High-resolution melting curve analysis, a rapid and affordable method for mutation analysis in childhood acute myeloid leukemia
- 1Department of Hematology/Oncology, Shanghai Children’s Medical Center, Shanghai Jiaotong University School of Medicine, Shanghai, China
- 2Division of Hematology, Oncology and Blood and Marrow Transplantation, Children’s Center for Cancer and Blood Disease, Children’s Hospital Los Angeles, Keck School of Medicine, University of Southern California, Los Angeles, CA, USA
Background: Molecular genetic alterations with prognostic significance have been described in childhood acute myeloid leukemia (AML). The aim of this study was to establish cost-effective techniques to detect mutations of FMS-like tyrosine kinase 3 (FLT3), nucleophosmin 1 (NPM1), and a partial tandem duplication within the mixed-lineage leukemia (MLL-PTD) genes in childhood AML.
Procedure: Ninety-nine children with newly diagnosed AML were included in this study. We developed a fluorescent dye SYTO-82 based high-resolution melting (HRM) curve analysis to detect FLT3 internal tandem duplication (FLT3-ITD), FLT3 tyrosine kinase domain (FLT3-TKD), and NPM1 mutations. MLL-PTD was screened by real-time quantitative PCR.
Results: The HRM methodology correlated well with gold standard Sanger sequencing with less cost. Among the 99 patients studied, the FLT3-ITD mutation was associated with significantly worse event-free survival (EFS). Patients with the NPM1 mutation had significantly better EFS and overall survival. However, HRM was not sensitive enough for minimal residual disease monitoring.
Conclusion: High-resolution melting was a rapid and efficient method for screening of FLT3 and NPM1 gene mutations. It was both affordable and accurate, especially in resource underprivileged regions. Our results indicated that HRM could be a useful clinical tool for rapid and cost-effective screening of the FLT3 and NPM1 mutations in AML patients.
Introduction
Acute myeloid leukemia (AML) is a group of heterogeneous disease harboring different genetic alterations and with considerable diversity in clinical behavior and prognosis (1–3). Karyotype is the major prognostic factors in AML (1–3). Several molecular alterations have been previously identified in children with AML, especially in cases with cytogenetic normal AML (CN-AML) (4–15).
FMS-like tyrosine kinase receptor (FLT3) internal tandem duplications (ITDs) are caused by the duplication of the exon 14 sequence at juxtamembrane domain, leading to constitutive activation of downstream signaling (16, 17). FLT3-ITD has been described in 4–27% of patients with childhood AML and a high FLT3-ITD allelic ratio is associated with worse outcome (4–7). Point mutations in codon 835 or 836 of the FLT3 gene tyrosine kinase domain (FLT3-TKD) occur in 3–11% of patients (4–7, 18, 19), but the prognostic relevance remains to be defined.
Nucleophosmin 1 (NPM1) is a nuclear and cytoplasmic protein, which functions as a molecular chaperone to prevent protein aggregation in the nucleolus and to regulate p53 levels (9). Point mutations within the C-terminal region of the NPM1 gene result in cytoplasmic accumulation of the mutated NPM protein (9, 11, 12). This occurs in ~25% of pediatric CN-AML cases and is associated with a favorable clinical outcome (8, 10).
Partial tandem duplication (PTD) of the myeloid/lymphoid or mixed-lineage leukemia (MLL) gene without major chromosome structural aberrations involving band 11q23 has been identified in 5–13% of patients with de novo AML (13–15). Event-free survival (EFS) and complete remission (CR) duration were shorter in adult patients with CN-AML bearing MLL-PTD mutation than patients with the wild-type MLL gene (13–15).
In the United States and Europe, mutation detections of FLT3-ITD, FLT3-TKD, and NPM1 have been established as conventional diagnostic tests in clinical practice. However, they have not been applied to routine clinical care in China. Part of the reason has been cost (20, 21); when facing a catastrophic event such as cancer, it remains a significant challenge for Chinese pediatric oncologists to provide valuable and affordable clinical diagnostic tests.
