1
Neuroscience Institute, Georgia State University, Atlanta, GA, USA
2
Department of Pyschology, Dominican University, River Forest, IL, USA
3
Department of Computer Science, Georgia State University, Atlanta, GA, USA
The basic unit of any nervous system is the neuron. Therefore, understanding the operation of nervous systems ultimately requires an inventory of their constituent neurons and synaptic connectivity, which form neural circuits. The presence of uniquely identifiable neurons or classes of neurons in many invertebrates has facilitated the construction of cellular-level connectivity diagrams that can be generalized across individuals within a species. Homologous neurons can also be recognized across species. Here we describe NeuronBank.org, a web-based tool that we are developing for cataloging, searching, and analyzing neuronal circuitry within and across species. Information from a single species is represented in an individual branch of NeuronBank. Users can search within a branch or perform queries across branches to look for similarities in neuronal circuits across species. The branches allow for an extensible ontology so that additional characteristics can be added as knowledge grows. Each entry in NeuronBank generates a unique accession ID, allowing it to be easily cited. There is also an automatic link to a Wiki page allowing an encyclopedic explanation of the entry. All of the 44 previously published neurons plus one previously unpublished neuron from the mollusc, Tritonia diomedea, have been entered into a branch of NeuronBank as have 4 previously published neurons from the mollusc, Melibe leonina. The ability to organize information about neuronal circuits will make this information more accessible, ultimately aiding research on these important models.
The goal of brain atlases is to provide a common structure upon which individual observations can be laid. The finest anatomical scale of a nervous system atlas would be the neurons and synapses, i.e. a cellular level wiring diagram. Yet, despite over a hundred years of studying neurons and neuronal interactions, there is no consistent means of representing neurons and neural circuitry. Furthermore, there is no universal electronic repository for that knowledge and thus there is no efficient means to search for information about neuronal circuits across species. Here, the problems associated with creating a neural circuit atlas are defined and a tool called NeuronBank.org is introduced as a step towards addressing these problems.
One challenge with creating an atlas of neuronal wiring is determining what constitutes a particular type of neuron (Masland, 2004
; Bota and Swanson, 2007
). Mammalian brains have billions of neurons that are thought to fall into a few thousand classes (Bullock, 1993a
). It has been known since the work of Cajal that classes or types of neurons form stereotyped neural circuits (Ramon y Cajal, 1995
). For example, in the cerebellum, Purkinje neurons receive synaptic input from hundreds of thousands of granule cells and one climbing fiber. However, not all Purkinje neurons are equivalent; they project to different deep cerebellar nuclei and receive input from non-equivalent sets of climbing fibers and granule cells. For a discussion of “equivalent sets” of neurons see chapter 7 in Bullock (1993b)
.
The quandary of how to classify neurons in mammalian nervous systems is a serious one for understanding the basic wiring of the brain (Masland, 2004
; Bota and Swanson, 2007
). The problem is becoming more acute because molecular characterization of mammalian neurons is increasing the possible number of neuronal types (Van Hooser et al., 2005
; Nelson et al., 2006a
,b
). Furthermore, new imaging technologies are allowing neuronal wiring diagrams to be very finely specified (Briggman and Denk, 2006
; Livet et al., 2007
; Smith, 2007
; Lichtman et al., 2008
; Helmstaedter et al., 2008
; Jurrus et al., 2009
).
This problem is lessened in some invertebrate nervous systems where individual neurons can be uniquely identified and have similar properties from animal to animal (Croll, 1987
; Bullock, 2000
). In some invertebrate phyla such as gastropod molluscs and decapod crustaceans, the large size of neurons has facilitated simultaneous intracellular recording from multiple neurons. This has allowed detailed neuronal circuitry to be experimentally determined and related back to behavioral function (Kandel et al., 1967
; Brodfuehrer and Thorogood, 2001
; Comer and Robertson, 2001
; Selverston, 2008
; Katz, 2009
). Having atlases of the neuronal connectivity in these species is of interest to keep track of the increasingly complex circuitry and cellular properties. Furthermore, creating an atlas of identified neurons in such species can serve as a proving ground for nervous systems with less well-defined neurons.
Homologous neurons and neural circuits can be recognized across species. (Weiss and Kupfermann, 1976
; Croll, 1987
; Paul, 1991
; Newcomb and Katz, 2007
). Comparisons of the similarities and differences between species are often difficult to extract from the literature. Yet, there are important reasons why one would want to be able to use information from one species to guide research on neural circuits in other species. For example, the large size of grasshopper neurons allowed their early development to be mapped out and then applied to the much smaller nervous system of the fruit fly (Thomas et al., 1984
). A registry of neurons could help sort out homologies and thus help extend findings across taxa.
