- 1Department of Molecular Biology and Genetics, Democritus University of Thrace, Alexandroupoli, Greece
- 2Applied Human Molecular Genetics, Kennedy Center, Department of Clinical Genetics, Copenhagen University Hospital, Rigshospitalet, Glostrup, Denmark
- 3Danish Headache Center and Department of Neurology, Faculty of Health Sciences, University of Copenhagen, Glostrup Hospital, Glostrup, Denmark
- 4Institute of Biological Psychiatry, Mental Health Center Sct. Hans, Copenhagen University Hospital, Roskilde, Denmark
- 5Department of Biological Sciences, Purdue University, West Lafayette, IN, USA
Gilles de la Tourette syndrome (GTS) is a neuropsychiatric disorder characterized by multiple motor and vocal tics. GTS is a complex disorder, with environmental factors and several genes involved. Although variations within a few genes such as AADAC, NRXN1, SLITRK1, HDC, and IMMP2L have been tentatively associated with GTS (in a small number of patients), the causative genes underlying GTS pathophysiology remain unknown. In a previous genome-wide association study (GWAS) a single nucleotide polymorphism (SNP, rs2060546) near the Netrin-4 (NTN4 - MIM 610401) gene was shown to be associated with GTS [odds ratio (OR) = 1.7; p-value = 5.8 × 10-7] thus warranting further investigations. As NTN4 is one of the axon guidance molecules expressed in the central nervous system and it interacts with the encoded proteins of SLIT and WNT genes guiding the growth cone toward its target, it is an attractive candidate susceptibility gene for GTS. In this study we attempted to replicate the association of rs2060546 with GTS by genotyping a Danish cohort of 240 GTS patients and 1006 healthy controls. Our results did not reveal an association (OR = 1.363; p-value = 0.3329) in the Danish cohort alone, which may be due to the small sample size. However, a meta-analysis including the present cohort and a total of 1316 GTS patients and 5023 controls from the GTS GWAS Replication Initiative (GGRI) and the first GTS-GWAS yielded a significant signal (OR = 3.74; p-value = 0.00018) and same direction of effect in the three cohorts. Thus, our study strengthens the evidence of the possible involvement of NTN4 in GTS etiology, suggesting that further studies in even larger samples and functional studies are warranted to investigate the role of this region in GTS pathogenesis.
Introduction
Gilles de la Tourette syndrome (GTS) is a complex juvenile-onset neuro-developmental disorder characterized by the occurrence of multiple motor and vocal tics (Nag et al., 2013). Recent epidemiological studies estimated the worldwide prevalence of GTS to be approximately 1% with a male:female ratio of 4:1 (Robertson et al., 2009). GTS is often associated with co-morbidities such as attention-deficit hyperactivity disorder and obsessive-compulsive disorder (Dietrich et al., 2014). Additional co-occurring conditions are behavioral and emotional difficulties (hyperactivity, anxiety, and depression), sleeping disturbances, intellectual disabilities and autism spectrum disorder (Singer and Rosenberg, 1988; Wood et al., 2003). Studies so far conducted on GTS showed that there is a complex interplay of environmental and genetic factors, confirming the notion that GTS is a highly complex disorder (Davis et al., 2013; Dietrich et al., 2014).
To identify the causative genes and the biological pathways involved in GTS several approaches have been pursued, including candidate gene studies, family studies using linkage analysis, analysis of chromosomal abnormalities including copy number variants (CNV) and hypothesis-free genome-wide association studies (GWAS) (Paschou, 2013). For instance, chromosomal abnormalities have proven useful for identifying new candidate genes in GTS affected patients (Bertelsen et al., 2013). Chromosomal abnormalities have probed new candidate regions containing susceptible genes such as Slit- and Ntrk-Like Family, Member 1 (SLITRK1—MIM 609678), Neuroligin 4 (NLGN4—MIM 300427) and Contactin-Associated Protein-Like2 (CNTNAP2—MIM 604569) (Verkerk et al., 2003; Abelson et al., 2005; Lawson-Yuen et al., 2008; Patel et al., 2011). Specific CNV analyses have resulted in the identification of several other candidate genes, including Arylacetamide Deacetylase (AADAC—MIM 600338), Collagen, Type VIII, Alpha-1 (COL8A1—MIM 120251), Neurexin I (NRXN1—MIM 00565), Catenin Alpha-3 (CTNNA3—MIM 607667) and Inner Mitochondrial Membrane Peptidase, Subunit 2, (IMMP2L—MIM 605977) (Sundaram et al., 2010; Nag et al., 2013; Bertelsen et al., 2014, 2015). The first large GWAS for GTS did not identify any genome-wide significance SNP (Scharf et al., 2013). A multinational consortium, GTS GWAS Replication Initiative (GGRI), followed up on the results of the initial GWAS in an independent cohort of 636 cases and 641 controls, showing an association between GTS and rs2060546 (OR = 2.41; p-value = 5.8 × 10−7) on chromosome 12q23.1, ~32 kb upstream to the transcriptional start site of Netrin-4 (NTN4); a gene encoding an axon guidance protein expressed in the central nervous system (Paschou et al., 2014). As NTN4 is an attractive candidate susceptibility gene, we investigated the association of rs2060546 with GTS in a Danish cohort of 240 GTS patients and 1006 healthy controls.
