Genetically encoded Ca2+ indicators; expanded affinity range, color hue and compatibility with optogenetics
- 1Department of Biomolecular Science and Engineering, The Institute of Scientific and Industrial Research, Osaka University, Osaka, Japan
- 2PRESTO, Japan Science and Technology Agency, Tokyo, Japan
- 3Division for Bioimaging, Institute of Health Biosciences, The University of Tokushima Graduate School, Tokushima, Japan
- 4Department of Systems Neuroscience, Centre for Brain Integration Research, Tokyo Medical and Dental University Graduate School of Medical and Dental Sciences, Bunkyo-ku, Tokyo, Japan
Introduction
Fluorescent protein-based indicators are invaluable tools for functional imaging of living cells and organisms. Genetically encoded calcium indicators (GECIs) such as derivatives of yellow cameleons (YCs) and GCaMPs/pericams (Miyawaki et al., 1997; Nagai et al., 2001; Nakai et al., 2001) are a highly advanced class of indicators. Continued efforts for improvement of the performance of GECIs have resulted in brighter indicators with better photo-stability and expanded dynamic range, thus improving the sensitivity of detection. Fine-tuning of other properties, including Ca2+ affinity and Hill constant, have also contributed to increase the detectability of Ca2+ dynamics. Emerging optogenetic technology has forced the spectrally compatible GECI color variants. In this opinion, we highlight the recent development of GECIs including photo-switchable Ca2+ indicators and bioluminescence-based Ca2+ indicator, mainly invented in our group, focusing especially on the parameters determining their performance in order to provide a guideline for the selection of appropriate GECI for a given experiment.
Affinity Variant
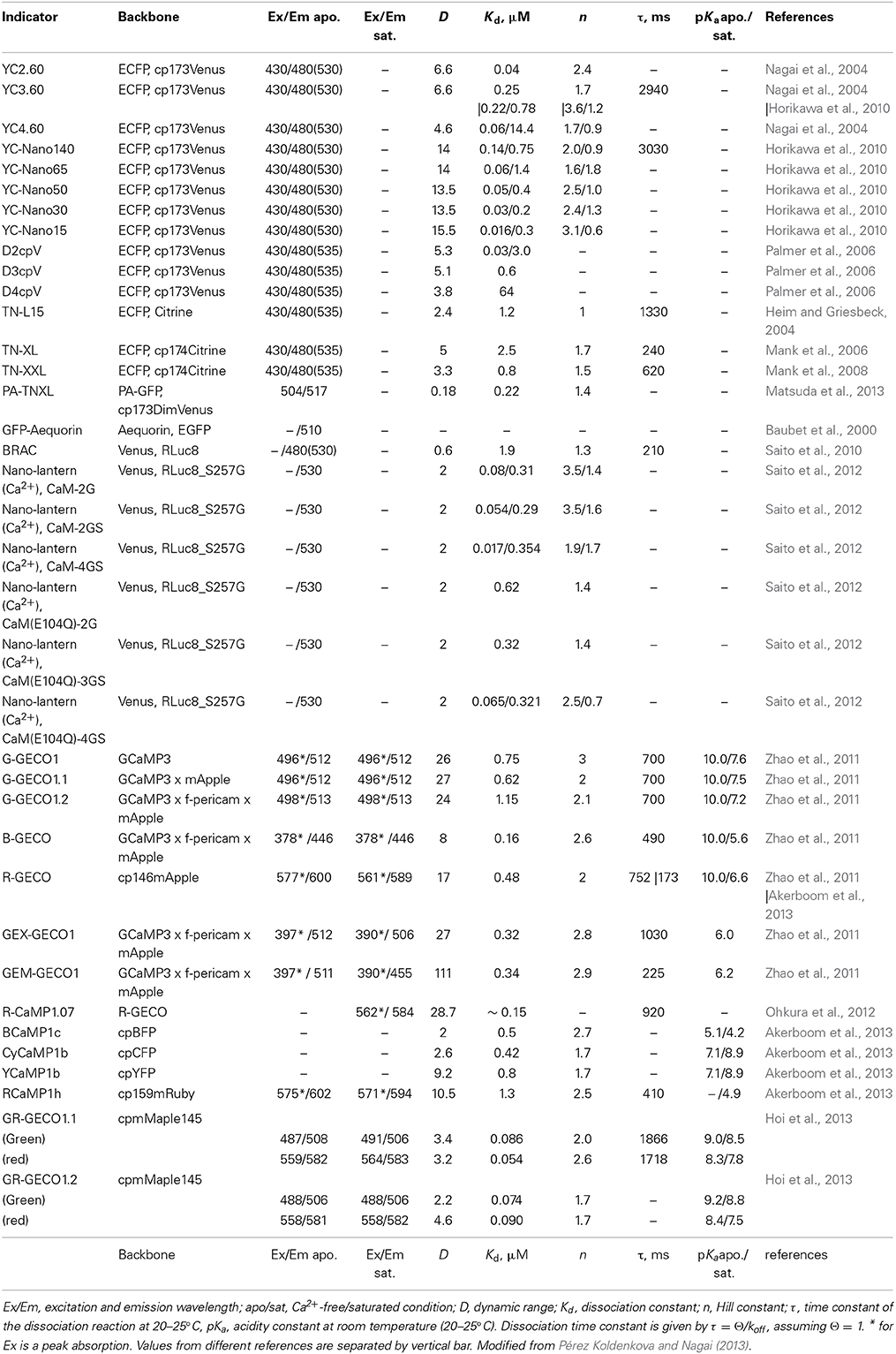
