure 2F) also differed across social conditions (ANOVA F = 3.598, p = 0.026), and post hoc Tukey’s tests indicated females performed significantly fewer glides when exposed to the LL stimuli pairing than the LS social exposure group (p = 0.042). In contrast to the up-down bias measure, total up-down swims were not different across groups (log total up-downs ANOVA F = 1.163, p = 0.342). In looking at overall activity, total transits were significantly different across treatment groups (log total transits ANOVA F = 4.10, p = 0.016; Figure 2B), and females performed significantly more transits when exposed to the SS group than the FF group (post hoc Tukey’s SS vs. FF, p = 0.011). We repeated the across-group ANOVAs on the reduced version of this data set used for our gene expression analysis (two females removed as outliers for a total n = 30; see Materials and Methods), and obtained identical patterns for all behaviors (data not shown). There were no relationships between pre-testing circulating E2 levels and any behavior either across or within treatment groups (data not shown).
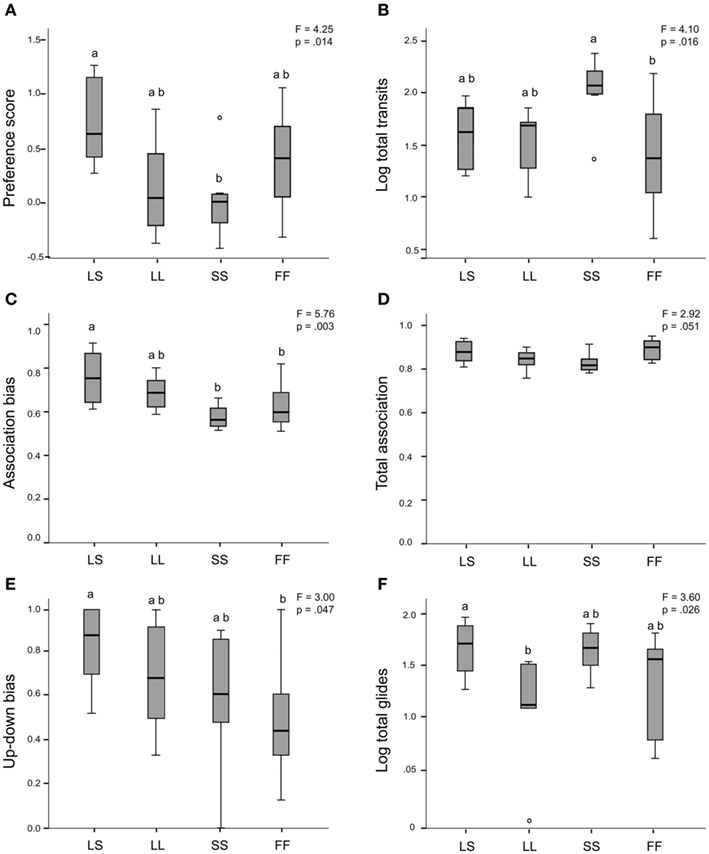
Figure 2. Mate preference behaviors and locomotor activity differ by social exposure. ANOVA results indicate significant group differences in the expression of preference score (A), association bias (C), and up-down bias swim behaviors (E), and a trend for group differences in proportion of total association with stimuli (D). Locomotor activity (log total transits) and log total glide swims were also significantly different between groups (B,F). Box plots indicate first and third quartiles, the line indicates the median, and the whiskers indicate high and low values. Circles indicate outliers. LS, large male/small male exposure; LL, large male/large male exposure; SS, small male/small male exposure; and FF, female/female-exposure. Glide bias and total up-down swims not shown.
Social Exposure Category Predicts Multivariate Behavior Patterns
We next tested if female behaviors differed across groups on a multivariate level. MANOVA analysis on our eight input behaviors identified significant group-wise differences in female behavior across the different social exposures (MANOVA eight input behaviors, F = 3.53, p = 9.6E−03). Female mate preference is often assessed using a single measure (association bias), so to justify the inclusion of multiple preference measures we repeated the MANOVA with association bias removed. We found significant behavioral differences between social exposure groups even when association bias was eliminated from the analysis (MANOVA seven input behaviors, F = 3.72, p = 8.36E−03).
We then analyzed all eight measured behaviors to assess (1) if multivariate behavioral patterns could successfully predict social group exposure, and (2) how these input behaviors are related in multivariate space (Figure 3A). MDA using social exposure group as the categorical dependent variable revealed two primary axes (Figure 3A), Behavior 1 (accounting for 50.2% between-group variation) and Behavior 2 (capturing 31.8% between-group variation), as well as a minor third axis (Behavior 3) that represented 18% of the between variance (data not shown). Overall classification rate was 69%, but classification prediction rates varied by exposure group. LS exposure was correctly predicted 62.5%, LL 50%, SS 86%, while FF exposure was correctly assigned 78% of the time. We estimated the precision of our MDA classification rates using jackknife resampling. As with the original MDA permutation, resampling classification rates were variable by social exposure group. For the behavior MDA, our overall rate was 53%, with rates ranging from a high of 71% for SS-exposed females and a low of 17% for LL-exposed females.
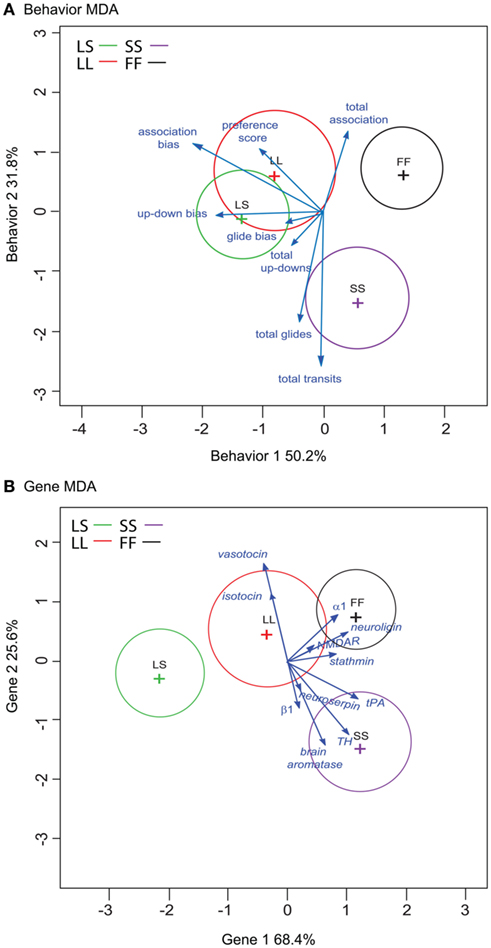
Figure 3. Spatial relationship of eight behaviors (A) and 11 input gene variables (B) onto MDA Axes. Circles represent the 95% confidence ellipses around each multivariate centroid (multivariate least squares mean for each treatment group), and the vectors represent the orientation in multidimensional space along the two primary axes Axis 1 (x-axis) and Axis 2 (y-axis) for each behavior (A) or gene (B) MDA. Vectors are scaled for the plot space, and vector length indicates the potency of each variable in terms of discriminating between groups. For each input variable, the vector points toward groups with higher mean values, and away from groups with lower mean values. Groups are plotted by color, LS = green, LL = red, SS = purple, and FF = black.