Traditionally, FLT3-ITD, FLT3-TKD, and NPM1 mutations are detected by PCR followed by electrophoresis or Taqman RQ-PCR (17, 22–24). The traditional PCR technique is convenient and less expensive. However, it is an open tube system with increased possibility of false positive results due to amplification product carry-over contamination. Taqman RQ-PCR is a closed-tube system requiring no post-PCR processing, and it has increased sensitivity and higher resolution when compared to traditional PCR. However, this technique can only detect known mutations matching the designed probe. Thus, the search continues for a sensitive, reliable, and good manufacturing procedure (GMP) leveled detection system for both the known and potential unknown gene mutations in clinical diagnosis.
High-resolution melting (HRM) curve analysis is a homogenous, closed-tube, post-PCR technique for rapidly and efficiently discovering genetic variations in DNA fragments (25–27), based on the sequence dependent dissociation behavior of DNA when exposed to increasing temperature. It is a simple, powerful, and cost-effective method for a large scale genotyping project (28). The accuracy of the technique depends on appropriate PCR design and saturating DNA dyes.
In this study, we investigated using HRM analysis to evaluate the prevalence of FLT3-ITD, FTL3-TKD, and NPM1 in childhood AML. We also developed real-time quantitative PCR (RQ-PCR) methodology to detect MLL-PTD gene mutations. We validated the HRM technology and analyzed the clinical significance of gene mutations in a group of 99 children with AML who were enrolled in the AML-XH-99 protocol. The AML-XH-99 protocol was a single arm clinical study to examine homoharringtonine, a plant alkaloid that has been used to treat leukemia in China since the 1980s (29), in combination with chemotherapy to treat childhood AML. The outcome of this trial has been previously published (30).
Materials and Methods
Patients
From January 1999 to December 2008, a total of 99 newly diagnosed PML/RARA negative AML pediatric patients were included in this study. All 99 patients were already enrolled in the AML-XH-99 protocol and had available bone marrow (BM) samples frozen in the Biobank at Shanghai Children’s Medical Center.
At diagnosis, all patients underwent blood testing for complete blood counts and biochemistry panels. BM cells were aspirated for morphology, immunohistochemistry, karyotype, immunophenotyping, and molecular testings for RUNX1-RUNX1T1(AML1-ETO), CBFB-MYH11, or PML-RARA gene mutations (31). CN-AML is defined as leukemia without chromosomal abnormalities by karyotype and negative for RUNX1-RUNX1T1, CBFB-MYH11, or PML-RARA by PCR.
Additional BM samples were collected for research purposes at the time of diagnosis (after a legal guardian signed an informed consent). The mononuclear cells were isolated by Ficoll-Hypaque gradient and cryopreserved in the Biobank. This study was approved by the Institutional Review Board of Shanghai Children’s Medica Center.
Chemotherapy
All patients were treated on the AML-XH-99 protocol, which included induction, consolidation, and continuation therapies (30). Induction consisted two cycles of DAE (daunorubicin 40 mg/m2/day, days 1–3, cytarabine 200 mg/m2 every 12 h, days 1–7, and etoposide 100 mg/m2/day, days 1–3). Consolidation included two courses of chemotherapy: (A) DA (cytarabine 2 g/m2 every 12 h, days 1–3, and daunorubicin 30 mg/m2/day, day 1 and 2); and (B) EA (cytarabine 2 g/m2 every 12 h, days 1–3, and VP-16 160 mg/m2/day, day 1 and 2). For continuation therapy, DA alternating with EA, followed by two courses of HA (homoharringtonine 3–4 mg/m2/day, days 1–9, and cytarabine 75 mg/m2 every 12 h, days 1–7), were given for a total of 12 cycles (30).
Complete remission was defined as marrow blasts <5% with evidence of blood count recovery. Central nervous system leukemia prevention consisted of intrathecal (IT) chemotherapy with dexamethasone 5 mg/m2, methotrexate 12.5 mg/m2 (maximum 12.5 mg), and cytarabine 1 mg/kg (maximum 35 mg) at various time points (30). Allogeneic hematopoietic stem cell transplantation (Allo-HSCT) was performed in some relapsed or refractory patients with suitable donors who had achieved CR or at least a good partial response (PR, defined as a BM blast reduction of >50%) after salvage chemotherapy.
