Cortical gene expression in spinal cord injury and repair: insight into the functional complexity of the neural regeneration program
- 1 Molecular Neurobiology Laboratory, Department of Neurology, Heinrich-Heine-University, Düsseldorf, Germany
- 2 Translational Glial Cell Research, Department of Neurology, Heinrich-Heine-University, Düsseldorf, Germany
- 3 Biomedical Research Center, Heinrich-Heine-University, Düsseldorf, Germany
Traumatic spinal cord injury (SCI) results in the formation of a fibrous scar acting as a growth barrier for regenerating axons at the lesion site. We have previously shown (Klapka et al., 2005) that transient suppression of the inhibitory lesion scar in rat spinal cord leads to long distance axon regeneration, retrograde rescue of axotomized cortical motoneurons, and improvement of locomotor function. Here we applied a systemic approach to investigate for the first time specific and dynamic alterations in the cortical gene expression profile following both thoracic SCI and regeneration-promoting anti-scarring treatment (AST). In order to monitor cortical gene expression we carried out microarray analyses using total RNA isolated from layer V/VI of rat sensorimotor cortex at 1–60 days post-operation (dpo). We demonstrate that cortical neurons respond to injury by massive changes in gene expression, starting as early as 1 dpo. AST, in turn, results in profound modifications of the lesion-induced expression profile. The treatment attenuates SCI-triggered transcriptional changes of genes related to inhibition of axon growth and impairment of cell survival, while upregulating the expression of genes associated with axon outgrowth, cell protection, and neural development. Thus, AST not only modifies the local environment impeding spinal cord regeneration by reduction of fibrous scarring in the injured spinal cord, but, in addition, strikingly changes the intrinsic capacity of cortical pyramidal neurons toward enhanced cell maintenance and axonal regeneration.
Introduction
Corticospinal tract (CST) axotomy permanently deprives the motoneurons in layer V of the sensorimotor cortex of their functions in locomotor control. Possible reasons for regeneration failure of injured axons in the central nervous system (CNS) are the lack of growth-promoting factors (Houweling et al., 1998), the presence of inhibitory cues like the myelin proteins Nogo-A, myelin-associated glycoprotein (MAG), and oligodendrocyte-myelin glycoprotein (OMGP; McKerracher and Winton, 2002; Filbin, 2003), as well as the lesion scar (Fawcett and Asher, 1999; Klapka and Müller, 2006). The latter growth barrier consists of a collagen type IV network (basement membrane) which accumulates inhibitory molecules (Yurchenco and Schittny, 1990; Timpl, 1994) such as chondroitin sulfate proteoglycans (CSPG, Snow et al., 1990; Jones et al., 2002; Morgenstern et al., 2002; Davies et al., 2004), semaphorins (de Winter et al., 2002; Niclou et al., 2003), and ephrins (Bundesen et al., 2003) thereby preventing axonal regeneration across the lesion site.
Although several studies investigated the gene expression response in the CNS after brain injury (Salin and Chesselet, 1992; Abankwa et al., 2002; Bareyre et al., 2002; Raghavendra Rao et al., 2003; Küry et al., 2004; Poulsen et al., 2005; Israelsson et al., 2006), only little is known about the transcriptional response in affected brain regions following spinal cord injury (SCI). Most studies suggest that CNS neurons differ in their response to axotomy compared to PNS neurons as exemplified by analyses done on growth-associated protein 43 (Gap43) and α-tubulin (Mikucki and Oblinger, 1991; Tetzlaff et al., 1994; Fournier and McKerracher, 1997). Data of Mason et al. (2003) further suggest that cortical neurons show variations in axotomy-induced gene expression depending on the distance of the injury site to the cell body. The authors identified increased expression of a number of growth-associated genes such as the oncogene c-jun/ap-1, l1cam/ncaml1, atf3, and krox2-4/egr1 after intracortical but not after spinal axotomy of layer V pyramidal neurons. In addition a recent study revealed no changes in expression of the receptors for myelin-associated inhibitor proteins in any neuronal population of the mouse brain after severe T10 spinal cord contusion (Barrette et al., 2007).
Until now, neither a systematic investigation on cortical gene expression after SCI nor the molecular response to a regeneration-promoting treatment in the injured CNS has been performed. Here, for the first time, the questions (i) whether axotomized cortical cells display a regeneration-associated gene expression program upon therapeutic intervention and (ii) how this treatment-triggered transcriptional program differs from the lesion-induced response are addressed.
Previous work in our lab showed that transient scar suppression by local application of an anti-scarring treatment (AST) using an iron chelator and cyclic AMP enabled long distance axon growth and functional improvement after dorsal hemisection of the CST (Klapka et al., 2005; Brazda and Müller, 2009). In addition, AST treatment rescued pyramidal cells in layer V of the sensorimotor cortex that project into the lesioned CST, thus preventing the injury-induced loss or atrophy of up to 40% of this cell population as described by Hains et al. (2003). Hence, AST treatment not only promoted axon regeneration through the fibrous scar at the lesion site, but also exerted retrograde neuroprotective effects on lesioned cortical neurons (Klapka et al., 2005). The beneficial effect of scar suppression on functional recovery in SCI has independently been proven by other labs using different therapeutic approaches (Davies et al., 2006; Tian et al., 2006; Ito et al., 2009).
Affymetrix microarray technology was applied to characterize transcriptional responses of sensorimotor cortex to SCI and AST. We made use of adult rat in three experimental paradigms including (i) sham-operated, (ii) lesion-only, and (iii) AST-treated animals, at distinct time-points after lesion investigating the following stages: acute (1 day), early subacute (7 days), late subacute (21 days) and chronic injury (60 days). We identified a large number of lesion-triggered as well as AST-affected regulated genes as early as 1 day after CST-axotomy. Our data indicate dynamic and treatment-specific gene regulation patterns associated with distinct functional groups of genes involved in axonal outgrowth/guidance or apoptosis/cell survival. The data further demonstrate a remarkable AST-induced switch in the genetic program of the affected sensorimotor cortex toward a neuroprotective and growth-promoting state.
Materials and Methods
Treatment Paradigms and Animal Groups
To study changes in neocortical mRNA expression after axotomy of the CST in the thoracic spinal cord and to identify alterations in gene expression due to AST, three groups of adult rats were examined and compared (i) Sham-operated animals underwent laminectomy but did not receive a SCI, (ii) Lesion-only animals received a dorsal spinal cord hemisection including CST transection, (iii) AST-treated animals received a CST transection followed by AST resulting in regenerative reactions. The animals were sacrificed at 1, 7, 21, or 60 dpo. Groups of three to five animals per condition were analyzed. However, samples were not pooled so each animal was used to generate an individual probe that was hybridized to a separate microchip. Further considerations concerning replicate numbers and pooling strategies have previously been described in more detail (Kruse et al., 2008).
Spinal Cord Injury
Spinal cord injury was carried out as previously described (Klapka et al., 2005). Briefly, laminectomy of vertebrae Th8 and Th9 was performed under isoflurane–anesthesia. The dura was opened with spring scissors and the dorsal columns and dorsal CST were cut with a Scouten Wire Knife (Bilaney, Düsseldorf, Germany). The iron chelator 2,2′-bipyridine-5,5′-dicarboxylic acid (BPY-DCA, 40 mM in Tris-buffer) was injected both into the lesion site (four injections of 0.2 μl) and at 1 mm proximal to the lesion site (two injections of 0.2 μl). After injection, 2 mg of 8-Br-cAMP were applied onto the lesion area. Control animals received injections of Tris-buffer. Lesions of treated animals were covered with a piece of ELVAX (ethylene–vinyl-acetate) copolymer sheet loaded with BPY-DCA for slow release, whereas ELVAX copolymer in control animals contained Tris-buffer alone. The animals were treated with antibiotics (Baytril) for 1 week after surgery and, if necessary, the bladders were emptied manually. Over all locomotor behavior of the rats was monitored using the open field test. Analogous to the results recently described (Klapka et al., 2005) the AST-treated animals showed moderate but significant improvement in their BBB score compared to lesion-only animals at 60 days post lesion (data not shown). Animal experiments were all done in compliance with current German legislation on animal research and have previously been approved by the State Office for Nature, Environment and Consumer Protection of North-Rhine-Westphalia (LANUV NRW).
Cortical Tissue Preparation and Total RNA Isolation
Initially, the appropriate cortical area for tissue preparation of pyramidal cells in layer V of the sensorimotor cortex was determined by retrograde labeling with DiI (Figure 1A). For tissue preparation, the brains were removed in a cold room (∼4°C) immediately after sacrifice and directly frozen in isopropanol at −45 to −55°C. Using a cryotom, 40 serial coronal brain slices of 50 μm thickness were cut from sham-operated, spinal injured, and AST-treated spinal injured adult rats. The area of layer V containing the primary motor neurons and the subjacent part of layer VI were rapidly dissected at −20°C and collected in Qiazol. Total RNA was isolated using the RNeasy Lipid Tissue Mini Kit (Qiagen, Hilden, Germany) completed by an additional DNAse-I treatment to avoid genomic DNA contamination. Quality and integrity of total RNA preparation was finally confirmed using a 2100 Bioanalyzer System (Agilent Technologies, Santa Clara, CA, USA).
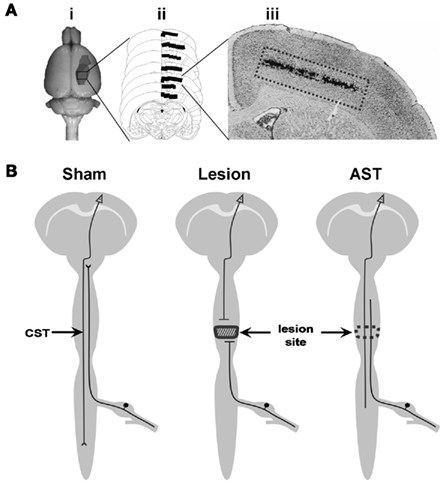
Figure 1. (A) Tissue preparation procedure: (i) representation of the cortical area labeled by the retrograde tracer DiI. This area represents the sensorimotor cortex containing neurons lesioned by the SCI paradigm. (ii) Schematic representation of frontal serial brain sections with the area indicated where DiI labeled neurons were found. (iii) Histological section of the cerebral cortex, showing DiI-traced neurons in layer V. (B) Schematic representation of rat CNS including the three experimental paradigms of the present investigation. Descending CST axons originating in pyramidal neurons of the cerebral cortex layer V are cut at spinal level Th8. Cortical tissue from sham-operated, CST-lesioned, and AST-treated animals was collected and analyzed.
Probe Labeling, Array Hybridization, and Data Analysis
GeneChip® Rat Expression 230A analysis was performed according to the manufacturer’s protocol (Affymetrix, Santa Clara CA, USA). Briefly, 2 μg of total RNA of each preparation were converted into labeled cRNA, subsequently fragmented, and hybridized to microarrays. Digitized signal intensities and raw data quality were independently evaluated and then the data were analyzed using ArrayAssist 4.2.0 software (Stratagene, Amsterdam, Netherlands) applying five different algorithms: MAS5.0 (Affymetrix, 2001), MBEI (Li and Wong, 2001), PLIER (Affymetrix, 2005), RMA (Irizarry et al., 2003a,b), and GCRMA (Wu and Irizarry, 2004). After variance stabilization (+16) and log-transformation (base 2) statistical analysis was carried out using ANOVA calculations. To specify the time point and experimental condition at which a particular gene is regulated post hoc tests were applied. The thresholds to assign genes to be regulated were >1.3 (fold-change) and <0.02 (p-values). Genes were considered significantly regulated if they fulfilled the given criteria by at least two algorithms, including either PLIER or GCRMA. Further detailed information on data analysis can be found in Kruse et al. (2008).