There are varying degrees to which a neuron can be individually identified (Bullock, 2000
). In the extreme, there is one neuron (or a bilaterally symmetric pair) that can be uniquely distinguished from all other neurons in the nervous system. In other cases, there is a small cluster of neurons that appear indistinguishable from each other. In animals that exhibit eutely (constancy of cell number), it is possible to identify every neuron in the nervous system, as has been done for the nematode, Caenorhabditis elegans, which has precisely 302 neurons (White et al., 1976
, 1986
). The small size of C. elegans has allowed the entire animal to be serially sectioned and reconstructed with electron microscopy, providing an unrivaled level of detail about the neurons and synapses. With a defined number of neurons, it is a relatively straightforward matter to create a database containing all neurons, i.e. its “neurome”. WormAtlas.org (Altun et al., 2009
) contains all of the neurons along with their synaptic connections, gene expression profile, anatomy, neurotransmitter, and developmental lineage. This type of comprehensive information is not available for any other nervous system. Such knowledge of the neurons and their connectivity is now allowing researchers to address questions related to how neuronal circuitry generates behavior in this animal (De Bono and Maricq, 2005
; Schafer, 2005
; Karbowski et al., 2007
).
The nervous systems in gastropod molluscs are more complex than that of C. elegans. There are approximately 8000–10,000 neurons in the central ganglia of opisthobranchs such as Aplysia californica and Tritonia diomedea (Coggeshall, 1967
; Willows et al., 1973
). This is two orders of magnitude larger than the nervous system of C. elegans, but many orders of magnitude smaller than vertebrate nervous systems. The number of neurons is also at least an order of magnitude less than the number found in insects and other arthropods. Among the 10,000 neurons, some of the larger ones have been individually identified. There are also clusters of neurons that appear similar and thus constitute a type of neuron, such as the sensory neurons in Aplysia (Emery and Audesirk, 1978
) or Tritonia (Getting, 1976
). Thus, the gastropod nervous systems provide an interesting test case for creating a neuronal circuitry atlas.
An atlas of neuronal circuitry must have certain features. These would include the ability to represent neurons from different parts of the nervous system and even from the nervous systems of different species. It would also need to be flexible enough to allow new characteristics to be added as our knowledge of neurons grows. Finally, it would need to allow for users to search this information within a species and across species.
One important step towards being able to catalogue and search neurons and neural circuitry is to develop a controlled vocabulary for the characters that apply to neurons. An example of this is the so-called “Petilla” terminology for classifying GABAergic neurons in cerebral cortex, which defines each of the characteristics that might be found in such neurons (Ascoli et al., 2008
). A broader approach has been used to create a Neuroscience Information Framework standardized ontology (Bug et al., 2008
). This has developed into NeuroLex.org, a semantic web wiki that organizes terms used in neuroscience. Any atlas needs to use terminology that is commonly agreed upon by the community of users.
Another step that is needed to unify knowledge about neuronal circuitry is the development of tools that allow neurons and neuronal connectivity to be catalogued and searched. There are a small number of such repositories in existence that are species-based or phylum-based. For example, as mentioned above, WormAtlas.org catalogues the neurons in the nematode, C. elegans. The Brain Architecture Management System (BAMS)
1
has a list of neurons from mammalian nervous systems (Bota et al., 2005
). However, the nervous system of C. elegans is sufficiently different from the mammalian nervous system that nematode neurons cannot be represented in BAMS and mammalian neurons cannot be represented in WormAtlas. Nematodes and rats are evolutionary distant, so it is not too surprising the ontology for one does not map onto the other in a systematic fashion. None-the-less, there are important commonalities such as molecular components (neurotransmitters, receptors, intracellular signaling molecules), physiological properties (plasticity, rhythmicity) and functional design (sensory systems, motor systems). Therefore, it would be of interest to be able to search across species to compare nervous system components and properties the way that one can currently search across species to compare genes and their functions in GenBank
2
There are several problems that one encounters in trying to represent knowledge about neurons and synapses that are not present for genes. The first is that whereas all genes, regardless of the organism, are strings of nucleotides, there is no uniform definition of what a neuron is (Moroz, 2009
). Furthermore, there is debate as to whether complex nervous systems arose independently in different phyla (Nielsen, 1999
; Miller, 2009
; Moroz, 2009
), which would make classification schemes based on homology invalid. However, even if there is deep homology of neuronal cell types (Arendt, 2008
; ito-Gutierrez and Arendt, 2009
), this does not aid in creating classification schemes because of problems of convergent evolution.
There are many features that can be used to characterize a neuron or a class of neurons: anatomical, electrophysiological, molecular, developmental, and functional. For most neurons, the information about these features is incomplete. New techniques continue to be developed that allow neurons to be categorized along additional dimensions, such as molecular expression patterns. It is virtually impossible at this point in history to fully list all of the possible characteristics of a neuron and its synapses. Therefore, any atlas of neurons and connectivity must by its nature be incomplete.
