- 1Department of Microbiology, Interdisciplinary Graduate School of Medicine and Engineering, University of Yamanashi, Chuo, Yamanashi, Japan
- 2Redox Response Cell Biology, Medical Research Institute, Tokyo Medical and Dental University, Tokyo, Japan
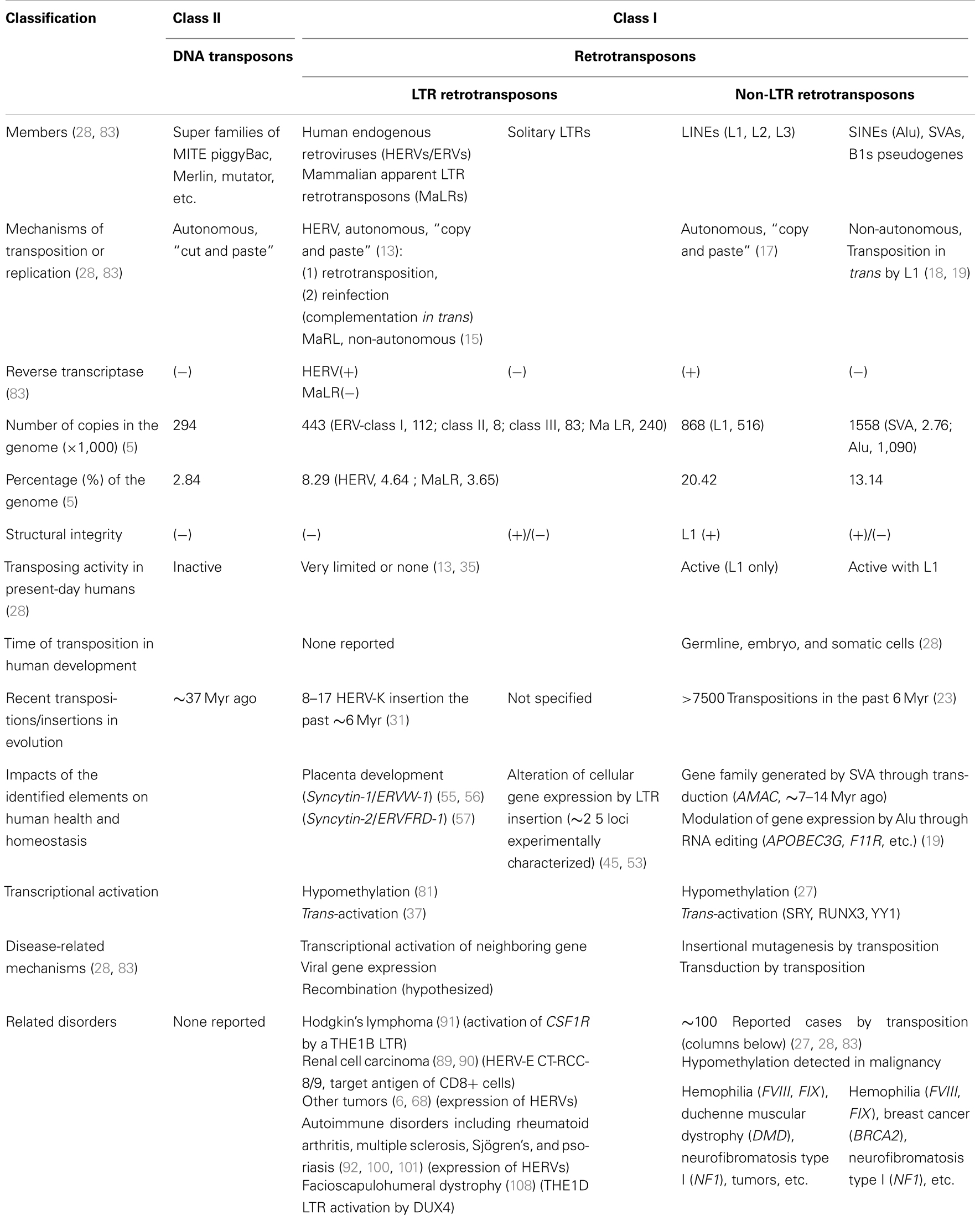
Since the human genome sequences became available in 2001, our knowledge about the human transposable elements which comprise ∼40% of the total nucleotides has been expanding. Non-long terminal repeat (non-LTR) retrotransposons are actively transposing in the present-day human genome, and have been found to cause ∼100 identified clinical cases of varied disorders. In contrast, almost all of the human endogenous retroviruses (HERVs) originating from ancient infectious retroviruses lost their infectivity and transposing activity at various times before the human-chimpanzee speciation (∼6 million years ago), and no known HERV is presently infectious. Insertion of HERVs and mammalian apparent LTR retrotransposons (MaLRs) into the chromosomal DNA influenced a number of host genes in various modes during human evolution. Apart from the aspect of genome evolution, HERVs and solitary LTRs being suppressed in normal biological processes can potentially act as extra transcriptional apparatuses of cellular genes by re-activation in individuals. There has been a reasonable prediction that aberrant LTR activation could trigger malignant disorders and autoimmune responses if epigenetic changes including DNA hypomethylation occur in somatic cells. Evidence supporting this hypothesis has begun to emerge only recently: a MaLR family LTR activation in the pathogenesis of Hodgkin’s lymphoma and a HERV-E antigen expression in an anti-renal cell carcinoma immune response. This mini review addresses the impacts of the remnant-form LTR retrotransposons on human pathogenesis.
Human Endogenous Retroviruses and Mammalian Apparent LTR Retrotransposons are Long Terminal Repeat-Retrotransposons in the Human Genome
Endogenous retroviruses (ERVs) stem from ancient infectious retroviruses that were reverse transcribed and integrated into the germ cell chromosomal DNA of ancestral host animals. The integrated viral genomes, termed proviruses, were subsequently fixed, and have been inherited for tens of millions of years (Myr). “Human endogenous retroviruses” (HERVs) refer to human ERVs, most of which appear to have entered the genome after the separation of Old and New World monkeys (∼35 Myr ago), and have increased in copy numbers through the human/primate evolution (1–4). The present-day human genome contains ∼400 thousand copies of HERVs and related sequences, accounting for ∼4.6% of the ∼3 billion nucleotide sequences (5, 6). In the eras of their copy number expansion, HERVs in the original form of 5′LTR-gag-pro-pol-env-3′LTR amplified themselves autonomously. However, all HERVs in the present-day humans are replication-incompetent, having varying degrees of mutations and deletions. Despite the absence of an obvious transposing activity, HERVs are classified as transposons based on the evidence of transposition during the human evolution and the existence of numerous animal ERVs presently mobilizing in their host genomes as represented by mouse intracisternal A particles (7, 8).