Materials and Methods
Samples
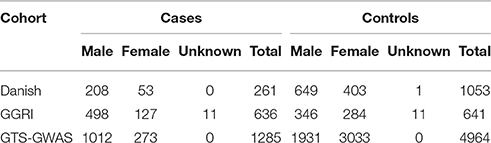
We investigated a Danish cohort and two previously published datasets (GGRI and GTS-GWAS) (Table 1). The first cohort (n = 1314) includes 261 deeply phenotyped GTS patients and 1053 healthy controls from Denmark. Danish ancestry was investigated using the genotype information. The second cohort from GGRI includes 1277 individuals (636 GTS cases and 641 healthy controls) from different European populations (Paschou et al., 2014). The third cohort (GTS-GWAS) includes 6249 individuals (1285 GTS cases and 4964 controls) of European ancestry (Scharf et al., 2013).
Genotyping
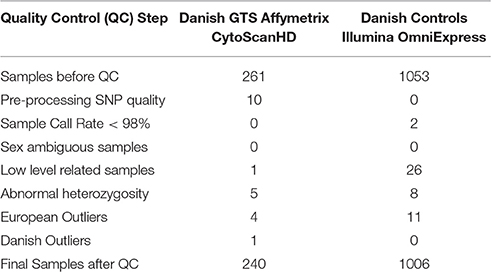
261 GTS cases from Denmark were genotyped on the Affymetrix CytoScanHD array (Affymetrix, Santa Clara, CA) covering around 750,000 single nucleotide polymorphisms (SNPs) and ~1.9 million single-locus copy number (CN) markers. 1053 healthy controls were genotyped on Illumina HumanOmniExpress arrays—18 on HumanOmniExpress12v1 and 1034 on HumanOmniExpress12v1-1 covering around 700,000 SNPs each. SNP calling and pre-processing of the raw data for Illumina controls were carried out using Illumina Genomestudio® software. For GTS cases genotyped on Affymetrix CytoScanHD SNP calling a pre-processing was carried out using default parameters for method apt-copynumber-cyto from Affymetrix Power Tools (APT, version 1.16) software.
Quality Control
A standard GWAS quality control measure (Scharf et al., 2013) was applied on both cases and controls from the Danish cohort to filter for population outliers and samples with low call rate; and to remove systematic bias using the software PLINK (Purcell et al., 2007). Quality control at SNP level includes removal of monomorphic SNPs, SNPs with genotyping rate <98%, SNPs with no information about chromosome location, SNPs with absolute minor allele frequency difference > 0.15, SNPs that fail Hardy-Weinberg Equilibrium and SNPs with AT/CG alleles. To check the quality of the Danish GTS cases metric values for CytoScanHD array recommended by the manufacture was used in the pre-processing step. Sample call rate calculated using PLINK (Purcell et al., 2007) was used to filter out samples that had more than 2% of SNPs missing. F statistic was calculated using SNPs from the X-chromosome. F <0.25 was assigned to female and F > 0.75 was assigned to male. Samples with an F-value between 0.25 and 0.75 were discarded due to sex ambiguity. Estimates of pairwise Identity-by-descent from PLINK (Purcell et al., 2007) were used to remove one of the samples from each pair which pass the relatedness metrics either pi-hat > 0.1 or Z1 > 0.2. Samples with high rates of heterozygosity Fhet > ± 0.05 are more likely to be result of contamination and were removed from the analysis. Principal Component Analysis (PCA) was applied using EIGENSTRAT (Price et al., 2006) and remaining samples were merged with the 1000 genomes cohort to remove European cluster and population outliers. Out of 261 GTS cases, four samples were removed as they did not lie inside the European cluster in Figure 1. Eleven healthy controls lied outside of the European cluster in Figure 2 and were removed from the association analysis. After removing the European outliers, Danish GTS cases and healthy controls were merged for 126,821 common SNPs. The PCA plot in Figure 3 was made using a LD-pruned set of ~90,000 SNPs to identify Danish outliers. One GTS case was removed as population outlier seen in Figure 3 which left in total 240 Danish GTS cases and 1006 healthy controls.