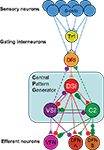
Yet despite these obstacles, researchers do create circuit diagrams of neurons and their connectivity. For example, Figure 1
shows the neural circuit mediating the escape swim response of the mollusc Tritonia (Katz, 2009
). The circuit illustrates the neurons that have been identified as playing a role in this behavior and their synaptic interactions (Getting and Dekin, 1985
; Getting, 1989
). Even though this neural circuit is relatively simple, it is difficult to represent all of the complexities in one circuit diagram. For example, sometimes neurons are represented as a class; there are three DSIs, but only one is shown on the diagram to make it simpler. The DFN-A and DFN-B neurons are functional classes, not identified neurons. The properties on the neurons and their synapses are not illustrated beyond the fact that some synapses are excitatory and others are inhibitory. Furthermore, by their nature, such circuit diagrams are static; they do not allow readers to explore the circuitry beyond what is presented; there are neurons that feed into this circuit that are not represented here.
Figure 1. Tritonia swim circuit. This image illustrates fundamental features of a neuronal wiring diagram used in publication. It represents neurons as balls and their synapses as sticks with triangles (excitatory), circles (inhibitory), or combinations of both. The names of the neurons are indicated on the balls.
In an effort to create a tool that could begin to address some of the needs mentioned, we embarked on the creation of a neural circuit atlas called NeuronBank (Calin-Jageman et al., 2007a
). Here, we will explain the fundamental organization of NeuronBank and demonstrate some of its functionality using the Tritonia nervous system as an example.
Basic architecture of the Neuronbank System
The primary challenge in designing NeuronBank was to accommodate species-diversity and continued scientific progress. Meeting these needs required flexibility in both the data model and the user interface, presenting relatively unique challenges for system design. Most current bioinformatics efforts utilize a three-tier architecture: (1) a single relational database provides data to (2) an application layer, which builds an invariant user interface for (3) web clients. In this architecture, changes to the data model must be infrequent because they require developer intervention to update both the database backend and the application layer. Moreover, there is typically only a single data store, so data sharing is not a concern. This type of architecture would not have been suitable for NeuronBank for several reasons. First, different species require different data models and user interfaces. Second, within each species, changes to the data model and user interface are likely to be frequent, making it impractical to require developer intervention. On the other hand, completely separating each species into separate applications would also be untenable. The duplication of effort and the challenges of maintaining interoperability across multiple, independent, yet related systems would override any gain in flexibility in data model and interface.
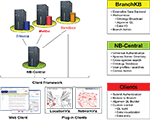
Our solution was to develop a service-oriented federation of customizable knowledgebases. Individual nodes within the federation (BranchKBs – Branch Knowledgebases) are specialized for knowledge of neural circuitry in a single species. New branches can be established using BranchKB, a software package that we have developed for creating an online, collaborative knowledgebase of neuronal types and synaptic connectivity. Branches are united by a common framework (NB-Central – NeuronBank Central), which enables search and analysis of neural circuits across all species. In this way, we provide the specialization and autonomy necessary to share and analyze knowledge from different species. Figure 2
shows the overall distributed architecture of NeuronBank.
Figure 2. Architecture of NeuronBank.
NB-Central serves two primary roles. First, it creates an environment for invoking and collating services provided by the BranchKBs. This enables a mechanism for searching and analyzing data across all the species represented. Second, it provides a set of web services to the BranchKBs and the clients. These services establish common core functionality, presenting a consistent experience across all branches. One of the major motivations for pursuing a service-based design for both NB-Central and the branches is that this allows for interoperability. Consistent interfaces allow us to support the development of third-party clients and allow for different implementations of the BranchKB using different technologies in the future.
Users interact with this federation via an integrated web client, which consumes resources from both NB-Central and various BranchKBs. Thus, the architectural complexity is masked and the user can seamlessly browse, search, and analyze knowledge from various species that is stored across the federation of NeuronBank branches. The web client features a plug-in architecture, making it possible to utilize different visualization and analysis tools on data from different BranchKBs (Figure 2
). Moreover, the web service interfaces to BranchKB and NB-Central are open, enabling the development of alternative clients that provide specialized functionality.
BranchKB
BranchKB is the storage system for each species-specific Branch within NeuronBank. In this respect, BranchKB is similar to the ‘database backend’ of a traditional web application. However, BranchKB differs from a traditional database in three important respects. First, BranchKB is designed to have a somewhat flexible data model, so that it can be adapted for representing knowledge within a particular species. Second, BranchKB provides services for accessing the current data model, exposing the meta-data required to build a dynamic user interface for that branch. Finally, BranchKB is designed to integrate with NB-Central so that knowledge can be shared across diverse communities.
Flexible data model of neural circuitry
Across all domains, neurons are described by sets of attributes (e.g. neurotransmitter, spike shape, and location) and identified by delineating the subset of attributes necessary and sufficient to reliably identify that neuron across different specimens. There is variation, however, in the attributes used to describe neurons in different species or even in different regions of the nervous system of a single species. In addition, new techniques within the neurosciences are constantly adding new attributes that can be used to distinguish neurons. NeuronBank uses a two part hierarchical ontology to represent the knowledge about neurons and connections: (a) a core ontology of classes and sub-classes that are applicable to all branches, and (b) an extensible list of attributes that can be tailored for a specific branch. All instances have a unique Accession ID that is generated by the system and provides a unique URL.