Human endogenous retroviruses are divided into 50–200 families primarily depending on the primer binding site (PBS) sequences complementary to the 3′ end of a cellular tRNA (9–11). For example, HERV-K and -W indicate utilization of tRNALys and tRNATry as the primers for reverse transcription, respectively. This terminology has nothing to do with their histories or functions. Nevertheless, the HERV-F/H family represents the oldest HERVs which entered into our genome before the split of simians and prosimians ∼60–70 Myr ago (12), while the HERV-K (HML-2) family contains the youngest HERVs having the best-preserved retroviral open reading frames (ORFs) (13, 14).
In addition to HERVs, the long terminal repeat (LTR) retrotransposon class in humans have another big subclass of “mammalian apparent LTR retrotransposons” (MaLRs), which consists of ∼240,000 copies accounting for 3.65% of the genomic sequences (5) (Table 1). Despite the classification, MaLRs lack a PBS and gag-pro-pol genes shared by animal retroelements (Copia/Ty1, Gypsy/Ty3, etc.) and retroviruses, and are thought to have originated before the radiation of eutherian mammals, 80–100 Myr ago (15, 16). The simple 5′LTR-ORF-3′LTR structure without a pol gene encoding reverse transcriptase (RT) and integrase suggests non-autonomous transposition. The transposon-like human element-1 (THE1) is known as a most widespread MaLR family in primates.
Mobilization of Non-LTR Retrotransposons
Transposons currently mobilizing on the human chromosomes include the long interspersed elements (LINEs), short interspersed elements (SINEs), and “SINE-R, variable of number of tandem repeats (VNTRs) and Alu” (SVA) elements, all of which are in the “non-LTR retrotransposons” family. L1 elements in the LINE family, are the only active autonomous retrotransposons, and replicate by a simple “copy and paste” mechanism involving target-site primed reverse transcription (17). Alu elements, the youngest subtypes of SINEs, and SVAs are apparently mobilized in trans by L1s (18, 19). Retrotransposition of these elements occurs in germline, early embryonic development, and in somatic cells (20–22).
In the past 6 Myr, >7,500 transpositions occurred in the primate genome (23). One retrotransposition per 10–100 births is estimated (19, 24). Thus, L1, Alu, and SVA elements have substantially impacted the human genome organization in evolution (25). Furthermore, the non-LTR retrotransposons are causative agents of human genetic disorders (26, 27); ∼100 cases of disorders caused by non-LTR retrotransposon insertions have been reported (27, 28) (Table 1).
HERVs and MaLRs Exist as Non-Transposable Elements
In comparison with the intra-nuclear transposition mechanisms of non-LTR retrotransposons, both of the ERV amplification pathways, “retrotransposition,” and “reinfection,” are tricky. The former comprises multiple steps including particle-like assembly in the cytosol, reverse transcription, nuclear translocation, and chromosomal insertion. The latter requires infectious virus particle production followed by germ-line infection ultimately leading to chromosomal integration and fixation (13, 29, 30). Defective ERVs are also able to increase their copy numbers by “complementation in trans,” where a matching element at a different locus serves as the helper (13, 29). These mechanisms underlay the HERV amplification in the past.
However, no evidence of HERV transposition has been detected in present-day humans, either in a scientific or clinical background. Studies suggested 8–17 full-length HERV-K insertions after the divergence of humans from chimpanzees ∼6 Myr ago (31, 32). The insertion of HERV-K113 and -K115 were estimated to be a minimum 0.8–1 Myr ago, and that of HERV-K106 to be ∼150,000 years ago (33, 34). Even though the possibility of the youngest HERV-K mobilization still remains (35), HERVs as transposable elements do not impose a “clear and present danger” to humans. In addition, transposition of the older resident MaLRs is the most improbable.
Hundreds of Thousands of LTRs in the Genome Can Provide Binding Sites for Transcription Factors
Solitary LTRs, or solo LTRs, were generated from existing HERVs and MaLRs by the loss of internal sequences upon recombination between the 5′ and 3′ LTRs within the same copies or between separate LTRs. The human genome contains ∼50 copies of ERV-9 provirus forms and 3,000–4,000 copies of ERV-9 solitary LTRs (36). A recent study identified 944 HERV-K (HML-2) solitary LTRs versus 91 copies of full-length and nearly full-length HERV-K (HML-2) (14). In general, 85% of the HERV copies consist of solitary LTRs (5). The majority of MaLRs also exist as solitary LTRs (15).
Retrovirus LTRs (300–1300 bp) in the U3–R–U5 architecture are essential for high-level expression of the viral sequences (37). The U3 region contains a TATA box and transcription regulatory sequences which govern ubiquitous or tissue-specific gene expression. For instance, the U3 regions of human immunodeficiency viruses (HIVs) and the group of human T-cell leukemia viruses (HTLVs) have NF-κB and CREB/ATF binding sequences, respectively (38–41). Regulatory factors bind to the cognate cis-acting sequences to induce transcription from the initiation site corresponding to the 5′ terminus of the R region. In fact, the HERV-K (HML-2) LTR has Sp1/Sp3 and microphthalmia-associated transcription factor-M (MITF-M) binding sites in U3 (42, 43).
Therefore, the ∼400,000 copies of the LTR-retroelements scattered throughout the human chromosomes potentially provide extra enhancer-promoter sequences and initiation sites for neighboring cellular genes. By utilization of a nearby LTR as its alternative transcription machinery, one gene can give rise to new isoforms, enhance its transcription, and alter the tissue-specificity. In this context, HERVs and MaLRs could have significantly influenced the human genome evolution (44).