After the first reports regarding design concept of YCs and GCaMPs/pericams (Miyawaki et al., 1997; Nagai et al., 2001; Nakai et al., 2001), their properties have been modified in term of dynamic range of signal change, pH sensitivity and color hue, and so on. However, application of these GECIs had been still limited in certain experimental targets. One of the critical limitations of these GECIs was their relatively poor repertoire of affinity variants. Because the Ca2+ concentration ([Ca2+]) at resting state and the amplitude of [Ca2+] change differ significantly within the subcellular locations, cell types, and organisms, a diverse set of affinity variants of GECIs covering dissociation constants (Kds) from nM to mM would be needed for studying a wide range of research targets.
While moderate- and low-affinity variants of YCs (Kd > 0.1 μM) were developed successfully by either site-directed mutagenesis of the Ca2+ binding domain in the indicator (Miyawaki et al., 1997) or by the rearrangement of the overall molecular structure of the indicator (Truong et al., 2001) (Table 1), there was no systematic way for engineering a high-affinity variant. In vitro analysis revealed that free calmodulin (CaM) and its binding peptide M13 had much higher Ca2+ affinity (Kd of 20 nM) than that of the CaM and M13 fusion protein linked with two amino acid linkers (Kd of 80 nM) (Porumb et al., 1994). This suggested that steric hindrance might prevent efficient interaction of Ca2+-CaM with M13 in YCs. This possibility was examined by serial increment of the length of the linker from 2 to 5 amino acids. Flexible linkers with 3, 4, and 5 amino acids yielded Kds of 60 nM, 30 nM, and 15 nM, respectively. Linker elongation also worked for YC 3.60, yielding five YC variants covering Kds from 15 nM to 140 nM (Table 1). These affinity variants of YCs called YC-Nano showed increased sensitivity and could detect subtle changes in [Ca2+] in pyramidal neurons (Horikawa et al., 2010; Yamada et al., 2011) (Table 1), becoming an ideal toolbox to efficiently monitor the novel Ca2+ dynamics in cases where the concentration range of Ca2+ is poorly described (Table 1). Recent identification of Ca2+ twinkle, which is a localized Ca2+ transient in the fine astrocytic processes, is one of the examples (Kanemaru et al., 2014).