Quantitative Polymerase Chain Reaction Procedures
Amplification primers for real-time PCR analysis of the transcripts of gm-csf, bag1, kalrn, prdx2, vdr, diablo, egr2, stxbp1, robo1, unc5A, nrp1, gal, and gdnf were designed by using Primer Express 2.0 Software (Applied Biosystems, Foster City, CA, USA) and subsequently tested for efficiency and specificity. Real-time quantitative polymerase chain reaction (qPCR) was performed by using SYBR green chemistry (Applied Biosystems) and relative changes in gene expression were determined using the ΔΔCt method (Livak and Schmittgen, 2001) with ornithine decarboxylase1 (odc1) and glyceraldehyde-3-phosphate dehydrogenase (gapdh) as reference genes. Primer sequences are available upon request.
Over- and Under-Representation in Functional Groups and Overlaps
To extract relevant functional information from our high throughput analysis we used the gene ontology (GO) annotation system (Ashburner et al., 2000) implemented in PathwayArchitect (Stratagene) to identify GO-terms with unbalanced distribution of a query list compared to a computed background, e.g., all probe sets on the rat 230A-microarray. This enrichment analysis was used to determine over- and under-represented functional groups of regulated genes. The same procedure was also applied to detect over- and under-representation of functional groups in overlaps between time point-, treatment- and direction of regulation-specific sets of genes. The level of significance was calculated via the hypergeometric distribution and corrected for multiple testing.
Retrograde Tracing of Pyramidal Neurons in Primary Sensorimotor Cortex
Cortical pyramidal neurons were identified by injection of 2 × 0.5 μL of Fluoro-Gold (Fluorochrome LLC, Denver, CO, USA; 3% in ddH2O) lateral to the CST at the most proximal end of segment Th7 using a 10-μL Hamilton syringe with a 32-gage canula. After 7 days the brains of traced animals were dissected either for calibration of the cortical tissue preparation technique or for cell identification in combination with immunohistochemistry.
Immunohistochemistry
Rats were transcardially perfused with 4% paraformaldehyde and brains were subsequently post-fixed in 4% paraformaldehyde, cryoprotected in 30% sucrose and 20 μm thick sections were cut on a cryostat. After microwave antigen retrieval in 10 mM citrate buffer (600W for 3 × 2 min) and blocking with 5% donkey-serum (Sigma-Aldrich, St. Louis, MO, USA) in PBS-T (PBS with 0.1% TritonX-100), the sections were incubated at 4°C over-night with either rabbit anti-galanin (1:100; Affinity BioReagents, Rockford, IL, USA), rabbit anti-VDR (1:750; Affinity BioReagents, Rockford, IL, USA), rabbit anti-GM-CSF (1:25; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), rabbit anti-Diablo (1:100; Calbiochem, Darmstadt, Germany), rabbit anti-EGR2 (1:25; Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA), or mouse anti-claudin-11 (1:1000; Abcam plc, Cambridge, UK) diluted in blocking solution. Alexa488-conjugated donkey anti-rabbit or donkey anti-mouse secondary antibodies (1:200 each; Invitrogen, Carlsbad, CA, USA) were used for visualization. After two washing steps and elimination of lipofuscin autofluorescence with 0.3% Sudan Black B (in 70% ethanol, 7 min at room temperature), the sections were mounted with FluoromountG (Southern Biotech, Birmingham, AL, USA). Since the Fluoro-Gold signal in retrogradely traced pyramidal neurons of sensorimotor cortex (see above) gets lost during the subsequent immunohistochemical procedures, the sections were photographed before and after immune staining at identical positions using a BZ-8000 digital microscope (Keyence, Osaka, Japan).
Results
Cortical Gene Expression Profiles Following Spinal Cord Injury
It has been shown previously (Klapka et al., 2005; Schiwy et al., 2009) that the CST, among other descending motor fiber tracts in the injured rat spinal cord, responds to local AST by enhanced regenerative axon growth. In order to characterize the temporal changes in cortical mRNA expression profile after thoracic CST transection we compared the transcriptomes from cortical layer V of AST-treated rats to those of sham-operated animals (Figure 1B). We systematically identified significantly regulated genes comprising specific molecular responses over a period of 1, 7, 21, and 60 dpo (Table 1). Out of a total of ∼11,000 rat genes represented on the microchip, approximately 3,100 genes were significantly regulated, at least at one of the time-points examined. Despite of the distance between the lesion site at Th8 and the corresponding sensorimotor cortex (several centimeters), 521 genes underwent significant regulation in cortical layer V as early as 1 day after transection. Later (7–60 dpo), the number of regulated genes further increased and a maximum of 1,199 genes was reached at 21 days after injury. Notably, the proportion of up- and down-regulations varied over time. While at 1, 7, and 60 dpo almost equal proportions of the genes were up- or down-regulated, at 21 dpo the majority (73%) of the regulated genes were repressed (Table 1).
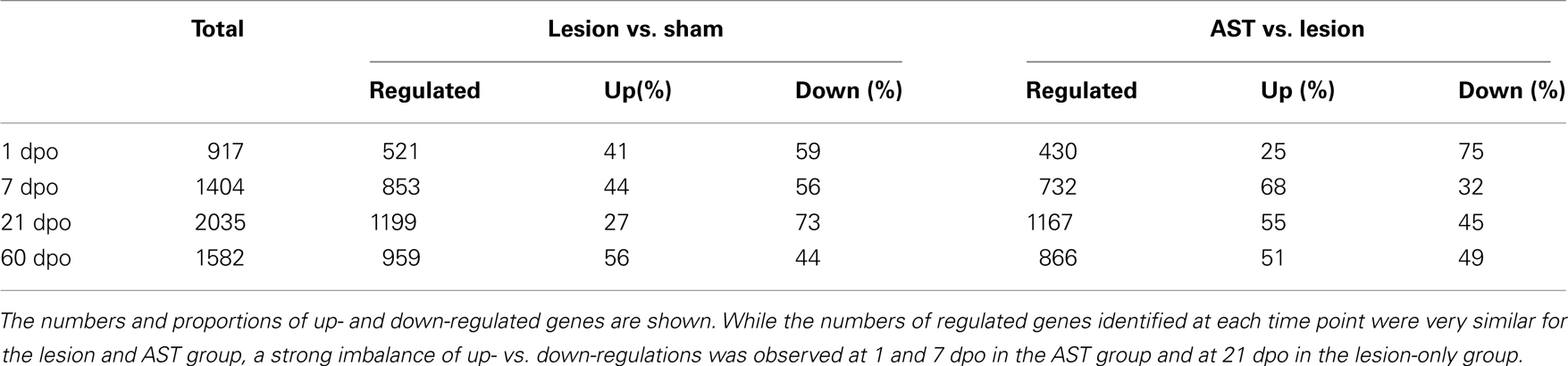
Table 1. Numbers of regulated genes in sensorimotor cortex of sham, lesioned-only and AST-treated rats at four different time points (1, 7, 21, 60 days) after surgical treatment.
The Lesion-Triggered Gene Expression Profile in Sensorimotor Cortex is Altered by Local Spinal Anti-Scarring Treatment
In order to determine AST-induced cortical gene regulations we compared (i) the transcriptomes of cortical layer V of injured but untreated (lesion-only) rats to that of sham animals (Lesion vs. Sham comparison), and (ii) the transcriptome of AST-treated rats to that of lesion-only animals (AST vs. Lesion comparison). Direct comparison of gene expression in AST-treated animals with lesion-only rats revealed a strong modulation of lesion-triggered gene expression profiles by AST (Table 1). Similar to lesion-only animals, the number of significantly regulated genes in AST-treated animals increased over time reaching a peak at 21 dpo (1167 genes, Table 1). During the entire period of 1–60 dpo approximately 2800 genes were significantly altered in treated rats when compared to lesioned animals. While at 1 dpo only 25% of the 430 significantly regulated genes in AST animals were up-regulated, nearly 70% of the 732 regulated genes were up-regulated at 7 dpo. Interestingly, the total number of regulated genes at each time point was similar in lesion-only and AST animals.
Time- and treatment-specific changes in gene expression patterns
We observed four different types of AST-triggered alterations of the lesion-induced gene expression profile (Figure 2). These included gene regulations in AST animals showing (i) no significant differences compared to lesion-only animals (injury-specific gene regulation), (ii) an additional effect of the AST treatment into the same direction of regulation as observed in lesion-only animals (gene expression boosted by AST), (iii) a counter-regulation back to or beyond the basal sham expression level (genes counter-regulated by AST), and (iv) AST-specific alterations not observed in lesion-only animals (AST-specific regulation). Interestingly, the group of AST-boosted probe sets comprised only a very small proportion (∼1%) of the regulated transcripts at all time-points examined. In contrast, the proportion of AST-counter-regulated transcripts clearly increased over time to more than 15% (Figure 2). The proportion of AST-specific transcripts of the overall detected regulated probe sets was rather high (40–46%, Figure 2). At each time point the numbers of injury-specific and of AST-specific regulated transcripts were very similar.
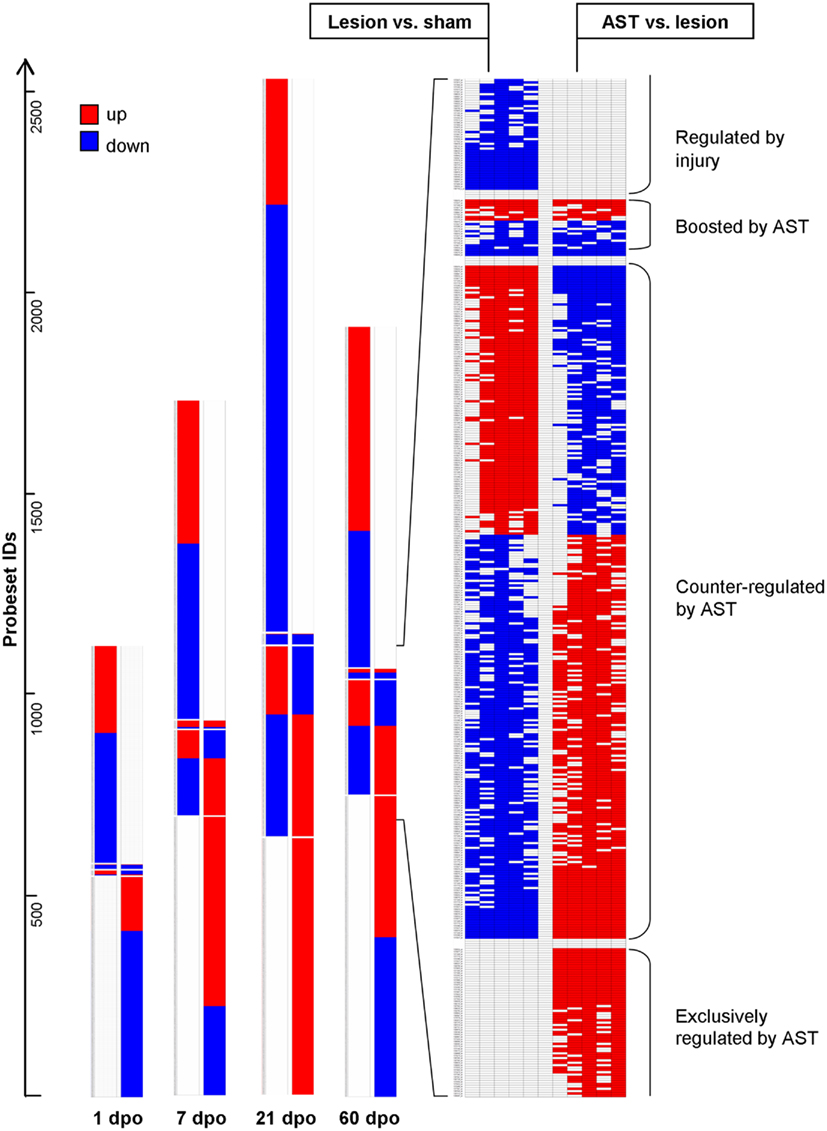
Figure 2. Schematic representation of expression changes in lesion-only animals compared to sham animals and in AST animals compared to lesion-only animals illustrates the various regulation patterns. In comparison to lesion-induced regulations, AST-induced regulations showed four different responses: (i) probesets “Regulated by injury” are strictly altered by SCI regardless of an additional AST treatment (no significant regulation in the comparison AST vs. Lesion); (ii) “Boosted by AST” are probesets with altered expression by injury that become further up- or down-regulated by AST; (iii) “Counter-regulated by AST” represents probesets which are regulated back to or beyond sham-level by AST; (iv) probesets “Exclusively regulated by AST” show no significant Lesion vs. Sham regulation. The detailed view on the right illustrates the results of all the five implemented algorithms (Mas5, LiWong, Plier, RMA, and GCRMA) for each comparison (Kruse et al., 2008).
