Molecular codes for neuronal individuality and cell assembly in the brain
- 1KOKORO-Biology Group, Graduate School of Frontier Biosciences, Laboratories for Integrated Biology, Osaka University, Yamadaoka, Suita, Osaka, Japan
- 2CREST, Osaka University, Suita, Japan
The brain contains an enormous, but finite, number of neurons. The ability of this limited number of neurons to produce nearly limitless neural information over a lifetime is typically explained by combinatorial explosion; that is, by the exponential amplification of each neuron's contribution through its incorporation into “cell assemblies” and neural networks. In development, each neuron expresses diverse cellular recognition molecules that permit the formation of the appropriate neural cell assemblies to elicit various brain functions. The mechanism for generating neuronal assemblies and networks must involve molecular codes that give neurons individuality and allow them to recognize one another and join appropriate networks. The extensive molecular diversity of cell-surface proteins on neurons is likely to contribute to their individual identities. The clustered protocadherins (Pcdh) is a large subfamily within the diverse cadherin superfamily. The clustered Pcdh genes are encoded in tandem by three gene clusters, and are present in all known vertebrate genomes. The set of clustered Pcdh genes is expressed in a random and combinatorial manner in each neuron. In addition, cis-tetramers composed of heteromultimeric clustered PCDH isoforms represent selective binding units for cell-cell interactions. Here I present the mathematical probabilities for neuronal individuality based on the random and combinatorial expression of clustered Pcdh isoforms and their formation of cis-tetramers in each neuron. Notably, clustered Pcdh gene products are known to play crucial roles in correct axonal projections, synaptic formation, and neuronal survival. Their molecular and biological features induce a hypothesis that the diverse clustered Pcdh molecules provide the molecular code by which neuronal individuality and cell assembly permit the combinatorial explosion of networks that supports enormous processing capability and plasticity of the brain.
Introduction
The mammalian brain is a complex multi-cellular system composed of an enormous number of cells, including neurons and glia. In the brain, the individual neurons are highly differentiated and well organized into neural networks that generate various brain functions, and the activity of each neuron reflects the encoded information.
Recent progress in neuroscience has revealed mechanisms by which many brain functions are controlled, but essential questions remain concerning the precise nature of information processing in the brain (reviewed by Buzsaki, 2010). How can nearly limitless number of information be processed by a finite number of neurons? How can such information be integrated with other information in the brain? How are different sets of information processed in parallel? The answers to these “how” questions require the existence of a basic neuronal code for information processing in the brain (reviewed by Sakurai, 1999). An individual neuron, the basic functional unit of the brain, has a specific firing activity, and is uniquely coordinated in a circuit with many other neurons in response to specific stimuli. A single neuron can have several to 10,000 synaptic contacts on it, and therefore receive several to ten thousand inputs.
Donald Hebb hypothesized that a discrete interconnected group of active neurons, a “cell assembly,” represents a distinct cognitive entity (Hebb, 1949). Although the experimental identification of these hypothesized cell assemblies proved difficult for decades, recent rapid progress in the large-scale recording from individual neurons has experimentally defined putative cell assemblies (reviewed by Buzsaki, 2010). Under Hebb's cell assembly hypothesis, a nearly limitless number of combinatorial neuronal groups can be theoretically produced from the limited number of neurons by combinatorial explosion. Thus, the “how” questions posed above can be solved, at least theoretically, by the cell assembly hypothesis. Furthermore, recent reports show that predictive neuronal activity by spontaneous firing is observed even before an event or experience happens (Kenet et al., 2003; Dragoi and Tonegawa, 2011). These findings might mean that each “cell assembly” is intrinsically predetermined before experiences are processed in the brain.
The immune system is a genetically predetermined system for recognizing external antigens (Tonegawa, 1983; Lieber, 1992). Enormous numbers of diverse immune cells are produced developmentally by the nearly random DNA rearrangement of immunoglobulin and T-cell receptor genes; these cells include the proper immune cells for responding to certain antigens (refer Figure 7). This system can learn and memorize a nearly limitless number of antigens, against which it produces antibodies when an animal is attacked again by the same antigen. The molecular mechanism for the predetermined immune memory system was solved decades ago, when only limited genomic information was available with random combinations. The identification of similar molecular mechanisms may explain the “how” questions of the enormous information processing capability of the brain. In particular, the molecular codes for neuronal individuality and interconnectivity are likely to be important; for example, the discovery of thousands of odorant receptors opened new avenues of investigation in the field of odorant sensory system biology (Buck and Axel, 1991).
By analyzing nerve regeneration, Langley and Sperry similarly hypothesized that there was some type of special chemical relationship between each class of nerve fiber and each class of nerve target cell (Sperry, 1963; Langley, 1895). Sperry's chemoaffinity hypothesis proposed the existence of individual identification tags that linked each axon to only specific target cells. Recent efforts to find “molecular tags” have led to the identification of “gradient molecules.” Complementary gradients of Eph kinases and their ligands, ephrins, play significant roles in establishing topographically organized maps, i.e., the retinotectal map (Cheng et al., 1995; Drescher et al., 1995; McLaughlin and O'Leary, 2005). In addition, axonal guidance molecules and receptors, which guide each axon to its target cells by contact-mediated and diffusible mechanisms, have been identified, and include ephrins, semaphorins, netrins, plexins, robos, slits, and others. The guidance cues act as both attractants and repellents (Dickson, 2002). In addition, specific adhesion and adhesion-inducing proteins are expressed differentially in specific neuronal populations. These include the cadherins and non-clustered Pcdh (∼20 genes, Takeichi, 2007), the neurexins and neuroligins, which have a large number of alternative splicing forms (Sudhof, 2008), and the olfactory receptors (∼1000 genes, Buck and Axel, 1991), which have all been proposed as supporting evidence for (and likely contributors to) the “area code hypothesis” (Dreyer, 1998).
Recent studies reported that two large protein families, Dscam1 in insects and clustered protocadherin (Pcdh) in vertebrates, are promising candidates for the molecular code that stamps individuality and specific interconnectivity on a given neuron (reviewed by Zipursky and Sanes, 2010). In both cases, a large diversity of proteins encoded in a complex genome structure is expressed in combinatorial and random patterns by individual neurons. These proteins mediate homophilic binding and play critical roles in neural development. In particular, the clustered Pcdh family is proposed to provide the molecular basis for neuronal individuality through their combinatorial and random expression, which is conserved in vertebrates, including humans (Yagi, 2008). In this paper, I summarize recent findings about the clustered Pcdh molecules and suggest a hypothesis of candidates for the molecular code for neuronal individuality and cell assembly in the brain.
Clustered Pcdh Molecules
In 1998, the identification of a group of eight homologous transmembrane proteins, called cadherin-related neuronal receptors (CNRs) has been reported (Kohmura et al., 1998). In 1999, Wu and Maniatis found a large gene cluster in the human genome project data by performing a BLAST search for CNRs (Wu and Maniatis, 1999). A total of 52 genes, called clustered Pcdh, are encoded in the human genome at 5q31. Exons encoding extracellular, transmembrane, and short intracellular domains are arranged in three groups called Pcdh-α, Pcdh-β, and Pcdh-γ, which have 15, 15, and 22 members, respectively. The Pcdh-α genes include the 8 CNR genes discovered in mice. The Pcdh-α and Pcdh-γ genes have very large first exons that encode almost an entire molecule, and that the 3 constant exons (exons 2–4) are very small and encode only the last 125–150 amino acids, which are shared by all Pcdh-α and Pcdh-γ genes. Their large exons have multipule promoters and are cis-spliced to the constant exons (Wang et al., 2002a). In addition, there are alternative splicing (A and B) forms in the constant exons of Pcdh-α genes (Sugino et al., 2000). The Pcdh-β cluster has no constant exons. Their cytoplasmic tails are distinct sequences, but highly conserved. In mice, a total of 58 genes are arranged in Pcdh-α, -β, and -γ, which have 14, 22, and 22 members, respectively, (Wu et al., 2001).