Samples
In this study, BM samples at diagnosis were analyzed for gene mutations. Control BM samples came from patients with idiopathic thrombocytopenia who underwent BM aspiration to rule out malignancy.
Total RNA was extracted from 1 × 107 BM mononuclear cells (BMMC) using the phenol–chloroform–isopropanol method (TRIzol®, Invitrogen). One microgram of total RNA was reverse transcribed according to the manufacturer’s protocols (Takara, Japan). RNA quality and quantity were assessed relative to the GAPDH housekeeping gene on the LightCycler® 480 System (Roche Diagnostics, Penzberg, Germany). Samples with CtGAPDH values over 30 were considered uninterpretable.
High-Resolution Melting Analysis of FLT3-ITD, FLT3-TKD, and NPM1 Mutation
The PCR and melting analysis for FLT3 and NPM1 mutations was performed on the LightCycler® 480, a real-time PCR machine with HRM and 96/384 well capacity. All samples were tested in triplicates. One positive control and one negative control with each gene mutation, as well as one blank control with distilled water only, were included in triplets on each run of unknowns. PCR was performed from ~10 ng of cDNA, using 0.3 μM each of the relevant forward and reverse primers in a total volume of 10 μl containing 5 μl ES Taq HS premix (Takara) and 5 μM SYTO-82 (Invitrogen) (32). PCR was performed at 94°C for 2 min, followed by 45 cycles at 94°C for 10 s, 56°C for 10 s, 72°C for 20 s, and a final extension step at 72°C for 5 min. The cycling conditions were the same for all three amplicons, allowing them to be performed in one run. The program ran for 40 cycles at 95°C for 30 s, 40°C for 30 s, then 60°C −95°C (5 s, 1°C/s). Upon completion of the run, analysis was performed using the software supplied with the LightCycler® 480. The melting curves were normalized, and temperature shifted to allow samples to be directly compared. Difference plots were generated by selecting a negative control as the baseline. The fluorescence of all other samples was plotted relative to this sample. Significant differences in fluorescence were indicative of mutations.
The primers used were: (1) FLT3-ITD: exon 11 (ITD-F) GCAATTTAGGTATGAAAGCCAGC, exon 15 (ITD-R) GTTGCGTTCATCACTTTTCCAA; (2) FLT3-TKD: TKD-F ACGTGCTTGTCACCC, TKD-R TTAATGGTGTAGATGCCTTCAAAC; and (3) NPM1: NPM-F TGACTGACCAAGAGGCTATTC, NPM-R GACAGCCAGATATCAACTGTTAC.
RQ-PCR Analysis of MLL-PTD Mutation
Real-time quantitative PCRs were performed on the LightCycler® 480 using ~20 ng of cDNA and 0.5 μM each of the relevant forward and reverse primers: MLL-F3: AAGCAGCCTCCACCACCAGAAT and MLL-R3: CCACGAGGTTTTCGAGGACTA in a total volume of 10 μl containing with 5 μl SYBR Green I premix (Takara) at 94°C for 2 min, followed by 35 cycles at 94°C for 10 s, 56°C for 20 s. Samples with CT values below 35 and with satisfying melting peak at 82–84°C were considered as MLL-PTD mutants.
Sanger Sequencing
For wild-type samples, PCR products were purified and sent to outside facilities for Sanger sequencing. For samples with mutations, PCR products were purified and cloned into TA vectors (Takara, Japan). Plasmids extracted from clones that were successfully inserted with target genes were sent for Sanger sequencing.
Statistical Analysis
Event-free survival was defined as the time elapsed from study enrollment to induction failure, relapse, or death. Overall survival (OS) was defined as the time from study enrollment until death from any cause. EFS and OS were calculated using the Kaplan–Meier method, and the log-rank test was used for comparisons of Kaplan–Meier curves. All p-values were two-sided and p < 0.05 was considered as statistically significant. Statistical calculations were performed using SAS version 9.2 (SAS Institute Inc., Cary, NC, USA).