Identification of time- and treatment-specific clusters of regulated genes
Temporal comparison revealed a rather low overlap (<20%) of regulated genes between consecutive survival time-points indicating dynamic changes in temporal regulation profiles which specify different acute and chronic stages. For further insight into the complexity of stage-specific gene regulation patterns, we investigated significant temporal overlaps between genes regulated by injury and/or AST throughout all time-points examined. If the numbers of genes that were co-regulated at certain experimental conditions significantly exceeded or fell short the number of overlaps expected merely by chance, those co-regulated clusters of genes were considered to be over- or under-represented in the given experimental condition. For better visualization of distinct responses showing high temporal overlaps of sets of regulated genes we converted the data (Table S1 in Supplementary Material) into a topographic map (Figure 3). Certain of these over- or under-representations might be expected, as, e.g., in one experimental condition genes cannot be up- and down-regulated at the same time. In such a case the overlaps will be zero as indicated by a dark blue pit. Additional dark blue areas denote the low amount of AST-boosted regulations. On the other hand, AST-triggered counter-regulations were very common and occurred at all time points, leading to ridge-like structures. For example, the lesion-triggered up-regulations at 7 and 21 dpo which are counter-regulated in AST animals are shown as very prominent ridges in the lower left corner of Figure 3. A similar strong overlap between counter-regulated gene clusters is observed between genes that are down-regulated in lesioned animals but up-regulated by AST at 7 and 60 dpo. Many of the AST-counter-regulated genes at 7 dpo could be attributed to the significantly enriched functional category of developmental processes including, e.g., mammalian achaete-scute homolog 1 (mash1), homeobox gene hoxA1, granulocyte-macrophage-colony stimulating factor (csf2/gm-csf), and others.
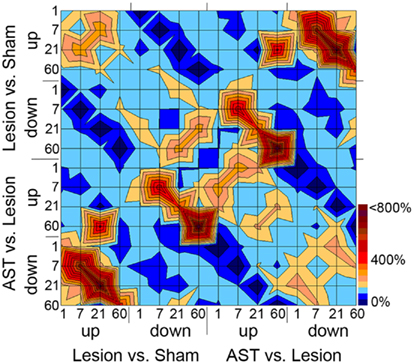
Figure 3. “Topographic” map illustrating overlaps between sets of regulated genes with respect to kind of treatment, direction of regulation, and time point of the respective gene regulation. Overlaps between groups of regulated genes for each time point, direction of regulation, and the comparison of Lesion vs. Sham and AST vs. Lesion have been calculated. The given number of overlaps is shown as percentage of the expected number of overlaps. Overlaps smaller than 150% of the expectancy value are illustrated in light to dark blue, over-representations in overlaps higher than 150% of the expectancy value are illustrated in light to dark red. For example AST-counter-regulations of lesion-induced genes at 7 and 21 dpo lead to the ridge structure in the lower left and in mirror image in the upper right corner of the topographic map. The corresponding values of the over- or under-representations including the level of significance are shown in Table S1 in Supplementary Material.
Another notable observation was the singular sharp peak representing a significant cluster of genes up-regulated at 21 dpo in lesioned animals that showed a delayed upregulation in AST animals at 60 dpo. Among the genes that showed this AST-triggered temporal shift in expression we found, e.g., fas apoptotic inhibitory molecule 2 (faim2), claudin-11 (cldn11), peroxisome proliferative activated receptor delta (ppard), dihydropyrimidinase-like 5 (dpysl5/crmp5), bcl2-like 1 (bcl2l1/Bclx), statin-like (stnl/eef1a2), syntaxin binding protein 1 (stxbp1), unc-5 homolog A (unc5A), and the neuronal zinc-finger protein neuro-d4 (neuD4). The observed clusters of counter-regulated genes and the temporal shift of co-regulated gene cluster demonstrate remarkable differences in the regulatory control of repressed and induced genes between the experimental conditions (lesion-only vs. AST-treated).
Differential Regulation of Biological Processes
To reveal biological processes that were activated or repressed following spinal cord lesion we assigned the regulated genes to functional categories using the DAVID Bioinformatics Resources (Dennis et al., 2003; Huang et al., 2009a). The stage-specific enrichment of several functional groups of regulated genes indicated the dynamic modulation of distinct cell physiological responses following SCI (Table 2, upper half). At 1 dpo affected processes included, among others, wounding responses, cytoskeletal reorganization, and cell survival, whereas at 7 dpo genes involved in metabolic and transport processes, in oxidative stress responses and brain development were predominantly changed. At 21 and 60 dpo enriched regulation of functional groups involved in protein biosynthesis and synaptic reorganization was observed. Likewise, apoptotic processes were strongly modulated at 60 dpo.
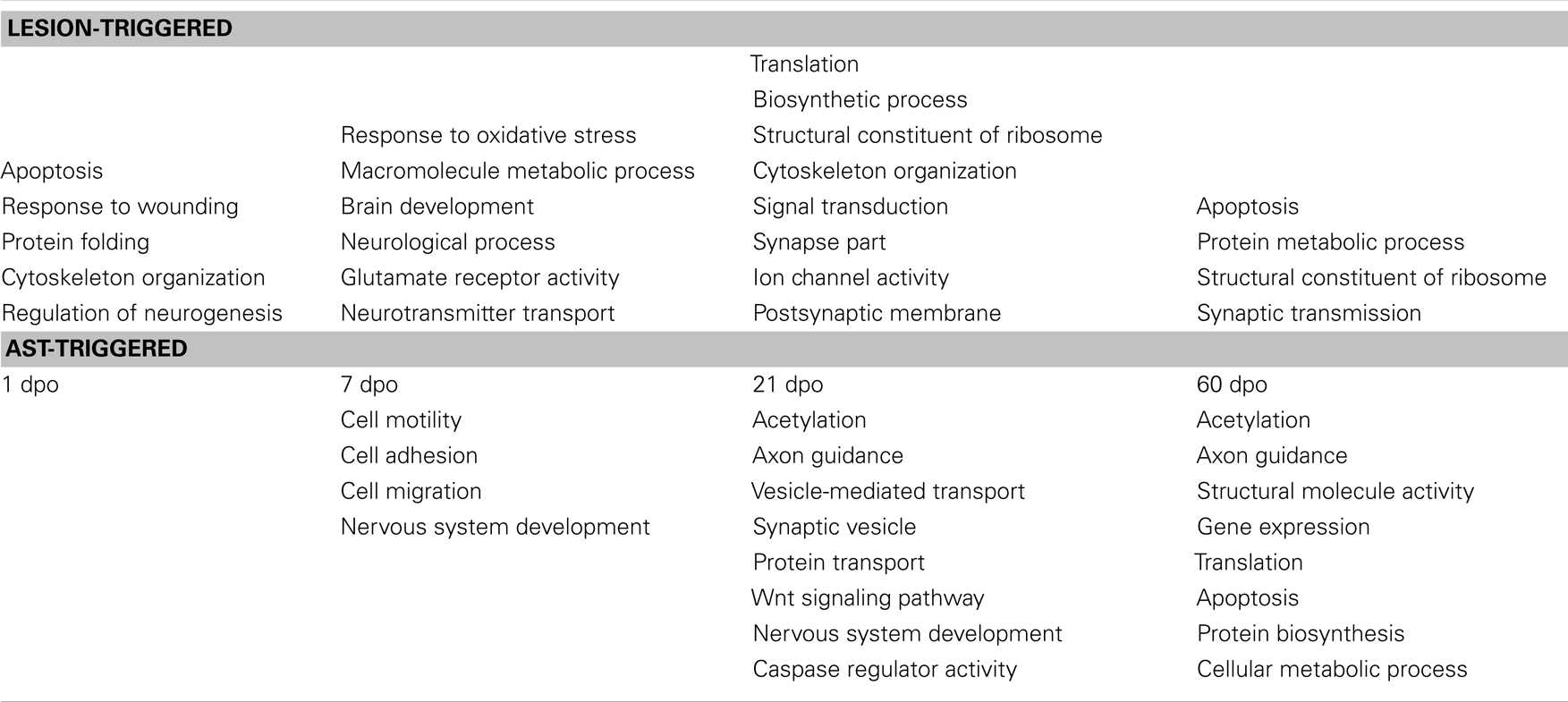
Table 2. Significantly enriched functional groups of regulated genes after SCI (upper panel) and after AST (lower panel).
The variety of biological processes that were predominant in AST animals compared to lesion-only rats are summarized in Table 2 (lower half). As in lesion-only animals the activation/repression of significantly enriched functional groups of regulated genes appears to be stage-dependent in AST animals. While genes involved in cell adhesion, motility, migration and nervous system development were predominant at 7 dpo, affected processes like, e.g., axon guidance, protein transport and vesicle transport, Wnt signaling as well as acetylation/deacetylation (including epigenetic responses), and caspase regulatory activity (apoptosis/cell survival) were enriched at 21 dpo. At 60 dpo processes comprising transcription, protein biosynthesis/translation and, again, axon guidance, acetylation/deacetylation as well as apoptosis/cell survival were among the significantly enriched functional categories. Some important functional categories including nervous system development, axon guidance, caspase regulatory activity/apoptosis, and acetylation/deacetylation showed significant enrichment in more than one stage. Although AST rats markedly differed from lesioned animals in the expression of 430 regulated cortical genes already at 1 dpo, no particular functional process was significantly enriched at this early time point. This indicates that AST treatment regulated genes participating largely in the same biological processes as those found in lesion-only animals.