The Pcdhs are fascinating for several reasons (Figure 1). First, their ectodomains have cadherin motifs. They belong to the cadherin superfamily, many other members of which play critical roles in developmental processes including synapse formation (Yagi and Takeichi, 2000). Mice lacking Pcdh-α are viable and fertile but have axon projection defects (Hasegawa et al., 2008; Katori et al., 2009). The loss of Pcdh-γ leads to neonatal death with neurological defects, including cell death and decreased numbers of synapses (Wang et al., 2002b). Thus, the Pcdhs are important for building proper neural networks in the brain. Second, they have a remarkable genomic organization, similar to that of immunoglobulin and T-cell receptor gene clusters. The N-terminal extracellular, transmembrane, and short cytoplasmic domains are encoded by a distinct and large exon, while the C-terminal cytoplasmic domain of each protein is identical among the α or γ members (Wu and Maniatis, 1999). Third, Pcdhs are expressed predominantly in the nervous system. Almost all of their isoforms are expressed in a scattered pattern over wide regions of the brain (Esumi et al., 2005; Kaneko et al., 2006; Noguchi et al., 2009; Yokota et al., 2011). In addition, at the single-cell level, individual family members are randomly expressed in combinatorial patterns (Esumi et al., 2005; Kaneko et al., 2006). Fourth, the gene regulation of Pcdhs is epigenetically controlled independently and monoallelically (Tasic et al., 2002; Kawaguchi et al., 2008). Their random expression in each neuron depends on the structure of the gene cluster (Figure 2) (Noguchi et al., 2009), and is controlled by cis-regulatory elements that independently influence the α and β gene clusters (Figure 3) (Ribich et al., 2006; Yokota et al., 2011). Fifth, the Pcdh proteins form heteromultimeric protein oligomers. The heterotetramer formed by the Pcdh-γ proteins is a homophilic binding unit that induces cell-cell adhesion and interaction (Figure 4) (Schreiner and Weiner, 2010). Finally, Pcdh orthologs are present in vertebrates but not in invertebrates (Hill et al., 2001; Noonan et al., 2004b; Hirayama and Yagi, 2006).
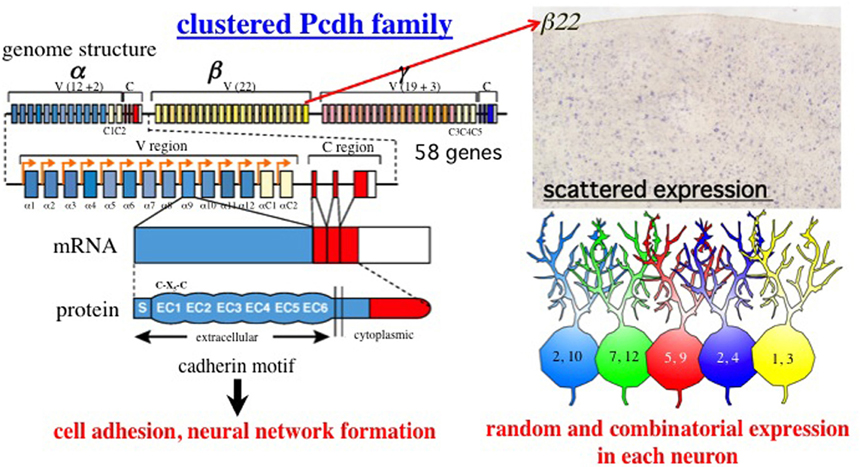
Figure 1. Summary of the clustered Pcdh family. Genomic organization of the Pcdh-α, Pcdh-β, and Pcdh-γ gene clusters in mouse chromosome 18. A total of 58 isoforms are encoded in these gene clusters. The mouse Pcdh-α gene consists of 14 exons (12 randomly and two constitutively expressed) in the variable (V) region and a set of three constant (C) region exons (A-type alterative splicing), Not shown here, B-type is derived from four constant region exons (Kohmura et al., 1998). Similar to Pcdh-α, the Pcdh-γ cluster consists of 22 variable exons (19 randomly and three constitutively expressed) and a set of three constant region exons. Mature mRNAs of the Pcdh-α and Pcdh-γ isoforms are produced from one of these variable exons and either the α or γ constant exons. The αC1, αC2, γC3, γC4, and γC5 exons are closely related in homology and gene regulation. The Pcdh-β cluster does not have constant exons; instead, 22 mature isoforms are produced from large single exons. All the Pcdh-α, Pcdh-β, and Pcdh-γ isoforms consist of a signal peptide (S) with six extracellular cadherin (EC) domains in the extracellular region, followed by a single transmembrane (TM) domain and cytoplasmic region. Interestingly, a Cys-(X)5-Cys (C-X5-C) motif in the EC1 domain is completely conserved in the vertebrate clustered Pcdh family (Morishita and Yagi, 2007). Loss-of-function analyzes have revealed that the Pcdh family has homologous cell adhesion activity, and critical roles in building neural networks, including axonal targeting, synapse formation, cell death, and dendritic arborization. Each of the 12 α, 22 β, and 19 γ isoforms exhibits random and combinatorial expression in individual neurons at the allelic level. Thus, they exhibit a scattered expression pattern in wide regions of the brain. The photograph shows the expression pattern of the β22 isoform in the cerebral cortex (provided by K. Hirano). The figures in the neurons are the a isoforms, illustrating the random and combinatorial expression in each individual neuron. Different colors represent different combinations.
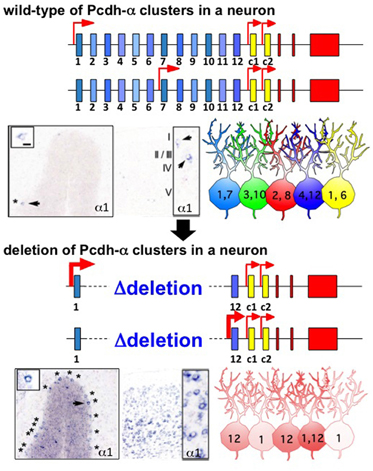
Figure 2. Random regulation of the Pcdh-α1 to α12 isoforms from the gene cluster in individual neurons. In wild-type, one (or two) is randomly chosen from 12 variable exons in a monoallelic manner. As a result, a random and a combinatorial expression of a isoforms are established in each individual neuron. The photographs show the representative, scattered expression patterns of the α1 isoform in Purkinje neurons and the cerebral cortex, by in situ hybridization. The numbers in the illustrated neurons give the number of a isoforms expressed in individual neurons. In a gene cluster in which variable exons α2–α11 are deleted, one (or two) is randomly chosen from the remaining two exons α1 and α12 in the monoallelic. As a result, either α1 or α12 is always expressed in individual neurons. The expression frequencies of α1 and α12 are therefore increased in the deletion mutants. The photographs show the expression patterns of the α1 isoform in the Purkinje cells and cortex of this mutant.
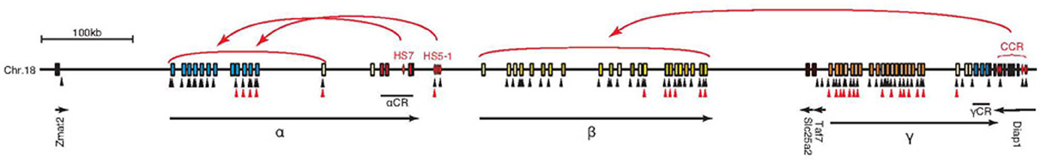
Figure 3. Schematic diagram of the independent regulation of the Pcdh-α, β, and γ gene clusters by cis-regulatory elements. The HS5–1 element controls the expression of α3–αC1 depending on the distance. The HS-7 element controls the whole Pcdh-α cluster. The cis-regulatory elements, called the cluster control region (CCR), control Pcdh-β and are located downstream from the Pcdh-γ cluster. The control elements for the Pcdh-γ cluster have not been identified yet. Several CTCF-binding and cohesin-SA1-binding sites exist in the large clustered Pcdh gene cluster locus. Black and red triangles indicate CTCF-binding and cohesin-SA1-binding sites, respectively.