Results
HRM Analysis
Results were analyzed using the LightCycler® 480 associated software (release 1.5.0). Data were presented in two formats: the normalized plot (Figures 1A–C), in which the amount of intercalating dye remaining at any temperature point was expressed as a fraction of the amount prior to data acquisition; and a difference plot (Figures 1D–F), where the average HRM profile of the control samples was used by the genotype function of the machine software as the standard wild-type profile for subsequent comparison to each of the test samples. Each mutant allele had its own distinctive melting curve when compared to the wild-type allele. The distinct melting curves of the mutant became more apparent when data were represented in a difference plot format than in a normalized plot.
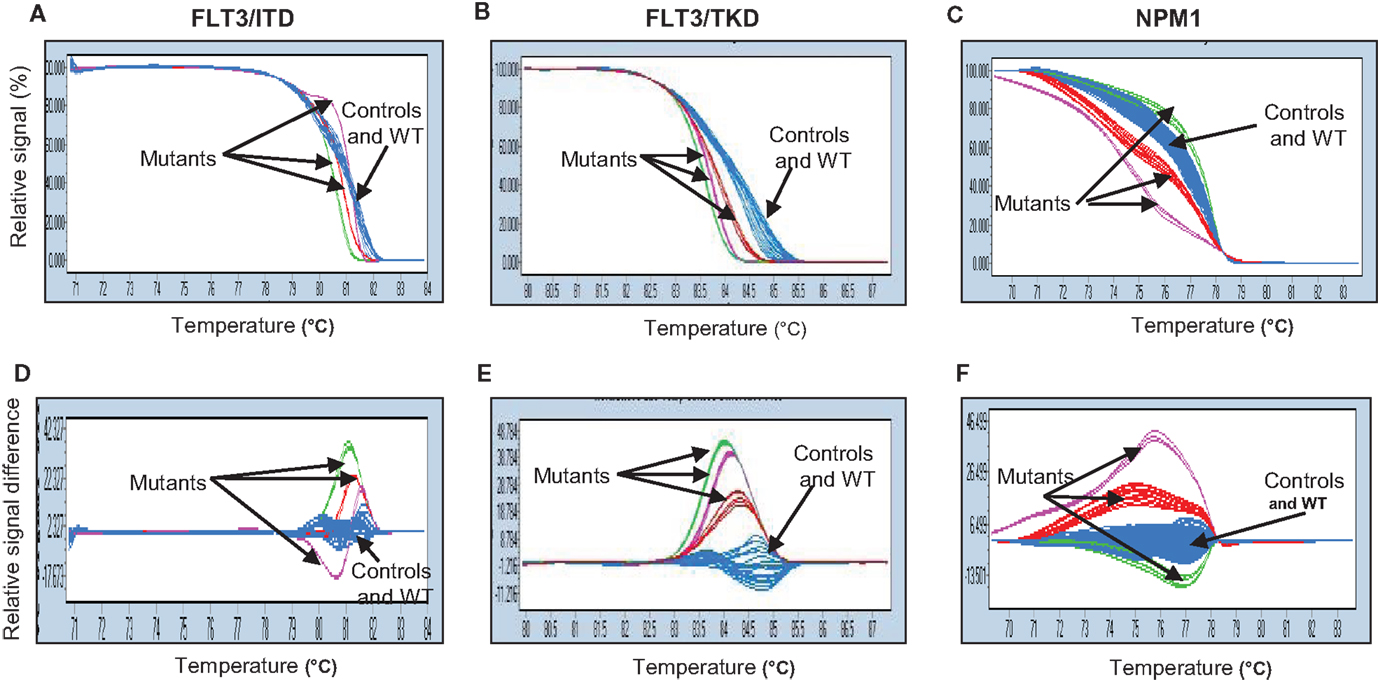
Figure 1. Genetic variation of FLT3-ITD (A,D), FLT3-TKD (B,E), and NPM1 (C,F) genes analyzed by high-resolution melting (HRM) curve analysis. Fragments of three genes amplified by PCR and HRM analysis from cDNA isolated from patients and normal BM samples were shown in two different plots: normalized plot (A–C) and difference plot (D–F). Both plots enable differentiation between control and wild-type samples (blue) and mutants (red, purple, and green). WT, wild-type.
