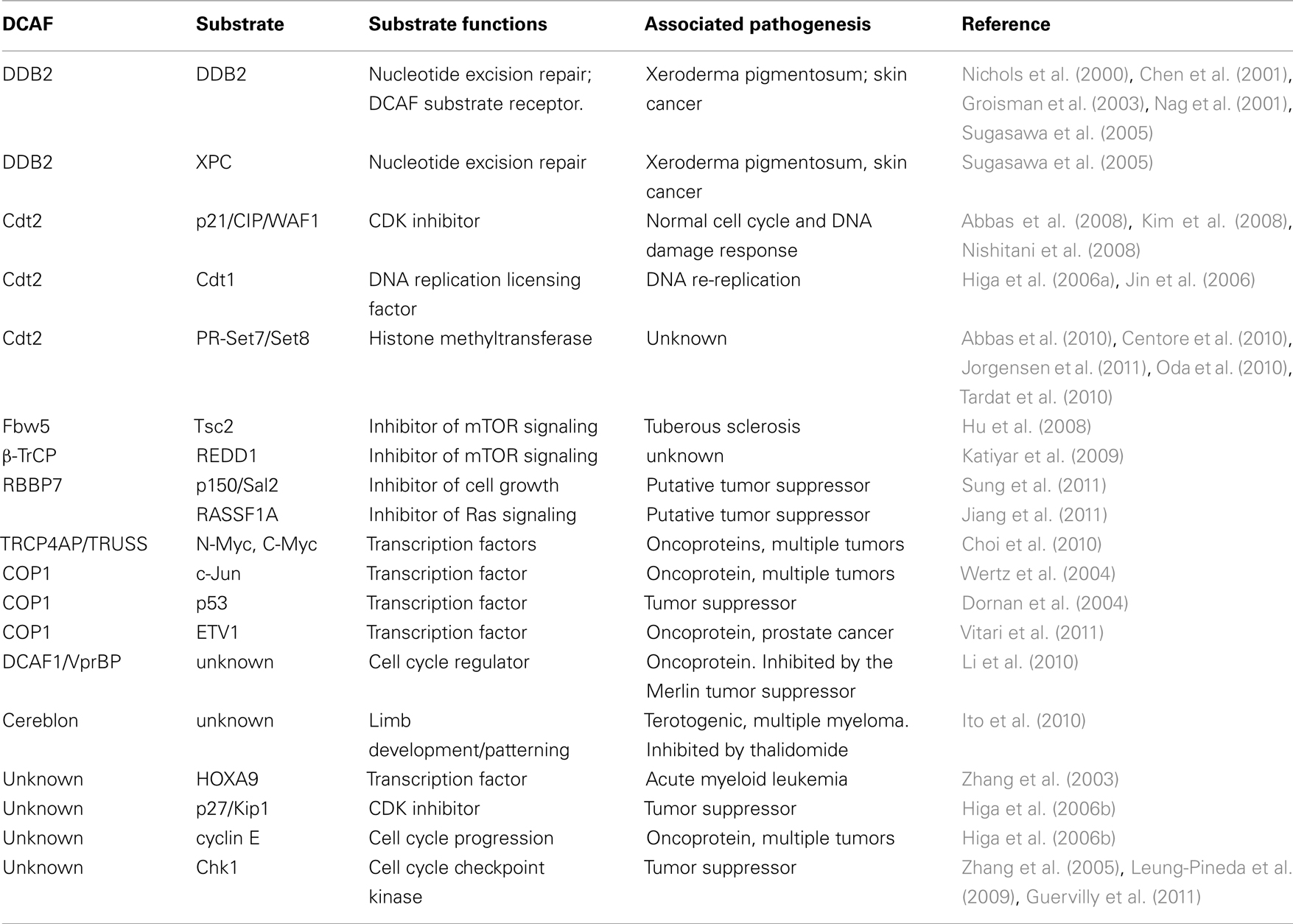
- Department of Pathology and Laboratory Medicine, Weill Cornell Medical College and Weill Graduate School of Medical Sciences of Cornell University, New York, NY, USA
The cullin 4-RING ubiquitin ligase (CRL4) family employs multiple DDB1–CUL4 associated factors substrate receptors to direct the degradation of proteins involved in a wide spectrum of cellular functions. Aberrant expression of the cullin 4A (CUL4A) gene is found in many tumor types, while mutations of the cullin 4B (CUL4B) gene are causally associated with human X-linked mental retardation. This focused review will summarize our current knowledge of the two CUL4 family members in the pathogenesis of human malignancy and neuronal disease, and discuss their potential as new targets for cancer prevention and therapeutic intervention.
Introduction
The cullin-RING ubiquitin ligases (CRLs) are the largest E3 ligase family in eukaryotes, and ubiquitinate a wide array of substrates involved in cell cycle, signaling, DNA damage response, gene expression, chromatin remodeling, and embryonic development. Cullins serve as elongated scaffolds that assemble functional E3 complexes by utilizing distinct adaptors to recruit substrate receptors and the ubiquitin-charged E2 conjugating enzyme (Figure 1). Within the E2–E3 complex, the RING domain-containing Rbx1/ROC1/Hrt1 or Rbx2/SAG adaptor bridges E2 binding to the cullin carboxyl terminus, which is necessary for transfer of ubiquitin to all cullin substrates. The cullin amino terminus binds cullin-specific adaptors, which in turn recruit distinct classes of substrate receptors that target substrates to the E2–CRL complex for ubiquitination and subsequent degradation by the 26S proteasome. CRL activity is regulated by the Nedd8 post-translational modification: the ubiquitin-like Nedd8 protein is conjugated to cullins in a manner highly analogous to ubiquitination. In a cascade of events designated the neddylation cycle, neddylation promotes CRL activity, while deneddylation by the COP9 signalosome inhibits cullins (reviewed in Petroski and Deshaies, 2005; Sarikas et al., 2011).
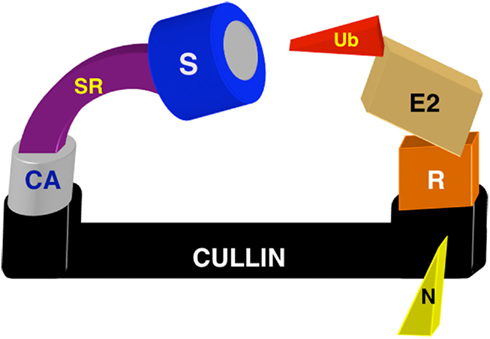
Figure 1. Schematic diagram of the multimeric CRL ubiquitin ligase complex. The elongated cullin scaffold binds cullin-specific adaptors (CA) at the N-terminus to recruit substrates (S). All cullins except CUL3 require separate substrate receptors (SR) to bridge the adaptor–substrate interaction. RING protein adaptors (R) bind the ubiquitin (Ub)-charged E2 conjugating enzyme at the cullin C-terminus. Nedd8 post-translational modification (N) promotes cullin activity.