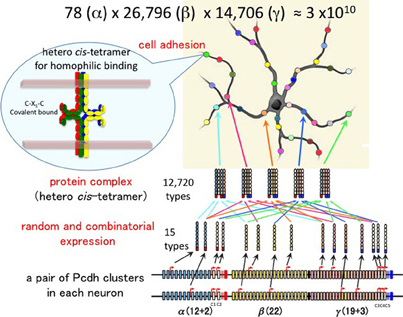
Figure 4. Individuality of neurons determined by the random expression of clustered Pcdh isoforms and the random production of heteromultimeric cis-tetramers. From each clustered Pcdh cluster allele, 1 of the α, 2 of the β, and 2 of the γ isoforms are randomly expressed in combination. From the two alleles in each neuron, 2 of the α, 4 of the β, and 4 of the γ isoforms are randomly expressed in combination and with repetition, e.g., sometimes the same isoform is chosen from both alleles. Thus, the 78 possible combinations from 12 (α), 26,796 from 22 (β), and 14,706 from 19 (β) yield approximately 3 × 1010 variations that could be expressed by individual neurons. The five C-type isoforms are constitutively expressed from both alleles in each neuron. Therefore, a total of 15 isoforms is expressed in each neuron, 10 (2α + 4β + 4γ) random and five constitutive isoforms. From 15 isoforms, 12,720 types of cis-tetramers are possible by combination with repetition and considering the topological variations (see Figure 5). The protein structure of the heteromultimeric cis-tetramers has not been revealed yet. The C-X5-C motif is conserved among all clustered Pcdh isoforms and is important for forming the cis-tetramers Schreiner and Weiner (2010). Actually the clustered Pcdh proteins are localized as their protein dots in neuritis Phillips et al. (2003); Murata et al. (2004), and also shown that over expression of intact and truncated Pcdh-γ isoform can inhibit synaptogenesis Femandez-Monreal et al. (2009).
Interestingly, there are many nucleotide polymorphisms among the clustered Pcdh genes of mouse subspecies (Taguchi et al., 2005) and individual humans (Noonan et al., 2003; Miki et al., 2005). Evolutionarily, the clustered Pcdh gene clusters are conserved and homogenized (appeared similar sequences specified in species) within each vertebrate species (Noonan et al., 2004a; Ishii et al., 2004; Schmutz et al., 2004; Yagi, 2008). Together, these molecular features suggest the clustered Pcdhs as possible candidates for producing complex neural networks at the individual neuron level in vertebrates.
Gene Regulation of Clustered Pcdhs at the Individual Neuron Level
The clustered Pcdhs are candidates for the molecular code for neuronal individuality. Single-cell RT-PCR analysis of Purkinje cells, which contain a large amount of mRNA, revealed strong evidence for the stochastic, combinatorial expression of clustered Pcdhs in individual neurons (Esumi et al., 2005; Kaneko et al., 2006). Each Purkinje cell expresses ∼2 of the 5′ members of the 12 Pcdh-α isoforms and ∼4 of the 5′ members of the 19 Pcdh-γ isoforms. In addition, ∼4 of the 22 Pcdh-β isoforms are expressed (Hirano et al, unpublished data; their scattered expression referring in Yokota et al., 2011). These expressions are stochastically regulated monoallelically. Interestingly, their random expressions depend on the number of variable exons in the cluster. When a deletion allele of exons Pcdh-α2 to α11, which spares only exons α1 and α12, was used to make a transgenic knock-in mouse, the expression frequencies of the α1 and α12 isoforms differed from those of the wild-type allele (Figure 2) (Noguchi et al., 2009). Namely, each individual neuron always expressed α1, α12, or both isoforms from the deletion allele, whereas the α1 and α12 isoforms are only sometimes expressed from among the 12 variable exons of the wild-type allele. Thus, the expressions of the variable exons are random or stochastic, like the results of throwing dice.
The random and scattered expression of variable exons is found in Purkinje neurons (Esumi et al., 2005; Kaneko et al., 2006), suggesting almost all the neurons in the brain have random and scattered expression pattern of variable exons of clustered Pcdh (Noguchi et al., 2009; Yokota et al., 2011). In contrast, the 3′ members (“C” isoforms) of each cluster, αC1 and αC2 in Pcdh-α and γC3, γC4, and γC5 in Pcdh-γ, are expressed constitutively and biallelically by Purkinje neurons (Kaneko et al., 2006). Their biallelic expressions also depend on the position of the C exon in the gene cluster; when a deletion construct that removes Pcdh-α11–the αC2 exon is knocked-in, the nearest exon from the constant region, α10, is expressed constitutively and biallelically (Noguchi et al., 2009). Thus, the monoallelic and biallelic expressions of the Pcdh isoforms are regulated by the structure of the gene cluster.
From each allele in individual neurons, 1, 2, and 2 isoforms, respectively, are randomly expressed from among the total 12 in the α, 22 in the β, and 19 in the γ cluster (Figure 4). The calculation of the number of possible combinations in each allele is represented as 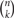 , where n is the number of total isoforms, and k is the number expressed in a cell, calculated by a formula of n!/(n − k)!k!.
, where n is the number of total isoforms, and k is the number expressed in a cell, calculated by a formula of n!/(n − k)!k!.
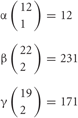
Thus, the number of combinations with repetition from both alleles, is 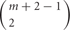 , where m is the number of permutations from each monoallelice, and 2 is the number of alleles.
, where m is the number of permutations from each monoallelice, and 2 is the number of alleles.
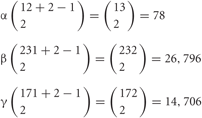
Therefore, a total of 78 × 26,796 × 14,706 = 30,736,834,128 (approximate 3 × 1010) variations are possible for each neuron. In addition, the five “C” isoforms αC1, αC2, γC3, γC4, and γC5, which are constitutively expressed in neurons, increase the total number of isoforms expressed per neuron but does not contribute to variation. It is estimated that the cerebral cortex of the human brain contains 1010 neurons. Therefore, these calculations suggest that the variations caused by the random expression of clustered Pcdh isoforms could account for the individuality of all the neurons in the brain.
All the variable exons of clustered Pcdh have promoters that contain a conserved sequence element (CGCT) (Figure 3). Therefore, their isoform expressions are regulated by a mechanism of promoter choice in individual neurons (Tasic et al., 2002). The expression of clustered Pcdh isoforms is epigenetically controlled. Cell lines expressing specific clustered Pcdh isoforms have differential DNA-methylation patterns in their promoter regions: the active promoters are hypomethylated, and silent ones are methylated (Kawaguchi et al., 2008). In vivo, Purkinje neurons have distinct and variable DNA-methylation patterns in the clustered Pcdh promoter regions. In addition, the cis-regulatory elements HS7 and HS5–1 control Pcdh-α (Ribich et al., 2006; Kehayova et al., 2011) and CCR controls Pcdh-β (Yokota et al., 2011), respectively, (Figure 3). Interestingly, the zinc finger DNA-binding protein CTCF binds to almost all the variable exons and cis-elements (Handoko et al., 2011), and regulates the expression of clustered Pcdh isoforms (Golan-Mashiach et al., 2011; Kehayova et al., 2011). The regulator of chromatin conformation, cohesin-SA1, also binds to several variable exons and regulates the expression of clustered Pcdh isoforms (Remeserio et al., 2012) (Figure 3). The Pcdh cluster is also modified by histone methylation and acetylation (Mikkelsen et al., 2007), and is enriched in binding sites for the demethylation factor Tet1 (Xu et al., 2011). Thus, the stochastic expression of clustered Pcdh isoforms in individual neurons appears to be regulated by epigenetic factors and by interactions between each promoter and cis-elements within the gene clusters.