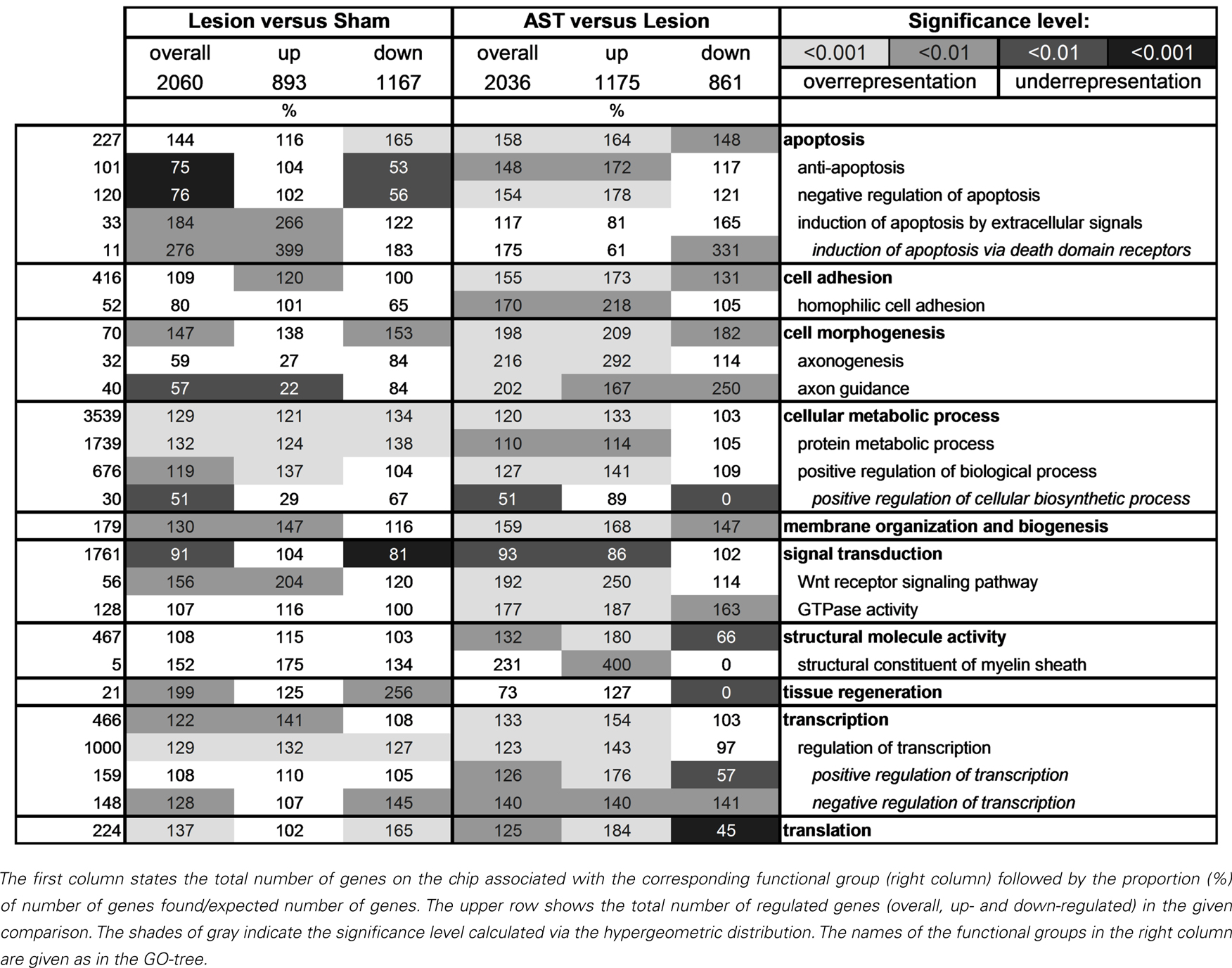
To reveal the differential activation or repression of physiological functions in response to regeneration-promoting AST treatment as compared to lesion-only animals we analyzed which GO-categories (Ashburner et al., 2000) were over- or under-represented. Ontologies that did not exceed their stochastic expectation were not taken into account. A selection of the most affected GO-categories is shown in Table 3. Many genes are part of several GO-classes and may thus be involved in multiple functional processes.
Interestingly, we observed clear differences between lesioned and AST-treated animals, when we focused on genes linked to apoptosis. While in lesion-only animals numerous genes responsible for induction of apoptosis by extracellular signals via death domain receptors were significantly enriched in the group of up-regulated genes, in AST-treated animals a large number of genes of this subcategory was significantly down-regulated. In addition, we observed enhanced representation of anti-apoptotic or neuroprotective genes in treated animals. In these animals the number of up-regulated anti-apoptotic or neuroprotective genes increased over time (Figure 4).
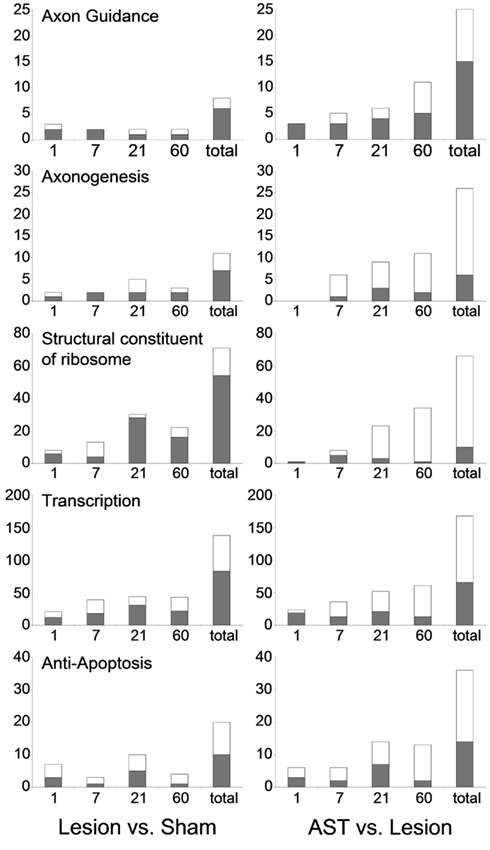
Figure 4. Selected functional groups of genes that were strongly modulated by AST treatment. The number of significant regulations associated with the GO-terms axon guidance, axonogenesis, structural constituent of ribosome, transcription, and anti-apoptosis throughout the entire time period examined are shown (up-regulation: white bar; down-regulation: gray bar). Note that the number of regulations related to axon guidance which are altered by AST compared to lesion at 60 dpo alone is higher than the total number of axon guidance genes regulated after lesion-only over all time points.
The GO-term cell morphogenesis also displayed striking differences between lesioned and AST-treated animals. The subcategories axonogenesis and axon guidance were of particular interest. Both groups were under-represented among lesion-induced genes (+ 3) at all time-points examined (Figure 4). In contrast, in AST-treated animals genes related to axonogenesis and axon guidance were increasingly over-represented after treatment (Table 3; Figure 4) indicating a regulatory functional switch in cortical cells. The numbers of up-regulated genes in these two functional subcategories were found to rise continuously. Throughout the entire period from 1 to 60 dpo the temporal regulation profile of axon guidance associated genes changed from solely down-regulation to predominant up-regulation in treated animals whereas genes related to axonogenesis showed AST-induced up-regulation between 7 and 60 dpo (Figure 4).
Genes associated with membrane organization and biogenesis were identified as over-represented in the up-regulated gene pools of both lesioned and AST-treated animals (Table 4). Surprisingly, we also found a significant over-representation of up-regulated structural constituents of myelin in AST-treated animals at 21 and 60 dpo (data not shown).
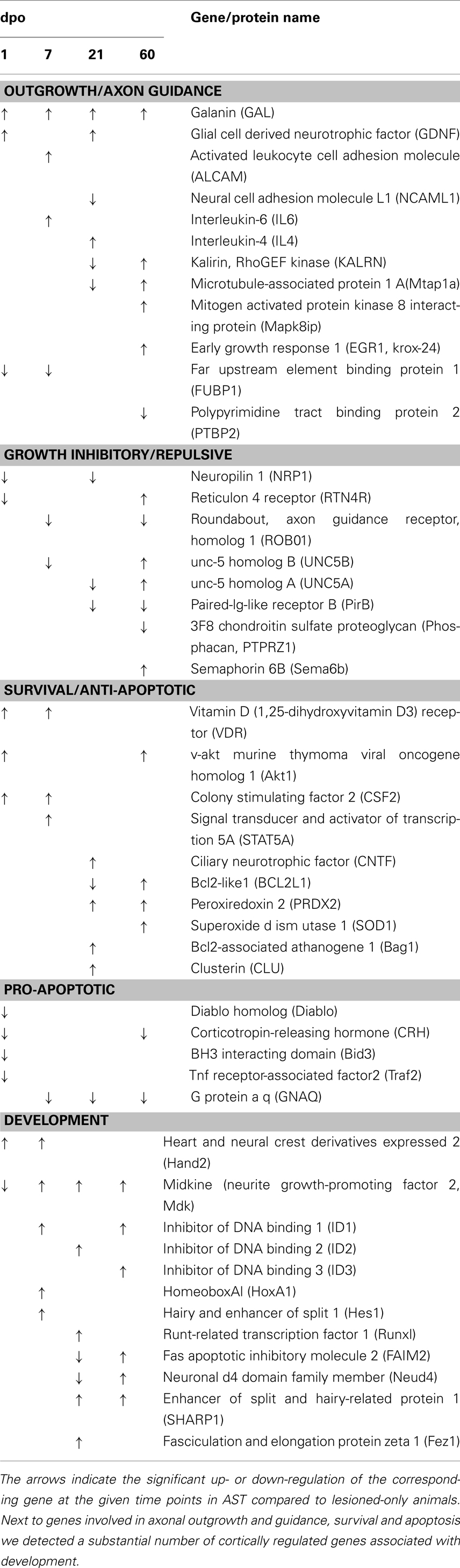
Table 4. Sensorimotor cortical regulations of selected genes in the AST vs. Lesion comparison are displayed for selected functional groups.
We further observed altered transcriptional and translational activities in sensorimotor cortex after SCI, which were substantially enhanced by AST treatment (Table 3). As shown in Figure 4 a high number of genes linked to the GO-term structural constituent of ribosome was significantly enriched in the group of lesion-triggered down-regulations reaching a maximum number at 21 dpo. In contrast, AST-triggered up-regulation of genes related to this category increased steadily up to 60 dpo. Interestingly, at 21 dpo the majority of the down-regulated genes of this functional category in lesioned animals was, at the same time, counter-regulated in AST animals. A very similar temporal profile as shown for structural constituent of ribosome was observed for the category transcription (Figure 4).
Identification of Regulated Genes Associated with AST-Dependent Responses
While one goal of this study was to characterize novel transcripts in sensorimotor cortex that were regulated as a response to traumatic SCI, the second aim was to identify regulated genes that could play a role in successful AST-promoted axonal regeneration.
Axon growth
Numerous known genes involved in axon growth in regenerating peripheral neurons were up-regulated in sensorimotor cortex of AST-treated animals (Table 4) such as galanin (GAL), glial cell derived neurotrophic factor (GDNF), kalirin (KALRN), LIM domain only 4 (LMO4), galectin 1 (LGALS1), paternally expressed 3 (PEG3), serum/glucocorticoid regulated kinase (SGK), mitogen activated protein kinase 8 interacting protein (MAPK8IP), actin beta (ACTB), interleukins 4 and 6 (IL4, IL6), vitamin D3 receptor (VDR), enabled homolog (ENAH), and S100 calcium binding protein beta (S100B).
Important genes involved in axonal guidance also underwent AST-triggered changes in expression. Among those genes were the roundabout homolog 1 (robo1), reticulon 4 receptor (rtn4r/nogor), unc-5 homolog A, B (unc5A, 5B), dihydropyrimidinase-like 5 (dpysl5/crmp5), paired-Ig-like receptor B (pirb), neuropilin 1 (nrp1), receptor-like tyrosine kinase (ryk), cyclin-dependent kinase 5 (cdk5), semaphorin 6B (sema6b/semaZ), contactin 2 (cntn2/tax1), neural cell adhesion molecule L1 (l1cam/ncaml1), fasciculation and elongation protein zeta 1 (fez1), and activated leukocyte cell adhesion molecule (alcam). Several of these genes which are known to be involved in growth cone repulsion and/or collapse, like robo1, unc5A and unc5B, dpysl5/crmp5, RTN4R, pirb and nrp1, were down-regulated in AST-treated compared to lesion-only animals in at least one of the observed time points (Table 4). On the other hand, genes like cntn2, fez1, and alcam, which are positively involved in outgrowth and axonal pathfinding, were up-regulated in the AST group. Sema6b was up-regulated in the cortex in AST-treated rats at 60 dpo.
Surprisingly, we neither found significant cortical up-regulation of gap43 in lesioned-only nor in AST-treated rats. However, in treated animals we detected the sequential down-regulation of the two transcripts fubp1 (far upstream element binding protein 1) at 1 and 7 dpo and ptbp2 (polypyrimidine tract binding protein 2) at 60 dpo (Table 4). These proteins are known to interact with gap43 mRNA, thereby influencing its stability (Irwin et al., 1997).