To assess the specificity of the HRM test, we selected nine samples, seven of which tested positive and two negative for FLT3-ITD. Direct Sanger sequencing confirmed 100% consistency. Furthermore, we performed similar validation for the NPM1 gene mutation in six mutants and five wild-type samples. Results were confirmed with 100% consistency when compared to Sanger sequencing (Table 1). Sanger sequencing verification showed that in patients with FLT3-ITD, the size of internal tandem duplication varied from 3 to 66 bp. No difference was observed between the size of the ITD and the results seen in HRM analysis in all the samples analyzed. Sequencing of samples with FLT3-TKD confirmed the most frequent D835Y mutation. The NPM1 mutation involved 4 bp insertions, which altered the tryptophan at amino acid position 288.
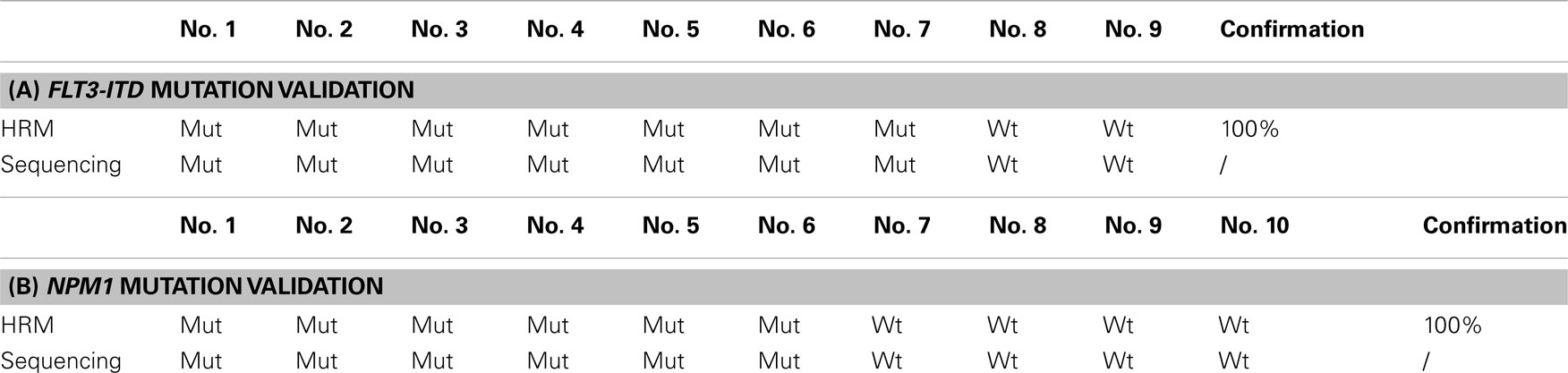
Table 1. Sanger sequencing validation of FLT3-ITD (A) and NPM1 (B) gene mutations detected by HRM analysis using fluorescent dye SYTO-82.
We also tested whether HRM analysis could be used as a method for MRD monitoring. The ability to detect low levels of FLT3-ITD, FLT3-TKD, and NPM1 gene mutations in a background of non-mutated DNA was evaluated by titrating each of the mutant alleles with wild-type DNA to produce a range of the mutant allele dilutions. Unfortunately, the lower limit of detection was 1/10 ~ 1/100 for FLT3-ITD and 1/100 ~ 1/1000 for both FLT3-TKD and NPM1. This suggested that HRM analysis was not sensitive enough for MRD monitoring in this study (Figure 2).