Heteromultimeric Protein Complex
The clustered Pcdh proteins have a punctate localization (Phillips et al., 2003; Murata et al., 2004; Femandez-Monreal et al., 2009), and may function in complexes: Pcdh-α and Pcdh-γ may form heteromultimers (Figure 4). The Pcdh-γ proteins induce the membrane surface expression of Pcdh-α proteins (Murata et al., 2004). In addition, Pcdh-β proteins associate with Pcdh-α and Pcdh-γ proteins (Han et al., 2010), and locate in synapses (Junghans et al., 2008). Schreiner and Weiner (2010) showed that 7 Pcdh-γ members exhibit isoform-specific homophilic binding, and that heteromultimeric cis-tetramers function as a homophilic binding unit (Schreiner and Weiner, 2010). The binding behavior of the cis-tetramers is very different from that of classical cadherins, which do not form multimers, and mediate cell-cell interactions by binding an identical cadherin on a different cell (Figure 5A). The clustered Pcdh cis-tetramers are formed before they engage in cell-cell interactions. As shown in Figure 5A, if two cells express two Pcdh isoforms, and only one of them is expressed in common, only one type of cis-tetramer on each cell is capable of cell-cell homophilic binding. In fact, cells that express only 1 or 2 of the four isoforms in common bind very poorly, whereas those expressing three or four of the four isoforms in common bind well, which supports the proposed cis-tetramer binding activity (Schreiner and Weiner, 2010).
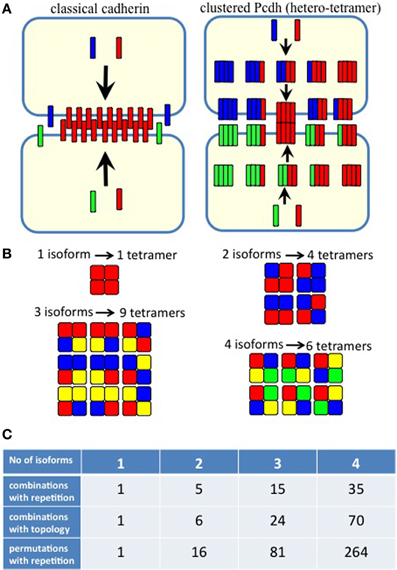
Figure 5. (A) Homophilic cell adhesion as achieved by classical cadherins versus the cis-tetramers of clustered Pcdh isoforms. Red bars represent the common type of cadherin or clustered isoform molecules expressed on and binding between two interacting cells. Blue and green bars show additional cadherins or clustered Pcdh isoforms that are differentially expressed in the interacting cells. From two clustered Pcdh isoforms, five types of cis-tetramers can be produced in combination with repetition. In this example, only the red cis-tetramers can bind homophilically. (B) Variations of heteromultimeric cis-tetramers from each combination. One, two, three, and four isoforms can form 1, 4, 9, and 6 possible combinations, respectively, with repetition and topological variation. (C) Table shows the number of possible cis-tetramers from i isoforms calculated by combinations with repetition 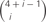 , combinations with topological variations
, combinations with topological variations 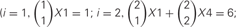
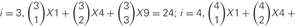
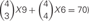 and permutations with repetition i4.
and permutations with repetition i4.
Combinations with repetition are calculated as 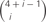 , where 4 is the number of protein isoforms for a cis-tetramer, and i is the number of different protein isoforms expressed in an individual neuron. When 4 different isoforms are expressed in cells, the number of distinct cis-tetramers is
, where 4 is the number of protein isoforms for a cis-tetramer, and i is the number of different protein isoforms expressed in an individual neuron. When 4 different isoforms are expressed in cells, the number of distinct cis-tetramers is 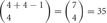 . One, two and three isoforms expressing cells have
. One, two and three isoforms expressing cells have 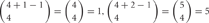 and
and 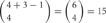 distinct cis-tetramers, respectively. Therefore, in cells that express four isoforms with 1, 2, and 3 isoforms in common, 1/35 (2.8%), 4/35 (11.4%), and 15/35 (42.9%) cis-tetramers will match, respectively.
distinct cis-tetramers, respectively. Therefore, in cells that express four isoforms with 1, 2, and 3 isoforms in common, 1/35 (2.8%), 4/35 (11.4%), and 15/35 (42.9%) cis-tetramers will match, respectively.
However, the cis-tetramers also have possible topological variations. Figure 5B shows the topological variations of cis-tetramers. Therefore, considering the topological variations of cis-tetramers, one, two, three, and four kinds of isoforms produce 1, 4, 9, and 6 distinct cis-tetramers, respectively. If there are 4 different isoforms expressed in cells, the total number of distinctive topological cis-tetramers is 70 (70 = 4 + 24 + 36 + 6; 1 choice from 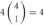 , 4 × 1 = 4; 2 choices from
, 4 × 1 = 4; 2 choices from 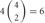 , 6 × 4 = 24; 3 choices from
, 6 × 4 = 24; 3 choices from 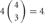 , 4 × 9 = 36; 4 choices from
, 4 × 9 = 36; 4 choices from 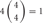 , 1 × 6 = 6). Calculating the topological variation, cells expressing 1, 2, and 3 isoforms have 1, 6, and 24 distinct topological cis-tetramers, respectively. Therefore, in cells sharing one, two, or three isoforms versus cells expressing four types of isoforms, 1/70 (1.4%), 6/70 (8.6%), or 24/70 (34.3%) cis-tetramers are matched.
, 1 × 6 = 6). Calculating the topological variation, cells expressing 1, 2, and 3 isoforms have 1, 6, and 24 distinct topological cis-tetramers, respectively. Therefore, in cells sharing one, two, or three isoforms versus cells expressing four types of isoforms, 1/70 (1.4%), 6/70 (8.6%), or 24/70 (34.3%) cis-tetramers are matched.
On the other hand, this calculation does not consider the molecular amounts of each type of cis-tetramer. If i types of isoforms are expressed in equal amounts in cells, the total amount of cis-tetramers can be represented by a permutation with a repetition of i4. Therefore, although repetitions of the same type of cis-tetramer exist, cells sharing one, two, and three isoforms versus cells expressing four types of isoforms are calculated as 14/44 = 1/264 (0.4%), 24/44 = 16/264 (6.1%), and 34/44 = 81/264 (30.1%), respectively. These calculations contain several simplifications and assumptions for equal transcription and translation of each isoform (summarized in Figure 5C). In any cases, these calculations support the above-described experimental results of poor cell adhesion in cells expressing different isoform combinations, and together these findings suggest that the heteromultimeric cis-tetramer of clustered Pcdh protein isoforms could serve as the specific binding unit for cell adhesion and neuronal interconnections (Schreiner and Weiner, 2010).
In addition to Pcdh-γ isoforms, the heteromultimeric cis-tetramers may contain a combination of Pcdh-α, Pcdh-β, and Pcdh-γ isoforms. The evidence is as follows. First, α and γ isoforms are immunoprecipitated with each other's specific antibody (Murata et al., 2004), and β proteins associate with Pcdh-α and Pcdh-γ proteins (Han et al., 2010). Second, various Pcdh-α isoforms translocate to the cell- surface upon the expression of various Pcdh-γ isoforms, and various combinations of Pcdh-α and Pcdh-γ isoforms have been confirmed (Murata et al., 2004). In addition, the Cys-(X)5-Cys (C-X5-C) motif was found to be important for the formation and cell-surface expression of covalently bound cis-tetramers (Schreiner and Weiner, 2010) (Figure 4), and the C-X5-C motif in the first cadherin domain (EC1) is completely conserved among all clustered Pcdh proteins in vertebrates. Furthermore, analysis of the protein structure of the EC1 domain of Pcdh-α4 indicated that the motif is located at the protein's surface (Morishita et al., 2006), and the C-X5-C motif of the EC1 domain is also conserved in the solitary Pcdh-δ2 proteins (Morishita and Yagi, 2007).