Among the eight cullins (CUL1-7 and PARC) in higher organisms, the CUL4 subfamily of CRLs is uniquely comprised of two members, CUL4A and CUL4B, that share extensive sequence homology and functional redundancy. While most cullins recruit substrate receptors using BTB domain-containing adaptors, the CUL4 family employs the structurally distinct triple WD40 β-propeller domain-containing DDB1 adaptor to recruit members of the DDB1–CUL4 associated factors (DCAF) family of substrate receptors (Angers et al., 2006; He et al., 2006; Higa et al., 2006a; Jin et al., 2006; reviewed in Lee and Zhou, 2007). Genetic approaches have been utilized to dissect the physiological relevance and unique functions of the CUL4 family members, as well as the individual components of the CRL4 ubiquitin ligase complex (Yoon et al., 2004; Kopanja et al., 2009, 2011; Liu et al., 2009; Yin et al., 2011). This focused review will summarize recent findings that have shed light on the role of CUL4 activity in human disease.
CUL4A and Cancer
CUL4A was initially identified as an amplified or overexpressed gene in primary human breast cancers (Chen et al., 1998). Genome-wide analysis of human cancers revealed CUL4A amplification in 5% of familial and sporadic breast cancers, and as high as 20% in the basal-like breast cancer subtype that is associated with aggressive growth and poor prognosis (Melchor et al., 2009). CUL4A amplification has also been found in squamous cell carcinomas (Shinomiya et al., 1999), adrenocortical carcinomas (Dohna et al., 2000), childhood medulloblastoma (Michiels et al., 2002), hepatocellular carcinomas (Yasui et al., 2002), and primary malignant pleural mesotheliomas (Hung et al., 2011). Recently, genome-wide high-density SNP arrays further revealed high CUL4A gene copy number in a subset of lung and ovarian carcinomas, as well as other solid tumor types (Beroukhim et al., 2010). Moreover, high CUL4A expression correlates with significantly shorter overall and disease-free survival (Schindl et al., 2007), indicating that dysregulation of CUL4A may play a role in promoting oncogenesis. Mouse models support this hypothesis, as skin-specific Cul4a knockout mice showed marked resistance to UV-induced carcinogenesis compared to wild-type and heterozygous mice (Liu et al., 2009). Transgenic mice with inducible expression of exogenous CUL4A developed pulmonary hyperplasia, which is consistent with a role for dysregulated CUL4A in driving uncontrolled proliferation (Li et al., 2011a). The role of CUL4B in carcinogenesis remains to be determined.
The damaged DNA binding proteins DDB1 and DDB2 were first characterized as DNA damage sensors that initiate the nucleotide excision repair (NER) pathway following UV irradiation (reviewed in Tang and Chu, 2002). Earlier studies identified the DDB1–DDB2 heterodimer as both a target and component of the CRL4 ubiquitin ligase complex (Shiyanov et al., 1999; Chen et al., 2001; Nag et al., 2001; Groisman et al., 2003). DDB2 mutations that impair the recognition of UV-induced DNA lesions are causal for the photosensitivity and early onset of skin cancer found in xeroderma pigmentosum group E (XPE) patients (Nichols et al., 2000), and were recapitulated in the Ddb2 knockout mouse model (Itoh et al., 2004; Yoon et al., 2004; Alekseev et al., 2005). Conversely, enforced expression of DDB2 in transgenic mice delayed the onset of UV-induced squamous cell carcinomas (Alekseev et al., 2005), further highlighting the significance of DDB2 activity in DNA repair and cancer prevention. The physiological relevance of CUL4A-mediated degradation of DDB2 was determined in the Cul4a knockout mouse, as skin-specific deletion of CUL4A significantly enhanced resistance to UV-induced skin carcinogenesis (Liu et al., 2009). Protein levels of DDB2 and XPC, another NER damage sensor and CRL4DDB2 substrate (Sugasawa et al., 2005), were found to accumulate, thus augmenting NER activity and decreasing tumorigenic potential.
In addition to DNA repair, CRL4 also plays a significant role in cell cycle regulation by targeting the Cdt1 DNA replication licensing factor, the p21 cyclin-dependent kinase inhibitor, and the PR-Set7/Set8 histone H4K20 methyltransferase for ubiquitin-proteolytic degradation in a Cdt2 (DCAF)- and PCNA-dependent manner (Higa et al., 2003; Zhong et al., 2003; Hu et al., 2004; Jin et al., 2006; Nishitani et al., 2006, 2008; Abbas et al., 2008, 2010; Kim et al., 2008; Centore et al., 2010; Oda et al., 2010; Tardat et al., 2010; Jorgensen et al., 2011). Knockdown of Cdt2 resulted in G2 arrest and DNA re-replication of the genome (Jin et al., 2006), indicating a critical role for CRL4Cdt2 in limiting the replication of DNA during S phase. In response to UV or ionizing radiation, Cdt1, p21, and PR-Set7/Set8 were rapidly degraded in a CRL4-dependent manner (Higa et al., 2003; Hu et al., 2004; Abbas et al., 2008, 2010; Centore et al., 2010; Jorgensen et al., 2011). S phase arrest is also triggered by CRL4-mediated degradation of Chk1 in a phosphorylation-dependent manner under normal conditions and in the presence of genotoxic stress (Zhang et al., 2005; Leung-Pineda et al., 2009). While these proteins are targeted by both CUL4 family members, p21 protein levels were found to accumulate in primary Cul4a−/− mouse embryonic fibroblasts (MEFs) following UV irradiation, resulting in prolonged G1/S arrest (Liu et al., 2009). Higher p21 levels enforced the G1/S cell cycle checkpoint post-UV, thus allowing additional time for NER activities to conclude prior to the initiation of DNA replication. The absence of G2 arrest or DNA re-replication in the Cul4a knockout mouse model indicates that CUL4B at least partially compensates for the loss of CUL4A activity. Simultaneous inactivation of both CUL4A and CUL4B in primary MEFs led to growth arrest (Liu et al., 2009), which recapitulates the rapid G1 arrest observed in DDB1 knockout MEF cells (Cang et al., 2006).