(a) The core ontology currently consists of root classes for Neurons, Connections, and Annotations. Additional root classes can be added if needed. The core ontology has deliberately been designed to be as simple as possible so that it will be applicable to neural circuits from any species. For example, all neurons have the following four core attributes: Name, Inputs, Outputs, and Annotations. All other properties are represented in the extensible list of attributes. The Inputs, Outputs, and Annotations classes are further sub-classified in a hierarchical manner to represent commonly agreed upon classifications. But, this sub-classification can be changed in the future; it is not a fixed component of NeuronBank. The current core ontology is shown at http://neuronbank.org/help/index.php/Core_Ontology
.
(b) The extensible list of attributes allows individual branches of NeuronBank to customize the properties of objects. The NeuronBank application is not affected by any changes made to the extensible list of attributes and provides the ability for schema evolution that is typically not available with a relational database system. Changes to the extensible attribute list are made with the attribute editor (Figure 3
). An attribute can be either a classification or a measurement. Classification is for attributes with enumerated values, whereas measurement is for attributes that have non-discrete numerical values, sometimes with a standard error.
Figure 3. Screenshot showing editing of extensible attributes in the web client. (A). The list of extensible attributes for a neuron is editable. This screenshot shows the first 6 attributes of Neurons in the Tritonia extensible list. A new attribute can be added by filling in the name (here “My_Attribute”) and choosing whether the attribute is a Measurement or a Classification. Clicking on the attribute’s name allows further editing. (B) This is an example of editing the Attribute “Activity_Resting”. This is a classification with the values Silent, Spiking, Bursting, Irregular. Additional values can be added if they are needed for describing neuronal resting activity.
The administrative editing capability of BranchKB allows each branch to be customized to provide the attributes necessary to describe neurons in that species. As new techniques enable the measurement of additional neural attributes, the data model can be further extended. We envision that communities of neuroscientists interested in a given species will work together to devise the attribute set appropriate for that species.
Meta-data for dynamic user interfaces
Although a dynamic interface was a design requirement for web and plug-in clients, it has significant implications for the design of BranchKB. Specifically, BranchKB must enable clients to retrieve not only data but also the current data model. Thus, each BranchKB exposes functions that enable client applications to retrieve the current class structure, the attributes that apply to a particular class, and the attribute values that are valid for a particular instance. These functions are made available through a set of web services so that each customized instance of BranchKB maintains a consistent set of interfaces for various clients (see below).
NB-Central
NB-Central is a hub that unifies the diverse branches into a coherent federation. First, it provides a common set of web services to the BranchKBs. These services impart common core functionality, eliminating duplication of effort and generating a consistent experience across all branches. Second, NB-Central creates an environment for invoking and collating services provided by the BranchKBs. This enables a mechanism for searching and analyzing data across all of the species represented. By drawing on existing work in the fields of web services and distributed architectures, we have designed NB-Central as a unifying mechanism for managing heterogeneous, dispersed resources. The specific functions of NB-Central are as follows:
(1) Branch Directory. NB-Central maintains a master list of online BranchKBs, periodically polls their status, and provides information to clients about each branch.
(2) User Authentication and profiles. NB-Central provides a single-point authentication system; each BranchKB invokes this service to check a user’s credentials and establish his/her privileges. Thus, a logon can be executed from any point in the federation. In addition to Branch-level permissions, NB-Central stores all user-profile information and provides secure access to this information via a web service. This enables a user’s preferences and privileges to be available to all BranchKBs.
(3) Cross-branch searching. Despite the heterogeneity across BranchKBs, it is essential to enable comparative work across species. NB-Central works in concert with the federation of BranchKBs to provide this function by exposing a web service for cross-species search. This service accepts a query and then passes it on to each available BranchKB. NB-Central collates the results and returns them back to the user. Thus, NB-Central provides a single point to access data throughout the federation.
User Roles
NeuronBank has just four types of users: Branch Administrators, Branch Editors, Registered Users, and Unregistered Users. All users have full access to read the knowledge in the federation. They can search, browse, and export this knowledge. They can be anonymous or register with NB-Central to enable stored searches, favorites, and other user services. Registered users can modify the Help Wiki and the NeuronBank Wiki (see below). Branch Editors curate the customizations, which specialize the Branches. Specifically, Branch Editors create, edit, and document the specialized data model, i.e. the extensible list of attributes for describing neurons in that species. They can add new neurons, annotate existing knowledge, elucidate new properties of existing neurons, etc. Branch Administrators have all of the access of Branch editors, but also perform user management for their Branch.
Web Client
NeuronBank web client is the primary user interface for the NeuronBank federation. It invokes web services of both NB-Central and the various BranchKBs. The primary functions of the web client are to enable knowledge access (browsing and searching), knowledge entry (create, edit, and delete), knowledge analysis (visualization) ontology curation (customizing species-specific neuron attributes), and user administration.