Influence of LTR/HERV Insertions on Cellular Gene Expression in Human Evolution
More than 25 experimentally characterized cellular genes show LTR-mediated evolutionary changes, in which inserted LTRs act as alternative promoters to provide new tissue-specificity, play as the major promoters, or exert only minor effects (45). For example, an ERV-L LTR plays as an alternative promoter of β1,3-galactosyltransferase 5 gene (B3GALT5) to induce colon-specific transcription (46). A HERV-K(HML-5) LTR serves as the major promoter of INSL4, a primate-specific insulin-like growth factor gene expressed in placenta (47). A HERV-E family LTR plays as an alternative tissue-specific promoter of the endothelin B receptor (EDNRB) gene, by which the gene expression increased ∼15% in placenta (48). LTR-derived promoters often increase placenta-specific gene expression, although the overall effect of the LTR insertion appears moderate in many cases (45). Notably, the 5′ terminus of a HERV-E insert in reversed orientation provides a salivary-specific enhancer for the human-only amylase gene AMY1C (49, 50).
The LTR of ERV-9 located in the β-globin locus control region (β-LCR) serves as an additional enhancer region for the downstream globin genes (36, 51). The U3 region has 5–17 tandem repeats of 37–41 bases with GAGA, CCAAT, and CCACC motifs, in which the strong affinity of NF-Y, a ubiquitous transcriptional regulatory factor, to the CCAAT motif allows a robust enhancer complex formation with other proteins (52). An ERV-9 LTR upstream of TP63 (p63), a member of the tumor suppressor TP53 (p53) gene family, acts as a strong promoter to express novel p63 isoforms in the testis of hominidae (53). Thus, some of the 5,000 copies of ERV-9 LTRs, 15–38 Myr after their integration (54), are involved in adjacent host gene control.
A HERV-W-derived Env glycoprotein encoded by ERVW-1 (7q21.2) (also termed syncytin-1 or ERVWE1) causes cytotrophoblast fusion, suggesting an essential function in syncytiotrophoblast formation in placenta tissue development (55). In addition, a HERV-FRD Env protein encoded by ERVFRD-1, or syncytin-2 (6p24), was found to have an immunosuppressive activity (56), while a role in placenta morphogenesis was also suggested (57). In conjunction with the upstream regulatory sequences, the HERV-W LTR drives the strong ERVW-1 expression in placenta (58).
The Hypothesis of HERV-Mediated Oncogenesis
Animal exogenous retroviruses carrying oncogenes can trigger transformation of the host cells by expression of the viral oncogenes (59). The virus env gene can act like an oncogene (60, 61). Retroviruses without an oncogene are also oncogenic. If retroviral integration occurs in the vicinity of a proto-oncogene which typically encodes a growth factor receptor, signaling molecule, or transcriptional activator, the LTR can activate it. “Transduction” of oncogenes and “insertional mutagenesis” by the LTRs represent major retroviral tumorigenesis mechanisms (59, 62). Based on the knowledge of retroviral functions, contribution of HERVs and solitary LTRs to human tumorigenesis has long been hypothesized.
The Np9 and Rec genes overlapping the env gene of HERV-K are particularly intriguing as candidate oncogenes. HERV-K was found as an endogenous retrovirus expressed in teratocarcinoma cell lines in an earlier study (63). Nuclear protein Np9 interacts with E3 ubiquitin ligase Ligand of Numb Protein-X (LNX), and thereby possibly influences the Notch pathway (64). Rec transgenic mice manifest neoplastic changes in the testes which correspond to the precursor phenotype of seminoma in humans (65). The Rec protein enhances androgen receptor-mediated transcription (66). High-level expression of Rec and Np9 also occurs in melanoma (67).
Activation of specific or selective HERVs occurs in various tumors and cell lines (68). HERV-K has been most extensively analyzed in cancers from different tissue origins. For example, HERV-K mRNA and proteins were detected in melanoma tissues and cell lines (69, 70). A recent study identified 24 HERV-K (HML-2) loci being transcribed in melanoma tissues (67). The RT and its enzyme activity increase in breast cancer tissues and hormone-treated cell lines (71). A large population (>85%) of breast cancer patients express HERV-K Env proteins which elicit both serologic and cell-mediated immune responses (72). HERV-K virus-like particle production was observed in teratocarcinoma cell lines (73, 74). HERV-K Gag and Env proteins are expressed in germ cell tumors, and the patients produce antibodies against these viral proteins (75, 76). Forward and reverse transcripts of several HERV-K loci are detected in prostate cancer cell lines (77). Furthermore, the gag gene of a HERV-H locus on chromosome Xp22 is frequently expressed in colon cancer tissues (78). Ovarian cancer tissues produce the Env proteins of HERV-K, ERV3, and HERV-E (79).
Transcriptional Activation of HERVs by Epigenetic Changes in Cancers
Transposons undergo long-term epigenetic silencing by DNA methylation, which is stably maintained from gametes to early embryos and the adult tissues (80). Cancer-associated hypomethylation, or derepression, frequently occurs in repeated DNA sequences including heterochromatic DNA repeats, dispersed retrotransposons, and endogenous retroviral elements (81). Although genome wide hypomethylation in cancers seems to be a side effect of carcinogenesis rather than the cause (82), site-specific hypomethylation may possibly cause re-activation of tumor-related genes leading to initiation and progression of malignancies and other diseases (83).
Recent results show that increased HERV-K (22q11.23) expression in prostate and ovarian cancers is associated with hypomethylation of the HERV-K locus (84, 85). Reduced methylation of CpG dinucleotides within the promoter region corresponds to HERV-W expression in ovarian cancer (86) and HERV-H (Xp22.3) expression in colon, gastric, and pancreatic cancers (87). Hypomethylation of the U3 region of several HERV-W loci seems to be critical for the HERV-W activation in testicular cancer (88).