In the isoform-specific binding activity, both the EC2 and EC3 domains are important for homophilic binding specificity (Schreiner and Weiner, 2010). Notably, among all the clustered Pcdh isoforms, the EC2 and EC3 domains are the most divergent (Kohmura et al., 1998; Wu and Maniatis, 1999).
If 15 isoforms of clustered Pcdhs are expressed in an individual neuron, this number of isoforms could generate 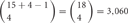 possible combinations with repetition and
possible combinations with repetition and 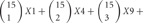
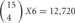 types of cis-tetramers in an individual neuron (Figure 4). However, in these cells, 5 “C” isoforms are constitutively expressed, and the remaining 10 isoforms are randomly chosen and expressed. The expression of 15 isoforms in an individual neuron contains assumptions of randomly chosen 4 Pcdh-β isoforms by our unpublished data (Hirano et al. in preparation). An individual neuron is estimated to form several to tens of thousands of synapses, suggesting that the variation created by cis-tetramers of cluster Pcdh isoforms could cover the number of synapses in a neuron.
types of cis-tetramers in an individual neuron (Figure 4). However, in these cells, 5 “C” isoforms are constitutively expressed, and the remaining 10 isoforms are randomly chosen and expressed. The expression of 15 isoforms in an individual neuron contains assumptions of randomly chosen 4 Pcdh-β isoforms by our unpublished data (Hirano et al. in preparation). An individual neuron is estimated to form several to tens of thousands of synapses, suggesting that the variation created by cis-tetramers of cluster Pcdh isoforms could cover the number of synapses in a neuron.
Next, I calculated the number of kinds of cis-tetramers that could be generated from the number of distinct isoforms (Figure 6A), and the probability of matching cis-tetramers (the matching probability) occurring between a pair of neurons, each of which expresses 15 clustered Pcdh isoforms, when the number of different isoforms between them changes (Figure 6B). The matching probabilities (P) of the isoforms decrease exponentially with as the number of different (d) isoforms increases.
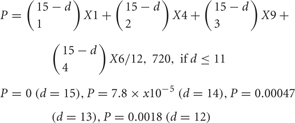
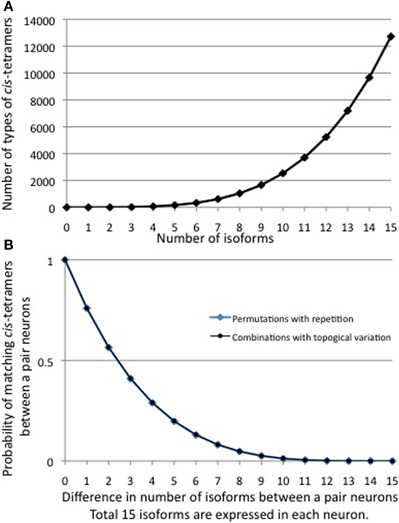
Figure 6. (A) The number of combinations of heteromultimeric cis-tetramers increases as the number of isoforms increases, as a combination function. If 15 isoforms are expressed in individual neurons, the number of possible cis-tetramer combinations is 12,720. (B) The probability that matching cis-tetramers will be expressed on a pair of neurons, as a function of the number of isomers that are different between the two neurons, if each neuron expresses 15 isoforms. Calculations are done by two methods: combinations with topological variations (black thin line), and permutations with repetitions (blue bold line). A small number of different isoforms expressed between a pair of neurons will sharply decrease the matching probability of cis-tetramers, e.g., a difference of only 3 of a total of 15 isoforms leads to 0.41 (41%) cis-tetramers matching between a pair neurons. However, 10 differences in a total of 15 isoforms (5 isoforms in common between a pair of neurons) yields a score of 0.013, meaning that only 1.3% of the cis-tetramers match between the pair of neurons.
Surprisingly, the matching probability of the types of cis-tetramers decreases rapidly with small differences in the number of different isoforms between the two cells; for example, a difference of only 3 isoforms yields a matching probability of 41.1% (below 50%). On the other hand, these calculations do not consider the molecular amount of each type of cis-tetramer. If i types of isoforms are expressed at equal amounts in each cell, the total number of possible cis-tetramers is represented by i4 of permutation with repetition though including the same type of cis-tetramers. Considering the total amount of cis-tetramers, the amounts of different cis-tetramers can be shown as i4-(i−d)4. Then, i4-(i−d)4/i4 represents the probability of the total difference in the amounts of cis-tetramers between a pair of neurons expressing different numbers of isoforms. In our analysis with Purkinje neurons, we estimated that i = 15 in individual neurons. Here I hypothesize that the total number of possible cis-tetramers is 154, when every isoform has the same propensity for producing cis-tetramers. If 1 of the 15 isoforms is different (14 isoforms shared) between a pair of neurons, 144 (38,416) of the total 154 (50,625) are the same types of cis-tetramers, and thus 154—144 = 12,209 are different cis-tetramers. The function curve of the permutation with repetition is similar to the calculation curve of the differences of cis-tetramers considering the variations of their combinations with repetition and topology (Figure 6B).
In any case, these calculations demonstrate that a few distinctly expressed clustered Pcdh isoforms can lead to distinct neuronal individuality by virtue of their heteromultimeric cis-tetramers. In addition, interestingly, the common expression of several clustered Pcdh isoforms has little effect on the amount of variation between a pair of neurons. For example, a difference of 10 isoforms among a total of 15 (5 isoforms expressed in common) is calculated as generating only 1.3% matching cis-tetramers. Thus, even if the five “C” type clustered Pcdh isoforms are constitutively expressed in each neuron, the individuality of the neurons can be robustly maintained with 98% different cis-tetramers by the random expression of clustered Pcdh isoforms in each neuron. Thus, the stochastic expression of clustered Pcdh isoforms may provide a molecular code capable of stamping a high degree of individuality on every neuron in the brain. To examine this possibility, we need to study the function of the homophilic activity of the heteromultimeric cis-tetramers of clustered Pcdh isoforms in the brain.
Similar stochastic expressions have been reported for Dscam1 isoforms in insect neurons, and these molecules might serve as molecular codes for neuronal individuality in the insect brain. In Drosophila, alternative splicing of the single gene Dscam1 can generate 19,008 isoforms. The homophilic binding of the isoforms results in the repulsion of self-neurites. Individual neurons randomly express multiple isoforms; the number of Dscam isoforms expressed by each neuron is estimated to be 10 to 50 (Hattori et al., 2009). The Dscam1 protein isoforms have homophilic activity at the single isoform level. Calculation using a Monte Carlo simulation (Hattori et al., 2009) and combinatorics by closed-form solutions (Forbes et al., 2011) indicated a 4.4% chance that a pair of neurons shares at least one isoform, from 30 random expressions of 20,000 isoforms. Similar probabilities are estimated for Dscam1 in insects and clustered Pcdhs in vertebrates, even though the mechanism for randomness is different; that is, alternative splicing of Dscam1 or promoter choice and cis-tetramers for clustered Pcdh. Thus, neuronal individuality could be important in both vertebrates and invertebrates for developing complex neural networks.
Cell Assembly and Clustered Pcdhs
The functions of the clustered Pcdhs have been examined by producing loss-of-function mice. Mice lacking Pcdh-α are viable and fertile, but they have defects in contextual learning and special working memory (Fukuda et al., 2008). The olfactory sensory neurons and serotonergic neurons of these mutants have projection errors (Hasegawa et al., 2008; Katori et al., 2009). In wild-type mice, the axons of olfactory neurons that express the same olfactory receptor converge to innervate the proper glomeruli of the olfactory bulb. However, in the mutants, abnormal ectopic convergence is observed, even in adults (Hasegawa et al., 2008). Similarly, serotonergic fibers are abnormally distributed and condensed in several brain areas of serotonergic targeting (Katori et al., 2009). These axonal targeting phenotypes are also detected in the cytoplasmic deletion mutants, suggesting that the constant cytoplasmic tail of the Pcdh-α proteins is important for correct axonal targeting. In addition, loss of Pcdh-α in mice has functional impairments of cortico-cortical pathways between both hemispheres of primary somatosensory cortex by different mechanism on NMDA receptor (Yamashita et al., 2012).