Apoptosis and protection
Numerous apoptosis-associated genes were regulated by AST treatment (Table 4). Notably, genes known to be involved in pro-apoptotic processes such as bcl2-like proteins 1 (bcl2l1/Bclx) and 13 (bcl2l13), Nucleus accumbens-1 (nac1/btbd14b), direct inhibitor of apoptosis-binding protein with low pI (diablo), corticotropin-releasing hormone (crh), TNF receptor-associated factor 2 (traf2), and neuronal death protein Dp5 (bid3/dp5) were exclusively down-regulated in treated animals. On the other hand, the group of genes encoding proteins with anti-apoptotic and/or neuroprotective properties like protein kinase B (akt1), Bcl2-associated athanogene 1 (bag1), peroxiredoxin 2 (prdx2), superoxide dismutase (sod1), colony stimulating factor 2 (csf2/gm-csf), and signal transducer and activator of transcription 5A (stat5A/mgf), were found to be up-regulated in AST animals compared to lesion controls.
Development
Regeneration-promoting AST provoked induction of numerous genes with known functions during development (Table 4), including, amongst others, genes encoding for the DNA binding proteins hand2 and hoxA1, the helix-loop-helix (HLH) inhibitors of DNA binding 1–3 (id1,2,3), the transcription factors runx1, hairy and enhancer of split 1 (hes1) as well as the related repressor sharp1, the neurite growth-promoting factor 2 alias midkine (mdk), the component of the neuron-specific chromatin remodeling nBAF-complex Neuro-D4 (neuD4), and the fasciculation and elongation protein zeta1 (fez1).
Validation of Microarray Expression Data by QPCR
We performed real-time qPCR to verify the validity of our microarray data. For most genes tested the qPCR data confirmed our microarray results, both in temporal pattern as well as in magnitude of transcript regulation. The fold-change of regulation detected by qPCR was, in general, about 1.5–2 times higher than in the array-based data. Genes from several relevant functional groups were chosen for validation (Figure 5). Csf2/gm-csf, bag1, kalrn, prdx2, vdr, egr2/krox20, robo1, unc5A, and gdnf showed the same significant regulation patterns with qPCR as with the arrays. Galanin 1 (gal) and syntaxin binding protein (stxbp1) even showed qPCR-based regulations at more time points than originally detected by microarrays. In the case of diablo and nrp1, the significance of the array-based regulation could be reproduced for some but not all time points. In general, the qPCR analysis confirmed gene regulations observed by microarray analysis.
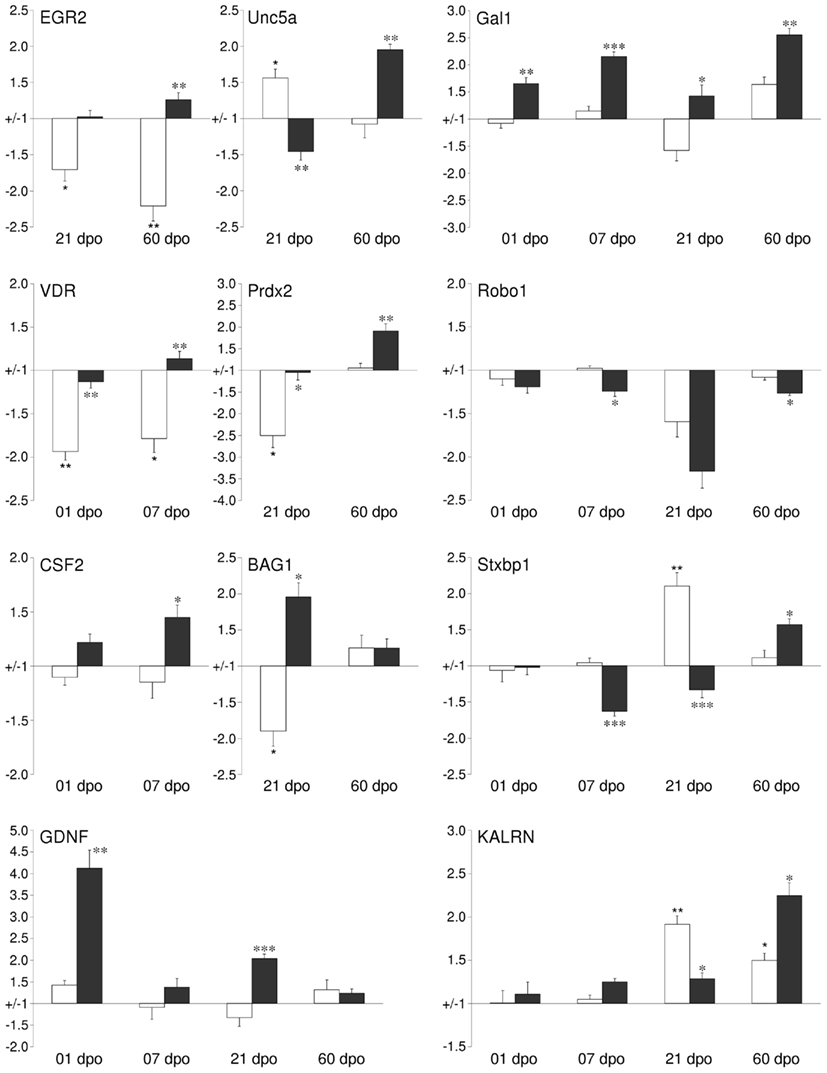
Figure 5. Validation of changes in mRNA expression by quantitative qPCR for a subset of regulated cortical genes identified by the GeneChip analysis. For selected time points the mRNA expression levels of cortical tissues from either lesion-only (white bars) or AST- treated (black bars) were compared to sham-operated animals. Abbreviations: early growth response 2 (EGR2), unc-5 homolog A (UNC5A), galanin (GAL), vitamin D receptor (VDR), peroxiredoxin 2 (PRDX2), roundabout homolog 1 (ROBO1), colony stimulating factor 2 (GM-CSF), Bcl2-associated athanogene 1 (BAG1), syntaxin binding protein 1 (STXBP1), glial cell derived neurotrophic factor (GDNF), kalirin (KALRN). Note: Significance was calculated for the comparisons Lesion-only vs. Sham-operated (corrected p-value <0.05*, <0.01**, <0.001***) and AST-treated vs. Lesion-only (corrected p-value < 0.05*, <0.01**, <0.001***).
Cellular Localization of Transcripts and Proteins
In order to reveal cellular origins of transcripts of interest, we consulted the public Allan database (Allen Brain Atlas; Lein et al., 2007; http://mouse.brain-map.org/welcome.do). Fifty-nine transcripts out of the 60 significantly regulated genes mentioned in this paper have been localized by in situ hybridization in sensorimotor cortex. Of these transcripts, 49 could be classified as of neuronal origin, and 44 of those were clearly found in pyramidal cells of cortical layer V (Table S4 in Supplementary Material). However, non-neuronal gene regulation from neighboring glial and/or immune cells is also likely to be expected in spinal injury- and treatment-affected sensorimotor cortex. We further performed immunohistochemical localization of selected proteins with different putative functions in regeneration, namely proteins promoting axon outgrowth (Gal), neuronal maturation and plasticity (Egr2/Krox20), neuroprotection (GM-CSF, VDR), myelination (Egr2/Krox20, Cldn11), or, e.g., cell death (Diablo). As shown in Figure 6 almost all of these proteins could be localized in retrogradely labeled corticospinal neurons in layer V of sensorimotor cortex. For GM-CSF and Galanin, localization in pyramidal layer V neurons has not been described before. Depending on the particular intracellular distribution of these proteins, we observed differential staining patterns. While the cytokine GM-CSF (Figures 6A–C) and the neuropeptide Galanin (Figures 6G–I) showed a homogeneous cytoplasmic localization in pyramidal neurons, the spot-like distribution of Diablo in neuronal cytoplasm (Figures 6D–F) fits well with its known mitochondrial localization (Martinez-Ruiz et al., 2008). The immunostaining of the transcription modulator vitamin D receptor (VDR), on the other hand, was predominantly observed in the nucleus of the pyramidal projection neurons (Figures 6M–O). Staining for the multifunctional C(2)H(2)-type zinc-finger transcription factor Egr2/Krox20 revealed expression in somata and neurites of cortical neurons, but failed to demonstrate additional glial cell expression (Figures 6J–L). However, immunostaining of the tight junction-related Claudin-11 protein revealed a non-neuronal localization (Figures 6P–R) in cortical layer V and, therefore, is an example for the contribution of non-neuronal cells to spinal injury and treatment-triggered cortical gene expression. The Claudin-11 protein is part of the radial component of CNS myelin which determines the permeability between layers of compact myelin (Bronstein et al., 1997; Morita et al., 1999).
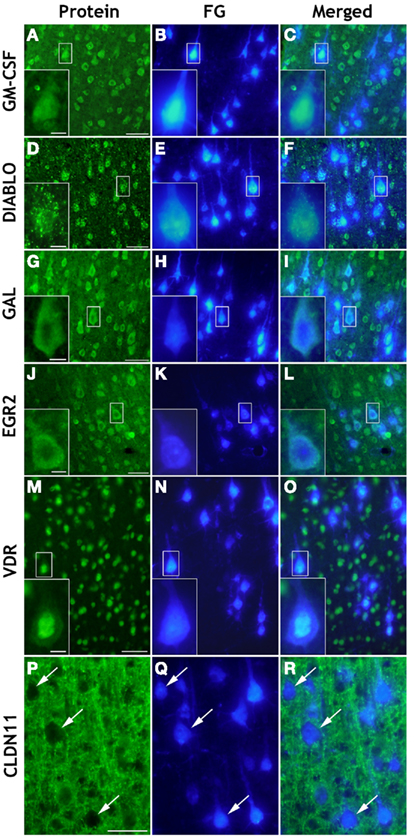
Figure 6. Cellular localization by immunohistochemistry of selected proteins in coronal brain cryosections (20 μm) of sensorimotor cortex layer V. Retrogradely Fluoro-Gold (FG) labeled pyramidal neurons [FG/blue: (B,E,H,K,N) and arrows in (Q)] showed positive immunoreactivity (green) of GM-CSF (A,C), DIABLO/SMAC (D,F), GAL (G,I), EGR2 (J,L), and VDR (M,O). While GM-CSF, GAL, and EGR2/KROX20 were homogeneously distributed in the cytoplasm of retrogradely labeled injured pyramidal neurons, Diablo showed a spot-like cytoplasmic distribution, whereas VDR was mainly detected in the pyramidal cell nucleus. As expected, Claudin-11 (CLDN11) could not be localized in retrogradely FG-labeled cell bodies of pyramidal neurons [see arrows in (P,Q,R)] but it was detected in extraneuronal membranous structures [green: (P,R)]. Scale bars = 50 μm in (A,D,G,J,M,P); 10 μm in the insets.
Discussion
Axotomy of the CST Leads to Specific Temporal Gene Expression Profiles in Cortical Layer V
To gain insight into cortical responses following SCI and regeneration-promoting AST we have investigated gene expression profiles and their temporal changes in layer V of the sensorimotor cortex of rat. We were able to demonstrate the highly complex and extensive dynamic transcriptional reactions in sensorimotor cortex following SCI. Furthermore, this analysis revealed extensive neuroprotective and regeneration-promoting cortical responses to local application of AST (Klapka et al., 2005).