Identification of LTRs that Contribute to Tumor Immunity and Oncogenesis
A pioneering study identified a HERV-encoded peptide as a tumor-specific antigen that works in the successful hematopoietic stem cell transplantation for the therapy of metastatic renal cell carcinoma (RCC) (89). The tumor antigen, CT-RCC-1, recognized by RCC-specific CD8+ T cells is encoded by novel spliced variants of the HERV-E transcript from chromosome 6q. Furthermore, this antigen expression results from hypomethylation of the LTR sequences, as well as hypoxia-inducible transcription factor (HIF-2α) binding to the HIF responsive element (HRE) in the LTR (90). HIF-2α is stabilized in many cases of RCC due to the von Hippel–Lindau (VHL) tumor suppressor gene inactivation. This whole scenario enlightens the relevance of HERVs in cancer biology and treatment.
A study on tumorigenesis of Hodgkin’s lymphoma provided the first and so far only evidence that endogenous LTR activation can be oncogenic (91). These malignant cells strongly express CSF1R, the colony stimulating factor 1 receptor gene, to allow CSF1/CSF1R-dependent cell growth. Intriguingly, a LTR of THE1B (a MaLR family LTR retrotransposon) is located 6.2 kb upstream of the CSF1R coding sequences, and is activated by the loss of CpG methylation to drive the aberrant CSF1R gene expression from the initiation site within the LTR. The regulatory region contains Sp1, GATA, AP-1, and NF-κB binding motifs. Anaplastic large cell lymphomas also have the same CSF1R transcript. Thus, hypomethylation of the THE1B LTR causes the CSF1R oncogene activation, reminiscent of “insertional mutagenesis” by exogenous retroviruses.
Possible Contribution of HERVs to Autoimmune Diseases
Human endogenous retroviruses have also been implicated in autoimmune diseases including rheumatoid arthritis (RA), multiple sclerosis (MS), and systemic lupus erythematosus (SLE). Expression of multiple ERVs was detected in RA patients (92), and that of HERV-K10 in juvenile RA (93). A recent report indicated a strong linkage between the MS-associated retroviral element (MRSV)-type HERV-W and MS: the env RNA and protein expression significantly increases in the blood and brain cells of MS patients (94). HERV-H/F family HERV-Fc1 expression in T lymphocytes and the plasma HERV-Fc1 RNA level also increase in patients with MS (95). HERV-K18 (chromosome 1) was found to be a risk factor of MS (96) and other autoimmune disorders (97, 98). Expression of HERV-W, K, E, and a newly identified ERV-9/HERV-W variant in psoriasis was also observed (99).
Three mechanisms have been proposed for HERV-related pathology of autoimmune diseases: molecular mimicry, superantigen production, and LTR-mediated alterations of gene expression (100). The Gag and Env proteins of HERV-K10 share the peptide sequences with the rheumatoid factor epitopes on IgG1Fc, implying molecular mimicry (101). The env genes of defective HERV-K alleles at 1q21.2–q22 encode superantigens, and are transcriptionally activated by the interferon α signaling to cause polyclonal T-cell activation (102, 103). There was an interesting hypothesis that solitary LTRs in the 5′-flanking region of the HLA-DQB1 gene (encoding major histocompatibility complex, class II, DQ beta 1) influence the susceptibility to autoimmune diseases (104–106). However, these results remain ambiguous, and need to be more critically examined to determine whether or not the LTR/HERV activation is the true cause of the clinical consequences (101, 107).
Furthermore, the LTR of MaLR family THE1D is activated by transcription factor DUX4 which is specifically expressed in facioscapulohumeral dystrophy, implying involvement of DUX4 in the pathophysiology (108). Hypomethylation of HERV-E and HERV-K was also observed in SLE patients (109).
Concluding Remarks
Human endogenous retroviruses are remnant forms of infectious retroviruses that integrated into the chromosomal DNA of germ-line cells of human ancestors, increased their copy numbers and have been inherited by present-day humans. None of the HERVs poses an immediate risk as a transposable element. However, solitary LTRs derived from HERVs and MaLRs dominate the provirus forms in the copy numbers, and can serve as redundant enhancer-promoter sequences for nearby cellular genes. When the DNA methylation-mediated suppression system becomes compromised, HERVs and LTRs can cause detrimental and/or self-protecting effects. Two prominent examples of the clinically significant HERV/LTR activation have been reported: CSF1R oncogene activation by a MaLR LTR in Hodgkin’s lymphoma and RCC-specific novel HERV-E antigen expression facilitating the immunotherapy. Future researches in oncology and immunogenetics will unveil more details about the endogenous LTR functions in human pathogenesis.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The work was partially supported by Grant-in-Aid for Scientific Research (C) 25460475 from MEXT, Japan (to Iyoko Katoh).
References
1. Sverdlov ED. Retroviruses and primate evolution. Bioessays (2000) 22:161–71. doi:10.1002/(SICI)1521-1878(200002)22:2<161::AID-BIES7>3.0.CO;2-X
2. Bannert N, Kurth R. Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci U S A (2004) 101:14572–9. doi:10.1073/pnas.0404838101
3. Hughes JF, Coffin JM. Human endogenous retroviral elements as indicators of ectopic recombination events in the primate genome. Genetics (2005) 171:1183–94. doi:10.1534/genetics.105.043976
4. Moyes DL, Martin A, Sawcer S, Temperton N, Worthington J, Griffiths DJ, et al. The distribution of the endogenous retroviruses HERV-K113 and HERV-K115 in health and disease. Genomics (2005) 86:337–41. doi:10.1016/j.ygeno.2005.06.004
5. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature (2001) 409:860–921. doi:10.1038/35057062
6. Paces J, Pavlícek A, Paces V. HERVd: database of human endogenous retroviruses. Nucleic Acids Res (2002) 30:205–6. doi:10.1093/nar/30.1.205
7. Ribet D, Harper F, Dupressoir A, Dewannieux M, Pierron G, Heidmann T. An infectious progenitor for the murine IAP retrotransposon: emergence of an intracellular genetic parasite from an ancient retrovirus. Genome Res (2008) 18:597–609. doi:10.1101/gr.073486.107
8. Gifford WD, Pfaff SL, Macfarlan TS. Transposable elements as genetic regulatory substrates in early development. Trends Cell Biol (2013) 23:218–26. doi:10.1016/j.tcb.2013.01.001
9. Jurka J. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet (2000) 16:418–20. doi:10.1016/S0168-9525(00)02093-X
10. Tristem M. Identification and characterization of novel human endogenous retrovirus families by phylogenetic screening of the human genome mapping project database. J Virol (2000) 74:3715–30. doi:10.1128/JVI.74.8.3715-3730.2000
11. Mager DL, Medstrand P. Retroviral repeat sequences. In: Cooper D, editor. Nature Encyclopedia of the Human Genome. Hampshire: MacMillan Publishers (2003). p. 291–315.