The loss of Pcdh-γ in mice leads to neonatal lethality with neurological defects involving apoptosis and decreased synapses (Wang et al., 2002b). The increased apoptosis occurs during the period of naturally occurring neuronal cell death (Lefebvre et al., 2008; Prasad et al., 2008). Even when the apoptosis defects are eliminated using Bax mutants, the Pcdh-γ mutants still show decreased synapses in the spinal cord (Weiner et al., 2005). In the retina, the Pcdh-γs are also indispensable for neuronal survival, and decreased synapses are seen in the Pcdh-γ mutants, although these are not rescued by Bax deletion, unlike in the spinal cord. Therefore it is not yet clear that in the retina there is a clear circuit formation role for the Pcdh-γs (Lefebvre et al., 2008).
There are no data to date that demonstrate the clustered Pcdh diversity is required in vivo. Genetic studies of the clustered Pcdhs, however, have gradually revealed their functions for building the correct neural networks. Also diversity of Pcdh-γ proteins has crucial roles for their selective homophilic adhesion activity in cultured K562 cells. The clustered Pcdhs are randomly expressed in every individual neuron and form an enormous number of variable cis-tetramers, speculating their function for building neural networks at an individual neuron level in the brain.
Recent physiological approaches have revealed that local neural networks form complex networks with neuronal ensembles at an individual neuron level (Song et al., 2005; Yoshimura et al., 2005). In addition, specific local connectivity develops preferentially among sister excitatory cortical neurons (Yu et al., 2009). Theoretical analyzes analyzes of neural networks suggest that complex networks exist in the brain (Sporns, 2011). Interestingly, Watts and Strogatz showed that “small-world” networks [by analogy with the small-world phenomenon known as six degrees of separation (Guare, 1990)] with high clustering coefficients and short characteristic path lengths emerge as a consequence of both random interactions and highly regulated ones (Watts and Strogatz, 1998).
To understand how complex brain networks form and function, we must first understand the mechanisms for creating randomness and regularity in the brain. In addition, considering the recent physiological results on spontaneous neural assembly and predetermined neural activity (Buzsaki, 2010), we need to examine the intrinsic and individual mechanisms for generating neural networks with randomness and regularity during brain development. In this line, the random expression of the clustered Pcdh family molecules in individual neurons during development and their specific cell adhesion activities for neural network formation make them intriguing candidates for molecules that enable intrinsic neural network formation; they could provide both the “small-world” cell assembly feature and account for the nearly limitless neural information processed within the limited brain mass. As shown in Figure 7, both the immune system and the brain might be similarly predetermined systems involving diverse individual cells created randomly before being exposed to external experiences for acquiring nearly limitless memories. In the immune system, antigens serve as the “external experiences.” In the brain system, the mechanisms that serve as the “external experiences” that assemble the predetermined neural circuits in the context of developmental programs and that generate functional networks by means of synaptic plasticity have not been fully elucidated, but the continued examination of the clustered Pcdh family may uncover some of the answers.
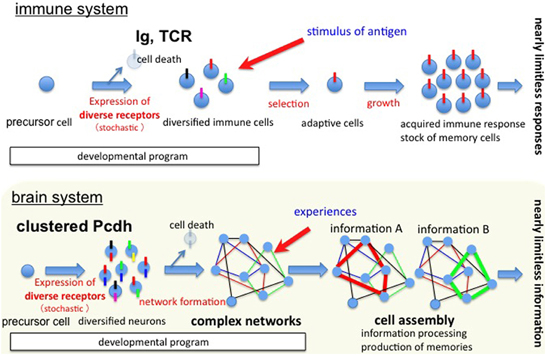
Figure 7. Diagrams of memory systems for both the immune system and the brain. (A) In the immune system, an enormous number of diverse immune cells are developmentally produced; thus a set of pre-functional immune cells prepared early in life. The mechanism for diversity is the stochastic DNA rearrangement of the immunoglobulin (Ig) and T-cell receptor (TCR) genes. After infection with external antigens, the appropriate immune cells for responding to the antigen are selected, expanded, and stored in memory in the form of memory cells. In this way, numerous and nearly limitless adaptive immune responses and antigen memories are generated. Thus, the functional immune cells are predetermined by developmental programming, including a stochastic mechanism. (B) In the brain system, clustered Pcdh isoforms from the α, β, and γ gene clusters are stochastically expressed in neurons to produce individual neuronal identities. The expressed clustered Pcdh isoforms produce functional cis-tetramers. At the individual neuron level, each neuron is incorporated into neural networks via the affinity of its cellular interactions. The randomly expressed clustered Pcdh isoforms in an individual neuron form cis-tetramers that specifically bind the matching cis-tetramers on other neurons, generating a complex neural network that is determined by randomness and by high cluster coefficients during development. Thus, the process network formation during development results in numerous cell assemblies. As a result of experiences, the cell assemblies that respond to a specific experience are selected, strengthened, and the experience is memorized in the form of the strengthened cell assembly. In this way, nearly limitless neural information processing and memories can be generated. Thus, functional cell assemblies might be predetermined by developmental programs that involve stochastic expression and specific cellular interactions to form neural networks in the brain.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I thank T. Kitsukawa, A. Uchimura, and S. Hasegawa for discussion and calculation of the combinations of cis-tetramers. I also thank members of our laboratories, especially K. Hirano, R. Kaneko, T. Hirayama, and T. Hirabayashi for the expression analysis for Pcdh-β and discussion. I thank K. Shibuki, E. Tarusawa, and Y. Yoshimura for discussion and colaboration of the cell assembly hypothesis in the brain. This work was supported in part by Grant-in-Aid for Scientific Research (S), a Grant-in-Aid for Scientific Research on Innovative Area “Mesoscopic Neurocircuitry” from the Ministry of Education, Science, Sports, and Culture of Japan (MEXT), and CREST from Japan Science and Technology Agency (JST) (Takeshi Yagi).
References
Buck, L., and Axel, R. (1991). A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65, 175–187.
Cheng, H. J., Nakamoto, M., Bergemann, A. D., and Flanagan, J. G. (1995). Complementary gradients in expression and binding of ELF-1 and Mek4 in development of the topographic retinotectal projection map. Cell 82, 371–381.
Dragoi, G., and Tonegawa, S. (2011). Preplay of future place cell sequences by hippocampal cellular assemblies. Nature 469, 397–401.
Drescher, U., Kremoser, C., Handwerker, C., Loschinger, J., Noda, M., and Bonhoeffer, F. (1995). In vitro guidance of retinal ganglion cell axons by RAGS, a 25 kDa tectal protein related to ligands for Eph receptor tyrosine kinases. Cell 82, 359–370.
Dreyer, W. J. (1998). The area code hypothesis revisited: olfactory receptors and other related transmembrane receptors may function as the last digits in a cell surface code for assembling embryos. Proc. Natl. Acad. Sci. U.S.A. 95, 9072–9077.
Esumi, S., Kakazu, N., Taguchi, Y., Hirayama, T., Sasaki, A., Hirabayashi, T., Koide, T., Kitsukawa, T., Hamada, S., and Yagi, T. (2005). Monoallelic yet combinatorial expression of variable exons of the protocadherin-alpha gene cluster in single neurons. Nat. Genet. 37, 171–176.
Femandez-Monreal, M., Kang, S., and Phillips, G. R. (2009). Gamma-protocadherin homophilic interaction and intracellular trafficking is controlled by the cytoplasmic domain in neurons. Mol. Cell. Neurosci. 40, 344–353.
Forbes, E. M., Hunt, J. J., and Goodhill, G. J. (2011). The combinatorics of neurite self-avoidance. Neural Comput. 23, 2746–2769.