The multivariate centroids for the four social exposure groups are plotted in Figure 3A, where the circles represent the 95% confidence ellipses around each centroid and the vectors indicate the direction and relative discriminating power (potency index sensu Perrault et al., 1979) of each behavioral measure’s loading onto the two major axes. The behavior MDA axes scores differed by social exposure group (MANOVA F = 7.60, p = 2.75E−07). Treatment group scores were significantly different for both of the major axes (Behavior 1 ANOVA F = 11.9, p = 4.44E−05; Behavior 2 ANOVA F = 7.50, p = 8.97E−04). In Behavior 1, post hoc Tukey’s tests revealed significant differences between females exposed to large males (LS and LL) from other groupings. Specifically, there were significant pairwise differences between LS vs. SS (p = 0.006) and FF (p = 0.6.25E−05) exposure groups, and LL vs. FF (p = 0.002) and a trend for LL vs. SS (p = 0.091) pairings. Within Behavior 2, post hoc Tukey’s test indicated significant differences between females exposed to pairings that included a small male (SS) from other groupings. In the Behavior 2 axis, SS-females were distinct from the other groupings: LL (p = 0.004), FF (p = 0.001), with a trend for LS (p = 0.053). There was also a significant treatment difference within the minor Behavior 3 axis (ANOVA F = 4.25, p = 0.014), and this axis distinguished between females exposed to the large male groupings (LL vs. LS p = 0.012).
Inspection of the input behaviors onto the axes (Figure 3A; Table 1) indicates preference measures tended to load more heavily onto the Behavior 1 axis, and that these behaviors increased in groupings that contained a large male (LS and LL). Of the four preference measures, association bias (r = −0.7241, p = 6.71E−05) and up-down bias (r = −0.5985, p = 0.003) loaded most strongly onto the Behavior 1 axis scores. In contrast, the general sociality and activity measures loaded more heavily onto the Behavior 2 axis. Total association loaded toward groupings without a small male (LL, FF; r = 0.4513, p = 0.045) while the more active total glide and total transit measures loaded toward the small male grouping (SS; total glides r = −0.6200, p = 0.001; total transits r = −0.8618, p = 1.05E−08). There was no relationship between pre-testing E2 levels and behavior MDA axes scores (data not shown).
Social Exposure Category Predicts Gene Expression Patterns
When assessed individually, gene expression did not vary by social or control HT conditions across groups (data not shown). However, MDA analysis revealed that exposure to different social conditions was reflected in the underlying gene expression structure and identified three axes. The Gene 1 and Gene 2 axes accounted for 68.4%, 25.6% of the between-class variation (Figure 3B), respectively, and the minor Gene 3 axis captured 6.0% (data not shown). MDA modeling correctly predicted group assignment 64% overall, but prediction rates differed when examined by group. Modeling correctly predicted the LS 87.5%, FF 78%, and SS group exposure 57%. In contrast, LL-group exposure was correctly assigned only 33% of the time. As with the Behavior MDA jackknife test, resampling classification rates for the Gene MDA were variable across groups. For the gene MDA, our overall resampling precision was 33%, ranging from a high of 56% for FF-exposed females and a low of 14% for SS-exposed females.
As with the Behavior MDA, gene MDA axes scores were significantly different across social exposure groups (MANOVA F = 8.24, p = 8.25E−08). Treatment groups were significantly different within the two major Gene axes (Gene 1 ANOVA F = 19.8, p = 6.83E−07; Gene 2 ANOVA F = 7.41, p = 0.9.58E−04; Figure 3B). Post hoc Tukey’s analysis on Gene 1 axis scores revealed pairwise differences between groups containing large males relative to other exposure groups (LS vs. SS p = 0.4.20E−06, LS vs. FF p = 2.00E−06, LL vs. SS p = 0.046, LL vs. FF p = 0.042), including a discrimination between large male-containing groups (LS vs. LL p = 0.013). Within Gene 2, post hoc Tukey’s identified significant differences between SS-females and those exposed to groupings without a small male (SS vs. LL p = 0.010, SS vs. FF p = 7.34E−04). There were no significant treatment differences within the minor Gene 3 axis scores (ANOVA F-1.75, p = 0.182).