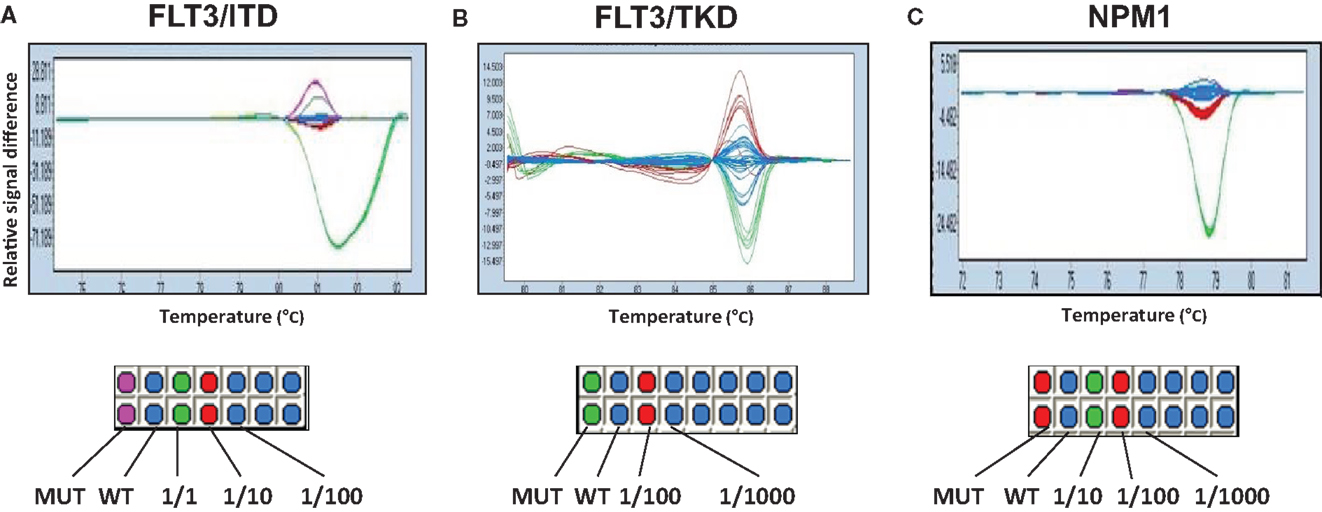
Figure 2. Evaluation of the ability of HRM for MRD detection. Mutant cDNA of FLT3-ITD (A), FLT3-TKD (B), and NPM1 (C) genes was titrated and mixed with different ratio of wild-type cDNA to produce a range of mutant allele dilutions. Fractions show the proportion of mutated/wild-type genes. Color differences demonstrate the identification of sample types. Duplications were made in columns.
Patient Characteristics and Genetic Findings
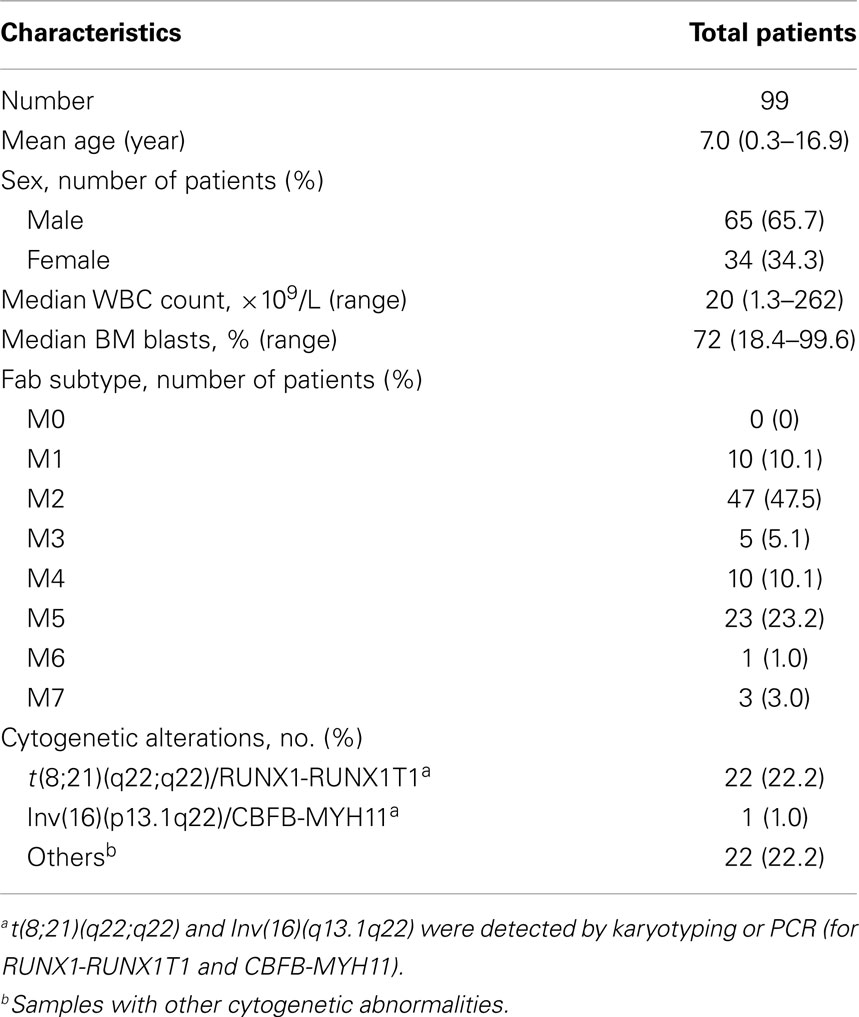
Patient characteristics were summarized in Table 2. The mean age of patients was 7 years (range 0.3–16.9 years). Slightly more males than females were included in the study. Five patients had M3 morphology. However, they were all negative for t(15;17) and PML-RARA. Therefore, they were all treated with AML-XH-99 protocol and were included in the analysis. Among all 99 PML-RARA negative AML patients, 22 (24.2%) were positive for RUNX1-RUNX1T1, and 1 (1.0%) was positive for CBFB–MYH11. Fifty four patients (54.5%) had normal karyotype and were defined as CN-AML.