12. Bénit L, Calteau A, Heidmann T. Characterization of the low-copy HERV-Fc family: evidence for recent integrations in primates of elements with coding envelope genes. Virology (2003) 312:159–68. doi:10.1016/S0042-6822(03)00163-6
13. Bannert N, Kurth R. The evolutionary dynamics of human endogenous retroviral families. Annu Rev Genomics Hum Genet (2006) 7:149–73. doi:10.1146/annurev.genom.7.080505.115700
14. Subramanian R, Wildschutte J, Russo C, Coffin J. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology (2011) 8:90. doi:10.1186/1742-4690-8-90
15. Smit AFA. Identification of a new, abundant superfamily of mammalian LTR-transposons. Nucleic Acids Res (1993) 21:1863–72. doi:10.1093/nar/21.8.1863
16. Lynch C, Tristem M. A co-opted gypsy-type LTR-retrotransposon is conserved in the genomes of humans, sheep, mice, and rats. Curr Biol (2003) 13:1518–23. doi:10.1016/S0960-9822(03)00618-3
17. Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. EMBO J (2002) 21:5899–910. doi:10.1093/emboj/cdf592
18. Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet (2003) 35:41–8. doi:10.1038/ng1223
19. Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet (2009) 10:691–703. doi:10.1038/nrg2640
20. Kano H, Godoy I, Courtney C, Vetter MR, Gerton GL, Ostertag EM, et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Gene Dev (2009) 23:1303–12. doi:10.1101/gad.1803909
21. Hancks DC, Kazazian HH Jr. SVA retrotransposons: evolution and genetic instability. Semin Cancer Biol (2010) 20:234–45. doi:10.1016/j.semcancer.2010.04.001
22. Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature (2011) 479:534–7. doi:10.1038/nature10531
23. Mills RE, Bennett EA, Iskow RC, Luttig CT, Tsui C, Pittard WS, et al. Recently mobilized transposons in the human and chimpanzee genomes. Am J Hum Genet (2006) 78:671–9. doi:10.1086/501028
24. Li X, Scaringe WA, Hill KA, Roberts S, Mengos A, Careri D, et al. Frequency of recent retrotransposition events in the human factor IX gene. Hum Mutat (2001) 17:511–9. doi:10.1002/humu.1134
25. Deininger PL, Moran JV, Batzer MA, Kazazian HH Jr. Mobile elements and mammalian genome evolution. Curr Opin Genet Dev (2003) 13:651–8.
26. Chen JM, Stenson PD, Cooper DN, Ferec C. A systematic analysis of LINE-1 endonuclease-dependent retrotranspositional events causing human genetic disease. Hum Genet (2005) 117:411–27. doi:10.1007/s00439-005-1321-0
27. Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell (2010) 141:1253–61. doi:10.1016/j.cell.2010.05.020
28. Hancks DC, Kazazian HH Jr. Active human retrotransposons: variation and disease. Curr Opin Genet Dev (2012) 22:191–203. doi:10.1016/j.gde.2012.02.006
29. Belshaw R, Katzourakis A, Paces J, Burt A, Tristem M. High copy number in human endogenous retrovirus families is associated with copying mechanisms in addition to reinfection. Mol Biol Evol (2005) 22:814–7. doi:10.1093/molbev/msi088
30. Dewannieux M, Harper F, Richaud A, Letzelter C, Ribet D, Pierron G, et al. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res (2006) 16:1548–56. doi:10.1101/gr.5565706
31. Barbulescu M, Turner G, Seaman MI, Deinard AS, Kidd KK, Lenz J. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr Biol (1999) 9:861–S861. doi:10.1016/S0960-9822(99)80390-X
32. Shin W, Lee J, Son SY, Ahn K, Kim HS, Han K. Human-specific HERV-K insertion causes genomic variations in the human genome. PLoS One (2013) 8:e60605. doi:10.1371/journal.pone.0060605
33. Jha AR, Pillai SK, York VA, Sharp ER, Storm EC, Wachter DJ, et al. Cross-sectional dating of novel haplotypes of HERV-K 113 and HERV-K 115 indicate these proviruses originated in Africa before homo sapiens. Mol Biol Evol (2009) 26:2617–26. doi:10.1093/molbev/msp180
34. Jha AR, Nixon DF, Rosenberg MG, Martin JN, Deeks SG, Hudson RR, et al. Human endogenous retrovirus K106 (HERV-K106) was infectious after the emergence of anatomically modern humans. PLoS One (2011) 6:e20234. doi:10.1371/journal.pone.0020234
35. Turner G, Barbulescu M, Su M, Jensen-Seaman MI, Kidd KK, Lenz J. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr Biol (2001) 11:1531–5. doi:10.1016/S0960-9822(01)00455-9
36. Ling J, Pi W, Bollag R, Zeng S, Keskintepe M, Saliman H, et al. The solitary long terminal repeats of ERV-9 endogenous retrovirus are conserved during primate evolution and possess enhancer activities in embryonic and hematopoietic cells. J Virol (2002) 76:2410–23. doi:10.1128/jvi.76.5.2410-2423.2002
37. Vogt VM. Retroviral virions and genomes. In: Coffin JM, Hughes JF, Varmus HE, editors. Retroviruses. New York: Cold Spring Harbor Laboratory Press (1997). p. 27–69.
38. Kawakami K, Scheidereit C, Roeder RG. Identification and purification of a human immunoglobulin-enhancer-binding protein (NF-kappa B) that activates transcription from a human immunodeficiency virus type 1 promoter in vitro. Proc Natl Acad Sci U S A (1988) 85:4700–4. doi:10.1073/pnas.85.13.4700
39. Katoh I, Yoshinaka Y, Ikawa Y. Bovine leukemia virus trans-activator p38tax activates heterologous promoters with a common sequence known as a cAMP-responsive element or the binding site of a cellular transcription factor ATF. EMBO J (1989) 8:497–503.