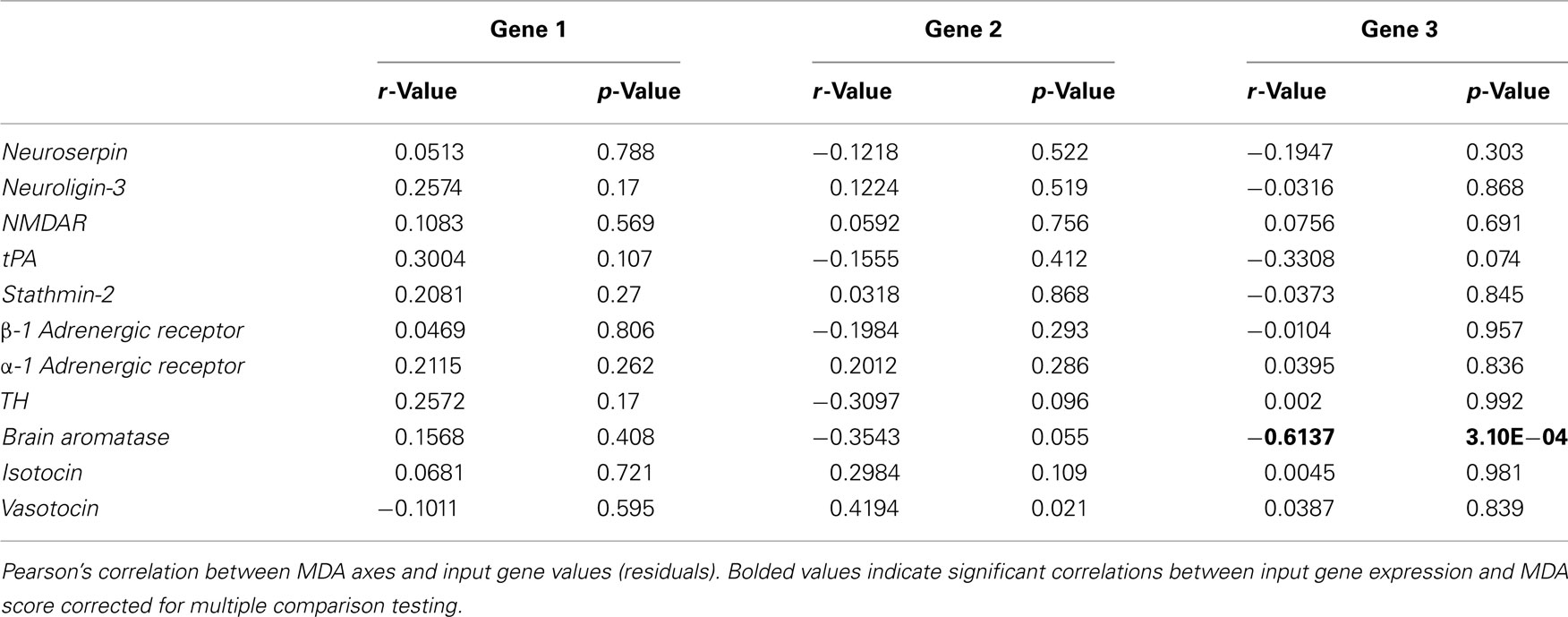
We assessed the relative importance of the input gene variables by analyzing the spatial orientation and discriminating potency of the 11 gene vectors (Figure 3B). No single gene was significantly associated with either of the major Gene axes (Table 2). However, the gene vector orientations do highlight two clusters of mostly mate preference- and reproduction-associated genes loading mostly onto Gene 1 axis (Figure 3B). Both clusters were directed away from large male-containing groups (LS, LL), with one cluster generally loading toward FF-females (α-1 adrenergic receptor, neuroligin-3, NMDAR, stathmin-2) and the other loading toward SS-females (tPA, TH, neuroserpin, brain aromatase, β-1 adrenergic receptor). In contrast, two genes associated with affiliative behaviors (isotocin, vasotocin) exhibited a distinct orientation onto the Gene 2 axis that was almost directly opposed to two preference-associated genes (neuroserpin, β-1 adrenergic receptor) and brain aromatase (Figure 3B). To more directly compare the behavior and gene data sets, we did not include the HT group in our Gene MDA analysis, however inclusion of the HT group resulted in nearly identical group centroid and input gene vector positioning in multivariate space, with the HT centroid falling between the LL and FF groups along the Gene 1 axis (data not shown). There was no relationship between pre-testing circulating E2 levels and gene expression for any individual input gene (including brain aromatase r = 0.2376, p = 0.215) or gene MDA axes scores (data not shown).
Context-Specific Preference Behavior and Gene Expression Patterns are Correlated in Multivariate Space
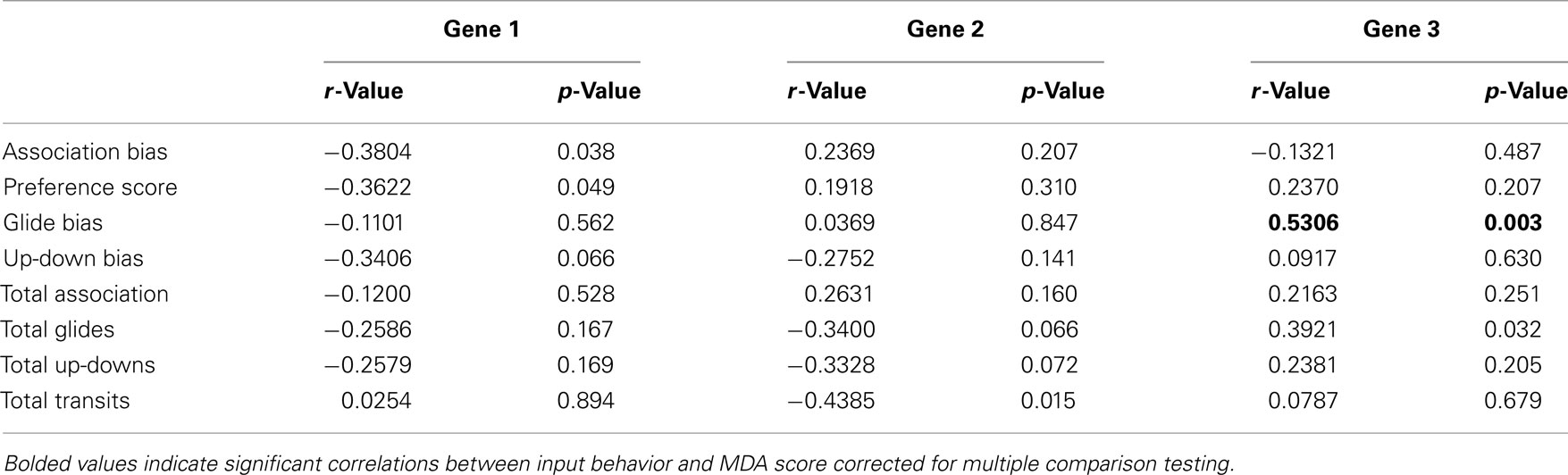
Initial examinations of individual input behaviors and gene MDA scores produced significant correlations wherein two of the four preference measures were correlated with Gene 1 (Association bias, r = −0.3804; p = 0.038; Preference Score r = −0.3622, p = 0.049; Table 3), and the overall activity measure was associated with Gene 2 (r = −0.4385, p = 0.015, Table 3). However, no relationships between individual behaviors and the two major Gene MDA axes were significant following multiple test corrections.