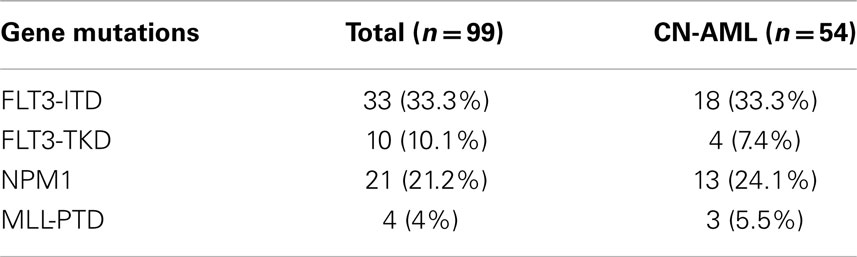
Of the total 99 patients, HRM analysis detected 33 patients with FLT3-ITD mutations, 10 with FLT3-TKD mutations, and 21 with NPM1 mutations. Four patients were positive for MLL-PTD mutations (Table 3) by RQ-PCR. The presence of the mutations was not mutually exclusive. Among the 33 patients with the FLT3-ITD mutations, 4 had NPM1 mutations, and 2 patients had the MLL-PTD mutations. Similarly, among the 10 patients with the FLT3-TKD mutations, 2 of them had NPM1 mutations and 1 had the MLL-PTD mutation. One patient was found to have concurrent FLT3-ITD, FLT3-TKD, and NPM1 mutations. In total, 66% patients had at least one gene mutation.
Of the 54 CN-AML patients, there were 18 (33.3%) FLT3-ITD mutants, 4 (7.4%) FLT3-TKD mutants, 13 (24.1%) NPM1 mutants, and 3 (5.56%) MLL-PTD mutants (Table 3).
Clinical Outcome
The EFS and OS at the median follow-up time of 49.6 months for all patients were 45.2 and 50.0%, respectively. The EFS of patients with FLT3-ITD mutations were significantly worse when compared to that of patients without FLT3-ITD mutations (p = 0.038, Figure 3A). The OS for patients with and without FLT3-ITD mutations were 38 and 55%, respectively. However, the difference was not statistically significant (Figure 3D). There was no significant difference in EFS and OS for patients with or without FLT3-TKD mutations (Figures 3B,E). The EFS and OS for all patients with NPM1 mutations were significantly better than patients without NPM1 mutations (p = 0.01, Figures 3C,F). All four patients with MLL-PTD mutations died from disease.