Conflicting reports indicate that additional cell cycle regulators may be targeted by CRL4-based ubiquitin ligases for proteasome-mediated degradation. CUL4A overexpression reduced the steady-state levels of the CDK inhibitor p27Kip1 in 293T cells, while CUL4A shRNA or dominant-negative CUL4A resulted in the accumulation of p27Kip1 in mouse mammary epithelial cells or the human MCF-7 breast cancer cell, respectively (Miranda-Carboni et al., 2008). However, the turnover rate of p27Kip1 was not directly measured under these conditions. Using primary MEF cells, Cang et al. (2006) showed that deletion of DDB1 resulted in G1/S cell cycle arrest and p27Kip1 accumulation. However, the half-life of p27 was not prolonged in the absence of DDB1, arguing against p27Kip1 as a direct substrate of the CRL4 ubiquitin ligase. Future studies should determine whether a CRL4-based E3 ligase directly or indirectly regulates p27Kip1 protein stability. Interestingly, silencing of CUL4B in primary MEF cells had little effect on cell proliferation (Liu et al., 2009), but CUL4B knockdown in HeLa cells resulted in S phase cell cycle arrest (Zou et al., 2007) as well as cyclin E accumulation (Higa et al., 2006b; Zou et al., 2009). Given the stimulatory role of cyclin E in S phase progression and cell proliferation, it remains to be determined how increased cyclin E levels would result in growth inhibition, and whether additional CUL4B substrates are also involved in triggering S phase arrest.
The amplification or overexpression of CUL4A observed in various cancers likely corresponds with diminished post-translational stability of its substrates, many of which are tumor suppressors (Table 1). CRL4Fbw5 may promote oncogenesis by targeting the mTOR inhibitor tuberous sclerosis protein 2 (Tsc2) for ubiquitination (Hu et al., 2008). Mutations in the Tsc1 and Tsc2 tumor suppressors are causal for the synonymous autosomal dominant disease that is marked by the formation of benign growths on the skin, nervous system, kidneys, and heart. REDD1, another inhibitor of mTOR signaling, is targeted for degradation by the CRL4βTrCP ubiquitin ligase (Katiyar et al., 2009). Additional tumor suppressors that are subject to CUL4A-mediated ubiquitination include p150/Sal2, which is degraded as cells transition from quiescence to an actively dividing state (Sung et al., 2011), and RASSF1A (Jiang et al., 2011), a negative Ras effector that was previously identified as a target of the CRL1/SCFSkp2 (Skp1, CUL1, F-box-containing substrate receptor, and Rbx1) ubiquitin ligase (Song et al., 2008). Contrary to the trend of CUL4A-mediated degradation of tumor suppressors, the CRL4 substrate receptor TRCP4AP/TRUSS targets both N-Myc and C-Myc transcription factors for degradation (Choi et al., 2010). Stabilization of Myc protein in many cancer cell lines corresponds with TRCP4AP/TRUSS downregulation, indicating the potential significance of CUL4-mediated post-translational regulation in restricting Myc protein levels. COP1, acting as a CRL4 substrate receptor, functions as a tumor suppressor by targeting the c-Jun proto-oncogene for ubiquitination (Wertz et al., 2004). However, tissue-specific differences have been reported in CRL4-independent COP1 ubiquitin ligase activity. The p53 tumor suppressor is targeted for degradation by COP1 (Dornan et al., 2004), which may provide mechanistic insight into COP1 overexpression observed in ovarian and breast cancer. Conversely, the oncogenic ETS transcription factors are also degraded in a COP1-dependent manner, and loss of COP1 activity results in ETV1 accumulation that promotes prostate epithelial cell proliferation (Vitari et al., 2011). Finally, ectopic expression of CUL4A in PC12 rat pheochromocytoma cells suppresses apoptosis and promotes cell survival (Tan et al., 2011), further indicating that CUL4A activity supports cell growth.
The oncogenic effects of CUL4A are highlighted by the finding that Merlin, a tumor suppressor encoded by NF2, inhibits the ubiquitin ligase activity of CUL4A in complex with VprBP/DCAF1 (Li et al., 2010). Moreover, mutations in Merlin that ablate the enzymatic inhibition of CRL4VprBP have been found in patients with the neurofibromatosis type 2 familial cancer syndrome (Li et al., 2010). VprBP silencing compromises tumorigenesis in Merlin-deficient mesothelioma cell lines, indicating the significance of CRL4VprBP signaling in promoting cellular proliferation (Li et al., 2010). Huang and Chen showed that Merlin was targeted for degradation by the CRL4VprBP ubiquitin ligase (Huang and Chen, 2008). However, Li et al. demonstrated that Merlin was not a substrate of CRL4VprBP, but rather served as a negative regulator of the CRL4VprBP ubiquitin ligase. Further biochemical and genetic studies are required to determine the functional relationship between Merlin and CRL4VprBP. VprBP is also required for cell cycle progression into S phase (Belzile et al., 2007; Hrecka et al., 2007; Le Rouzic et al., 2007; Tan et al., 2007; Wen et al., 2007), but the mechanism of cell cycle regulation and the cellular targets of CRL4VprBP have yet to be determined. Nevertheless, these findings indicate a role for the CRL4VprBP ubiquitin ligase in promoting cell cycle progression and oncogenic transformation.
Despite the numerous growth-promoting pathways that are amplified by CUL4A activity, the degradation of other identified CUL4A substrates reveals a more complex effect on growth regulation. CUL4A plays a critical role in granulopoiesis by degrading the HOXA9 homeodomain protein, which may also restrict HOXA9-induced leukemogenesis (Zhang et al., 2003). The leukemogenic NUP98–HOXA9 fusion protein, which is derived from the t(9;11)(p15;p15) chromosomal translocation in acute myeloid leukemia patients, is resistant to CUL4A-mediated degradation, further indicating that CUL4A may play a role in the proper differentiation of hematopoietic cells (Chung et al., 2006). In addition to restricting the NER threshold, CRL4 also responds to DNA damage through histone H3 and H4 ubiquitination, which facilitates the recruitment of repair proteins (e.g., XPC) to damaged DNA (Wang et al., 2006). Thus, the CUL4A ubiquitin ligase may play distinct roles in tumorigenesis that are dictated by cellular context or environmental conditions.
CUL4B and Human X-Linked Mental Retardation
CUL4B mutations in human patients have been found to be causal for X-linked mental retardation (XLMR) syndrome (Tarpey et al., 2007; Zou et al., 2007; Badura-Stronka et al., 2010; Isidor et al., 2010). Patient-derived cells also display increased camptothecin-induced topoisomerase I-dependent DNA breaks, which are associated with the peripheral neuropathy spinocerebellar ataxia with axonal neuropathy-1 (SCAN-1; Kerzendorfer et al., 2010). The unique CUL4B N-terminus may mediate the recruitment of distinct substrates for degradation, and their accumulation likely contributes to the CUL4B phenotype. WDR5, a subunit of the H3K4 methyltransferase complex, was initially identified as a DCAF and more recently characterized as a CUL4B substrate (Nakagawa and Xiong, 2011). XLMR-derived CUL4B mutations resulted in the accumulation of WDR5 and subsequent activation of neuronal genes that promote neurite extension. Peroxiredoxin III is another unique CUL4B substrate that may affect neural development through the regulation of reactive oxygen species (ROS) levels (Li et al., 2011b). Finally, the arylhydrocarbon receptor (AhR) acts as a unique CUL4B substrate receptor that targets estrogen and androgen receptors for proteasome-mediated degradation (Ohtake et al., 2007), further highlighting the role of CUL4B as a transcriptional regulator.