However, the components of both social behaviors and gene expression are multi-faceted, therefore we next utilized multivariate approaches to assess the relationship between behavior and gene expression. We first tested the input data sets without considering social context. A Mantel test comparing the Euclidean distance matrices for behavior and gene data sets found no significant group-level relationships (matrix correlation r = 0.4618, p = 0.294), indicating that the input matrices are not correlated when treatment is not factored in the analysis. Next, a global canonical correlation analysis (CCA) between the behavioral and gene expression data sets produced a first canonical root that captured significant relationships between the two data sets (r = 0.9837, p = 0.007), however this relationship did not hold following removal of a statistical outlier (r = 0.9172, p = 0.800).
We next retained social exposure as an explicit factor in our modeling by directly comparing the behavior and gene MDA’s. Looking at the scores by axis, Behavior 1 and Gene 1 (r = 0.4811, p = 0.007; Figure 4A), and Behavior 2 and Gene 2 (r = 0.4909, p = 0.006; Figure 4B) were positively correlated, while the minor Behavior 3 and Gene 3 axes were negatively correlated (r = −0.5140, p = 0.004).
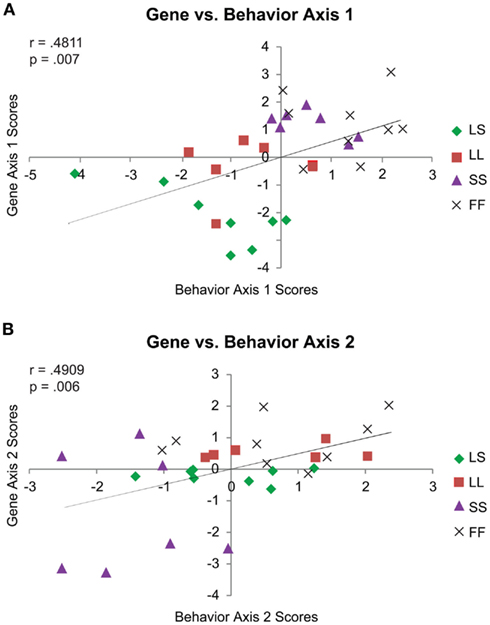
Figure 4. Context-specific correlations for behavior vs. gene MDA scores. Behavior 1 and Gene 1 Pearson’s correlation (A). Behavior 2 and Gene 2 Pearson’s correlation (B). For both panels, behavior MDA scores are depicted on the x-axis and gene MDA scores on the y-axis. MDA score generation explicitly accounts for treatment group, and so although group membership is denoted by symbol (see panel legends), the correlations are calculated including all MDA scores for each axis. Only the correlations between the two main axes are shown.
Discussion
Social behaviors and the genomic pathways that help coordinate them are complex, and multiple facets of social behavior and gene expression co-express across quite different social contexts. Mate preference is one subcategory of more general social behaviors, and the neurological pathways associated with female preference behaviors may draw from those associated with general sociality and female sexual behavior in addition to mate discrimination. In the current experiment, we sampled behaviors and genes representing general female social affiliation, mate preference, and reproduction (Figure 1). Our goal was to characterize mate preference gene and behavior relationships both in relevance to differing social environments (i.e., specific social context) as well as in relationship to other molecular pathways involved in female social behaviors. We found that examining both behavior and gene expression within a multidimensional context provided greater power to characterize differences between social contexts than more traditional approaches that apply multidimensional approaches only to gene expression (cf. Table 3 with Figure 4). With this approach, we identified distinct behavioral and gene expression patterns under mate choice contexts (LL, LS) from those associated with no choice (SS) or a general social affiliation context (FF; Figure 3). Furthermore, suites of genes associated with mate preference clustered independently from those associated with general social affiliation (vasotocin, isotocin) across these different social contexts.
Differential Social Exposure Elicits Distinct Behavior Patterns
Female swordtails demonstrated strong behavioral differences across social exposure groups, whether between general social conditions (females only) vs. mate choice conditions or between mate choice groupings of varying complexity (Figure 3). The existence of discrete male phenotypes in X. nigrensis allowed us to vary the composition of the stimulus males, thereby creating three separate mate choice contexts. For example, females presented with the large male–small male pair (LS), were exposed to a simple or salient choice environment with one male potentially displaying multiple visual cues (e.g., large size, courtship behavior, ornamentation) and the other male lacking those features. In contrast, the two large male pairing was presumably more complex as females could discriminate between two stimuli similarly matched in female-preferred traits, while the pairing of two small males (two coercive stimuli) represented a minimal choice environment where the suite of female-preferred traits was absent in both males.
On a behavioral level, multivariate group differences were primarily driven by the presence or absence of a large male (i.e., choice vs. no choice; Figure 3A) with the primary Behavior 1 axis driven by the higher expression of preference behaviors in large male social contexts. These preference measures (whether measured as a time or activity bias) formed a cluster distinct from non-specific time or activity measures, suggesting that mate preference and general social affiliation measures are substantially different behavioral suites in this species. Behavior 2 discriminated between the coercive small male pairing vs. all other social contexts containing more favorable social stimuli, and was driven by the overall sociality and movement measures. Females tended to spend less overall time associating with stimuli when small males were present, and were more active (more transits and glide swims). Interestingly, despite having the major axis driven by the expression of preference behaviors, multidimensional classification was most successful in the non-mate choice categories (SS 86%, FF 78%), and less predictive in the mate choice contexts (LS 62.5%, LL 50%).
Differential Social Exposure Elicits Distinct Gene Expression Patterns
Multivariate differences in gene expression patterns were also driven by differential compositions of social exposure groups (Figure 3B), although inherent pre-existing individual variation in expression patterns may also contribute to these differences. Whereas the Behavior 1 axis split apart groups roughly based on presence or absence of a large male in the pairing (and did not discriminate between LL and LS), the Gene 1 axis was more precise in discriminating amongst social environments and included differentiation reflecting scaling levels of the mate choice environment (e.g., significant distinctions between LL vs. LS vs. SS). Furthermore, the gene MDA showed a marked improvement relative to the behavior MDA in classifying females exposed to the most salient mate choice context (LS) with a near-perfect classification rate (87.5%).