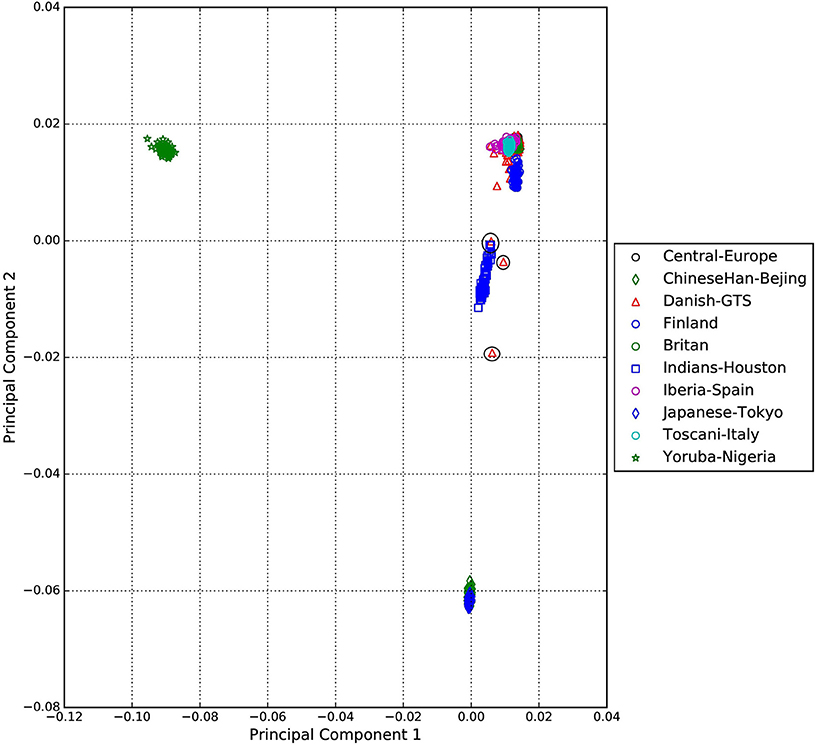
Figure 1. Two dimensional PCA plot of Danish GTS cases with selected populations from 1000genomes cohort.
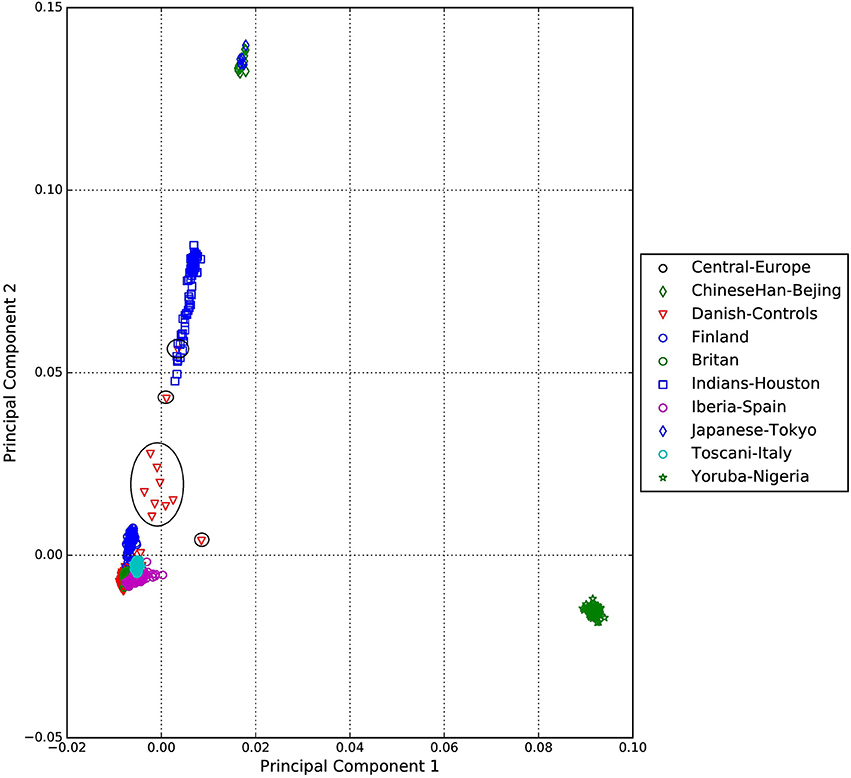
Figure 2. Two dimensional PCA plots of Danish controls and selected populations of 1000genomes cohort.