So far, high-affinity variants of the GCaMP and pericam families are not available (smallest Kd of 160 nM for B-GECO) (Zhao et al., 2011) (Table 1). Although these single FP-based Ca2+ indicators have distinct structural design unlike YCs, it will be interesting to examine whether elongation of linkers, which connect the sensor modules, contributes to increased Ca2+ affinity, as in the case of YC-Nano.
Possible Side Effects of GECI
One might worry that the strong Ca2+ chelating effect of YC-Nano would affect endogenous Ca2+ homeostasis. Depletion of target molecules or ions by loaded indicators is often problematic for imaging of non-buffered signaling molecules, such as cyclic nucleotides and NO, but this is not the case for Ca2+. Like H+, cytosolic free [Ca2+] is maintained dynamically through the balancing action of Ca2+ buffers (i.e., Ca2+ binding proteins), which exist in abundance within the cell. Of course, excessive loading of Ca2+ indicator/chelator beyond the buffering capacity of these buffers does affect cellular Ca2+ homeostasis. In cases where more than mM concentrations of EGTA were loaded to observe subcellularly localized Ca2+, Ca2+ puffs and blips were generated (Cheng and Lederer, 2008). Moderate loading/expression at sub-μM concentration of YC-Nano never affected the viability of fish embryos including a set of neurons (Horikawa et al., 2010).
On the other hand, functional interference of GECIs with endogenous Ca2+ binding proteins and their targets could pose a problem. The CaM of YCs potentially trans-activate endogenous CaM targets. In vitro analysis reported that excessive amounts of CaM affect the dynamic range of conventional YCs in a dose-dependent manner (Palmer et al., 2006). To avoid these side effects, computational re-design of Ca2+ sensing motifs was performed. Modified binding interface of the synthetic CaM and its target prevented intermolecular interaction. The resulting YCs, named D2/3/4cpV, have been demonstrated to be insensitive to large excesses of CaM, while maintaining a Kd of 0.03–64 μM and a large dynamic range of 3.8- to 5.3-fold (Palmer et al., 2006) (Table 1).
An alternate way to avoid uncontrolled interaction of GECI with endogenous proteins is to employ a different Ca2+-binding motif. While CaM has a variety of downstream targets, troponin C (TnC), a skeletal and cardiac muscle-specific Ca2+-binding protein, is known to limit its interaction to just troponin I and troponin T. Indicators incorporating TnC from avian skeletal muscle or human cardiac muscle were generated, based on molecular design similar to that of cameleon. The resulting TN-L15 and TN-hTnC displayed a moderate Ca2+ affinity but a lower Ca2+ specificity (due to its cross reactivity with Mg2+) and a small signal change (due to the lack of its binding peptide which enhances the conformational change of sensor motif) (Heim and Griesbeck, 2004) (Table 1). As with YCs, further improvements have been introduced to TN-L15. Mg2+ reactivity was eliminated by site directed mutagenesis on TnC, and the dynamic range was increased by replacing the Citrine acceptor with its cp174 variant, eventually yielding TN-XL (Mank et al., 2006). Low affinity of TN-XL was improved in TN-XXL by replacing TnC moiety with a concatenate of its high affinity C-lobe (Table 1). Although the in vitro dynamic range of TN-XXL was small, its in vivo performance was acceptable, suggesting the advantages of using TnC with reduced interference (Mank et al., 2008) (Table 1).