The gene MDA was extremely effective at discriminating amongst social exposure groups, however this discrimination was not driven by differential expression of any one particular gene on the major axes (Figure 3B; Table 2). Most striking, instead, was the directionality of the gene vectors in multivariate space. The mate preference- and reproduction-associated genes formed two clusters bisected by the Gene 1 axis. Both clusters loaded heavily toward non-mate choice environments (one toward conspecific females, and the other toward small male pairs) and so were down-regulated in the presence of favored large males, a pattern consistent with many of the genes in mate choice conditions from our earlier microarray results (Cummings et al., 2008). Gene by behavior relationships are complex, however, and can vary based on context and the level of analysis. For example, while we consistently find this male- vs. female-exposure pattern when looking between social groups, analyses of some of these “preference-associated genes” (e.g., neuroserpin, neuroligin-3) within male-exposed groups have instead shown positive correlations with preference behavior (e.g., Cummings et al., 2008; Lynch et al., 2012).
One striking feature of some of these preference-associated genes identified in our initial swordtail microarray experiment (e.g., neuroserpin, neuroligin-3, NMDAR, stathmin-2), as well as tPA, a gene functionally linked with both neuroserpin and NMDAR (Yepes and Lawrence, 2004; Samson and Medcalf, 2006), is their association with neural plasticity and learning in other taxa (Clayton, 2000; Reidel et al., 2003; Miranda and Lomas, 2006; Sudhof, 2008; Biswas et al., 2010; Borges et al., 2010). These genes share functional similarities as they are associated with synaptic plasticity and have been linked to social behavior, exploration, learning, and memory (Madani et al., 2003; Pawlak et al., 2003; Reidel et al., 2003; Tabuchi et al., 2007; Martel et al., 2008; Biswas et al., 2010). As such, these genes may represent components of an assessment/learning module that may help integrate incoming social/environmental information with an appropriate behavioral response. Social learning and decision-making have been implicated in mate choice behavior (Witte and Noltemeier, 2002; Ophir and Galef, 2004; White, 2004), and the association between mate preference context and synaptic plasticity-associated genes is particularly intriguing in light of evidence that female preference behaviors are age-dependent in X. nigrensis (Wong et al., 2011).
Whereas the primary axis in the gene MDA was dominated by preference-associated genes and characterized differences across mate choice contexts, the secondary axis was dominated by social affiliation-associated gene expression and maximized the variance along a social affiliation continuum (favorable to non-favorable contexts). Similar to the secondary behavior axis pattern, Gene 2 differentiated females exposed to the small male pairing from groups containing favorable stimuli (large males or conspecific females), and may capture aspects of an expression-level social affiliation – aversion axis. Supporting this hypothesis, genes associated with sociality and affiliative behavior loaded more prominently onto the Gene 2 axis (vasotocin, isotocin, Figure 3B).
Sociality is a critical component of female preference behavior, and the neurogenetics of social affiliation has been a particularly exciting area of research in recent years (Goodson and Bass, 2001; Donaldson and Young, 2008; Kelly et al., 2011). The neuropeptides vasopressin and oxytocin have been well-studied in mediating pair bonding in voles (Young and Wang, 2004), and the fish homologs vasotocin and isotocin have been implicated in social approach and reproductive behavior in fish (Goodson and Bass, 2000; Thompson and Walton, 2004). Social behavior and affiliation are also linked with reward circuitry (Young and Wang, 2004) and have a strong neuroendocrine component (Adkins-Regan, 2009). Therefore we categorized vasotocin, isotocin, tyrosine hydroxylase (TH; a rate-limiting enzyme in catecholamine synthesis often used as a marker for dopamine synthesis; Masserano and Weiner, 1983), and the highly expressed teleost-specific brain isoform of aromatase [the enzyme that converts testosterone to estradiol (brain aromatase (cyp19a1b); Pasmanik and Callad, 1985)] as being associated with the social/affiliation components of mate choice behavior (Figure 1).
In the current experiment, TH and brain aromatase (also categorized under the female receptivity/reproduction subunit of social behavior) fell into the preference and reproduction cluster associated with SS-exposure. Meanwhile, vasotocin and isotocin loaded almost exclusively along the Gene 2 axis, and oriented toward the group containing favorable sexual (large males) stimuli. Furthermore, isotocin and vasotocin and the SS gene cluster orient along a continuum between attractive (LL) and unattractive (SS) males. The clear distinction in multidirectional space of these two “social affiliation” genes from the mate preference/plasticity gene suites suggests that general social affiliation pathways may differ from those modulating a more specific form of social behavior–mate preference.
This differential expression of isotocin and vasotocin vs. the bulk of the preference-associated gene clusters is intriguing. Both are known to modulate social behaviors, and in particular alterations in vasotocin mRNA abundance and localization have been associated with the rapid behavioral responses associated with changes in social status (e.g., dominant/subordinate transitions in cichlids; Greenwood et al., 2008; Renn et al., 2008), or socially induced sex-change in some fish species (for review see Godwin, 2010). However, to our knowledge, neither vasotocin nor isotocin transcriptional abundance has been directly implicated in a cascade modulating social behavior on this sort of time scale (30 min). Future studies should directly test dynamic transcriptional regulation of these two peptides in female preference behavior.
In contrast to the mate preference- and affiliation-associated genes, there was an absence of spatial patterning for genes categorized under the receptivity/reproduction aspect of social behavior. We looked at four genes representing aspects of neurological pathways underlying female receptivity (Figure 1). Best-studied in rodents, the catecholaminergic system is critical for female sexual behavior in several vertebrate taxa (Everitt et al., 1975; Meston et al., 1996; Etgen et al., 1999, 2001; Becker et al., 2001; Riters et al., 2007), and β-1 adrenergic receptor was previously identified with male exposure and female receptivity displays in X. nigrensis (Cummings et al., 2008). Therefore, we utilized TH and representatives from the two main adrenergic receptor groups, β-1 adrenergic receptor and α-1 adrenergic receptor, as representatives of genes involved in female sexual behavior. We also cross-categorized brain aromatase. Female reproductive behavior and neurophysiology is closely linked with steroid hormones, particularly estrogen (for a review see Flanagan-Cato, 2011), and in fish the brain isoform of aromatase is associated with sexual behaviors and arousal (Forlano et al., 2006; Forlano and Bass, 2011).