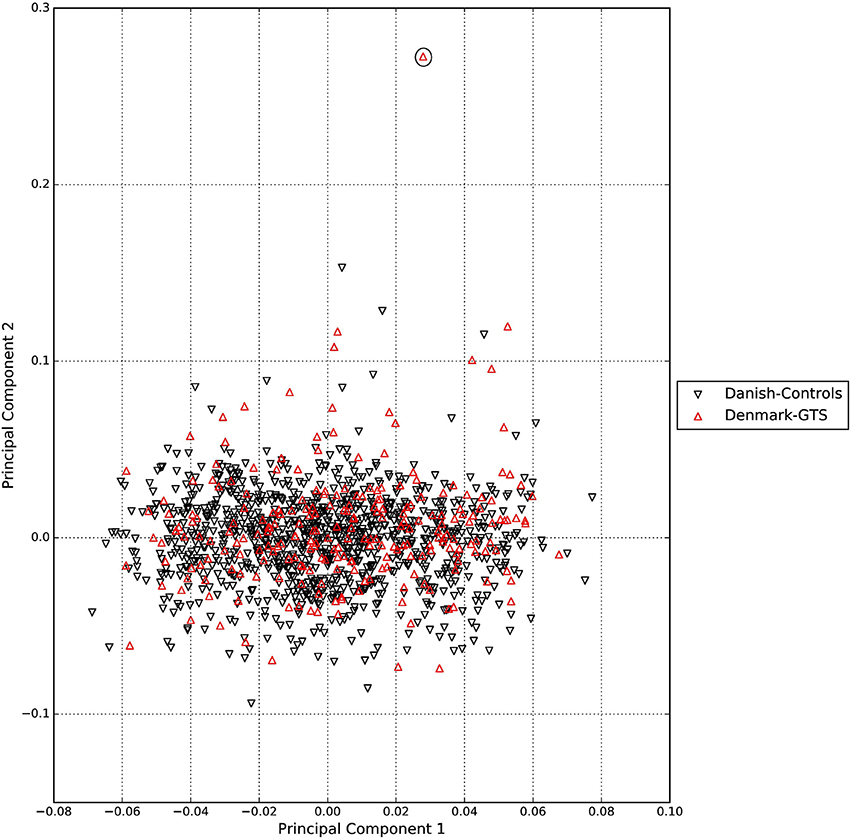
Figure 3. Two dimensional PCA plot of Danish GTS cases and controls to check for population outliers.
Association Analysis
Standard case control association analysis using PLINK (Purcell et al., 2007) was performed using χ2 test comparing SNP frequency between cases and controls.
Meta-Analysis
Metal (Willer et al., 2010) was used to perform sample weighted meta-analysis of association results of the SNP rs2060546 from all three cohorts. Association results for the GGRI and GTS-GWAS cohorts were taken from the original studies (Scharf et al., 2013; Paschou et al., 2014).