Fukuda, E., Hamada, S., Hasegawa, S., Katori, S., Sanbo, M., Miyakawa, T., Yamamoto, T., Yamamoto, H., Hirabayashi, T., and Yagi, T. (2008). Down-regulation of protocadherin-alpha A isoforms in mice changes contextual fear conditioning and spatial working memory. Eur. J. Neurosci. 28, 1362–1376.
Golan-Mashiach, M., Grunspan, M., Emmanuel, R., Gibbs-Bar, L., Dikstein, R., and Shapiro, E. (2011). Identification of CTCF as a master regulator of the clustered protocadherin genes. Nucleic Acids Res.
Han, M. H., Lin, C., Meng, S., and Wang, X. (2010). Proteomics analysis reveals overlapping functions of clustered protocadherins. Mol. Cell Proteomics 9, 71–83.
Handoko, L., Xu, H., Li, G., Ngan, C. Y., Chew, E., Schnapp, M., Lee, C. W., Ye, C., Ping, J. L., Mulawadi, F., Wong, E., Sheng, J., Zhang, Y., Poh, T., Chan, C. S., Kunarso, G., Shahab, A., Bourque, G., Cacheux-Rataboul, V., Sung, W. K., Ruan, Y., and Wei, C. L. (2011). CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 43, 630–638.
Hasegawa, S., Hamada, S., Kumode, Y., Esumi, S., Katori, S., Fukuda, E., Uchiyama, Y., Hirabayashi, T., Mombaerts, P., and Yagi, T. (2008). The protocadherin-alpha family is involved in axonal coalescence of olfactory sensory neurons into glomeruli of the olfactory bulb in mouse. Mol. Cell. Neurosci. 38, 66–79.
Hattori, D., Chen, Y., Matthews, B. J., Salwinski, L., Sabatti, C., Grueber, W. B., and Zipursky, S. L. (2009). Robust discrimination between self and non-self neurites requires thousands of Dscam1 isoforms. Nature 461, 644–648.
Hill, E., Broadbent, I. D., Chothia, C., and Pettitt, J. (2001). Cadherin superfamily proteins in Caenorhabditis elegans and Drosophila melanogaster. J. Mol. Biol. 305, 1011–1024.
Hirayama, T., and Yagi, T. (2006). The role and expression of the protocadherin-alpha clusters in the CNS. Curr. Opin. Neurobiol. 16, 336–342.
Ishii, Y., Asakawa, S., Taguchi, Y., Ishibashi, S., Yagi, T., and Shimizu, N. (2004). Construction of BAC library for the amphibian Xenopus tropicalis. Genes Genet. Syst. 79, 49–51.
Junghans, D., Heidenreich, M., Hack, I., Taylor, V., Frotscher, M., and Kemler, R. (2008). Postsynaptic and differential localization to neuronal subtypes of protocadherin beta16 in the mammalian central nervous system. Eur. J. Neurosci. 27, 559–571.
Kaneko, R., Kato, H., Kawamura, Y., Esumi, S., Hirayama, T., Hirabayashi, T., and Yagi, T. (2006). Allelic gene regulation of Pcdh-αlpha and Pcdh-gamma clusters involving both monoallelic and biallelic expression in single Purkinje cells. J. Biol. Chem. 281, 30551–30560.
Katori, S., Hamada, S., Noguchi, Y., Fukuda, E., Yamamoto, T., Yamamoto, H., Hasegawa, S., and Yagi, T. (2009). Protocadherin-alpha family is required for serotonergic projections to appropriately innervate target brain areas. J. Neurosci. 29, 9137–9147.
Kawaguchi, M., Toyama, T., Kaneko, R., Hirayama, T., Kawamura, Y., and Yagi, T. (2008). Relationship between DNA methylation states and transcription of individual isoforms encoded by the protocadherin-alpha gene cluster. J. Biol. Chem. 283, 12064–12075.
Kehayova, P., Monahan, K., Chen, W., and Maniatis, T. (2011). Regulatory elements required for the activation and repression of the protocadherin-alpha gene cluster. Proc. Natl. Acad. Sci. U.S.A. 108, 17195–17200.
Kenet, T., Bibitchkov, D., Tsodyks, M., Grinvald, A., and Arieli, A. (2003). Spontaneously emerging cortical representations of visual attributes. Nature 425, 954–956.
Kohmura, N., Senzaki, K., Hamada, S., Kai, N., Yasuda, R., Watanabe, M., Ishii, H., Yasuda, M., Mishina, M., and Yagi, T. (1998). Diversity revealed by a novel family of cadherins expressed in neurons at a synaptic complex. Neuron 20, 1137–1151.
Langley, J. N. (1895). Note on regeneration of prae-ganglionic fibres of the sympathetic. J. Physiol. 18, 280–284.
Lefebvre, J. L., Zhang, Y., Meister, M., Wang, X., and Sanes, J. R. (2008). gamma-Protocadherins regulate neuronal survival but are dispensable for circuit formation in retina. Development 135, 4141–4151.
Lieber, M. R. (1992). The mechanism of V(D)J recombination: a balance of diversity, specificity, and stability. Cell 70, 873–876.
McLaughlin, T., and O'Leary, D. D. (2005). Molecular gradients and development of retinotopic maps. Annu. Rev. Neurosci. 28, 327–355.
Miki, R., Hattori, K., Taguchi, Y., Tada, M. N., Isosaka, T., Hidaka, Y., Hirabayashi, T., Hashimoto, R., Fukuzako, H., and Yagi, T. (2005). Identification and characterization of coding single-nucleotide polymorphisms within human protocadherin-alpha and -beta gene clusters. Gene 349, 1–14.
Mikkelsen, T. S., Ku, M., Jaffe, D. B., Issac, B., Lieberman, E., Giannoukos, G., Alvarez, P., Brockman, W., Kim, T. K., Koche, R. P., Lee, W., Mendenhall, E., O'Donovan, A., Presser, A., Russ, C., Xie, X., Meissner, A., Wernig, M., Jaenisch, R., Nusbaum, C., Lander, E. S., and Bernstein, B. E. (2007). Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560.
Morishita, H., Umitsu, M., Murata, Y., Shibata, N., Udaka, K., Higuchi, Y., Akutsu, H., Yamaguchi, T., Yagi, T., and Ikegami, T. (2006). Structure of the cadherin-related neuronal receptor/protocadherin-alpha first extracellular cadherin domain reveals diversity across cadherin families. J. Biol. Chem. 281, 33650–33663.
Morishita, H., and Yagi, T. (2007). Protocadherin family: diversity, structure, and function. Curr. Opin. Cell Biol. 19, 584–592.
Murata, Y., Hamada, S., Morishita, H., Mutoh, T., and Yagi, T. (2004). Interaction with protocadherin-gamma regulates the cell surface expression of protocadherin-alpha. J. Biol. Chem. 279, 49508–49516.
Noguchi, Y., Hirabayashi, T., Katori, S., Kawamura, Y., Sanbo, M., Hirabayashi, M., Kiyonari, H., Nakao, K., Uchimura, A., and Yagi, T. (2009). Total expression and dual gene-regulatory mechanisms maintained in deletions and duplications of the Pcdha cluster. J. Biol. Chem. 284, 32002–32014.
Noonan, J. P., Grimwood, J., Danke, J., Schmutz, J., Dickson, M., Amemiya, C. T., and Myers, R. M. (2004a). Coelacanth genome sequence reveals the evolutionary history of vertebrate genes. Genome Res. 14, 2397–2405.
Noonan, J. P., Grimwood, J., Schmutz, J., Dickson, M., and Myers, R. M. (2004b). Gene conversion and the evolution of protocadherin gene cluster diversity. Genome Res. 14, 354–366.
Noonan, J. P., Li, J., Nguyen, L., Caoile, C., Dickson, M., Grimwood, J., Schmutz, J., Feldman, M. W., and Myers, R. M. (2003). Extensive linkage disequilibrium, a common 16.7-kilobase deletion, and evidence of balancing selection in the human protocadherin alpha cluster. Am. J. Hum. Genet. 72, 621–635.