Distinct expression patterns may also account for the phenotypes observed in the Cul4a knockout mouse model. In addition to enhanced resistance against UV-induced skin carcinogenesis, CUL4A knockout also resulted in male infertility (Kopanja et al., 2011; Yin et al., 2011). These studies revealed non-overlapping expression patterns between CUL4A and CUL4B during the pachytene to diplotene stages in adult testes, thus accounting for the physiological requirement for CUL4A activity in spermatogenesis (Yin et al., 2011). The disease syndrome manifested in humans with CUL4B mutations may be attributed to exclusive substrate targeting and/or differential expression patterns of the CUL4 family members. In vivo interrogation of CUL4B activity using knockout mouse models would shed insight into the mechanism of pathology, and may identify avenues for therapeutic intervention.
CRL4 as a Candidate for Therapeutic Intervention
Manipulation of the post-translational stability of cellular proteins has been demonstrated to be a new and effective cancer therapeutic strategy. Proteasome inhibitors, such as bortezomib, non-specifically block the overall degradation of poly-ubiquitinated proteins, but preferentially sensitize rapidly dividing cells to apoptosis (Richardson et al., 2003; Kane et al., 2007; Orlowski and Kuhn, 2008). MLN4924, a small molecule inhibitor of the Nedd8 E1 activating enzyme, specifically blocks cullin activity, which leads to the accumulation of their substrates (Soucy et al., 2009). However, more precise targeting of CRL complexes has been shown to be possible with the discovery that the teratogenic agent thalidomide specifically inhibits Cereblon (CRBN) (Ito et al., 2010), an identified DCAF for the CRL4 ubiquitin ligase family. Substrates for CRL4CRBN have yet to be determined, but aberrant Fgf8 expression and signaling were observed following Cereblon inhibition (Ito et al., 2010). Thalidomide is currently used to treat multiple myeloma, indicating that Cereblon activity likely promotes cell growth.
Viral hijack of CRL4 activity may provide further insight into possible mechanisms of CRL4 intervention. Parainfluenza virus 5 (PIV5, formerly SV5) V protein binds DDB1, which recruits STAT2 to target STAT1 for ubiquitination and proteasome-mediated degradation, thus inhibiting the cellular interferon-induced response to viral infection (Precious et al., 2005, 2007). Additional rubulaviruses, such as human parainfluenza virus 2 and mumps virus, also target STAT proteins for degradation by redirecting DDB1 activity through their V proteins (Ulane and Horvath, 2002). DDB1 binding by the hepatitis B virus X protein (HBx) resulted in S phase arrest (Martin-Lluesma et al., 2008), and the subsequent deleterious effects contributed to hepatocellular carcinomas in a cell-non-autonomous manner that was recapitulated in hepatocyte-specific Ddb1 knockout mice (Yamaji et al., 2010). S phase arrest is also triggered by HIV-1 Vpr and HIV-2 Vpx proteins through CRL4VprBP binding, which may promote macrophage infection (Sharifi et al., 2012). Thus, the modification of DDB1 substrate binding represents another method of altering CRL4 activity for therapeutic purposes.
Concluding Remarks
Despite the significant sequence conservation and functional redundancy of the CUL4 family members, CUL4A and CUL4B play strikingly diverse roles in human disease. CUL4A activity promotes oncogenesis, as demonstrated by the overexpression and/or amplification of CUL4A in several tumor types and the resistance to UV-induced skin carcinogenesis in the Cul4a knockout mouse model, while loss-of-function mutations in CUL4B are causal for human X-linked mental retardation syndrome. Surprisingly, gene-trapped inactivation of Cul4b in mice resulted in embryonic lethality (Cox et al., 2010). The genetic interrogation of CUL4B awaits the generation of a conditional Cul4b knockout mouse model to identify the unique in vivo contributions of CUL4B activity, and the major substrates that are responsible for CUL4B-dependent disease phenotypes. Furthermore, additional mouse models are required to gain a comprehensive understanding of the role of CRL4 in pathogenesis through the functional delineation of the DCAF substrate receptors, their substrates (Emanuele et al., 2011), and the associated cellular pathways whose dysregulation contribute to pathogenesis.
CUL4A represents an ideal target for therapeutic intervention, as genetic ablation in mice resulted in a marked resistance to carcinogenesis. Additionally, the recent structural determination of CRL4 assembly and activation provides a molecular platform for the design of inhibitors (Angers et al., 2006; Li et al., 2006; Fischer et al., 2011). Although the extensive protein–protein interface between CUL4 and DDB1 may present a formidable challenge for direct intervention by small molecule inhibitors, allosteric inhibitors are an attractive alternative to attenuate CRL4 activity, especially given the stringent requirements of proximity and E2–E3 orientation for effective ubiquitin transfer. Finally, the striking efficacy of thalidomide in treating multiple myeloma and its inhibition of Cereblon activity indicate that developing specific inhibitors for multiple subunits or interfaces, e.g., DCAF binding to either CRL4 or substrates, may prove to be feasible therapeutic strategies.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jeffrey Hannah and Jae Yong Eom for critical reading of the manuscript. We apologize for not being able to comprehensively cite references related to CRL4 due to focused topics discussed in this review and space constraints. This work is supported by National Institute of Health grants 5R01CA098210, 1R01CA159925, and the Irma T. Hirschl Career Scientist Award to Pengbo Zhou.
References
Abbas, T., Shibata, E., Park, J., Jha, S., Karnani, N., and Dutta, A. (2010). CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol. Cell 40, 9–21.
Abbas, T., Sivaprasad, U., Terai, K., Amador, V., Pagano, M., and Dutta, A. (2008). PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 22, 2496–2506.
Alekseev, S., Kool, H., Rebel, H., Fousteri, M., Moser, J., Backendorf, C., de Gruijl, F. R., Vrieling, H., and Mullenders, L. H. (2005). Enhanced DDB2 expression protects mice from carcinogenic effects of chronic UV-B irradiation. Cancer Res. 65, 10298–10306.
Angers, S., Li, T., Yi, X., MacCoss, M. J., Moon, R. T., and Zheng, N. (2006). Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 443, 590–593.