To accommodate the heterogeneity in data models across different BranchKBs, the interfaces for accessing, editing, and analyzing knowledge must be dynamic and built on-the-fly from meta-data retrieved from the relevant BranchKB. Moreover, visualization and analysis needs vary across branches. To achieve a high level of flexibility, the web client utilizes a plug-in system. A software plug-in is a program that extends the capability of the main program. The plug-in programs interact with the main program through services such as Application Programming Interfaces (API) or data exchange protocols. The plug-in system provides much flexibility and extensibility, allowing third parties to develop small programs for their specific applications. Different plug-ins can be invoked on data from different BranchKBs. Additional plug-ins can be developed for general use or to meet specialized needs for a particular branch. The current NeuronBank includes two plug-in programs: LocationViz and NetworkViz. These two programs retrieve neuron data from the knowledgebase through a standard data exchange protocol, and then display the neuron location or circuit information in 2D maps.
Implementation Technologies
BranchKB
We implemented BranchKB as a set of Java-based web-services and servlets. As a knowledge store, we used the Protégé knowledge-management system
3
. Protégé is a mature, open-source Java-based knowledgebase system (Musen et al., 1994
; Noy et al., 2003
). It provides a class/instance/attribute data store that can be accessed via both a Java API and an extensible query language called Algernon. Protégé knowledgebases can be stored in a number of formats, including MySQL database tables. Finally, Protégé knowledgebases can be imported and exported to a variety of formats, including RDF and XML. Import and export formats can also be extended via a plug-in architecture. Protégé provides native support for both a dynamic data model and meta-data access. This enabled us to rapidly prototype BranchKB.
NB-Central
Similar to BranchKB, we implemented NB-Central as a set of Java-based web services and servlets. As the data model for user and branch information is relatively stable and simple, we used a conventional relational database backend (mySQL 5.0).
Web and plug-in clients
We implemented our web client as a Java Server Faces (JSF) web application hosted on Apache Tomcat application server. JSF is a stable, well-supported web-application technology. It features a free and full-featured IDE (Sun Developer Studio). The Visualization clients were implemented as Java applets.
Servers
We use Dell PowerEdge servers with dual Xeon processors running Fedora Red Hat Operating System. We currently maintain both a test server and a production server. Each runs Apache web server, Apache Tomcat application server, mySQL, Protégé and PHP. NB-Central currently resides on the same server as our two BranchKBs, along with our help system, public website, code versioning repository, and developer documentation system. The public website is implemented using Exponent, which requires a mySQL database and PHP scripting system. The Wikis use the popular MediaWiki package.
To illustrate the interactions between system components and provide more insight into operation of our federated architecture, we will walk through the use of NeuronBank. Currently, we have implemented most of the functionality envisioned for general users, but have only prototyped the functions for Branch Editors and Administrators, which we grant for the Sandbox branch upon request. Our progress has been sufficient, however, to deploy our first Branches for public testing and to populate them with all the known neurons and connection in two species, Tritonia diomedea, which now has 45 neurons and Melibe leonina, which has only 4 published neurons.
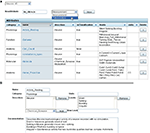
Branch Listing
This interface from the web client presents a directory of branches, allowing users to quickly identify the species catalogued in the federation, the status of each branch, and their permissions on each branch (Figure 4
). From here, users can jump directly into a branch to view, edit, or analyze knowledge within that species. This simple interface masks considerable complexity. The web client dynamically builds this interface by invoking the Branch-directory web service on NB-central.
Figure 4. Branch List. Screen shot showing the Branch List and the navigation bar at the top. The branch list is dynamically generated to show the active branches, the number of neurons in each branch and the permissions of the user for each branch. Text searches can be initiated from the navigation bar (i).
Dynamic Search Interfaces
One of the most important aspects for any knowledge management system is an effective search interface. We have implemented five types of search functions in NeuronBank v1.0:
(1) Text searches. Text strings can be searched for from any page in NeuronBank using the “Find” function, which is located on the navigation bar (Figures 4
i, 5
i).
(2) Browse neurons. A simple browse function allows users in one click to view all of the neurons in that branch (Figure 5
ii).
(3) Form-based searches. We have implemented a more powerful querying system using Algernon
4
, a powerful LISP-based query language (Figure 5
iii), which enables path-based searches. (e.g., find all the neurons with chemical synapses where the presynaptic neuron is involved with the behavior “swimming” Figure 5
). Creating queries directly in Algernon is relatively complex and would not be suitable for most users. Therefore, we developed a form-based interface for the web that allows users to construct ad-hoc Algernon queries. The primary novelty of this interface is that it builds the search interface dynamically to match the current data model of a selected branch. Specifically, the web client retrieves the current class structure from a selected BranchKB. When a class is selected as part of a search, the client retrieves from the BranchKB the attributes that can apply to that class and the paths leading away from that class. Thus, a single search interface on the client can generate queries for any BranchKB, despite substantial heterogeneity in their data models. Once the user has designed a query, the client constructs an equivalent Algernon statement and executes the search.