Using microarray hybridization we identified 521 significantly regulated genes which showed a lesion-induced regulation as early as 24 h after injury (see Table 1) indicating early cortical responses to distant SCI. Each of the four pathophysiological stages following SCI (acute, early, and late subacute, chronic) are characterized by a distinct set of significantly enriched functional groups of regulated cortical genes (see Table 2). Interestingly, many of the early affected genes were related to wounding, apoptosis, neurogenesis, and cytoskeletal reorganization suggesting their critical roles in the initial response of the sensorimotor cortex to axotomy of the CST. Over time, not only the numbers of regulated genes (see Table 1) but also the number of enriched functional groups reflecting stage-specific lesion-associated processes (see Table 2) further increased in lesion-only animals reaching a maximum at 3 weeks after injury. Even after 60 dpo the cortical gene expression profile in chronically spinal injured animals still showed persisting alterations in more than 900 genes compared to sham controls. The data obtained from the Allan database (Table S4 in Supplementary Material) as well as the immunostainings presented here (Figure 6) indicate predominant recordings of neuronal rather than non-neuronal expression profiles in the examined spinal injury- or AST-affected cortical layer V. Nevertheless, our data also revealed additional non-neuronal gene regulations executed by neighboring cells, presumably glia, that contribute to the recorded cortical expression profiles (see Figures 6P–R).
AST Treatment Strongly Modifies the Lesion-Induced Cortical Gene Expression Profiles
Until now, studies on gene expression after CNS axotomy focused on lesioned animals lacking therapeutic treatment. AST provides an excellent opportunity to investigate cortical gene expression under regeneration-promoting conditions. As shown here, AST-induced rapid as well as long-lasting massive changes in the lesion-triggered cortical gene expression profile. Already at 1 dpo, 430 genes were differentially regulated in AST rats as compared to the lesion-only control animals (see Table 1). This AST-triggered modification of the lesion response increased over time peaking at 21 dpo. As outlined in Figure 2, we observed several interesting responses. First, a substantial number of regulated transcripts in lesion-only control animals were not changed by AST, indicating that these regulations were triggered by SCI regardless of treatment. Second, there was a group of injury-triggered regulations that were boosted by AST, indicating that some of the injury responses were already pointing into a supportive direction, but needed to be reinforced in order to stimulate the regenerative process. Third, we noticed a very interesting group of counter-regulated genes that increased with time. The latter group of genes indicates that AST was able to reverse numerous of the injury-induced regulations that presumably could be harmful or detrimental for the regenerative process. Finally, a surprisingly large proportion of transcript regulations was AST-specific (∼42%) demonstrating a treatment-dependent fundamental change toward a cortical regeneration-associated program, rather than a slight modulation of the lesion-induced gene expression profile.
Functional Classification Suggests a Modification of Gene Expression Profiles toward Survival and Axon Growth-Promoting Processes by AST.
Our study further reveals that numerous genes with known functions during development show an AST-mediated induction (see Table 4). For example, within the first week of AST we could demonstrate an early and sustained upregulation of hand2, encoding a bHLH transcription factor with crucial functions in neurogenesis and survival of sympathetic neurons as well as in neuronal differentiation (Doxakis et al., 2008; Schmidt et al., 2009). Interestingly, midkine (mdk) encoding a heparin-binding factor promoting growth, survival and/or migration of various target cells (Michikawa et al., 1993; Owada et al., 1999; Zou et al., 2006) and a critical factor in regeneration of injured peripheral nerves (Sakakima et al., 2009) showed a biphasic AST-induced expression pattern with significant upregulation starting at 7dpo. The ID HLH proteins act as transcriptional regulators during development and are generally considered as modulators of differentiation (Stewart et al., 1997; Norton et al., 1998). Similar to the AST-induced regulation described in Table 4 Id-1, -2, and -3 showed axotomy-induced up-regulation of expression also in peripheral nerves (Gillen et al., 1995; Stewart et al., 1997; Bosse et al., 2006). Furthermore, a very recent paper specified that id2 reprograms the intrinsic growth state of adult DRG neurons and promotes neuritogenesis following SCI (Yu et al., 2011). Even later, at 21dpo, we observed a distinct AST-mediated upregulation of developmental genes like fez1, a gene required for the generation, differentiation, and axon targeting of layer V corticospinal motor neurons (Chen et al., 2005; Molyneaux et al., 2005) and involved in neurite outgrowth (Miyoshi et al., 2003). The developmental and neuron-specific regulator of Fas/CD95-mediated apoptosis faim2, which exhibits neuroprotective potential in the context of neurological diseases (Reich et al., 2011) shows a late AST-induced upregulation. Furthermore, AST-induced developmental genes including, e.g., transcription factor mash1 and the homeobox gene hoxA1 were repressed at 7 dpo after injury when a concomitant regeneration-promoting treatment was lacking. Therefore, our analysis suggests that, at least, partial and selective reactivation and maintenance of specific developmental responses could be an important aspect of successful repair in both the PNS (Vogelaar et al., 2003; Bosse et al., 2006) and the CNS, although the repertoire of the re-induced developmental genes may be different.
Well-regulated interactions with attractive or repulsive guidance cues are important for successful axonal regeneration (for review see Koeberle and Bähr, 2004; Bolsover et al., 2008; Curinga et al., 2008). Accordingly, we observed an AST-triggered regulation of genes involved in axon guidance/repulsion (down-regulation of unc5A, unc5B, sema6b, reticulon 4 receptor) and axonogenesis (up-regulation of gal, kalrn, egr1, mtap1a) in treated animals (see Table 4). This group of genes became more prominent over time and is likely to participate in AST-promoted regenerative axon growth.
We further observed an AST-associated over-representation of anti-apoptotic and an under-representation of pro-apoptotic genes (see Tables 4) indicating that AST drives cortical gene expression toward protective as opposed to cell death-promoting pathways. These findings on the molecular level correlate well with the previously published neuroprotective effects of AST (Klapka et al., 2005). Interestingly, the AST-triggered changes in the expression of genes involved in apoptosis/cell survival occurred in a biphasic mode at early (1 dpo) as well as late time points (60 dpo), suggesting distinct phases of neural protection in the acute phase and during later stages after injury and treatment (see Table 2).
Regulated Genes with Proven Functions in Axonal Regeneration
On the level of individual genes we could demonstrate that genes positively involved in axon growth and guidance were predominantly up-regulated in AST animals throughout the entire period studied (see Table 4). Conversely, transcripts encoding growth inhibitory molecules were largely down-regulated. The same observations were made in the groups of genes involved in regulation of apoptosis/cell survival. While numerous survival-promoting and anti-apoptotic genes were up-regulated, many pro-apoptotic genes were down-regulated, especially at the early time-point (see Table 4). Our data indicate that the statistical calculations of over- and under-represented functional groups of regulated genes in AST animals can, indeed, be confirmed at the level of individual genes with known functions.
Some regeneration-associated genes (RAGs), which are well known in the context of peripheral nerve regeneration, were also found in this CNS study. Galanin, for example, is up-regulated in peripheral neurons after injury, and is regarded as a “regenerating axon” marker (Villar et al., 1989; Suarez et al., 2006). We found persisting up-regulation of galanin mRNA under AST conditions at all time-points studied (see Figure 5), suggesting long-term support of central axon regeneration, which was corroborated by its proven localization in pyramidal neurons of cortical layer V (see Figure 6).
Midkine (Mdk), a heparin-binding growth factor, is implicated in development and repair of various tissues, including the nervous system. It has been reported that Mdk promotes peripheral nerve regeneration (Sakakima et al., 2009), protects from focal brain ischemia and augments central neurogenesis (Ishikawa et al., 2009). Therefore, the long-lasting AST-triggered induction of mdk (7–60 dpo) as observed here may indicate a critical role of mdk in AST-induced neuroprotection and subsequent regeneration.
The relevance of vitamin D, a neuroactive seco-steroid, during CNS development is also receiving increasing recognition (Veenstra et al., 1998; Keisala et al., 2009). Its receptor (VDR) belongs to the family of trans-acting transcriptional regulatory factors, and genetic vdr ablation produces severe motor impairment (Kalueff et al., 2004) confirming the importance of the vitamin D system in motor functions. Thus, it is likely that the AST-mediated counter-regulation of vdr in pyramidal neurons (see Figure 6) compared to the diminished expression after lesion (see Figure 5) may also contribute to the beneficial AST effect after SCI.
GM-CSF is a cytokine that not only controls the production, differentiation, and function of granulocytes and macrophages, but also induces dose-dependent neuronal differentiation of neural stem cells (Krüger et al., 2007). GM-CSF and its receptors are broadly expressed throughout the brain and known to be up-regulated after ischemic insults counteracting programmed cell death (Schäbitz et al., 2008). Further, GM-CSF-administration in a rat SCI-model resulted in significantly reduced glial scar formation and enhanced integrity of axonal structure (Huang et al., 2009b). Thus, the early but sustained (1–21 dpo) AST-mediated induction of csf2/gm-csf (see Figure 5) suggests a role both in the rescue of pyramidal cells in cortical layer V where GM-CSF is expressed (see Figure 6) as well as in successful regeneration of CST axons as previously observed in AST animals (Klapka et al., 2005).
The transcription factor c-Jun/AP-1 has been identified to be differentially expressed after axotomy (Broude et al., 1997; Kenney and Kocsis, 1997; Vaudano et al., 1998) and associated with apoptosis in the nervous system (Herdegen et al., 1997). Consistently, the AST-triggered counter-regulation of c-jun/ap-1 mRNA in cortical neurons back down to sham levels as observed in the present investigation could reduce putative pro-apoptotic effects of c-Jun/AP-1 and thus support neuronal survival.
Conclusion
In this unique molecular systems investigation we observed that cortical neurons are capable of dynamic responses on the transcriptional level to SCI, and that they further modify their complex lesion-triggered gene expression profile in response to distant regeneration-promoting treatment (AST) in spinal cord. We were able to correlate AST-promoted effects, namely retrograde neuronal protection, enhanced regenerative axon growth and functional (locomotor) recovery, with significant changes in cortical gene expression profiles. Obviously, AST not only modifies the local environment impeding spinal cord regeneration by reduction of fibrous scarring in the injured spinal cord, but, in addition, the treatment strikingly changes the intrinsic capacity of cortical pyramidal neurons toward enhanced cell maintenance and axonal regeneration. The data suggest that combined administration of the iron chelator and cyclicAMP could successfully facilitate axonal regeneration by (i) reducing the accumulation of growth inhibitory factors at the lesion site, (ii) modulating the immune response, (iii) increasing growth factor availability, (iv) regulating cytoskeletal reorganization and protein synthesis, and (v) providing neuroprotection by sequestering redox-active iron and thereby inhibiting the Fenton reaction as well stabilizing the neuroprotective transcriptional activator HIF-1α. Further detailed studies will be required in order to elucidate the essential molecular players and participating pathways employed by AST treatment to improve axonal regeneration after traumatic SCI.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors gratefully acknowledge the critical reading of the manuscript and suggestions of Christian Stephan on the bioinformatic analysis. This work was supported by the DFG (Contract grant number: SFB 590/TP C2), a grant of the faculty of medicine of the Heinrich-Heine-University, the German Paraplegia Foundation (DSQ) and the International Institute for Research in Paraplegia (IRP, Switzerland).
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/molecular_neuroscience/10.3389/fnmol.2011.00026/abstract
Table S1. Overlaps between gene regulations in Lesion and AST paradigms at different time points. The given overlap between groups of regulated genes is shown as percentage of the number of overlaps merely expected by chance as calculated from the group sizes and the total number of genes on the chip. Each possible comparison for the time point-specific sets of up- and down-regulated genes in the groups Lesion vs. Sham and AST vs. Lesion is illustrated. Overlaps showing significant over- or under-representation are shown in orange (over-representation) and blue (under-representation), respectively. Interestingly, some of the overlapping effects were prolonged over more than one time point. For example a significant proportion of lesion-induced genes detected at 1 dpo was down-regulated by AST at 1 (259%) and also at 7 (331%) dpo.
Table S2. Significantly regulated genes showing AST-induced counter-regulation or a temporal shift according to Figure 3. The respective regulated Probeset IDs are shown.