Photoactivatable GECI
To visualize Ca2+ dynamics in specific cell types, tissues, or organs, targeted expression of GECI gene is imperative. Although many promoters for cell/tissue/organ-specific expression are available, they do not cover all types of cell/tissue/organ. Photoswitchable GECIs (PS-GECI) can help overcome this limitation. Fluorescence status of PS-GECIs can be switched through light irradiation in arbitrary cell/tissue/organ, enabling cell/tissue/organ-specific visualization of Ca2+ dynamics. This “highlighted Ca2+ imaging” is beneficial in elucidating the activity of a single cell in the convoluted cell population of the neuronal network. There are only two reported PS-GECIs: a photoactivatable GECI, PA-TNXL (Matsuda et al., 2013), and a photoconvertible GECI, GR-GECO (Hoi et al., 2013) (Supplementary Image 1). The PA-TNXL was developed by replacing the donor and the acceptor fluorescent proteins in the TN-XL with a PA-GFP (photoactivatable GFP) and a dim yellow fluorescent protein DimVenus, respectively (Supplementary Image 1A and Table 1). Fluorescence of PA-TNXL can be switched on by violet light (~ 400 nm) irradiation. The fluorescence of the photoactivated PA-TNXL dims upon Ca2+ binding. GR-GECO has a similar design as GCaMPs/pericams. It has an mMaple, which can change fluorescence wavelength from green to red on being irradiated with violet light (~ 400 nm) (McEvoy et al., 2012; Hoi et al., 2013) (Supplementary Image 1B and Table 1). The intensity of both green and red fluorescence gets brighter with increase in [Ca2+]. For a wider range of applications, new PS-GECIs showing larger change in fluorescence intensity on photostimulation, higher dynamic range, broader Ca2+ affinity range, and/or reversible photoswitching are required.
GECIs for Optogenetics
In neuroscience, a paradigm shift has been brought about by optogenetics. Channel rhodopsin (ChR), a light-gated ion channel, and halorhodopsin (HR), a pump, allow us to control the activity of neural circuits with fine spatio-temporal resolution (Boyden et al., 2005; Zhang et al., 2007). As ChR and HR are activated by blue (400–500 nm) and yellow (500–600 nm) light, respectively; spectrally separated GECIs are necessary for combinatorial application of optogenetics with Ca2+ imaging. GECOs are the first reported color variants of single-FP GECIs based on cpGFPs and cpmApple (Zhao et al., 2011). Large-scale screening carried out by utilizing bacterial periplasmic expression system helped identify blue and red color variants of GECOs, including green and ratio metric alternates. In addition to the expanded color pallet, GECOs are also show sizable signal change (111-fold for GEM-GECO1), sensitizing them for the detection of subtle Ca2+ response. Structure guided evolution of GCaMP yielded BCaMP1c, CyCaM1a, YCaMP1b, and a series of RCaMPs (Akerboom et al., 2013). Compatibility of GECI color variants with optogenetic control was demonstrated by using CA3 pyramidal neuron co-expressing ChR2 and RCaMP1.07, which was in turn developed by the site-directed mutagenesis of R-GECO1 (Ohkura et al., 2012).
Bioluminescence-based Ca2+ imaging is an alternate and ideal strategy that is highly compatible with optogenetics. As bioluminescent indicators do not require excitation with light, observation can be free from functional crosstalk between optogenetic actuators. The limitation of this indicator was their dim signal as in the case of Ca2+ sensitive Aequorin and its emission-enhanced variants such as GA (Baubet et al., 2000). A considerable increase of emission signal was, however, achieved in BRAC and Nano-lantern (Ca2+), the latest version of bioluminescence-based GECIs (Saito et al., 2010, 2012) (Supplementary Image 1C and Table 1). BRAC is the cameleon like fluorescence resonance energy transfer (FRET)-based indicator harboring CaM-M13 moiety fused with an improved luciferase (RLuc8) derived from Renilla reniformis, which acts as a donor, and Venus, which acts as an acceptor. BRAC displays Ca2+ dependent FRET emission change (Supplementary Image 1C, left). Using BRAC, Ca2+ signaling was successfully visualized in plant leaves, in which fluorescence-based Ca2+ indicators cannot be applied due to their strong auto-fluorescence and intrinsic photosensitivity.