(4) Cross-Branch searches. Each branch stores knowledge for a different species using a slightly different data model. Normally, this would make it difficult to conduct a search efficiently across multiple Branches. However, each BranchKB presents a consistent interface via web services. This enabled us to further generalize the dynamic search interface in the web client. Specifically, the same query can be submitted to all BranchKBs in the federation using the same interface. Cross branch searches can be conducted using the “Find” text search or the form-based query.
(5) Anatomical searches. One of the important distinguishing characteristics of a neuron is its location within a nervous system. However, given the incommensurable body plans of distantly related organisms–no single coordinate or mapping system could possibly represent anatomical locations across species. To overcome this problem, we have supplemented the web client with a LocationViz plug-in. This embedded JAVA applet visualizes the somatic locations of neurons in 2- dimensions on an anatomical plane. This representation system is sufficient for a variety of species in which soma location can be indicated relative to a set of standard brain views or slices. For example, in Tritonia, neurons can be located based on standardized views of the dorsal and ventral surface of the brain. The LocationViz plug-in can accept a different set of reference views for each BranchKB, thus enabling the same plug-in to visualize anatomical information over many branches. For species in which this form of representation is not sufficient (e.g. mammals), a different visualization plug-in will need to be developed.
First Public Branches
We have deployed the first branches of NeuronBank for public use: the Tritonia and Melibe branches. These branches contain the published information about the neurons and synaptic connectivity in the molluscs, Tritonia diomedea and Melibe leonina, which are model neuronal systems (Thompson and Watson, 2005
; Katz, 2009
). The branches serve as a convenient test for establishing a branch because the number of publications related to identification of neurons and connections is relatively small (<15 for both) and the community of researchers is also small (7 independent labs). It took less than 2 weeks for one person to enter all of the information about neurons and synaptic connections in Tritonia into our test server. We had a second person repeat the process and again found that it took less than 2 weeks of moderate work to input all 44 neurons and their connections. We then had students, post-docs, and other Tritonia researchers search the knowledgebase for errors in content. We immediately found one omission; a user noticed that we had neglected to include Neuron Pd14 (Tri0002792). This oversight illustrates the need for NeuronBank; the description of this neuron was buried in a paper (Murray and Willows, 1996
) and thus not apparent to most researchers. It was the author of that paper who noticed the omission from NeuronBank. We have also used the Tritonia branch as an outlet for publication of a neuron that we identified, but have not published in a journal, the LU neuron (Tri0002823).
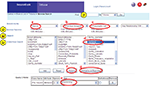
An Example Search
To illustrate the features of the system, we will walk through a form-based search. Let’s say you are on the Tritonia branch and want to search for all neurons that make a chemical synapse with a presynaptic neuron that is involved with the behavior “swimming”. Figure 5
shows the pull-menus that would be used to construct this query. One drawback of this system is that it takes some knowledge of the meaning of the attributes to construct appropriate queries. Future implementations will allow the user access to definitions of all of the attributes as well as their links in NeuroLex.org.
Figure 5. Screen shot of a search on the Tritonia branch. Four types of searching are available i. Text string, ii. Browsing, iii. Form-based search, iv. Anatomical search. In a form-based search shown, the user has created a query for a neuron (a) with a chemical synapse (b) where the presynaptic cell (c) has an attribute of “-behavior” (d) and the value of that attribute is “Swimming” (e). This query can be submitted to the Tritonia branch (f) or it can be submitted to all branches (g).
The query can be submitted to the current branch (Tritonia) or it can be submitted to all branches (Figure 5
f,g). Generalizing a search across the federation presents two problems. (1) How is the client to discover available BranchKBs? (2) How should the client collate results across the federation? These problems are overcome by NB-Central, which acting as a hub for the federation generates the Algernon query and invokes the search web services across all the branches. The results are then displayed in a table (Figure 6
), which shows how many hits were found in each branch. This query had 37 results in Tritonia, 4 in the Sandbox branch, and one in the Melibe branch.
Figure 6. Cross branch query results. The query from Figure 5
was submitted to all branches. The number of results obtained from each branch is displayed in the column on the right. Clicking on the name of the branch takes the user to the results for that branch.
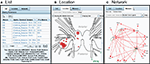
The results within a branch are displayed in one of three formats: List, Location, or Network (Figure 7
). The List format presents the components of the query, allowing users to easily trace the inference chain performed on the knowledgebase. For example, the results of the query shown in Figure 7
A include the Neuron, the Chemical synapse, and the Presynaptic Neuron. Clicking on any entry in this table will take the user to the respective object in the Branch.
Figure 7. Three ways to view the results: List, Location, and Network (tabs). (A) The List view shows all parts of the search result as clickable items. (B) The Location view displays the neuron cell bodies on a map of the nervous system. (C) The Network view displays the results in the form of a synaptic wiring diagram. Red arrows represent chemical synapses and green arrows represent electrical connections.