Badura-Stronka, M., Jamsheer, A., Materna-Kiryluk, A., Sowinska, A., Kiryluk, K., Budny, B., and Latos-Bielenska, A. (2010). A novel nonsense mutation in CUL4B gene in three brothers with X-linked mental retardation syndrome. Clin. Genet. 77, 141–144.
Belzile, J. P., Duisit, G., Rougeau, N., Mercier, J., Finzi, A., and Cohen, E. A. (2007). HIV-1 Vpr-mediated G2 arrest involves the DDB1-CUL4A-VPRBP E3 ubiquitin ligase. PLoS Pathog. 3, e85. doi:10.1371/journal.ppat.0030085
Beroukhim, R., Mermel, C. H., Porter, D., Wei, G., Raychaudhuri, S., Donovan, J., Barretina, J., Boehm, J. S., Dobson, J., Urashima, M., Mc Henry, K. T., Pinchback, R. M., Ligon, A. H., Cho, Y. J., Haery, L., Greulich, H., Reich, M., Winckler, W., Lawrence, M. S., Weir, B. A., Tanaka, K. E., Chiang, D. Y., Bass, A. J., Loo, A., Hoffman, C., Prensner, J., Liefeld, T., Gao, Q., Yecies, D., Signoretti, S., Maher, E., Kaye, F. J., Sasaki, H., Tepper, J. E., Fletcher, J. A., Tabernero, J., Baselga, J., Tsao, M. S., Demichelis, F., Rubin, M. A., Janne, P. A., Daly, M. J., Nucera, C., Levine, R. L., Ebert, B. L., Gabriel, S., Rustgi, A. K., Antonescu, C. R., Ladanyi, M., Letai, A., Garraway, L. A., Loda, M., Beer, D. G., True, L. D., Okamoto, A., Pomeroy, S. L., Singer, S., Golub, T. R., Lander, E. S., Getz, G., Sellers, W. R., and Meyerson, M. (2010). The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905.
Cang, Y., Zhang, J., Nicholas, S. A., Bastien, J., Li, B., Zhou, P., and Goff, S. P. (2006). Deletion of DDB1 in mouse brain and lens leads to p53-dependent elimination of proliferating cells. Cell 127, 929–940.
Centore, R. C., Havens, C. G., Manning, A. L., Li, J. M., Flynn, R. L., Tse, A., Jin, J., Dyson, N. J., Walter, J. C., and Zou, L. (2010). CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol. Cell 40, 22–33.
Chen, L. C., Manjeshwar, S., Lu, Y., Moore, D., Ljung, B. M., Kuo, W. L., Dairkee, S. H., Wernick, M., Collins, C., and Smith, H. S. (1998). The human homologue for the Caenorhabditis elegans cul-4 gene is amplified and overexpressed in primary breast cancers. Cancer Res. 58, 3677–3683.
Chen, X., Zhang, Y., Douglas, L., and Zhou, P. (2001). UV-damaged DNA-binding proteins are targets of CUL-4A-mediated ubiquitination and degradation. J. Biol. Chem. 276, 48175–48182.
Choi, S. H., Wright, J. B., Gerber, S. A., and Cole, M. D. (2010). Myc protein is stabilized by suppression of a novel E3 ligase complex in cancer cells. Genes Dev. 24, 1236–1241.
Chung, K. Y., Morrone, G., Schuringa, J. J., Plasilova, M., Shieh, J. H., Zhang, Y., Zhou, P., and Moore, M. A. (2006). Enforced expression of NUP98-HOXA9 in human CD34(+) cells enhances stem cell proliferation. Cancer Res. 66, 11781–11791.
Cox, B. J., Vollmer, M., Tamplin, O., Lu, M., Biechele, S., Gertsenstein, M., van Campenhout, C., Floss, T., Kuhn, R., Wurst, W., Lickert, H., and Rossant, J. (2010). Phenotypic annotation of the mouse X chromosome. Genome Res. 20, 1154–1164.
Dohna, M., Reincke, M., Mincheva, A., Allolio, B., Solinas-Toldo, S., and Lichter, P. (2000). Adrenocortical carcinoma is characterized by a high frequency of chromosomal gains and high-level amplifications. Genes Chromosomes Cancer 28, 145–152.
Dornan, D., Wertz, I., Shimizu, H., Arnott, D., Frantz, G. D., Dowd, P., O’Rourke, K., Koeppen, H., and Dixit, V. M. (2004). The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 429, 86–92.
Emanuele, M. J., Elia, A. E., Xu, Q., Thoma, C. R., Izhar, L., Leng, Y., Guo, A., Chen, Y. N., Rush, J., Hsu, P. W., Yen, H. C., and Elledge, S. J. (2011). Global identification of modular cullin-RING ligase substrates. Cell 147, 459–474.
Fischer, E. S., Scrima, A., Bohm, K., Matsumoto, S., Lingaraju, G. M., Faty, M., Yasuda, T., Cavadini, S., Wakasugi, M., Hanaoka, F., Iwai, S., Gut, H., Sugasawa, K., and Thomä, N. H. (2011). The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell 147, 1024–1039.
Groisman, R., Polanowska, J., Kuraoka, I., Sawada, J., Saijo, M., Drapkin, R., Kisselev, A. F., Tanaka, K., and Nakatani, Y. (2003). The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113, 357–367.
Guervilly, J. H., Renaud, E., Takata, M., and Rosselli, F. (2011). USP1 deubiquitinase maintains phosphorylated CHK1 by limiting its DDB1-dependent degradation. Hum. Mol. Genet. 20, 2171–2181.
He, Y. J., McCall, C. M., Hu, J., Zeng, Y., and Xiong, Y. (2006). DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev. 20, 2949–2954.
Higa, L. A., Mihaylov, I. S., Banks, D. P., Zheng, J., and Zhang, H. (2003). Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint. Nat. Cell Biol. 5, 1008–1015.
Higa, L. A., Wu, M., Ye, T., Kobayashi, R., Sun, H., and Zhang, H. (2006a). CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat. Cell Biol. 8, 1277–1283.
Higa, L. A., Yang, X., Zheng, J., Banks, D., Wu, M., Ghosh, P., Sun, H., and Zhang, H. (2006b). Involvement of CUL4 ubiquitin E3 ligases in regulating CDK inhibitors Dacapo/p27Kip1 and cyclin E degradation. Cell Cycle 5, 71–77.
Hrecka, K., Gierszewska, M., Srivastava, S., Kozaczkiewicz, L., Swanson, S. K., Florens, L., Washburn, M. P., and Skowronski, J. (2007). Lentiviral Vpr usurps Cul4-DDB1[VprBP] E3 ubiquitin ligase to modulate cell cycle. Proc. Natl. Acad. Sci. U.S.A. 104, 11778–11783.