Results
We attempted to replicate the association of SNP rs2060546 with GTS by investigating quality control passed genotyped data of 240 GTS patients and 1006 healthy controls from Denmark (Table 2). We did not find the SNP rs2060546 to be significantly associated in the Danish cohort (OR = 1.363, 95% CI 0.7188-2.675; p-value = 0.3329) (Table 3). This might be attributed to the small size of the cohort and we proceeded to perform a meta-analysis with previously published data (Paschou et al., 2014). However, meta-analysis of the Danish, the GGRI and the first GTS-GWAS cohorts, showed significant association to the studied SNP (OR = 3.74; p-value = 0.00018) and the same direction of effect in all three cohorts (Table 3). Thus, the results supported the involvement of this particular chromosomal region in GTS etiology, and potentially NTN4.
Discussion
Involvement of NTN4 in neurodevelopment makes it an attractive candidate protein as a contributing factor to GTS pathology. During the development of the nervous system, numerous dynamic guidance cues direct the trajectory of the migrating developed axon toward its suitable target (Killeen and Sybingco, 2008). Netrins are axon guidance cues which are composed of six members: Netrin 1-4 Netrin-related molecules, Netrin-G1 and G2 (Davis et al., 2013). Netrin-4 functions as guidance cue for axonal growth, neurite elongation, neuronal remodeling and plasticity (Zhang et al., 2004). The neuronal growth cones sense Netrin-4 as either attractant or repellent cues, depending on different receptors expressed on their surface or differences in the cellular signal transduction machinery (Koch et al., 2000; Qin et al., 2007). Netrin-4 might mediate axon outgrowth by attracting actions through deleted in colorectal carcinoma (DCC) or neogenin (Neo 1) receptors and repulsive effects through unc-5 homolog (UNC5A) receptors (Qin et al., 2007; Schubert et al., 2009). Previous imaging studies showed that changes in the thalamus and cerebral cortex volumes play a significant role in regulating the severity of tic symptoms implicated in GTS cases (Rueda et al., 2005; Rickards et al., 2008). Notably, expression of the NTN4 gene was detected in human brain regions such as cerebellum, thalamus, cerebral cortex and olfactory bulb, thus, supporting the role of this gene in GTS pathogenesis (Zhang et al., 2004; Amat et al., 2006; Lee et al., 2006).
In this study, we could not identify any association between rs2060546 SNP and GTS in a relatively small Danish cohort alone, however, meta-analyses with the GGRI and GTS-GWAS cohorts provides further support for the possible association of this SNP with GTS in European populations. The close localization of rs2060546 to NTN4 and the abundant expression of Netrin-4 in different brain regions, associated with GTS pathogenesis, rendered further support to the hypothesis that NTN4 might be a new candidate gene for GTS.
Author Contributions
SP, performed data analysis, interpretation, and participated in manuscript writing. ZT and PP supervised the study, interpretation of the results, and participated in manuscript writing. RH participated in biological relevance of result and manuscript writing. FT participated in data analysis and manuscript writing. AE, JO, TW, TH, ZT, BB provided the data and participated in manuscript writing. All authors read and approved the final version to be published and agreed to be accountable for all aspects of the work.
Funding
This project was supported by FP7-People-2012-ITN, project: TS-EUROTRAIN, grant number 316978; and Lundbeck Foundation, grant number R24-A2419 and R100-A9332.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank all individuals who participated and contributed to this work.