The Location format (Figure 7
B) is generated with the LocationViz plug-in, which is also used for Anatomical searches. Users can click on the plotted neurons to quickly access their detailed information. This plug-in is also used by Branch Editors to mark or edit the soma location of an identified neuron.
The Network format (Figure 7
C) is generated by another plug-in, NetworkViz. A major function of NeuronBank is to catalog synaptic connectivity. As most neurons have many synaptic partners, a simple list of inputs and outputs is overwhelming. Typically, a network diagram provides a more intuitive display of a neuron’s local circuitry. The NetworkViz plug-in provides a complete network graph of search results and also the local partners of individual neurons. Users can re-arrange the nodes in the network and link into the detail pages of displayed neurons and connections. We have not yet implemented search and edit modes for this plug-in. This plug-in was developed with the Prefuse.org visualization toolkit, a free (BSD license), Java-based toolkit that provides optimized data structures for graphs, a host of layout and visual encoding techniques, and support for dynamic queries, integrated search, and knowledgebase connectivity.
Each of the neurons is represented as a node connected by arrows to other neurons. This network diagram was dynamically generated from the information in the Tritonia branch. Thus, we have created a system that can automatically generate neuronal circuit diagrams. In this case, the entire swim central pattern generator and its immediate outputs were pulled from the knowledgebase. Right clicking on any neuron or synapse will open the detail page for that item.
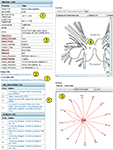
An Example Neuron
The detail page for one neuron (DSI) is shown in Figure 8
. This page demonstrates more of the features incorporated into both BranchKB and the web client. The top left panel shows the attributes of DSI and the values of those attributes. Some of the attributes (Figure 8
, #1
) are core attributes, such as the Accession ID, which is used in an automated Return URL (Figure 8
, #2
) that links directly to this record. This accession ID has now been cited in published papers (Calin-Jageman et al., 2007b
; Hill and Katz, 2007
; Katz, 2007
, 2009
; Sakurai et al., 2007
; Hill et al., 2008
; Sakurai and Katz, 2008
, 2009
), allowing readers of those papers access to the information about the neuron stored in NeuronBank. This table also shows a number of extensible attributes. (Figure 8
, #3
), such as soma coloration and behavior, which are specific to Tritonia. The location of the cell body indicated on a map, was dynamically generated by LocationViz (Figure 8
, #4
). All input and output synapses are graphically represented in a dynamic format by NetworkViz (Figure 8
, #5
). The detail page contains annotations that document the sources of information and provide additional comments (Figure 8
, #6
). Finally, a link is provided to the entry for this neuron in the NeuronBank Wiki (Figure 8
, #7
), which is explained below.
Figure 8. Detail page dynamically generated for DSI.
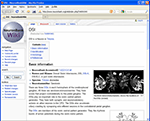
Neuronbank Wiki
In parallel to the NeuronBank Branches, we have created a Wikipedia-like NeuronBank Wiki. A link to the Wiki appears at the top of each page. All objects in NeuronBank have an Accession ID, which automatically reserves a unique wiki page URL. This wiki is used for a less constrained description of objects in NeuronBank. The wiki page for DSI is shown in Figure 9
.
Figure 9. Screen shot of the NeuronBank Wiki page for DSI.
The NeuronBank Wiki has several functions that are synergistic with the NeuronBank knowledgebase. It can be used for information that does not fit a class of NeuronBank objects. In fact, neurons from species that don’t have Branches yet can and do have NeuronBank Wiki entries
5
. Unlike the BranchKBs, the NeuronBank Wiki is open to be edited by any registered user of NeuronBank. It can serve as a Neuromics Encyclopedia. We have had success in using the NeuronBank Wiki as a means to gather information from researchers for ultimate incorporation into a NeuronBank branch. We have also used the wiki for educational purposes. Students can initiate pages about neurons, which can be read and released by instructors. Some of the student-initiated pages can be seen at http://neuronbank.org/wiki/index.php/Category:Biol6102_student_pages
.
NeuronBank is a first generation neural circuit atlas that fulfills many of the necessary criteria. NeuronBank users access a web interface that hides the complexity of the underlying structure of NeuronBank. The different branches are unified by NB-Central. The information about each species is housed in a separate data store, which is accessed through BranchKB. Each branch allows for a flexible representation of neurons; new attributes can be added at any time and incorporated in the extensible list of attributes. NeuronBank can represent neurons from different species even if those nervous systems have different characteristics. The extensible attribute list allows the different branches of NeuronBank to be customized. Plug-in architecture allows data stored in the database to be visualized differently. Currently, there are two plug-ins: one for cell soma location (LocationViz) and the other for neural circuit visualization (NetworkViz). Additional plug-ins can be developed as the need arises. Finally, NeuronBank provides a number of different means to search the information in the database within a species and across species.