Hu, J., McCall, C. M., Ohta, T., and Xiong, Y. (2004). Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat. Cell Biol. 6, 1003–1009.
Hu, J., Zacharek, S., He, Y. J., Lee, H., Shumway, S., Duronio, R. J., and Xiong, Y. (2008). WD40 protein FBW5 promotes ubiquitination of tumor suppressor TSC2 by DDB1-CUL4-ROC1 ligase. Genes Dev. 22, 866–871.
Huang, J., and Chen, J. (2008). VprBP targets Merlin to the Roc1-Cul4A-DDB1 E3 ligase complex for degradation. Oncogene 27, 4056–4064.
Hung, M. S., Mao, J. H., Xu, Z., Yang, C. T., Yu, J. S., Harvard, C., Lin, Y. C., Bravo, D. T., Jablons, D. M., and You, L. (2011). Cul4A is an oncogene in malignant pleural mesothelioma. J. Cell. Mol. Med. 15, 350–358.
Isidor, B., Pichon, O., Baron, S., David, A., and Le Caignec, C. (2010). Deletion of the CUL4B gene in a boy with mental retardation, minor facial anomalies, short stature, hypogonadism, and ataxia. Am. J. Med. Genet. A 152A, 175–180.
Ito, T., Ando, H., Suzuki, T., Ogura, T., Hotta, K., Imamura, Y., Yamaguchi, Y., and Handa, H. (2010). Identification of a primary target of thalidomide teratogenicity. Science 327, 1345–1350.
Itoh, T., Cado, D., Kamide, R., and Linn, S. (2004). DDB2 gene disruption leads to skin tumors and resistance to apoptosis after exposure to ultraviolet light but not a chemical carcinogen. Proc. Natl. Acad. Sci. U.S.A. 101, 2052–2057.
Jiang, L., Rong, R., Sheikh, M. S., and Huang, Y. (2011). Cullin-4A.DNA damage-binding protein 1 E3 ligase complex targets tumor suppressor RASSF1A for degradation during mitosis. J. Biol. Chem. 286, 6971–6978.
Jin, J., Arias, E. E., Chen, J., Harper, J. W., and Walter, J. C. (2006). A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell 23, 709–721.
Jorgensen, S., Eskildsen, M., Fugger, K., Hansen, L., Larsen, M. S., Kousholt, A. N., Syljuasen, R. G., Trelle, M. B., Jensen, O. N., Helin, K., and Sørensen, C. S. (2011). SET8 is degraded via PCNA-coupled CRL4(CDT2) ubiquitylation in S phase and after UV irradiation. J. Cell Biol. 192, 43–54.
Kane, R. C., Dagher, R., Farrell, A., Ko, C. W., Sridhara, R., Justice, R., and Pazdur, R. (2007). Bortezomib for the treatment of mantle cell lymphoma. Clin. Cancer Res. 13, 5291–5294.
Katiyar, S., Liu, E., Knutzen, C. A., Lang, E. S., Lombardo, C. R., Sankar, S., Toth, J. I., Petroski, M. D., Ronai, Z., and Chiang, G. G. (2009). REDD1, an inhibitor of mTOR signalling, is regulated by the CUL4A-DDB1 ubiquitin ligase. EMBO Rep. 10, 866–872.
Kerzendorfer, C., Whibley, A., Carpenter, G., Outwin, E., Chiang, S. C., Turner, G., Schwartz, C., El-Khamisy, S., Raymond, F. L., and O’Driscoll, M. (2010). Mutations in Cullin 4B result in a human syndrome associated with increased camptothecin-induced topoisomerase I-dependent DNA breaks. Hum. Mol. Genet. 19, 1324–1334.
Kim, Y., Starostina, N. G., and Kipreos, E. T. (2008). The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev. 22, 2507–2519.
Kopanja, D., Roy, N., Stoyanova, T., Hess, R. A., Bagchi, S., and Raychaudhuri, P. (2011). Cul4A is essential for spermatogenesis and male fertility. Dev. Biol. 352, 278–287.
Kopanja, D., Stoyanova, T., Okur, M. N., Huang, E., Bagchi, S., and Raychaudhuri, P. (2009). Proliferation defects and genome instability in cells lacking Cul4A. Oncogene 28, 2456–2465.
Le Rouzic, E., Belaidouni, N., Estrabaud, E., Morel, M., Rain, J. C., Transy, C., and Margottin-Goguet, F. (2007). HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4-DDB1 ubiquitin ligase. Cell Cycle 6, 182–188.
Lee, J., and Zhou, P. (2007). DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol. Cell 26, 775–780.
Leung-Pineda, V., Huh, J., and Piwnica-Worms, H. (2009). DDB1 targets Chk1 to the Cul4 E3 ligase complex in normal cycling cells and in cells experiencing replication stress. Cancer Res. 69, 2630–2637.
Li, T., Chen, X., Garbutt, K. C., Zhou, P., and Zheng, N. (2006). Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase. Cell 124, 105–117.
Li, T., Hung, M. S., Wang, Y., Mao, J. H., Tan, J. L., Jahan, K., Roos, H., Xu, Z., Jablons, D. M., and You, L. (2011a). Transgenic mice for cre-inducible overexpression of the Cul4A gene. Genesis 49, 134–141.
Li, X., Lu, D., He, F., Zhou, H., Liu, Q., Wang, Y., Shao, C., and Gong, Y. (2011b). Cullin 4B protein ubiquitin ligase targets peroxiredoxin III for degradation. J. Biol. Chem. 286, 32344–32354.
Li, W., You, L., Cooper, J., Schiavon, G., Pepe-Caprio, A., Zhou, L., Ishii, R., Giovannini, M., Hanemann, C. O., Long, S. B., Erdjument-Bromage, H., Zhou, P., Tempst, P., and Giancotti, F. G. (2010). Merlin/NF2 suppresses tumorigenesis by inhibiting the E3 ubiquitin ligase CRL4(DCAF1) in the nucleus. Cell 140, 477–490.
Liu, L., Lee, S., Zhang, J., Peters, S. B., Hannah, J., Zhang, Y., Yin, Y., Koff, A., Ma, L., and Zhou, P. (2009). CUL4A abrogation augments DNA damage response and protection against skin carcinogenesis. Mol. Cell 34, 451–460.
Martin-Lluesma, S., Schaeffer, C., Robert, E. I., van Breugel, P. C., Leupin, O., Hantz, O., and Strubin, M. (2008). Hepatitis B virus X protein affects S phase progression leading to chromosome segregation defects by binding to damaged DNA binding protein 1. Hepatology 48, 1467–1476.