Nano-lantern (Ca2+) was constructed by direct fusion of Venus with RLuc8, which is split by insertion of CaM-M13. Ca2+ binding to CaM-M13 induces reconstitution of the split Rluc8. This leads to FRET from reconstituted Rluc to Venus, resulting in a large increase in emission signal (Supplementary Image 1C, right and Table 1). Thus, Ca2+ dynamics is monitored as a total intensity change, from both RLuc8 and FRET-enhanced emission of Venus. In the state-of-the-art demonstration by using cultured hippocampal neurons, Ca2+, transiently triggered by photo-activated ChR2, were imaged at 10 Hz with high SNR, showing the good compatibility of bioluminescence imaging with optogenetics (Saito et al., 2012) (Supplementary Images 1D,E).
Conclusion and Perspective
GECIs are advantageous over synthetic Ca2+ dyes in their targetability and reliability for chronic imaging. However, there remains room for further improvement on several parameters. Suboptimal kinetic property, non-linearity due to cooperativity, and pH sensitivity of single-FP based indicator should be improved to perform reliable detection of Ca2+. The future development of GECI is, regardless of the faults, promising, because of its evolvability. As a result of the past and the current attempts, not only have the basic property of GECIs been optimized, but also of new family of GECIs have been successfully developed, including color variants and self-illuminating GECIs. Future experiments will focus on improving the compatibility of GECIs with optogenetic tools. GECIs, used in collaboration with latest imaging platform such as a deep tissue imaging and ultra-fast and large-scale recording systems, will pave way to deepening our understanding of the supple ability our brain to learn and to memorize.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to acknowledge funding from Grant-in-Aid for Scientific Research on Innovative Areas, “Spying minority in biological phenomena” (No. 3306) from MEXT, a grant from the Cooperative Research Program of “Network Joint Research Center for Medical Devices,” and PRESTO and SENTAN from JST.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fnmol.2014.00090/abstract
Supplementary Image S1. Schematic representation of photoactivatable, photoconvertible or bioluminescent Ca2+ indicators. (A) PA-TNXL is composed of a troponin C (TnC) linked to a photoactivatable GFP (PA-GFP) at the N-terminal and a dim variant of Venus (DimVenus) at the C-terminal. On violet light irradiation, TnC starts fluorescing green. As the Ca2+ concentration increases, the intensity of fluorescence decreases proportionately due to FRET from PA-GFP to DimVenus (B) GR-GECO was developed from a photoconvertible FP mMaple, whose fluorescence changes from green to red on violet light irradiation. Circular permutation was introduced into mMaple and then linked to calmodulin (CaM) and M13 to create GR-GECO. The fluorescence in both green and red state increases proportionally with an increase in Ca2+ concentration. (C) A schematic representation of bioluminescence-based GECIs BRAC and Nano-lantern (Ca2+) for detecting Ca2+: Calmodulin (CaM) and M13 are used as Ca2+ binding domain for both of them. (Left) BRAC has Venus and RLuc8 at N- and C- terminal of CaM-M13, respectively. It causes wavelength shift from cyan to yellow thorough FRET on Ca2+ biding. (Right) A split RLuc is used for Nano-lantern (Ca2+). Each half of RLuc is linked to the terminals of CaM-M13. Compaction of CaM-M13 by Ca2+ binding reconstitutes the whole structure of RLuc, and luminescence intensity increases. Luminescence from RLuc is enhanced by VenusΔ C10 located at N-terminal of the Nano-lantern (Ca2+). An increase in Ca2+ can therefore be observed as FRET results in an increase in the intensity of yellow fluorescence. (D, E) Ca2+ imaging in the rat hippocampal neuron co-expressing Nano-lantern (Ca2+) and ChR2. (D) Ratio image (L/L0) of the Nano-lantern (Ca2+) without (i and iii) and with (ii and iv) activation of ChR2. (E) Time course of the ratio change. ChR2 is activated during the time period marked in gray.