We found no distinct multidimensional clustering for genes associated with female sexual behaviors, presumably due to considerable overlap between sociality, mate preference and female reproduction. There are extensive neuroendocrine contributions to sensory modulation and synaptic plasticity (Etgen et al., 1999; Forlano et al., 2006; Hojo et al., 2008; Srivastava et al., 2008; Maney and Pinaud, 2011), and many genes associated with sociality/affiliation, preference, and sexual behaviors are steroid-sensitive (Witt, 1995; Adkins-Regan, 1998; Gimpl and Gahreholz, 2001; Riters et al., 2007). β-1 adrenergic receptor expression (and to a lesser extent TH) was closely associated with brain aromatase and the neural plasticity-associated neurosperpin along the Gene 2 axis while α-1 adrenergic receptor expression was not associated with brain aromatase and instead clustered along with other preference/synaptic plasticity-associated genes oriented away from the LS group along Gene 1. In the absence of sexual contact, female reproduction-associated gene expression may reflect a more diffuse, motivation toward sociality that helps to modulate context-specific expression of appropriate genes and behaviors.
Multivariate Correlations between Behavior and Gene Expression
While “mate preference” is a subset of “social affiliation,” it appears to evoke a more salient behavioral and genetic response in X. nigrensis females than more general social affiliation. The dominant axes for both the behavioral and gene MDA analyses captured the variance associated with preference or choice vs. no choice social contexts (Figure 3), and in this study the most prominent differences in gene expression were found between a high mate choice context (LS) and non-mate choice context (FF). This result is consistent with our previous microarray study that found significant genomic (>0300 genes) differences that were greatest between LS and FF social contexts across an LS, FF, SS, and asocial comparison (Cummings et al., 2008). Previous research has also revealed significant correlations between the expression patterns of several candidate preference-associated genes and individual variation in mate preference behaviors within male-exposed groups (Cummings et al., 2008; Lynch et al., 2012). In the current study, we explored the relationship between genes and behavior between social contexts and found correlations between multivariate patterns of social behaviors and gene expression that were identifiable only when evaluated by social context.
While individual behaviors showed some insight into the patterns of gene expression (Table 3), greater statistical power was found by examining the relationship between the multidimensional axes of behavior and gene expression (Figure 4). Multivariate behavior and gene expression scores clustered by social context (Figure 4), and reflect exposure to the four different social groups. The primary MDA axes, Behavior 1 and Gene 1, were positively correlated and dominated by preference behaviors toward attractive males coupled with lower expression of mate preference-associated gene suites in the presence of attractive males (Figures 3 and 4). This pattern is consistent with our earlier microarray results (Cummings et al., 2008), and may capture aspects of a release from inhibition in the presence of an attractive male. The secondary behavior and gene axes were also positively correlated, and were dominated by presence or absence of an aversive stimulus. The Behavior 2 and Gene 2 axes captured overall sociality measures (time or activity) that were consistent with an expression-level signature of affiliative and sociosexual-associated gene suites. In the absence of small males, females were more willing to spend time associating with social stimuli, and this tendency was reflected on a gene level with higher isotocin and vasotocin. On the aversive end of the axes (SS), females were more active, with higher brain aromatase, β-1 adrenergic receptor, and neuroserpin. The context-specific relationship between active behavior measures and neuroserpin expression is particularly interesting given its known association with exploratory behavior and neophobia in mice (Madani et al., 2003). Future studies comparing suites of candidate female social/sexual genes with randomly selected genes may provide further verification of these context-specific correlations, including the distinction between mate preference- and social affiliation-dominated axes.
Conclusion
Female behaviors were context-specific across a variety of social conditions that included both gradations of male stimulus pairs (male exposure groups of differing complexities) as well as female conspecific exposure. Multiple discriminant analysis (MDA) of 8 behaviors and 11 genes representing three spheres of female social behavior (mate preference, social/affiliative, and sexual behavior) parsed most of the between-group variation into primary axes linked to preference and social discrimination including discrimination based on mate choice complexity. Further, we found greater distinctions between social context at the molecular (genetic expression) level than at the behavioral level, suggesting that multivariate behavioral genomics approaches may be able to identify subtle differences in brain responses based on context that are not detectable at a behavioral level. Our prediction that mate preference would represent a distinct subset of a gene–behavior complex from that of general affiliative interactions was met as the mate preference axes were distinct from secondary axes that captured more general social and reproduction-associated gene–behavior suites. The distinction between mate preference and synaptic plasticity-associated genes from general affiliative measures suggests that precopulatory mate discrimination may be mediated by gene modules distinct from general social affiliation. Further, multivariate behavior and gene measure were significantly correlated only when differential social exposure was considered as an explicit factor, suggesting that gene–behavior suites covary in a socially relevant context.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank undergraduate students Katherine Chastain, Wendy Vu, Maneesh Anand, and Luis Betencourt for their invaluable contributions to this work. We thank members of the Cummings lab for helpful discussions, and wish to particularly acknowledge Kathleen Lynch for helpful comments on data interpretation and earlier versions of this manuscript. We also thank the Mexican government for collecting permits (DGOPA07311-13709-2261) and the Brackenridge Field Laboratory at the University of Texas at Austin for animal care facilities. This work was supported by NSF SGER IOS-0813742 and NSF IOS-0843000 to Molly E. Cummings.
Footnotes
References
Aubin-Horth, N., and Renn, S. C. P. (2009). Genomic reaction norms: using integrative biology to understand molecular mechanisms of phenotypic plasticity. Mol. Ecol. 18, 3763–3780.
Basolo, A. (2004). Variation between and within the sexes in body size preference. Anim. Behav. 68, 75–82.
Becker, J. B., Rudick, C. N., and Jenkins, W. J. (2001). The role of dopamine in the nucleus accumbens and striatum during sexual behavior in the female rat. J. Neurosci. 21, 3236–3241.
Benjamini, Y., Drai, D., Elmer, G., Kafkafi, N., and Golani, I. (2001). Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 125, 279–284.
Biswas, S., Reinhard, J., Oakeshott, J., Russell, R., Srinivasan, M. V., and Claudianos, C. (2010). Sensory regulation of neuroligins and neurexin 1 in the honeybee brain. PLoS ONE 5, e9133. doi:10.1371/journal.pone.0009133
Borges, V. M., Lee, T. W., Christie, D. L., and Birch, N. P. (2010). Neuroserpin regulates the density of dendritic protrusions and dendritic spine shape in cultured hippocampal neurons. J. Neurosci. Res. 88, 2610–2617.
Cummings, M. E., Garcia de Leon, F. J., Mollaghan, D. M., and Ryan, M. J. (2006). Is UV ornamentation an amplifier in swordtails? Zebrafish 3, 91–100.