Table S3. Lists of significantly regulated genes associated with the GO-terms axon guidance, axonogenesis, structural constituent of ribosome, transcription, and anti-apoptosis. The respective regulated Probeset IDs are shown. The number of regulations for the comparison of AST vs. Lesion are shown in Figure 4.
Table S4. List of significantly regulated transcripts identified in the present investigation, which had previously been localized by in situ hybridization in layer V cells of mouse sensorimotor cortex (see Allan Brain Atlas; http://mouse.brain-map.org/welcome.do)
Figure S1. Examples for cellular localization of selected transcripts by ISH in sensorimotor cortex of rat as found in the mouse brain atlas of the Allen Institute for Brain Science. Large pyramidal cell bodies in cortical layer V show strong signals for nrp1 and robo1, repectively. Scale bars = 100 μm.
References
Abankwa, D., Küry, P., and Müller, H. W. (2002). Dynamic changes in gene expression profiles following axotomy of projection fibers in the mammalian CNS. Mol. Cell. Neurosci. 21, 421–435.
Affymetrix. (2001). Statistical Algorithms Reference Guide [Technical Report]. Santa Clara, CA: Affymetrix. Available at: www.affymetrix.com/support/technical/technotes/statistical_reference_guide.pdf
Affymetrix. (2005). Guide to Probe Logarithmic Intensity Error (PLIER) Estimation, Affymetrix [published online].
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T., Harris, M. A., Hill, D. P., Issel-Tarver, L., Kasarskis, A., Lewis, S., Matese, J. C., Richardson, J. E., Ringwald, M., Rubin, G. M., and Sherlock, G. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29.
Bareyre, F. M., Haudenschild, B., and Schwab, M. E. (2002). Long-lasting sprouting and gene expression changes induced by the monoclonal antibody IN-1 in the adult spinal cord. J. Neurosci. 22, 7097–7110.
Barrette, B., Vallières, N., Dubé, M., and Lacroix, S. (2007). Expression profile of receptors for myelin-associated inhibitors of axonal regeneration in the intact and injured mouse central nervous system. Mol. Cell. Neurosci. 34, 519–538.
Bolsover, S., Fabes, J., and Anderson, P. N. (2008). Axonal guidance molecules and the failure of axonal regeneration in the adult mammalian spinal cord. Restor. Neurol. Neurosci. 26, 117–130.
Bosse, F., Hasenpusch-Theil, K., Küry, P., and Müller, H. W. (2006). Gene expression profiling reveals that peripheral nerve regeneration is a consequence of both novel injury-dependent and reactivated developmental processes. J. Neurochem. 96, 1441–1457.
Brazda, N., and Müller, H. W. (2009). Pharmacological modification of the extracellular matrix to promote regeneration of the injured brain and spinal cord. Prog. Brain Res. 175, 269–281.
Bronstein, J. M., Micevych, P. E., and Chen, K. (1997). Oligodendrocyte-specific protein (OSP) is a major component of CNS myelin. J. Neurosci. Res. 50, 713–720.
Broude, E., McAtee, M., Kelley, M. S., and Bregman, B. S. (1997). c-Jun expression in adult rat dorsal root ganglion neurons: differential response after central or peripheral axotomy. Exp. Neurol. 148, 367–377.
Bundesen, L. Q., Scheel, T. A., Bregman, B. S., and Kromer, L. F. (2003). Ephrin-B2 and EphB2 regulation of astrocyte-meningeal fibroblast interactions in response to spinal cord lesions in adult rats. J. Neurosci. 23, 7789–7800.
Chen, B., Schaevitz, L. R., and McConnell, S. K. (2005). Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc. Natl. Acad. Sci. U.S.A. 102, 17184–17189.
Curinga, G., Snow, D. M., and Smith, G. M. (2008). Mechanisms regulating interpretation of guidance cues during development, maturation, and following injury. Rev. Neurosci. 19, 213–226.
Davies, J. E., Tang, X., Bournat, J. C., and Davies, S. J. A. (2006). Decorin promotes plasminogen/plasmin expression within acute spinal cord injuries and by adult microglia in vitro. J. Neurotrauma 23, 397–408.
Davies, J. E., Tang, X., Denning, J. W., Archibald, S. J., and Davies, S. J. (2004). Decorin suppresses neurocan, brevican, phosphacan and NG2 expression and promotes axon growth across adult rat spinal cord injuries. Eur. J. Neurosci. 19, 1226–1242.
de Winter, F., Oudega, M., Lankhorst, A. J., Hamers, F. P., Blits, B., Ruitenberg, M. J., Pasterkamp, R. J., Gispen, W. H., and Verhaagen, J. (2002). Injury-induced class 3 semaphorin expression in the rat spinal cord. Exp. Neurol. 175, 61–75.
Dennis, G. Jr., Sherman, B. T., Hosack, D. A., Yang, J., Gao, W., Lane, H. C., and Lempicki, R. A. (2003). DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 4, P3.
Doxakis, E., Howard, L., Rohrer, H., and Davies, A. M. (2008). HAND transcription factors are required for neonatal sympathetic neuron survival. EMBO Rep. 9, 1041–1047.
Fawcett, J. W., and Asher, R. A. (1999). The glial scar and central nervous system repair. Brain Res. Bull. 49, 377–391.
Filbin, M. T. (2003). Myelin-associated inhibitors of axonal regeneration in the adult mammalian CNS. Nat. Rev. Neurosci. 4, 1–11.
Fournier, A. E., and McKerracher, L. (1997). Expression of specific tubulin isotypes increases during regeneration of injured CNS neurons, but not after the application of brain-derived neurotrophic factor (BDNF). J. Neurosci. 17, 4623–4632.
Gillen, C., Gleichmann, M., Spreyer, P., and Müller, H. W. (1995). Differentially expressed genes after peripheral nerve injury. J. Neurosci. Res. 42, 159–171.
Hains, B. C., Black, J. A., and Waxman, S. G. (2003). Primary cortical motor neurons undergo apoptosis after axotomizing spinal cord injury. J. Comp. Neurol. 462, 328–341.
Herdegen, T., Skene, P., and Bähr, M. (1997). The c-Jun transcription factor-bipotential mediator of neuronal death, survival and regeneration. Trends Neurosci. 20, 227–231.
Houweling, D. A., Bär, P. R., Gispen, W. H., and Joosten, E. A. (1998). Spinal cord injury: bridging the lesion and the role of neurotrophic factors in repair. Prog. Brain Res. 117, 455–471.
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2009a). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57.
Huang, X., Kim, J., Kong, T. H., Park, S. R., Ha, Y., Kim, M. H., Park, H., Yoon, S. H., Park, H. C., Park, J. O., Min, B. H., and Choi, B. H. (2009b). GM-CSF inhibits glial scar formation and shows long-term protective effect after spinal cord injury. J. Neurol. Sci. 277, 87–97.
Irizarry, R. A., Gautier, L., and Cope, L. (2003a). “An R package for analysis of Affymetrix oligonucleotide arrays,” in The Analysis of Gene Expression Data: Methods and Software, eds G. Parmigiani, E. Garrett, R. Irizarry, and S. Zeger (Berlin: Springer), 102–119.
Irizarry, R. A., Hobbs, B., Collin, F., Beazer-Barclay, Y. D., Antonellis, K. J., Scherf, U., and Speed, T. P. (2003b). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264.
Irwin, N., Baekelandt, V., Goritchenko, L., and Benowitz, L. I. (1997). Identification of two proteins that bind to a pyrimidine-rich sequence in the 3′-untranslated region of GAP-43 mRNA. Nucleic Acids Res. 25, 1281–1288.
Ishikawa, E., Ooboshi, H., Kumai, Y., Takada, J., Nakamura, K., Ago, T., Sugimori, H., Kamouchi, M., Kitazono, T., Ibayashi, S., and Iida, M. (2009). Midkine gene transfer protects against focal brain ischemia and augments neurogenesis. J. Neurol. Sci. 285, 78–84.
Israelsson, C., Lewén, A., Kylberg, A., Usoskin, D., Althini, S., Lindeberg, J., Deng, C. X., Fukuda, T., Wang, Y., Kaartinen, V., Mishina, Y., Hillered, L., and Ebendal, T. (2006). Genetically modified bone morphogenetic protein signalling alters traumatic brain injury-induced gene expression responses in the adult mouse. J. Neurosci. Res. 84, 47–57.
Ito, M., Natsume, A., Takeuchi, H., Shimato, S., Ohno, M., Wakabayashi, T., and Yoshida, J. (2009). Type I interferon inhibits astrocytic gliosis and promotes functional recovery after spinal cord injury by deactivation of the MEK/ERK pathway. J. Neurotrauma 26, 41–53.
Jones, L. L., Yamaguchi, Y., Stallcup, W. B., and Tuszynski, M. H. (2002). NG2 is a major chondroitin sulfate proteoglycan produced after spinal cord injury and is expressed by macrophages and oligodendrocyte progenitors. J. Neurosci. 22, 2792–2803.
Kalueff, A. V., Lou, Y., Laaksi, I., and Tuohimaa, P. (2004). Impaired motor performance in mice lacking neurosteroid vitamin D receptors. Brain Res. Bull. 64, 25–29.
Keisala, T., Minasyan, A., Lou, Y., Zou, J., Kalueff, A. V., Pyykkö, I., and Tuohimaa, P. (2009). Premature aging in vitamin D receptor mutant mice. J. Steroid Biochem. Mol. Biol. 115, 91–97.
Kenney, A. M., and Kocsis, J. D. (1997). Timing of c-jun protein induction in lumbar dorsal root ganglia after sciatic nerve transection varies with lesion distance. Brain Res. 751, 90–95.
Klapka, N., Hermanns, S., Straten, G., Masanneck, C., Duis, S., Hamers, F. P., Müller, D., Zuschratter, W., and Müller, H. W. (2005). Suppression of fibrous scarring in spinal cord injury of rat promotes long-distance regeneration of corticospinal tract axons, rescue of primary motoneurons in somatosensory cortex and significant functional recovery. Eur. J. Neurosci. 22, 3047–3058.
Klapka, N., and Müller, H. W. (2006). Collagen matrix in spinal cord injury. J. Neurotrauma 23, 422–436.
Koeberle, P. D., and Bähr, M. (2004). Growth and guidance cues for regenerating axons: where have they gone? J. Neurobiol. 59, 162–180.
Krüger, C., Laage, R., Pitzer, C., Schäbitz, W., and Schneider, A. (2007). The hematopoietic factor GM-CSF (granulocyte-macrophage colony-stimulating factor) promotes neuronal differentiation of adult neural stem cells in vitro. BMC Neurosci. 8, 88. doi: 10.1186/1471-2202-8-88
Kruse, F., Brazda, N., Küry, P., Bosse, F., and Müller, H. W. (2008). “Analyzing complex gene expression profiles in sensorimotor cortex following spinal cord injury and regeneration promoting treatment,” in Neural Degeneration and Repair. Gene Expression Profiling, Proteomics and Systems Biology, ed. H. W. Müller (Weinheim: Wiley-VCH), 61–89.
Küry, P., Abankwa, D., Kruse, F., and Müller, H. W. (2004). Gene expression profiling reveals multiple novel intrinsic and extrinsic factors associated with axonal regeneration failure. Eur. J. Neurosci. 19, 32–42.