40. Beimling P, Moelling K. Direct interaction of CREB protein with 21 bp tax-response elements of HTLV-ILTR. Oncogene (1992) 7:257–62.
41. Kurata S. Sensitization of the HIV-1-LTR upon long term low dose oxidative stress. J Biol Chem (1996) 271:21798–802.
42. Fuchs NV, Kraft M, Tondera C, Hanschmann KM, Lower J, Lower R. Expression of the human endogenous retrovirus (HERV) group HML-2/HERV-K does not depend on canonical promoter elements but is regulated by transcription factors Sp1 and Sp3. J Virol (2011) 85:3436–48. doi:10.1128/JVI.02539-10
43. Katoh I, Mirova A, Kurata S, Murakami Y, Horikawa K, Nakakuki N, et al. Activation of the long terminal repeat of human endogenous retrovirus K by melanoma-specific transcription factor MITF-M. Neoplasia (2011) 13:1081–92.
44. Griffiths DJ. Endogenous retroviruses in the human genome sequence. Genome Biol (2001) 2:REVIEWS1017. doi:10.1186/gb-2001-2-6-reviews1017
45. Cohen CJ, Lock WM, Mager DL. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene (2009) 448:105–14. doi:10.1016/j.gene.2009.06.020
46. Dunn CA, Medstrand P, Mager DL. An endogenous retroviral long terminal repeat is the dominant promoter for human beta1,3-galactosyltransferase 5 in the colon. Proc Natl Acad Sci U S A (2003) 100:12841–6. doi:10.1073/pnas.2134464100
47. Bièche I, Laurent A, Laurendeau I, Duret L, Giovangrandi Y, Frendo J-L, et al. Placenta-specific INSL4 expression is mediated by a human endogenous retrovirus element. Biol Reprod (2003) 68:1422–9. doi:10.1095/biolreprod.102.010322
48. Landry J-R, Mager DL. Functional analysis of the endogenous retroviral promoter of the human endothelin B receptor gene. J Virol (2003) 77:7459–66. doi:10.1128/JVI.77.13.7459-7466.2003
49. Ting CN, Rosenberg MP, Snow CM, Samuelson LC, Meisler MH. Endogenous retroviral sequences are required for tissue-specific expression of a human salivary amylase gene. Genes Dev (1992) 6:1457–65. doi:10.1101/gad.6.8.1457
50. Samuelson LC, Phillips RS, Swanberg LJ. Amylase gene structures in primates: retroposon insertions and promoter evolution. Mol Biol Evol (1996) 13:767–79. doi:10.1093/oxfordjournals.molbev.a025637
51. Pi W, Zhu X, Wu M, Wang Y, Fulzele S, Eroglu A, et al. Long-range function of an intergenic retrotransposon. Proc Natl Acad Sci U S A (2010) 107:12992–7. doi:10.1073/pnas.1004139107
52. Yu X, Zhu X, Pi W, Ling J, Ko L, Takeda Y, et al. The long terminal repeat (LTR) of ERV-9 human endogenous retrovirus binds to NF-Y in the assembly of an active LTR enhancer complex NF-Y/MZF1/GATA-2. J Biol Chem (2005) 280:35184–94. doi:10.1074/jbc.M508138200
53. Beyer U, Moll-Rocek J, Moll UM, Dobbelstein M. Endogenous retrovirus drives hitherto unknown proapoptotic p63 isoforms in the male germ line of humans and great apes. Proc Natl Acad Sci U S A (2011) 108:3624–9. doi:10.1073/pnas.1016201108
54. Costas J, Naveira H. Evolutionary history of the human endogenous retrovirus family ERV9. Mol Biol Evol (2000) 17:320–30. doi:10.1093/oxfordjournals.molbev.a026312
55. Mi S, Lee X, Li X-P, Veldman GM, Finnerty H, Racie L, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature (2000) 403:785–9. doi:10.1038/35001608
56. Mangeney M, Renard M, Schlecht-Louf G, Bouallaga I, Heidmann O, Letzelter C, et al. Placental syncytins: genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proc Natl Acad Sci U S A (2007) 104:20534–9. doi:10.1073/pnas.0707873105
57. Dupressoir A, Vernochet C, Harper F, Guégan J, Dessen P, Pierron G, et al. A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proc Natl Acad Sci U S A (2011) 108:E1164–73. doi:10.1073/pnas.1112304108
58. Prudhomme S, Oriol G, Mallet F. A retroviral promoter and a cellular enhancer define a bipartite element which controls env ERVWE1 placental expression. J Virol (2004) 78:12157–68. doi:10.1128/JVI.78.22.12157-12168.2004
59. Rosenberg N, Jolicoeur P. Retroviral pathogenesis. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. New York: Cold Spring Harbor Laboratory Press (1997). p. 475–585.
60. Aizawa S, Suda Y, Furuta Y, Yagi T, Takeda N, Watanabe N, et al. Env-derived gp55 gene of friend spleen focus-forming virus specifically induces neoplastic proliferation of erythroid progenitor cells. EMBO J (1990) 9:2107–16.