Cummings, M. E., Larkins-Ford, J., Reilly, C. R. L., Wong, R. Y., Ramsey, M. E., and Hofmann, H. A. (2008). Sexual and social stimuli elicit rapid and contrasting genomic responses. Proc. R. Soc. Lond. B Biol. Sci. 275, 393–402.
Cummings, M. E., and Mollaghan, D. (2006). Repeatability and consistency of female preference behaviors in a northern swordtail, Xiphophorus nigrensis. Anim. Behav. 72, 217–224.
Cummings, M. E., Rosenthal, G. G., and Ryan, M. J. (2003). A private ultravioloet channel in visual communication. Proc. R. Soc. Lond. B Biol. Sci. 270, 897–904.
Donaldson, Z. R., and Young, L. J. (2008). Oxytocin, vasopressin, and the neurogenetics of sociality. Science 322, 900–904.
Ellis, L. L., and Carney, G. E. (2009). Drosophila melanogaster males respond differently at the behavioural and genome-wide levels to Drosophila melanogaster and Drosophila simulans females. J. Evol. Biol. 22, 2183–2191.
Etgen, A. M., Ansonoff, M. A., and Quesada, A. (2001). Mechanisms of ovarian steroid regulation of norphinephrine receptor-mediated signal transduction in the hypothalamus: implications for female reproductive physiology. Horm. Behav. 40, 169–177.
Etgen, A. M., Chu, H.-P., Fiber, J. M., Karkanias, G. B., and Morales, J. M. (1999). Hormonal integration of neurochemical and sensory signals governing female reproductive behavior. Behav. Brain Res. 105, 93–103.
Everitt, B. J., Fuxe, K., Hokfelt, T., and Jonsso, G. (1975). Role of monoamines in the control by hormones of sexual receptivity in the female rat. J. Comp. Physiol. Psychol. 89, 556–572.
Filby, A. L., Paull, G. C., Hickmore, T. F. A., and Tyler, C. R. (2010). Unravelling the neurophysiological basis of aggression in a fish model. BMC Genomics 11, 498 doi:10.1186/1471-2164-11-498
Flanagan-Cato, L. M. (2011). Sex differences in the neural circuit that mediates female sexual receptivity. Front. Neuroendocrinol. 32, 124–136.
Forlano, P. M., and Bass, A. H. (2011). Neural and hormonal mechanisms of reproductive-related arousal in fishes. Horm. Behav. 59, 616–629.
Forlano, P. M., Schlinger, B. A., and Bass, A. H. (2006). Brain aromatase: new lessons from non-mammalian model systems. Front. Neuroendocrinol. 27, 247–274.
Gimpl, G., and Gahreholz, F. (2001). The oxytocin receptor system: structure, function, and regulation. Physiol. Rev. 81, 629–683.
Godwin, J. (2010). Neuroendocrinology of sexual plasticity in teleost fishes. Front. Neuroendocrinol. 31, 203–216.
Goodson, J. L., and Bass, A. H. (2000). Forebrain peptides modulate sexually polymorphic vocal circuitry. Nature 403, 769–772.
Goodson, J. L., and Bass, A. H. (2001). Social behavior functions and related anotomical characteristics of vasotocin/vasopressin systems in vertebrates. Brain Res. Rev. 35, 246–265.
Greenwood, A. K., Wark, A. R., Fernald, R. D., and Hofmann, H. A. (2008). Expression of arginine vasotocin in distinct preoptic regions is associated with dominant and subordinate behaviour in an African cichlid fish. Proc. R. Soc. Lond. B. Biol. Sci. 275, 2393–2402.
Hojo, Y., Murakami, G., Mukai, H., Higo, S., Hatanaka, Y., Ogiue-Ikeda, M., Ishii, H., Kimoto, T., and Kawato, S. (2008). Estrogen synthesis i the brain – Role in synaptic plasticity and memory. Mol. Cell. Endocrinol. 290, 31–43.
Hunt, G. J., Amdam, G. V., Schlipalius, D., Emore, C., Sardesai, N., Williams, C. E., Rueppel, O., Guzman-Novoa, E., Arechavaleta-Velasco, M., Chandra, S., Fondrk, M. K., Beye, M., and Page, R. E. (2007). Behavioral genomics of honeybee foraging and nest defense. Naturwissenschaften 94, 247–267.
Kelly, A. M., Kingsbury, M. A., Hoffbuhr, K., Schrock, S. E., Waxman, B., Kabelik, D., Thompson, R. R., and Goodson, J. L. (2011). Vasotocin neurons and septal V1a-like receptors potently modulate songbird flocking and responses to novelty. Horm. Behav. 60, 12–21.
Lynch, K. S., Ramsey, M. E., and Cummings, M. E. (2012). The mate choice brain: comparing gene profiles between female mate choice and male coercive poecillids. Genes Brain Behav. 11, 222–229.
Madani, R., Kozlov, S., Akhmedov, A., Cinelli, P., Kinter, J., Lipp, H.-P., Sonderegger, P., and Wolfer, D. P. (2003). Impaired explorative behavior and neophobia in genetically modified mice lacking or overexpressing the extracellular serine protease inhibitor neuroserpin. Mol. Cell. Neurosci. 23, 473–494.
Maney, D., and Pinaud, R. (2011). Estradiol-dependent modulation of auditory processing and selectivity in songbirds. Front. Neuroendocrinol. 32, 287–302.
Martel, G., Nishi, A., and Shumyatsky, G. P. (2008). Stathmin reveals dissociable roles of the basolateral amygdala in parental and social behaviors. Proc. Natl. Acad. Sci. U.S.A. 105, 14620–14625.
Masserano, J. M., and Weiner, N. (1983). Tyrosine hydroxylase regulation in the central nervous system. Mol. Cell. Biochem. 53/54, 129–152.
McLachlan, G. (2005). Discriminant Analysis and Statistical Pattern Recognition. Hoboken: John Wiley & Sons.
Meston, C., Moe, I., and Gorzalka, B. B. (1996). Effects of sympathetic inhibition on receptive, proceptive, and rejection behaviors in the female rat. Physiol. Behav. 59, 537–542.
Miranda, E., and Lomas, D. A. (2006). Neuroserpin: a serpin to think about. Cell. Mol. Life Sci. 63, 709–722.
Morris, M. R., Rios-Cardenas, O., and Tudor, M. S. (2006). Larger swordtail females prefer asymmetrical males. Biol. Lett. 2, 8–11.