Lein, E. S., Hawrylycz, M. J., Ao, N., Ayres, M., Bensinger, A., Bernard, A., Boe, A. F., Boguski, M. S., Brockway, K. S., Byrnes, E. J., Chen, L., Chen, L., Chen, T. M., Chin, M. C., Chong, J., Crook, B. E., Czaplinska, A., Dang, C. N., Datta, S., Dee, N. R., Desaki, A. L., Desta, T., Diep, E., Dolbeare, T. A., Donelan, M. J., Dong, H. W., Dougherty, J. G., Duncan, B. J., Ebbert, A. J., Eichele, G., Estin, L. K., Faber, C., Facer, B. A., Fields, R., Fischer, S. R., Fliss, T. P., Frensley, C., Gates, S. N., Glattfelder, K. J., Halverson, K. R., Hart, M. R., Hohmann, J. G., Howell, M. P., Jeung, D. P., Johnson, R. A., Karr, P. T., Kawal, R., Kidney, J. M., Knapik, R. H., Kuan, C. L., Lake, J. H., Laramee, A. R., Larsen, K. D., Lau, C., Lemon, T. A., Liang, A. J., Liu, Y., Luong, L. T., Michaels, J., Morgan, J. J., Morgan, R. J., Mortrud, M. T., Mosqueda, N. F., Ng, L. L., Ng, R., Orta, G. J., Overly, C. C., Pak, T. H., Parry, S. E., Pathak, S. D., Pearson, O. C., Puchalski, R. B., Riley, Z. L., Rockett, H. R., Rowland, S. A., Royall, J. J., Ruiz, M. J., Sarno, N. R., Schaffnit, K., Shapovalova, N. V., Sivisay, T., Slaughterbeck, C. R., Smith, S. C., Smith, K. A., Smith, B. I., Sodt, A. J., Stewart, N. N., Stumpf, K. R., Sunkin, S. M., Sutram, M., Tam, A., Teemer, C. D., Thaller, C., Thompson, C. L., Varnam, L. R., Visel, A., Whitlock, R. M., Wohnoutka, P. E., Wolkey, C. K., Wong, V. Y., Wood, M., Yaylaoglu, M. B., Young, R. C., Youngstrom, B. L., Yuan, X. F., Zhang, B., Zwingman, T. A., and Jones, A. R. (2007). Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176.
Li, C., and Wong, W. H. (2001). Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. U.S.A. 98, 31–36.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408.
Martinez-Ruiz, G., Maldonado, V., Ceballos-Cancino, G., Grajeda, J. P. R., and Melendez-Zajgla, J. (2008). Role of Smac/DIABLO in cancer progression. J. Exp. Clin. Cancer Res. 27, 48.
Mason, M. R. J., Lieberman, A. R., and Anderson, P. N. (2003). Corticospinal neurons up-regulate a range of growth-associated genes following intracortical, but not spinal, axotomy. Eur. J. Neurosci. 18, 789–802.
Michikawa, M., Xu, R. Y., Muramatsu, H., Muramatsu, T., and Kim, S. U. (1993). Midkine is a mediator of retinoic acid induced neuronal differentiation of embryonal carcinoma cells. Biochem. Biophys. Res. Commun. 192, 1312–1318.
Mikucki, S. A., and Oblinger, M. M. (1991). Corticospinal neurons exhibit a novel pattern of cytoskeletal gene expression after injury. J. Neurosci. Res. 30, 213–225.
Miyoshi, K., Honda, A., Baba, K., Taniguchi, M., Oono, K., Fujita, T., Kuroda, S., Katayama, T., and Tohyama, M. (2003). Disrupted-In-Schizophrenia 1, a candidate gene for schizophrenia, participates in neurite outgrowth. Mol Psychiatry 8, 685–694.
Molyneaux, B. J., Arlotta, P., Hirata, T., Hibi, M., and Macklis, J. D. (2005). Fezl is required for the birth and specification of corticospinal motor neurons. Neuron 47, 817–831.
Morgenstern, D. A., Asher, R. A., and Fawcett, J. W. (2002). Chondroitin sulphate proteoglycans in the CNS injury response. Prog. Brain Res. 137, 313–332.
Morita, K., Sasaki, H., Fujimoto, K., Furuse, M., and Tsukita, S. (1999). Claudin-11/OSP-based tight junctions of myelin sheaths in brain and Sertoli cells in testis. J. Cell Biol. 145, 579–588.
Niclou, S. P., Franssen, E. H., Ehlert, E. M., Taniguchi, M., and Verhaagen, J. (2003). Meningeal cell-derived semaphorin 3A inhibits neurite outgrowth. Mol. Cell. Neurosci. 24, 902–912.
Norton, J. D., Deed, R. W., Craggs, G., and Sablitzky, F. (1998). Id helix-loop-helix proteins in cell growth and differentiation. Trends Cell Biol. 8, 58–65.
Owada, K., Sanjyo, N., Kobayashi, T., Kamata, T., Mizusawa, H., Muramatsu, H., Muramatsu, T., and Michikawa, M. (1999). Midkine inhibits apoptosis via extracellular signal regulated kinase (ERK) activation in PC12 cells. J. Med. Dent. Sci. 46, 45–51.
Poulsen, C. B., Penkowa, M., Borup, R., Nielsen, F. C., Cáceres, M., Quintana, A., Molinero, A., Carrasco, J., Giralt, M., and Hidalgo, J. (2005). Brain response to traumatic brain injury in wild-type and interleukin-6 knockout mice: a microarray analysis. J. Neurochem. 92, 417–432.
Raghavendra Rao, V. L., Dhodda, V. K., Song, G., Bowen, K. K., and Dempsey, R. J. (2003). Traumatic brain injury-induced acute gene expression changes in rat cerebral cortex identified by GeneChip analysis. J. Neurosci. Res. 71, 208–219.
Reich, A., Spering, C., Gertz, K., Harms, C., Gerhardt, E., Kronenberg, G., Nave, K. A., Schwab, M., Tauber, S. C., Drinkut, A., Harms, K., Beier, C. P., Voigt, A., Göbbels, S., Endres, M., and Schulz, J. B. (2011). Fas/CD95 regulatory protein Faim2 is neuroprotective after transient brain ischemia. J. Neurosci. 31, 225–233.
Sakakima, H., Yoshida, Y., Yamazaki, Y., Matsuda, F., Ikutomo, M., Ijiri, K., Muramatsu, H., Muramatsu, T., and Kadomatsu, K. (2009). Disruption of the midkine gene (Mdk) delays degeneration and regeneration in injured peripheral nerve. J. Neurosci. Res. 87, 2908–2915.
Salin, P., and Chesselet, M. F. (1992). Paradoxical increase in striatal neuropeptide gene expression following ischemic lesions of the cerebral cortex. Proc. Natl. Acad. Sci. U.S.A. 89, 9954–9958.
Schäbitz, W., Krüger, C., Pitzer, C., Weber, D., Laage, R., Gassler, N., Aronowski, J., Mier, W., Kirsch, F., Dittgen, T., Bach, A., Sommer, C., and Schneider, A. (2008). A neuro-protective function for the hematopoietic protein granulocyte-macrophage colony stimulating factor (GM-CSF). J. Cereb. Blood Flow Metab. 28, 29–43.
Schiwy, N., Brazda, N., and Müller, H. W. (2009). Enhanced regenerative axon growth of multiple fiber populations in traumatic spinal cord injury following scar suppressing treatment. Eur. J. Neurosci. 30, 1544–1553.
Schmidt, M., Lin, S., Pape, M., Ernsberger, U., Stanke, M., Kobayashi, K., Howard, M. J., and Rohrer, H. (2009). The bHLH transcription factor Hand2 is essential for the maintenance of noradrenergic properties in differentiated sympathetic neurons. Dev. Biol. 329, 191–200.
Snow, D. M., Lemmon, V., Carrino, D. A., Caplan, A. I., and Silver, J. (1990). Sulfated proteoglycans in astroglial barriers inhibit neurite outgrowth in vitro. Exp. Neurol. 109, 111–130.
Stewart, H. J., Zoidl, G., Rossner, M., Brennan, A., Zoidl, C., Nave, K. A., Mirsky, R., and Jessen, K. R. (1997). Helix-loop-helix proteins in Schwann cells: a study of regulation and subcellular localization of Ids, REB, and E12/47 during embryonic and postnatal development. J. Neurosci. Res. 50, 684–701.
Suarez, V., Guntinas-Lichius, O., Streppel, M., Ingorokva, S., Grosheva, M., Neiss, W. F., Angelov, D. N., and Klimaschewski, L. (2006). The axotomy-induced neuropeptides galanin and pituitary adenylate cyclase-activating peptide promote axonal sprouting of primary afferent and cranial motor neurones. Eur. J. Neurosci. 24, 1555–1564.
Tetzlaff, W., Kobayashi, N. R., Giehl, K. M., Tsui, B. J., Cassar, S. L., and Bedard, A. M. (1994). Response of rubrospinal and corticospinal neurons to injury and neurotrophins. Prog. Brain Res. 103, 271–286.
Tian, D., Yu, Z., Xie, M., Bu, B., Witte, O. W., and Wang, W. (2006). Suppression of astroglial scar formation and enhanced axonal regeneration associated with functional recovery in a spinal cord injury rat model by the cell cycle inhibitor olomoucine. J. Neurosci. Res. 84, 1053–1063.
Vaudano, E., Campbell, G., Hunt, S. P., and Lieberman, A. R. (1998). Axonal injury and peripheral nerve grafting in the thalamus and cerebellum of the adult rat: up-regulation of c-jun and correlation with regenerative potential. Eur. J. Neurosci. 10, 2644–2656.
Veenstra, T. D., Prüfer, K., Koenigsberger, C., Brimijoin, S. W., Grande, J. P., and Kumar, R. (1998). 1,25-Dihydroxyvitamin D3 receptors in the central nervous system of the rat embryo. Brain Res. 804, 193–205.
Villar, M. J., Cortés, R., Theodorsson, E., Wiesenfeld-Hallin, Z., Schalling, M., Fahrenkrug, J., Emson, P. C., and Hökfelt, T. (1989). Neuropeptide expression in rat dorsal root ganglion cells and spinal cord after peripheral nerve injury with special reference to galanin. Neuroscience 33, 587–604.
Vogelaar, C. F., Hoekman, M. F. M., Gispen, W. H., and Burbach, J. P. H. (2003). Homeobox gene expression in adult dorsal root ganglia during sciatic nerve regeneration: is regeneration a recapitulation of development? Eur. J. Pharmacol. 480, 233–250.
Wu, Z., and Irizarry, R. A. (2004). Preprocessing of oligonucleotide array data. Nat. Biotechnol. 22, 656–658.
Yu, P., Zhang, Y. P., Shields, L. B., Zheng, Y., Hu, X., Hill, R., Howard, R., Gu, Z., Burke, D. A., Whittemore, S. R., Xu, X. M., and Shields, C. B. (2011). Inhibitor of DNA binding 2 promotes sensory axonal growth after SCI. Exp Neurol. 231, 38–44.
Yurchenco, P. D., and Schittny, J. C. (1990). Molecular architecture of basement membranes. FASEB J. 4, 1577–1590.
Keywords: anti-scarring treatment, axonal regeneration, axotomy, corticospinal tract, lesion scar, microarray, sensorimotor cortex, spinal cord injury
Citation: Kruse F, Bosse F, Vogelaar CF, Brazda N, Küry P, Gasis M and Müller HW (2011) Cortical gene expression in spinal cord injury and repair: insight into the functional complexity of the neural regeneration program. Front. Mol. Neurosci. 4:26. doi: 10.3389/fnmol.2011.00026
Received: 27 June 2011;
Paper pending published: 28 July 2011;
Accepted: 05 September 2011;
Published online: 29 September 2011.
Edited by:
Simone Di Giovanni, HIH, University of Tuebingen, GermanyReviewed by:
Paul Lingor, University Medicine Göttingen, GermanyJoost Verhaagen, Netherlands Institute fo Neuroscience, Netherlands
Copyright: © 2011 Kruse, Bosse, Vogelaar, Brazda, Küry, Gasis and Müller. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Hans W. Müller, Department of Neurology and Biomedical Research Center, Heinrich-Heine-University, Moorenstr. 5, 40225 Düsseldorf, Germany. e-mail: hanswerner.mueller@uni-duesseldorf.de