Phillips, G. R., Tanaka, H., Frank, M., Elste, A., Fidler, L., Benson, D. L., and Colman, D. R. (2003). Gamma-protocadherins are targeted to subsets of synapses and intracellular organelles in neurons. J. Neurosci. 23, 5096–5104.
Prasad, T., Wang, X., Gray, P. A., and Weiner, J. A. (2008). A differential developmental pattern of spinal interneuron apoptosis during synaptogenesis: insights from genetic analyses of the protocadherin-gamma gene cluster. Development 135, 4153–4164.
Remeserio, S., Cuadrado, A., Gomez-Lopez, G., Pisano, D. G., and Losada, A. (2012). A unique role of cohesin-SA1 in gene regulation and development. EMBO J.
Ribich, S., Tasic, B., and Maniatis, T. (2006). Identification of long-range regulatory elements in the protocadherin-alpha gene cluster. Proc. Natl. Acad. Sci. U.S.A. 103, 19719–19724.
Sakurai, Y. (1999). How do cell assemblies encode information in the brain? Neurosci. Biobehav. Rev. 23, 785–796.
Schmutz, J., Martin, J., Terry, A., Couronne, O., Grimwood, J., Lowry, S., Gordon, L. A., Scott, D., Xie, G., Huang, W., Hellston, U., Tran-Gyamfi, M., She, X., Prabhakar, S., Aerts, A., Altherr, M., Bajorek, E., Black, S., Branscomb, E., Caoile, C., Challacombe, J. F., Chan, Y. m., Denys, M., Detter, J. C., Escobar, J., Flowers, D., Fotopulos, D., Glavina, T., Gomez, M., Gonzales, E., Goodstein, D., Grigoriev, I., Groza, M., Hammon, N., Hawkins, T., Haydu, L., Israni, S., Jett, J., Kadner, K., Kimball, H., Kobabyashi, A., Lopez, G., Lou, Y., Martinez, D., Medina, C., Morgan, J., Nandkeshwar, R., Noonan, J. P., Pitluck, S., Pollard, M., Predki, P., Priest, J., Ramirez, L., Retterer, J., Rodriguez, A., Rogers, S., Salamov, A., Salazar, A., Thayer, N., Tice, H., Tsai, M., Ustaszewska, A., Vo, N., Wheeler, J., Wu, K., Yang, J., Dickson, M., Cheng, J. F., Eichler, E. E., Olsen, A., Pennacchio, L. A., Rokhsar, D. S., Richardon, P., Lucas, S. M., Myers, R. M., and Rubin, E. M. (2004). The DNA sequence and comparative analysis of human chromosome 5. Nature 431, 268–274.
Schreiner, D., and Weiner, J. A. (2010). Combinatorial homophilic interaction between gamma-protocadherin multimers greatly expands the molecular diversity of cell adhesion. Proc. Natl. Acad. Sci. U.S.A. 107, 14893–14898.
Song, S., Sjostrom, P. J., Reigl, M., Nelson, S., and Chklovskii, D. B. (2005). Highly nonrandom features of synaptic connectivity in local cortical circuits. PLoS Biol. 3:e68. doi: 10.1371/journal.pbio.0030068
Sperry, R. W. (1963). Chemoaffinity in the orderly growth of nerve fiber patterns and connections. Proc. Natl. Acad. Sci. U.S.A. 50, 703–710.
Sudhof, T. C. (2008). Neuroligins and neurexins link synaptic function to cognitive disease. Nature 455, 903–911.
Sugino, H., Hamada, S., Yasuda, R., Tuji, A., Matsuda, Y., Fujita, M., and Yagi, T. (2000). Genomic organization of the family of CNR cadherin genes in mice and humans. Genomics 63, 75–87.
Taguchi, Y., Koide, T., Shiroishi, T., and Yagi, T. (2005). Molecular evolution of cadherin-related neuronal receptor/protocadherin(alpha) [CNR/Pcdh(alpha)] gene cluster in Mus musculus subspecies. Mol. Biol. Evol. 22, 1433–1443.
Takeichi, M. (2007). The cadherin superfamily in neuronal connections and interactions. Nat. Rev. Neurosci. 8, 11–20.
Tasic, B., Nabholz, C. E., Baldwin, K. K., Kim, Y., Rueckert, E. H., Ribich, S. A., Cramer, P., Wu, Q., Axel, R., and Maniatis, T. (2002). Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol. Cell 10, 21–33.
Wang, X., Su, H., and Bradley, A. (2002a). Molecular mechanisms governing Pcdh-gamma gene expression: evidence for a multiple promoter and cis-alternative splicing model. Genes Dev. 16, 1890–1905.
Wang, X., Weiner, J. A., Levi, S., Craig, A. M., Bradley, A., and Sanes, J. R. (2002b). Gamma protocadherins are required for survival of spinal interneurons. Neuron 36, 843–854.
Watts, D. J., and Strogatz, S. H. (1998). Collective dynamics of “small-world” networks. Nature 393, 440–442.
Weiner, J. A., Wang, X., Tapia, J. C., and Sanes, J. R. (2005). Gamma protocadherins are required for synaptic development in the spinal cord. Proc. Natl. Acad. Sci. U.S.A. 102, 8–14.
Wu, Q., and Maniatis, T. (1999). A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell 97, 779–790.
Wu, Q., Zhang, T., Cheng, J. F., Kim, Y., Grimwood, J., Schmutz, J., Dickson, M., Noonan, J. P., Zhang, M. Q., Myers, R. M., and Maniatis, T. (2001). Comparative DNA sequence analysis of mouse and human protocadherin gene clusters. Genome Res. 11, 389–404.
Xu, Y., Wu, F., Tan, L., Kong, L., Xiong, L., Deng, J., Barbera, A. J., Zheng, L., Zhang, H., Huang, S., Min, J., Nicholson, T., Chen, T., Xu, G., Shi, Y., Zhang, K., and Shi, Y. G. (2011). Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol. Cell 42, 451–464.
Yagi, T., and Takeichi, M. (2000). Cadherin superfamily genes: functions, genomic organization, and neurologic diversity. Genes Dev. 14, 1169–1180.
Yamashita, H., Shanlin, C., Komagata, S., Hishida, R., Iwasato, T., Itohara, S., Yagi, T., Endo, N., Shibata, M., and Shibuki, K. (2012). Restoration of contralateral representation in the mouse somatosensory cortex after crossing nerve transfer. Plos One. (in press).
Yokota, S., Hirayama, T., Hirano, K., Kaneko, R., Toyoda, S., Kawamura, Y., Hirabayashi, M., Hirabayashi, T., and Yagi, T. (2011). Identification of the cluster control region for the protocadherin-beta genes located beyond the protocadherin-gamma cluster. J. Biol. Chem. 286, 31885–31895.
Yoshimura, Y., Dantzker, J. L., and Callaway, E. M. (2005). Excitatory cortical neurons form fine-scale functional networks. Nature 433, 868–873.
Yu, Y.-C., Bultje, R. S., Wang, X., and Shi, S.-H. (2009). Specific synapses develop preferentially among sister excitatory neurons in the neocortex. Nature 458, 501–505.
Keywords: protocadherin, Pcdh, CNR, neuron, neural circuits, complex networks, combinatorics, complexity
Citation: Yagi T (2012) Molecular codes for neuronal individuality and cell assembly in the brain. Front. Mol. Neurosci. 5:45. doi: 10.3389/fnmol.2012.00045
Received: 26 February 2012; Accepted: 22 March 2012;
Published online: 12 April 2012.
Edited by:
Joshua A. Weiner, The University of Iowa, USAReviewed by:
Joshua A. Weiner, The University of Iowa, USAWoj M. Wojtowicz, University of California, Berkeley, USA
Copyright: © 2012 Yagi. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Takeshi Yagi, Graduate School of Frontier Biosciences, Osaka University, 1–3 Yamadaoka, Suita, Osaka 565–0871, Japan. e-mail: yagi@fbs.oska-u.ac.jp