The detailed records for each neuron provide complete tables of attributes, all source information, and visualization of anatomical location and synaptic connectivity. This functionality can be readily tested on the Tritonia branch accessible from NeuronBank.org. One immediate advantage of having this system online is that information about Tritonia neurons is now accessible with a simple Google search, such as “DSI NeuronBank”, greatly increasing the accessibility of this information for both research and education.
The NeuronBank wiki is a unique contribution in that it acts to bridge the gap between the unstructured wiki, which is difficult to search, and the very structured database, which has a less familiar appearance to many users. Each entry in NeuronBank has a reserved wiki page. Similarly every object in the NeuronBank knowledgebase also has a unique AccessID, which defines a unique URL. This allows the wiki to link back to objects in the knowledgebase. Searching the knowledgebase can then link the user to information that is presented in an encyclopedic format on the wiki.
The wiki has the potential to become the source of input to a NeuronBank branch. Users can input information into the wiki, which can be monitored by editors who then incorporate that information into the formal knowledgebase.
NeuronBank and the NeuronBank Wiki have potential as an educational tool. Already there are several dozen pages that have been contributed to the NeuronBank wiki by students in courses and in laboratories. These pages can be edited by the community to provide encyclopedic references for neurons and neuronal features. Students who are learning about the neural circuits represented in NeuronBank can have a reference source for neuron identification.
NeuronBank also serves as a neural registry. As more neurons are identified, their characteristics can be compared against those in the registry to determine if they represent a new class of neurons. This may accelerate the investigation of research on neural circuits. Research involving the identification of neurons has slowed considerably since gastropod nervous systems were first examined with electrophysiology in the 1960s. In Tritonia, most of the neurons that are known today, were identified by 1973 (Willows et al., 1973
). Only three neuronal identifications were published in the last decade (Frost et al., 2001
, 2003
). However, lab books contain additional neurons that have been uniquely identified. Generally, unless a functional story is associated with the identification of a neuron, it is not possible to publish that identification in a journal. Therefore, having an outlet for publication of incremental knowledge is crucial for the continued advancement of the field.
Future Directions
The current version of NeuronBank should still be considered a prototype because the components supporting data entry, user management, and ontology curation are extremely rudimentary and have not yet been made available for public use. Moreover, some additional features remain to be implemented for general users. For example, the form based search, although powerful, is awkward to use and must be replaced by a more user-friendly search interface.
One issue to be resolved is how to maintain common ontologies across species to enable cross-species searches. One possible solution is to register all terms in a common lexicon, such as Neurolex.org. As long as the terms are consistent in their meaning and usage, then it ought to be possible to keep the various branches in alignment. Curation policies will need to be implemented.
A bigger problem with tools such as this is to get community buy in. There needs to be a critical mass of information already available before scientists will begin to use the tool. So, an initial investment needs to be made in creating branches and wiki pages. Recent initiatives by the International Neuroinformatics Coordinating Facility (INCF)
6
promise to increase the awareness and use of informatic tools such as NeuronBank.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Jason Pamplin, John Propper, Hao Tian, Piyaphol Phoungphol, Janaka Balasooriya, and Yan Chen for their contributions to NeuronBank. We thank Joshua Lillvis and Arianna Tomvacakis for their helpful suggestions on an earlier version of this manuscript. We are grateful to Akira Sakurai for helping with the figures. Supported by NIH R21 MH76753, NSF IIS-0827418, and seed grants from Georgia State University.
Altun, Z. F., Herndon, L. A., Crocker, C., Lints, R., and Hall, D. H. (2009). Available at: www.wormatlas.org
.
Ascoli, G. A., onso-Nanclares, L., Anderson, S. A., Barrionuevo, G., avides-Piccione, R., Burkhalter, A., Buzsaki, G., Cauli, B., Defelipe, J., Fairen, A., Feldmeyer, D., Fishell, G., Fregnac, Y., Freund, T. F., Gardner, D., Gardner, E. P., Goldberg, J. H., Helmstaedter, M., Hestrin, S., Karube, F., Kisvarday, Z. F., Lambolez, B., Lewis, D. A., Marin, O., Markram, H., Munoz, A., Packer, A., Petersen, C. C., Rockland, K. S., Rossier, J., Rudy, B., Somogyi, P., Staiger, J. F., Tamas, G., Thomson, A. M., Toledo-Rodriguez, M., Wang, Y., West, D. C., and Yuste, R. (2008). Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat. Rev. Neurosci. 9, 557–568.
Calin-Jageman, R. J., Dhawan, A., Yang, H., Wang, H.-C., Tian, H., Phounghol, P., Frederick, C., Balasooriya, J., Chen, Y., Prasad, S. K., Sunderraman, R., Zhu, Y., and Katz, P. S. (2007a). “Development of NeuronBank: A Federation of Customizable Knowledge Bases of Neuronal Circuitry,” in Proceedings of the Ist IEEE International Workshop on Service Oriented Technologies for Biological Databases and Tools, IEEE Services Computing Workshops (SOBDAT/SCW 2007). Salt Lake City, UT.