Melchor, L., Saucedo-Cuevas, L. P., Munoz-Repeto, I., Rodriguez-Pinilla, S. M., Honrado, E., Campoverde, A., Palacios, J., Nathanson, K. L., Garcia, M. J., and Benitez, J. (2009). Comprehensive characterization of the DNA amplification at 13q34 in human breast cancer reveals TFDP1 and CUL4A as likely candidate target genes. Breast Cancer Res. 11, R86.
Michiels, E. M., Weiss, M. M., Hoovers, J. M., Baak, J. P., Voute, P. A., Baas, F., and Hermsen, M. A. (2002). Genetic alterations in childhood medulloblastoma analyzed by comparative genomic hybridization. J. Pediatr. Hematol. Oncol. 24, 205–210.
Miranda-Carboni, G. A., Krum, S. A., Yee, K., Nava, M., Deng, Q. E., Pervin, S., Collado-Hidalgo, A., Galic, Z., Zack, J. A., Nakayama, K., Nakayama, K. I., and Lane, T. F. (2008). A functional link between Wnt signaling and SKP2-independent p27 turnover in mammary tumors. Genes Dev. 22, 3121–3134.
Nag, A., Bondar, T., Shiv, S., and Raychaudhuri, P. (2001). The xeroderma pigmentosum group e gene product ddb2 is a specific target of cullin 4a in mammalian cells. Mol. Cell. Biol. 21, 6738–6747.
Nakagawa, T., and Xiong, Y. (2011). X-linked mental retardation gene CUL4B targets ubiquitylation of H3K4 methyltransferase component WDR5 and regulates neuronal gene expression. Mol. Cell 43, 381–391.
Nichols, A. F., Itoh, T., Graham, J. A., Liu, W., Yamaizumi, M., and Linn, S. (2000). Human damage-specific DNA-binding protein p48. Characterization of XPE mutations and regulation following UV irradiation. J. Biol. Chem. 275, 21422–21428.
Nishitani, H., Shiomi, Y., Iida, H., Michishita, M., Takami, T., and Tsurimoto, T. (2008). CDK inhibitor p21 is degraded by a proliferating cell nuclear antigen-coupled Cul4-DDB1Cdt2 pathway during S phase and after UV irradiation. J. Biol. Chem. 283, 29045–29052.
Nishitani, H., Sugimoto, N., Roukos, V., Nakanishi, Y., Saijo, M., Obuse, C., Tsurimoto, T., Nakayama, K. I., Nakayama, K., Fujita, M., Lygerou, Z., and Nishimoto, T. (2006). Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 25, 1126–1136.
Oda, H., Hubner, M. R., Beck, D. B., Vermeulen, M., Hurwitz, J., Spector, D. L., and Reinberg, D. (2010). Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol. Cell 40, 364–376.
Ohtake, F., Baba, A., Takada, I., Okada, M., Iwasaki, K., Miki, H., Takahashi, S., Kouzmenko, A., Nohara, K., Chiba, T., Fujii-Kuriyama, Y., and Kato, S. (2007). Dioxin receptor is a ligand-dependent E3 ubiquitin ligase. Nature 446, 562–566.
Orlowski, R. Z., and Kuhn, D. J. (2008). Proteasome inhibitors in cancer therapy: lessons from the first decade. Clin. Cancer Res. 14, 1649–1657.
Petroski, M. D., and Deshaies, R. J. (2005). Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. 6, 9–20.
Precious, B., Young, D. F., Andrejeva, L., Goodbourn, S., and Randall, R. E. (2005). In vitro and in vivo specificity of ubiquitination and degradation of STAT1 and STAT2 by the V proteins of the paramyxoviruses simian virus 5 and human parainfluenza virus type 2. J. Gen. Virol. 86, 151–158.
Precious, B. L., Carlos, T. S., Goodbourn, S., and Randall, R. E. (2007). Catalytic turnover of STAT1 allows PIV5 to dismantle the interferon-induced anti-viral state of cells. Virology 368, 114–121.
Richardson, P. G., Barlogie, B., Berenson, J., Singhal, S., Jagannath, S., Irwin, D., Rajkumar, S. V., Srkalovic, G., Alsina, M., Alexanian, R., Siegel, D., Orlowski, R. Z., Kuter, D., Limentani, S. A., Lee, S., Hideshima, T., Esseltine, D. L., Kauffman, M., Adams, J., Schenkein, D. P., and Anderson, K. C. (2003). A phase 2 study of bortezomib in relapsed, refractory myeloma. N. Engl. J. Med. 348, 2609–2617.
Schindl, M., Gnant, M., Schoppmann, S. F., Horvat, R., and Birner, P. (2007). Overexpression of the human homologue for Caenorhabditis elegans cul-4 gene is associated with poor outcome in node-negative breast cancer. Anticancer Res. 27, 949–952.
Sharifi, H. J., Furuya, A. M., and de Noronha, C. M. (2012). The role of HIV-1 Vpr in promoting the infection of nondividing cells and in cell cycle arrest. Curr. Opin. HIV AIDS 7, 187–194.
Shinomiya, T., Mori, T., Ariyama, Y., Sakabe, T., Fukuda, Y., Murakami, Y., Nakamura, Y., and Inazawa, J. (1999). Comparative genomic hybridization of squamous cell carcinoma of the esophagus: the possible involvement of the DPI gene in the 13q34 amplicon. Genes Chromosomes Cancer 24, 337–344.
Shiyanov, P., Nag, A., and Raychaudhuri, P. (1999). Cullin 4A associates with the UV-damaged DNA-binding protein DDB. J. Biol. Chem. 274, 35309–35312.
Song, M. S., Song, S. J., Kim, S. J., Nakayama, K., Nakayama, K. I., and Lim, D. S. (2008). Skp2 regulates the antiproliferative function of the tumor suppressor RASSF1A via ubiquitin-mediated degradation at the G1-S transition. Oncogene 27, 3176–3185.
Soucy, T. A., Smith, P. G., Milhollen, M. A., Berger, A. J., Gavin, J. M., Adhikari, S., Brownell, J. E., Burke, K. E., Cardin, D. P., Critchley, S., Cullis, C. A., Doucette, A., Garnsey, J. J., Gaulin, J. L., Gershman, R. E., Lublinsky, A. R., McDonald, A., Mizutani, H., Narayanan, U., Olhava, E. J., Peluso, S., Rezaei, M., Sintchak, M. D., Talreja, T., Thomas, M. P., Traore, T., Vyskocil, S., Weatherhead, G. S., Yu, J., Zhang, J., Dick, L. R., Claiborne, C. F., Rolfe, M., Bolen, J. B., and Langston, S. P. (2009). An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736.
Sugasawa, K., Okuda, Y., Saijo, M., Nishi, R., Matsuda, N., Chu, G., Mori, T., Iwai, S., Tanaka, K., and Hanaoka, F. (2005). UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 121, 387–400.