References
Abelson, J. F., Kwan, K. Y., Roak, B. J. O., Baek, D. Y., Stillman, A. A., Morgan, T. M., et al. (2005). Sequence Variants in SLITRK1 are associated with Tourette' s Syndrome. Science 2126, 317–320. doi: 10.1126/science.1116502
Amat, J. A., Bronen, R. A., Saluja, S., Sato, N., Zhu, H., Gorman, D. A., et al. (2006). Increased number of subcortical hyperintensities on MRI in children and adolescents with Tourette's syndrome, obsessive-compulsive disorder, and attention deficit hyperactivity disorder. Am. J. Psychiatry 163, 1106–1108. doi: 10.1176/ajp.2006.163.6.1106
Bertelsen, B., Debes, N. M., Hjermind, L. E., Skov, L., Brøndum-Nielsen, K., and Tümer, Z. (2013). Chromosomal rearrangements in Tourette syndrome: implications for identification of candidate susceptibility genes and review of the literature. Neurogenetics 14, 197–203. doi: 10.1007/s10048-013-0372-y
Bertelsen, B., Melchior, L., Jensen, L. R., Groth, C., Glenthøj, B., Rizzo, R., et al. (2014). Intragenic deletions affecting two alternative transcripts of the IMMP2L gene in patients with Tourette syndrome. Eur. J. Hum. Genet. 22, 1283–1289. doi: 10.1038/ejhg.2014.24
Bertelsen, B., Stefánsson, H., Riff Jensen, L., Melchior, L., Mol Debes, N., Groth, C., et al. (2015). Association of AADAC Deletion and Gilles de la Tourette Syndrome in a Large European Cohort. Biol. Psychiatry 79, 383–391. doi: 10.1016/j.biopsych.2015.08.027
Davis, L. K., Yu, D., Keenan, C. L., Gamazon, E. R., Konkashbaev, A. I., Derks, E. M., et al. (2013). Partitioning the Heritability of Tourette Syndrome and obsessive compulsive disorder reveals differences in Genetic Architecture. PLoS Genet. 9:e1003864. doi: 10.1371/journal.pgen.1003864
Dietrich, A., Fernandez, T. V., King, R. A., State, M. W., Tischfield, J. A., Hoekstra, P. J., et al. (2014). The Tourette International Collaborative Genetics (TIC Genetics) study, finding the genes causing Tourette syndrome: objectives and methods. Eur. Child Adolesc. Psychiatry 24, 141–151. doi: 10.1007/s00787-014-0543-x
Killeen, M. T., and Sybingco, S. S. (2008). Netrin, Slit and Wnt receptors allow axons to choose the axis of migration. Dev. Biol. 323, 143–151. doi: 10.1016/j.ydbio.2008.08.027
Koch, M., Murrell, J. R., Hunter, D. D., Olson, P. F., Jin, W., Keene, D. R., et al. (2000). A novel member of the netrin family, beta-netrin, shares homology with the beta chain of laminin: identification, expression, and functional characterization. J. Cell Biol. 151, 221–234. doi: 10.1083/jcb.151.2.221
Lawson-Yuen, A., Saldivar, J.-S., Sommer, S., and Picker, J. (2008). Familial deletion within NLGN4 associated with autism and Tourette syndrome. Eur. J. Hum. Genet. 16, 614–618. doi: 10.1038/sj.ejhg.5202006
Lee, J. S., Yoo, S. S., Cho, S. Y., Ock, S. M., Lim, M. K., and Panych, L. P. (2006). Abnormal thalamic volume in treatment-naïve boys with Tourette syndrome. Acta Psychiatr. Scand. 113, 64–67. doi: 10.1111/j.1600-0447.2005.00666.x
Nag, A., Bochukova, E. G., Kremeyer, B., Campbell, D. D., Muller, H., Valencia-Duarte, A. V., et al. (2013). CNV analysis in Tourette syndrome implicates large genomic rearrangements in COL8A1 and NRXN1. PLoS ONE 8:e59061. doi: 10.1371/journal.pone.0059061
Paschou, P. (2013). The genetic basis of Gilles de la Tourette Syndrome. Neurosci. Biobehav. Rev. 37, 1026–1039. doi: 10.1016/j.neubiorev.2013.01.016
Paschou, P., Yu, D., Gerber, G., Evans, P., Tsetsos, F., Davis, L. K., et al. (2014). Genetic association signal near NTN4 in Tourette syndrome. Ann. Neurol. 76, 310–315. doi: 10.1002/ana.24215
Patel, C., Cooper-Charles, L., McMullan, D. J., Walker, J. M., Davison, V., and Morton, J. (2011). Translocation breakpoint at 7q31 associated with tics: further evidence for IMMP2L as a candidate gene for Tourette syndrome. Eur. J. Hum. Genet. 19, 634–639. doi: 10.1038/ejhg.2010.238
Price, A. L., Patterson, N. J., Plenge, R. M., Weinblatt, M. E., Shadick, N. A., and Reich, D. (2006). Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909. doi: 10.1038/ng1847