Mukai, M., Replogle, K., Drnevic, J., Wang, G., Wacker, D. W., Band, M., Clayton, D. F., and Wingfield, J. C. (2009). Seasonal differences of gene expression profiles in song sparrow (Melospiza melodia) hypothalamus in relation to territorial aggression. PLoS ONE 4, e8182. doi:10.1371/journal.pone.0008182
O’Connell, L. A., and Hofmann, H. A. (2011). Genes, hormones and circuits: an integrative approach to study the evolution of social behavior. Front. Neuroendocrinol. 32, 320–335.
Ophir, A. G., and Galef, B. G. (2004). Sexual experience can affect use of public information in mate choice. Anim. Behav. 68, 1221–1227.
Pasmanik, M., and Callad, G. V. (1985). Aromatase and 5a-reductase in the teleost brain, spinal cord, and pituitary gland. Gen. Comp. Endocrinol. 60, 244–251.
Pawlak, R., Magarinos, A. M., Melchor, J., McEwan, B., and Strickland, S. (2003). Tissue plasminogen activator in the amygdala is critical for stress-induced anxiety-like behavior. Nat. Neurosci. 6, 168–174.
Perrault, W. D., Behrman, D. N., and Armstrong, G. M. (1979). Alternative approaches for interpretation of multivariate discriminant analysis in marketing research. J. Bus. Res. 7, 343–346.
Ramsey, M. E., Wong, R. Y., and Cummings, M. E. (2011). Estradiol, reproductive cycle and preference behavior in a northern swordtail. Gen. Comp. Endocrinol. 170, 381–390.
Reidel, G., Platt, B., and Micheau, J. (2003). Glutamate receptor function in learnign and memory. Behav. Brain Res. 140, 1–47.
Renn, S. C. P., Aubin-Horth, N., and Hofmann, H. A. (2008). Fish and chips: functional genomics of social plasticity in an African cichlid fish. J. Exp. Biol. 211, 3041–3056.
Riters, L. V., Olesen, K. M., and Auger, C. J. (2007). Evidence that female endocrine state influences catecholamine responses to male courtship song in European starlings. Gen. Comp. Endocrinol. 154, 137–149.
Robinson, G. E., Fernald, R., and Clayton, D. F. (2008). Genes and social behavior. Science 322, 896–900.
Ryan, M. J., and Causey, B. A. (1989). “Alternative” mating behavior in the swordtails Xiphophorus nigrensis and Xiphophorus pygmaeus (pisces: poeciliidae). Behav. Ecol. Sociobiol. (Print) 24, 341–348.
Ryan, M. J., and Wagner, W. E. (1987). Asymmetries in mating preferences between species: female swordtails prefer heterospecific males. Science 236, 595–597.
Ryan, M. J., and Rosenthal, G. G. (2001). “Variation and selection in swordtails,” in Model Systems in Behavioral Ecology, ed. L. A. Dugatkin (Princeton, NJ: Princeton University Press), 133–148.
Samson, A. L., and Medcalf, R. L. (2006). Tissue-type plasminogen activator: a multifaceted modulator of neurotransmission and synaptic plasticity. Neuron 50, 673–678.
Srivastava, D. P., Woolfrey, K., Jones, K. A., Shum, C. Y., Lash, L. L., Swanson, G. T., and Penzes, P. (2008). Rapid enhancement of two-step wiring plasticity by estrogen and NMDA receptor activity. Proc. Natl. Acad. Sci. U.S.A. 105, 14650–14655.
Sudhof, T. C. (2008). Neuroligins and neurexins link synaptic function to cognitive disease. Nature 455, 903–911.
Tabuchi, K., Blundell, J., Etherton, M. R., Hammer, R. E., Liu, X., Powell, C. M., and Sudhof, T. (2007). A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science 318, 71–76.
Thompson, R. R., and Walton, J. C. (2004). Peptide effects on social behavior: effects of vasotocin and isotocin on social approach behavior in male goldfish (Carassius auratus). Behav. Neurosci. 118, 620–626.
Toth, A. L., Varala, K., Newman, T. C., Miguez, F. E., Hutchison, S. K., Willoughby, D. A., Simons, J. F., Egholm, M., Hunt, J. H., Hudson, M. E., and Robinson, G. E. (2007). Wasp gene expression supports an evolutionary link between maternal behavior and eusociality. Science 318, 441–444.
Watson, C. J. (1982). Approaches for the intrepetation of mulitple discriminant analysis in organizational research. Acad. Manage Rev. 7, 124–132.
White, D. J. (2004). Influences of social learning on mate-choice decisions. Learn. Behav. 32, 105–113.
Witt, D. (1995). Oxytocin and rodent sociosexual responses: from behavior to gene expression. Neurosci. Biobehav. Rev. 19, 315–324.
Witte, K., and Noltemeier, B. (2002). The role of information in mate-choice copying in female sailfin mollies (Poecilia latipinna). Behav. Ecol. Sociobiol. (Print) 52, 194–202.
Wong, R. Y., and Hofmann, H. A. (2010). Behavioural genomics: an organismic perspective. Encyclopedia Life Sci. doi: 10.1002/9780470015902.a0022554
Wong, R. Y., So, P., and Cummings, M. E. (2011). How female size and male displays influence mate preference in a swordtail. Anim. Behav. 82, 691–697.
Yepes, M., and Lawrence, D. A. (2004). Tissue-type plasminogen activator and neuroserpin: a well-balanced act in the nervous system? Trends Cardiovasc. Med. 14, 173–180.
Appendix
Keywords: mate choice, female preference, social behavior, poeciliid, sexual selection, multivariate
Citation: Ramsey ME, Maginnis TL, Wong RY, Brock C and Cummings ME (2012) Identifying context-specific gene profiles of social, reproductive, and mate preference behavior in a fish species with female mate choice. Front. Neurosci. 6:62. doi: 10.3389/fnins.2012.00062
Received: 19 January 2012; Accepted: 09 April 2012;
Published online: 01 May 2012.
Edited by:
David F. Clayton, University of Illinois, USAReviewed by:
Genevieve Konopka, University of Texas Southwestern Medical Center, USAKa Wan Li, VU University, Netherlands
Copyright: © 2012 Ramsey, Maginnis, Wong, Brock and Cummings. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Mary E. Ramsey, Section of Integrative Biology, University of Texas, 1 University Station C0930, Austin, TX 78712, USA. e-mail: mramsey@austin.utexas.edu