Sung, C. K., Dahl, J., Yim, H., Rodig, S., and Benjamin, T. L. (2011). Transcriptional and post-translational regulation of the quiescence factor and putative tumor suppressor p150(Sal2). FASEB J. 25, 1275–1283.
Tan, C., Zhang, L. Y., Chen, H., Xiao, L., Liu, X. P., and Zhang, J. X. (2011). Overexpression of the human ubiquitin E3 ligase CUL4A alleviates hypoxia-reoxygenation injury in pheochromocytoma (PC12) cells. Biochem. Biophys. Res. Commun. 416, 403–408.
Tan, L., Ehrlich, E., and Yu, X. F. (2007). DDB1 and Cul4A are required for human immunodeficiency virus type 1 Vpr-induced G2 arrest. J. Virol. 81, 10822–10830.
Tang, J., and Chu, G. (2002). Xeroderma pigmentosum complementation group E and UV-damaged DNA-binding protein. DNA Repair (Amst.) 1, 601–616.
Tardat, M., Brustel, J., Kirsh, O., Lefevbre, C., Callanan, M., Sardet, C., and Julien, E. (2010). The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat. Cell Biol. 12, 1086–1093.
Tarpey, P. S., Raymond, F. L., O’Meara, S., Edkins, S., Teague, J., Butler, A., Dicks, E., Stevens, C., Tofts, C., Avis, T., Barthorpe, S., Buck, G., Cole, J., Gray, K., Halliday, K., Harrison, R., Hills, K., Jenkinson, A., Jones, D., Menzies, A., Mironenko, T., Perry, J., Raine, K., Richardson, D., Shepherd, R., Small, A., Varian, J., West, S., Widaa, S., Mallya, U., Moon, J., Luo, Y., Holder, S., Smithson, S. F., Hurst, J. A., Clayton-Smith, J., Kerr, B., Boyle, J., Shaw, M., Vandeleur, L., Rodriguez, J., Slaugh, R., Easton, D. F., Wooster, R., Bobrow, M., Srivastava, A. K., Stevenson, R. E., Schwartz, C. E., Turner, G., Gecz, J., Futreal, P. A., Stratton, M. R., and Partington, M. (2007). Mutations in CUL4B, which encodes a ubiquitin E3 ligase subunit, cause an X-linked mental retardation syndrome associated with aggressive outbursts, seizures, relative macrocephaly, central obesity, hypogonadism, pes cavus, and tremor. Am. J. Hum. Genet. 80, 345–352.
Ulane, C. M., and Horvath, C. M. (2002). Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology 304, 160–166.
Vitari, A. C., Leong, K. G., Newton, K., Yee, C., O’Rourke, K., Liu, J., Phu, L., Vij, R., Ferrando, R., Couto, S. S., Mohan, S., Pandita, A., Hongo, J. A., Arnott, D., Wertz, I. E., Gao, W. Q., French, D. M., and Dixit, V. M. (2011). COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature 474, 403–406.
Wang, H., Zhai, L., Xu, J., Joo, H. Y., Jackson, S., Erdjument-Bromage, H., Tempst, P., Xiong, Y., and Zhang, Y. (2006). Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol. Cell 22, 383–394.
Wen, X., Duus, K. M., Friedrich, T. D., and de Noronha, C. M. (2007). The HIV1 protein Vpr acts to promote G2 cell cycle arrest by engaging a DDB1 and Cullin4A-containing ubiquitin ligase complex using VprBP/DCAF1 as an adaptor. J. Biol. Chem. 282, 27046–27057.
Wertz, I. E., O’Rourke, K. M., Zhang, Z., Dornan, D., Arnott, D., Deshaies, R. J., and Dixit, V. M. (2004). Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 22, 22.
Yamaji, S., Zhang, M., Zhang, J., Endo, Y., Bibikova, E., Goff, S. P., and Cang, Y. (2010). Hepatocyte-specific deletion of DDB1 induces liver regeneration and tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 22237–22242.
Yasui, K., Arii, S., Zhao, C., Imoto, I., Ueda, M., Nagai, H., Emi, M., and Inazawa, J. (2002). TFDP1, CUL4A, and CDC16 identified as targets for amplification at 13q34 in hepatocellular carcinomas. Hepatology 35, 1476–1484.
Yin, Y., Lin, C., Kim, S. T., Roig, I., Chen, H., Liu, L., Veith, G. M., Jin, R. U., Keeney, S., Jasin, M., Moley, K., Zhou, P., and Ma, L. (2011). The E3 ubiquitin ligase Cullin 4A regulates meiotic progression in mouse spermatogenesis. Dev. Biol. 356, 51–62.
Yoon, T., Chakrabortty, A., Franks, R., Valli, T., Kiyokawa, H., and Raychaudhuri, P. (2004). Tumor-prone phenotype of the DDB2-deficient mice. Oncogene 22, 22.
Zhang, Y., Morrone, G., Zhang, J., Chen, X., Lu, X., Ma, L., Moore, M., and Zhou, P. (2003). CUL-4A stimulates ubiquitylation and degradation of the HOXA9 homeodomain protein. EMBO J. 22, 6057–6067.
Zhang, Y. W., Otterness, D. M., Chiang, G. G., Xie, W., Liu, Y. C., Mercurio, F., and Abraham, R. T. (2005). Genotoxic stress targets human Chk1 for degradation by the ubiquitin-proteasome pathway. Mol. Cell 19, 607–618.
Zhong, W., Feng, H., Santiago, F. E., and Kipreos, E. T. (2003). CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature 423, 885–889.
Zou, Y., Liu, Q., Chen, B., Zhang, X., Guo, C., Zhou, H., Li, J., Gao, G., Guo, Y., Yan, C., Wei, J., Shao, C., and Gong, Y. (2007). Mutation in CUL4B, which encodes a member of cullin-RING ubiquitin ligase complex, causes X-linked mental retardation. Am. J. Hum. Genet. 80, 561–566.
Keywords: cullin, CUL4A, CUL4B, CRL, cancer, disease
Citation: Lee J and Zhou P (2012) Pathogenic role of the CRL4 ubiquitin ligase in human disease. Front. Oncol. 2:21. doi: 10.3389/fonc.2012.00021
Received: 13 January 2012; Paper pending published: 07 February 2012;
Accepted: 17 February 2012; Published online: 06 March 2012.
Edited by:
Hui-Kuan Lin, The University of Texas MD Anderson Cancer Center, USACopyright: © 2012 Lee and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Pengbo Zhou, Department of Pathology and Laboratory Medicine, Weill Cornell Medical College and Weill Graduate School of Medical Sciences of Cornell University, New York, NY 10065, USA. e-mail: pez2001@med.cornell.edu
 Jennifer Lee
Jennifer Lee