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A. R., Bender, D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi: 10.1086/519795
Qin, S., Yu, L., Gao, Y., Zhou, R., and Zhang, C. (2007). Characterization of the receptors for axon guidance factor netrin-4 and identification of the binding domains. Mol. Cell. Neurosci. 34, 243–250. doi: 10.1016/j.mcn.2006.11.002
Rickards, H., Wood, C., and Cavanna, A. E. (2008). Hassler and Dieckmann's seminal paper on stereotactic thalamotomy for Gilles de la Tourette syndrome: Translation and critical reappraisal. Mov. Disord. 23, 1966–1972. doi: 10.1002/mds.22238
Robertson, M. M., Eapen, V., and Cavanna, A. E. (2009). The international prevalence, epidemiology, and clinical phenomenology of Tourette syndrome: a cross-cultural perspective. J. Psychosom. Res. 67, 475–483. doi: 10.1016/j.jpsychores.2009.07.010
Rueda, M. R., Posner, M. I., and Rothbart, M. K. (2005). The development of executive attention: contributions to the emergence of self-regulation. Dev. Neuropsychol. 28, 573–594. doi: 10.1207/s15326942dn2802_2
Scharf, J. M., Yu, D., Mathews, C. A., Neale, B. M., Stewart, S. E., Fagerness, J. A., et al. (2013). Genome-wide association study of Tourette Syndrome. Mol. Psychiatry 18, 721–728. doi: 10.1038/mp.2012.69
Schubert, T., Denk, A., Mägdefrau, U., Kaufmann, S., Bastone, P., Lowin, T., et al. (2009). Role of the Netrin system of repellent factors on synovial fibroblasts in rheumatoid arthritis and osteoarthritis. Int. J. Immunopathol. Pharmacol. 22, 715–722. doi: 10.1177/039463200902200317
Singer, H. S., and Rosenberg, L. A. (1988). Development of behavioral and emotional problems in Tourette syndrome. Pediatr. Neurol. 5, 41–44. doi: 10.1016/0887-8994(89)90008-8
Sundaram, S. K., Huq, A. M., Wilson, B. J., and Chugani, H. T. (2010). Tourette syndrome is associated with recurrent exonic copy number variants. Neurology 74, 1583–1590. doi: 10.1212/WNL.0b013e3181e0f147
Verkerk, A. J. M. H., Mathews, C. A., Joosse, M., Eussen, B. H. J., Heutink, P., and Oostra, B. A. (2003). CNTNAP2 is disrupted in a family with Gilles de la Tourette syndrome and obsessive compulsive disorder. Genomics 82, 1–9. doi: 10.1016/S0888-7543(03)00097-1
Willer, C. J., Li, Y., and Abecasis, G. R. (2010). METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191. doi: 10.1093/bioinformatics/btq340
Wood, B. L., Klebba, K., Gbadebo, O., Lichter, D., Kurlan, R., Miller, B., et al. (2003). Pilot study of effect of emotional stimuli on tic severity in children with Tourette's syndrome. Mov. Disord. 18, 1392–1395. doi: 10.1002/mds.10552
Keywords: axon guidance, Gilles de la Tourette syndrome, GTS, Netrin-4, NTN4, single nucleotide polymorphism, SNP
Citation: Padmanabhuni SS, Houssari R, Esserlind A-L, Olesen J, Werge TM, Hansen TF, Bertelsen B, Tsetsos F, Paschou P and Tümer Z (2016) Investigation of SNP rs2060546 Immediately Upstream to NTN4 in a Danish Gilles de la Tourette Syndrome Cohort. Front. Neurosci. 10:531. doi: 10.3389/fnins.2016.00531
Received: 04 April 2016; Accepted: 31 October 2016;
Published: 22 November 2016.
Edited by:
John Vijay Sagar Kommu, National Institute of Mental Health and Neurosciences, IndiaReviewed by:
Munis Dundar, Erciyes University, TurkeySuhash Chakraborty, Hindustan Aeronautics Limited Hospital, India
Copyright © 2016 Padmanabhuni, Houssari, Esserlind, Olesen, Werge, Hansen, Bertelsen, Tsetsos, Paschou and Tümer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zeynep Tümer, zeynep.tumer@region.dk
 Shanmukha S. Padmanabhuni
Shanmukha S. Padmanabhuni Rayan Houssari
Rayan Houssari Ann-Louise Esserlind
Ann-Louise Esserlind Jes Olesen
Jes Olesen Thomas M. Werge4
Thomas M. Werge4 Thomas F. Hansen
Thomas F. Hansen Birgitte Bertelsen
Birgitte Bertelsen Fotis Tsetsos
Fotis Tsetsos Peristera Paschou
Peristera Paschou Zeynep Tümer
Zeynep Tümer