61. Yoshimura A, D’Andrea AD, Lodish HF. Friend spleen focus-forming virus glycoprotein gp55 interacts with the erythropoietin receptor in the endoplasmic reticulum and affects receptor metabolism. Proc Natl Acad Sci U S A (1990) 87:4139–43. doi:10.1073/pnas.87.11.4139
62. Bishop JM. Cellular oncogenes and retroviruses. Annu Rev Biochem (1983) 52:301–54. doi:10.1146/annurev.bi.52.070183.001505
63. Löwer R, Löwer J, Tondera-Koch C, Kurth R. A general method for the identification of transcribed retrovirus sequences (R-U5 PCR) reveals the expression of the human endogenous retrovirus loci HERV-H and HERV-K in teratocarcinoma cells. Virology (1993) 192:501–11. doi:10.1006/viro.1993.1066
64. Armbruester V, Sauter M, Roemer K, Best B, Hahn S, Nty A, et al. Np9 protein of human endogenous retrovirus K interacts with ligand of numb protein X. J Virol (2004) 78:10310–9. doi:10.1128/JVI.78.19.10310-10319.2004
65. Galli UM, Sauter M, Lecher B, Maurer S, Herbst H, Roemer K, et al. Human endogenous retrovirus rec interferes with germ cell development in mice and may cause carcinoma in situ, the predecessor lesion of germ cell tumors. Oncogene (2005) 24:3223–8. doi:10.1038/sj.onc.1208543
66. Hanke K, Chudak C, Kurth R, Bannert N. The Rec protein of HERV-K(HML-2) upregulates androgen receptor activity by binding to the human small glutamine-rich tetratricopeptide repeat protein (hSGT). Int J Cancer (2013) 132:556–67. doi:10.1002/ijc.27693
67. Schmitt K, Reichrath J, Roesch A, Meese E, Mayer J. Transcriptional profiling of human endogenous retrovirus group HERV-K(HML-2) loci in melanoma. Genome Biol Evol (2013) 5:307–28. doi:10.1093/gbe/evt010
68. Romanish MT, Cohen CJ, Mager DL. Potential mechanisms of endogenous retroviral-mediated genomic instability in human cancer. Semin Cancer Biol (2010) 20:246–53. doi:10.1016/j.semcancer.2010.05.005
69. Muster T, Waltenberger A, Grassauer A, Hirschl S, Caucig P, Romirer I, et al. An endogenous retrovirus derived from human melanoma cells. Cancer Res (2003) 63:8735–41.
70. Büscher K, Trefzer U, Hofmann M, Sterry W, Kurth R, Denner J. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res (2005) 65:4172–80. doi:10.1158/0008-5472.CAN-04-2983
71. Golan M, Hizi A, Resau JH, Yaal-Hahoshen N, Reichman H, Keydar I, et al. Human endogenous retrovirus (HERV-K) reverse transcriptase as a breast cancer prognostic marker. Neoplasia (2008) 10:521–33.
72. Wang-Johanning F, Radvanyi L, Rycaj K, Plummer JB, Yan P, Sastry KJ, et al. Human endogenous retrovirus K triggers an antigen-specific immune response in breast cancer patients. Cancer Res (2008) 68:5869–77. doi:10.1158/0008-5472.CAN-07-6838
73. Boller K, König H, Sauter M, Mueller-Lantzsch N, Löwer R, Löwer J, et al. Evidence that HERV-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma-derived retrovirus HTDV. Virology (1993) 196:349–53. doi:10.1006/viro.1993.1487
74. Lower R, Boller K, Hasenmaier B, Korbmacher C, Muller-Lantzsch N, Lower J, et al. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc Natl Acad Sci U S A (1993) 90:4480–4. doi:10.1073/pnas.90.10.4480
75. Sauter M, Schommer S, Kremmer E, Remberger K, Dolken G, Lemm I, et al. Human endogenous retrovirus K10: expression of Gag protein and detection of antibodies in patients with seminomas. J Virol (1995) 69:414–21.
76. Sauter M, Roemer K, Best B, Afting M, Schommer S, Seitz G, et al. Specificity of antibodies directed against Env protein of human endogenous retroviruses in patients with germ cell tumors. Cancer Res (1996) 56:4362–5.
77. Agoni L, Guha C, Lenz J. Detection of human endogenous retrovirus K (HERV-K) transcripts in human prostate cancer cell lines. Front Oncol (2013) 3:180. doi:10.3389/fonc.2013.00180
78. Alves PM, Levy N, Stevenson BJ, Bouzourene H, Theiler G, Bricard G, et al. Identification of tumor-associated antigens by large-scale analysis of genes expressed in human colorectal cancer. Cancer Immun (2008) 8:11.
79. Wang-Johanning F, Liu J, Rycaj K, Huang M, Tsai K, Rosen DG, et al. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int J Cancer (2007) 120:81–90. doi:10.1002/ijc.22256
80. Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature (2007) 447:425–32. doi:10.1038/nature05918
81. Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene (2002) 21:5400–13. doi:10.1038/sj.onc.1205651
82. Wild L, Flanagan JM. Genome-wide hypomethylation in cancer may be a passive consequence of transformation. Biochim Biophys Acta (2010) 1806:50–7. doi:10.1016/j.bbcan.2010.03.003
83. Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta (2007) 1775:138–62.
84. Goering W, Ribarska T, Schulz WA. Selective changes of retroelement expression in human prostate cancer. Carcinogenesis (2011) 32:1484–92. doi:10.1093/carcin/bgr181
85. Iramaneerat K, Rattanatunyong P, Khemapech N, Triratanachat S, Mutirangura A. HERV-K hypomethylation in ovarian clear cell carcinoma is associated with a poor prognosis and platinum resistance. Int J Gynecol Cancer (2011) 21:51–7. doi:10.1097/IGC.0b013e3182021c1a
86. Menendez L, Benigno BB, McDonald JF. L1 and HERV-W retrotransposons are hypomethylated in human ovarian carcinomas. Mol Cancer (2004) 3:12. doi:10.1186/1476-4598-3-12
87. Wentzensen N, Coy JF, Knaebel HP, Linnebacher M, Wilz B, Gebert J, et al. Expression of an endogenous retroviral sequence from the HERV-H group in gastrointestinal cancers. Int J Cancer (2007) 121:1417–23. doi:10.1002/ijc.22826
88. Gimenez J, Montgiraud C, Pichon J-P, Bonnaud B, Arsac M, Ruel K, et al. Custom human endogenous retroviruses dedicated microarray identifies self-induced HERV-W family elements reactivated in testicular cancer upon methylation control. Nucleic Acids Res (2010) 38:2229–46. doi:10.1093/nar/gkp1214
89. Takahashi Y, Harashima N, Kajigaya S, Yokoyama H, Cherkasova E, McCoy JP, et al. Regression of human kidney cancer following allogeneic stem cell transplantation is associated with recognition of an HERV-E antigen by T cells. J Clin Invest (2008) 118:1099–109. doi:10.1172/JCI34409
90. Cherkasova E, Malinzak E, Rao S, Takahashi Y, Senchenko VN, Kudryavtseva AV, et al. Inactivation of the von Hippel-Lindau tumor suppressor leads to selective expression of a human endogenous retrovirus in kidney cancer. Oncogene (2011) 30:4697–706. doi:10.1038/onc.2011.179
91. Lamprecht B, Walter K, Kreher S, Kumar R, Hummel M, Lenze D, et al. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat Med (2010) 16:571–9. doi:10.1038/nm.2129
92. Nakagawa K, Brusic V, McColl G, Harrison LC. Direct evidence for the expression of multiple endogenous retroviruses in the synovial compartment in rheumatoid arthritis. Arthritis Rheum (1997) 40:627–38. doi:10.1002/art.1780400407
93. Sicat J, Sutkowski N, Huber BT. Expression of human endogenous retrovirus HERV-K18 superantigen is elevated in juvenile rheumatoid arthritis. J Rheumatol (2005) 32:1821–31.