References
Akerboom, J., Carreras Calderón, N., Tian, L., Wabnig, S., Prigge, M., Tolö, J., et al. (2013). Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front. Mol. Neurosci. 6:2. doi: 10.3389/fnmol.2013.00002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Baubet, V., Le Mouellic, H., Campbell, A. K., Lucas-Meunier, E., Fossier, P., and Brúlet, P. (2000). Chimeric green fluorescent protein-aequorin as bioluminescent Ca2+ reporters at the single-cell level. Proc. Natl. Acad. Sci. U.S.A. 97, 7260–7265. doi: 10.1073/pnas.97.13.7260
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Boyden, E. S., Zhang, F., Bamberg, E., Nagel, G., and Deisseroth, K. (2005). Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8, 1263–1268. doi: 10.1038/nn1525
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cheng, H., and Lederer, W. J. (2008). Calcium sparks. Physiol. Rev. 88, 1491–1545. doi: 10.1152/physrev.00030.2007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Heim, N., and Griesbeck, O. (2004). Genetically encoded indicators of cellular calcium dynamics based on troponin C and green fluorescent protein. J. Biol. Chem. 279, 14280–14286. doi: 10.1074/jbc.M312751200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hoi, H., Matsuda, T., Nagai, T., and Campbell, R. E. (2013). Highlightable Ca2+ indicators for live cell imaging. J. Am. Chem. Soc. 135, 46–49. doi: 10.1021/ja310184a
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Horikawa, K., Yamada, Y., Matsuda, T., Kobayashi, K., Hashimoto, M., Matsu-ura, T., et al. (2010). Spontaneous network activity visualized by ultra-sensitive Ca2+ indicators, yellow Cameleon-Nano. Nat. Methods 7, 729–732. doi: 10.1038/nmeth.1488
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kanemaru, K., Sekiya, H., Xu, M., Satoh, K., Kitajima, N., Yoshida, K., et al. (2014). In vivo visualization of subtle, transient, and local activity of astrocytes using an ultrasensitive Ca2+ indicator. Cell Rep. 8, 311–318. doi: 10.1016/j.celrep.2014.05.056
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mank, M., Reiff, D. F., Heim, N., Friedrich, M. W., Borst, A., and Griesbeck, O. (2006). A FRET-based calcium biosensor with fast signal kinetics and high fluorescence change. Biophys. J. 90, 1790–1796. doi: 10.1529/biophysj.105.073536
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mank, M., Santos, A. F., Direnberger, S., Mrsic-Flogel, T. D., Hofer, S. B., Stein, V., et al. (2008). A genetically encoded calcium indicator for chronic in vivo two-photon imaging. Nat. Methods 5, 805–811. doi: 10.1038/nmeth.1243
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Matsuda, T., Horikawa, K., Saito, K., and Nagai, T. (2013). Highlighted Ca2+ imaging with a genetically encoded ‘caged’ indicator. Sci. Rep. 3:1398. doi: 10.1038/srep01398
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McEvoy, A. L., Hoi, H., Bates, M., Platonova, E., Cranfill, P. J., Baird, M. A., et al. (2012). mMaple: a photoconvertible fluorescent protein for use in multiple imaging modalities. PLoS ONE 7:e51314. doi: 10.1371/journal.pone.0051314
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Miyawaki, A., Llopis, J., Heim, R., McCaffery, J. M., Adams, J. A., Ikura, M., et al. (1997). Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388, 882–887. doi: 10.1038/42264
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nagai, T., Sawano, A., Park, E. S., and Miyawaki, A. (2001). Circularly permuted green fluorescent proteins engineered to sense Ca2+, Proc. Proc. Natl. Acad. Sci. U.S.A. 98, 3197–3202. doi: 10.1073/pnas.051636098
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nagai, T., Yamada, S., Tominaga, T., Ichikawa, M., and Miyawaki, A. (2004). Expanded dynamic range of fluorescent indicators for Ca2+ by circularly permuted yellow fluorescent proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 10554–10559. doi: 10.1073/pnas.0400417101
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nakai, J., Ohkura, M., and Imoto, K. (2001). A high signal-to-noise Ca2+ probe composed of a single green fluorescent protein. Nat. Biotechnol. 19, 137–141. doi: 10.1038/84397
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ohkura, M., Sasaki, T., Kobayashi, C., Ikegaya, Y., and Nakai, J. (2012). An improved genetically encoded red fluorescent Ca2+ indicator for detecting optically evoked action potentials. PLoS ONE 7:e39933. doi: 10.1371/journal.pone.0039933
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Palmer, A. E., Giacomello, M., Kortemme, T., Hires, S. A., Lev-Ram, V., Baker, D., et al. (2006). Ca2+ indicators based on computationally redesigned calmodulin-peptide pairs. Chem. Biol. 13, 521–530. doi: 10.1016/j.chembiol.2006.03.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pérez Koldenkova, V., and Nagai, T. (2013). Genetically encoded Ca2+ indicators: properties and evaluation. Biochim. Biophys. Acta 1833, 1787–1797. doi: 10.1016/j.bbamcr.2013.01.011
Porumb, T., Yau, P., Harvey, T. S., and Ikura, M. (1994). A calmodulin-target peptide hybrid molecule with unique calcium-binding properties. Protein Eng. 7, 109–115. doi: 10.1093/protein/7.1.109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Saito, K., Chang, Y. F., Horikawa, K., Hatsugai, N., Higuchi, Y., Hashida, M., et al. (2012). Luminescent proteins for high-speed single-cell and whole-body imaging. Nat. Commun. 3, 1262. doi: 10.1038/ncomms2248
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Saito, K., Hatsugai, N., Horikawa, K., Kobayashi, K., Matsu-Ura, T., Mikoshiba, K., et al. (2010). Auto-luminescent genetically-encoded ratiometric indicator for real-time Ca2+ imaging at the single cell level. PLoS ONE 5:e9935. doi: 10.1371/journal.pone.0009935
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Truong, K., Sawano, A., Mizuno, H., Hama, H., Tong, K. I., Mal, T. K., et al. (2001). FRET-based in vivo Ca2+ imaging by a new calmodulin-GFP fusion molecule. Nat. Struct. Biol. 8, 1069–1073. doi: 10.1038/nsb728
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yamada, Y., Michikawa, T., Hashimoto, M., Horikawa, K., Nagai, T., Miyawaki, A., et al. (2011). Quantitative comparison of genetically encoded Ca2+ indicators in cortical pyramidal cells and cerebellar purkinje cells. Front. Cell. Neurosci. 5:18. doi: 10.3389/fncel.2011.00018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, F., Wang, L. P., Brauner, M., Liewald, J. F., Kay, K., Watzke, N., et al. (2007). Multimodal fast optical interrogation of neural circuitry. Nature 446, 633–639. doi: 10.1038/nature05744
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhao, Y., Araki, S., Wu, J., Teramoto, T., Chang, Y. F., Nakano, M., et al. (2011). An expanded palette of genetically encoded Ca2+ indicators. Science 333, 1888–1891. doi: 10.1126/science.1208592
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: genetically encoded calcium ion indicator, Ca2+ imaging, FRET, fluorescence, bioluminescence
Citation: Nagai T, Horikawa K, Saito K and Matsuda T (2014) Genetically encoded Ca2+ indicators; expanded affinity range, color hue and compatibility with optogenetics. Front. Mol. Neurosci. 7:90. doi: 10.3389/fnmol.2014.00090
Received: 15 September 2014; Accepted: 30 October 2014;
Published online: 25 November 2014.
Edited by:
Katsuhiko Mikoshiba, RIKEN Brain Science Institute, JapanCopyright © 2014 Nagai, Horikawa, Saito and Matsuda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: ng1@sanken.osaka-u.ac.jp
 Takeharu Nagai
Takeharu Nagai Kazuki Horikawa
Kazuki Horikawa Kenta Saito
Kenta Saito Tomoki Matsuda
Tomoki Matsuda