94. Perron H, Germi R, Bernard C, Garcia-Montojo M, Deluen C, Farinelli L, et al. Human endogenous retrovirus type W envelope expression in blood and brain cells provides new insights into multiple sclerosis disease. Mult Scler (2012) 18:1721–36. doi:10.1177/1352458512441381
95. Laska MJ, Brudek T, Nissen KK, Christensen T, Møller-Larsen A, Petersen T, et al. Expression of HERV-Fc1, a human endogenous retrovirus, is increased in patients with active multiple sclerosis. J Virol (2012) 86:3713–22. doi:10.1128/JVI.06723-11
96. Tai A, O’Reilly E, Alroy K, Simon K, Munger K, Huber B, et al. Human endogenous retrovirus-K18 Env as a risk factor in multiple sclerosis. Mult Scler (2008) 14:1175–80. doi:10.1177/1352458508094641
97. de la Hera B, Varadé J, García-Montojo M, Lamas JR, de la Encarnación A, Arroyo R, et al. Role of the human endogenous retrovirus HERV-K18 in autoimmune disease susceptibility: study in the Spanish population and meta-analysis. PLoS One (2013) 8:e62090. doi:10.1371/journal.pone.0062090
98. Garcia-Montojo M, Varade J, Villafuertes E, Hera BD, Hoyas-Fernandez J, Dominguez-Mozo M, et al. Expression of human endogenous retrovirus HERV-K18 is associated with clinical severity in osteoarthritis patients. Scand J Rheumatol (2013). doi:10.3109/03009742.2013.779021
99. Molès JP, Tesniere A, Guilhou JJ. A new endogenous retroviral sequence is expressed in skin of patients with psoriasis. Br J Dermatol (2005) 153:83–9. doi:10.1111/j.1365-2133.2005.06555.x
100. Nelson PN, Hooley P, Roden D, Davari Ejtehadi H, Rylance P, Warren P, et al. Human endogenous retroviruses: transposable elements with potential? Clin Exp Immunol (2004) 138:1–9.
101. Tugnet N, Rylance P, Roden D, Trela M, Nelson P. Human endogenous retroviruses (HERVs) and autoimmune rheumatic disease: is there a link? Open Rheumatol J (2013) 7:13–21. doi:10.2174/1874312901307010013
102. Stauffer Y, Marguerat S, Meylan F, Ucla C, Sutkowski N, Huber B, et al. Interferon-α-induced endogenous superantigen: a model linking environment and autoimmunity. Immunity (2001) 15:591–601. doi:10.1016/S1074-7613(01)00212-6
103. Sutkowski N, Conrad B, Thorley-Lawson DA, Huber BT. Epstein-Barr virus transactivates the human endogenous retrovirus HERV-K18 that encodes a superantigen. Immunity (2001) 15:579–89. doi:10.1016/S1074-7613(01)00210-2
104. Seidl C, Donner H, Petershofen E, Usadel KH, Seifried E, Kaltwasser JP, et al. An endogenous retroviral long terminal repeat at the HLA-DQB1 gene locus confers susceptibility to rheumatoid arthritis. Hum Immunol (1999) 60:63–8. doi:10.1016/S0198-8859(98)00095-0
105. Pascual M, Martin J, Nieto A, Giphart MJ, van der Slik AR, de Vries RR, et al. Distribution of HERV-LTR elements in the 5’-flanking region of HLA-DQB1 and association with autoimmunity. Immunogenetics (2001) 53:114–8. doi:10.1007/s002510100307
106. Pani MA, Seidl C, Bieda K, Seissler J, Krause M, Seifried E, et al. Preliminary evidence that an endogenous retroviral long-terminal repeat (LTR13) at the HLA-DQB1 gene locus confers susceptibility to Addison’s disease. Clin Endocrinol (2002) 56:773–7. doi:10.1046/j.1365-2265.2002.t01-1-01548.x
107. Freimanis G, Hooley P, Ejtehadi HD, Ali HA, Veitch A, Rylance PB, et al. A role for human endogenous retrovirus-K (HML-2) in rheumatoid arthritis: investigating mechanisms of pathogenesis. Clin Exp Immunol (2010) 160:340–7. doi:10.1111/j.1365-2249.2010.04110.x
108. Geng LN, Yao Z, Snider L, Fong AP, Cech JN, Young JM, et al. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev Cell (2012) 22:38–51. doi:10.1016/j.devcel.2011.11.013
Keywords: human endogenous retrovirus, HERV, long terminal repeat, LTR, cancer, retrotransposon, autoimmune
Citation: Katoh I and Kurata S (2013) Association of endogenous retroviruses and long terminal repeats with human disorders. Front. Oncol. 3:234. doi: 10.3389/fonc.2013.00234
Received: 30 June 2013; Accepted: 27 August 2013;
Published online: 11 September 2013.
Edited by:
Peter K. Vogt, The Scripps Research Institute, USAReviewed by:
Joachim Denner, Robert Koch Institute, GermanyNorbert Bannert, Robert Koch Institute, Germany
Copyright: © 2013 Katoh and Kurata. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Iyoko Katoh, Department of Microbiology, Interdisciplinary Graduate School of Medicine and Engineering, University of Yamanashi, 1110 Shimokato, Chuo, Yamanashi 409-3898, Japan e-mail: iyoko@yamanashi.ac.jp