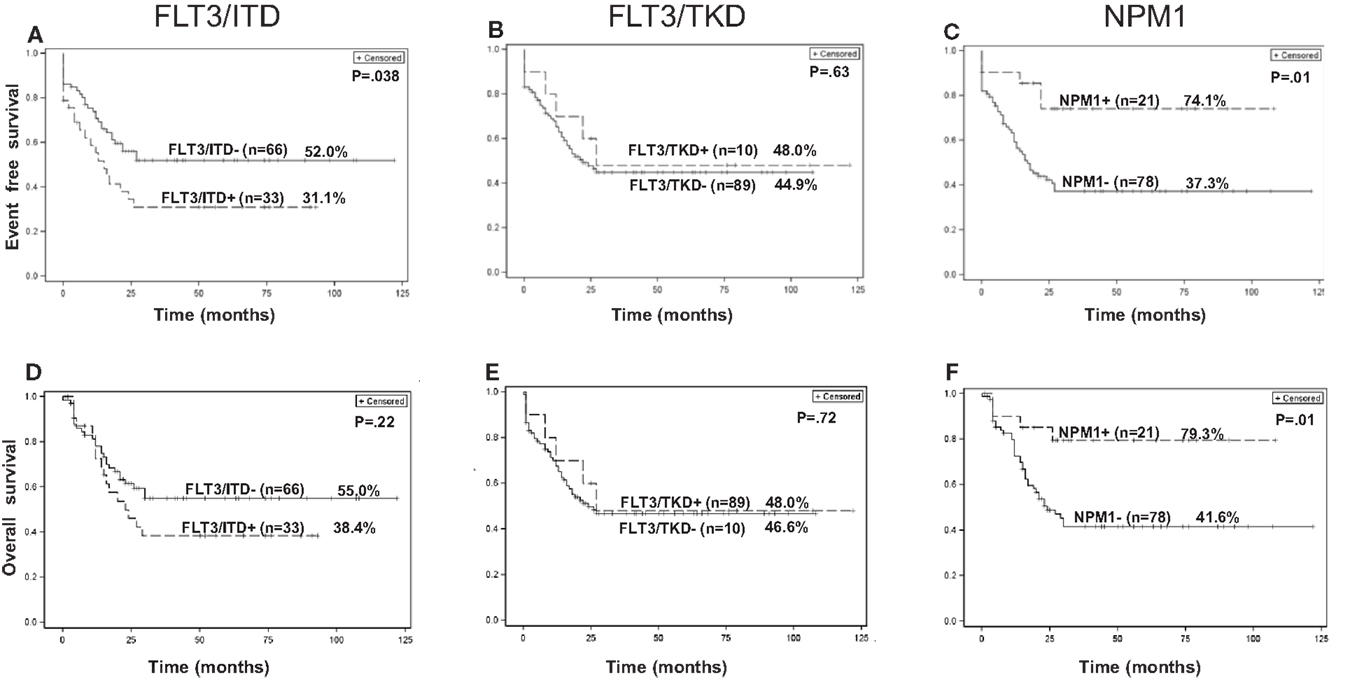
Figure 3. Kaplan–Meier curves of EFS (A–C) and OS (D–F) for all 99 patients with or without FLT3-ITD, FLT3-TKD, and NPM1 gene mutations.
In the CN-AML group of 54 patients, the EFS and OS time were 50 and 53%, respectively. We found no significant difference in EFS and OS for patients with or without FLT3-ITD mutations (Figures S1A,D in Supplementary Material) or FLT3-TKD mutations (Figures S1B,E in Supplementary Material). There is a trend toward better EFS in patients with the NPM1 mutation when compared to patients without the mutation (p = 0.07, Figure S1C in Supplementary Material); OS was significantly better (p = 0.05, Figure S1F in Supplementary Material). All three patients with MLL-PTD mutations in the CN-AML group died of disease progression.
Discussion
In this study, we developed HRM assays for assessment of FLT3-ITD, FLT3-TKD, and NPM1 gene mutations. As previously described, HRM allows closed-tube identification of mutations using a real-time PCR machine now common to most labs. Using the LightCycler® 480 Real-Time PCR System, PCR and analysis can be performed in one instrument. There is also less cDNA processing required, compared to non-homogeneous (gradient or gel-based) mutation screening methods. The PCR was easy to set up and the turn-around time was about 24 h. Fluorescent dye SYTO-82 did not inhibit the PCR, did not affect melting temperature, and was accurate for mutation detection. More importantly, it only cost $0.011 USD/test, a fraction of the cost of Sanger sequencing (25–27, 33).
Previously, Tan et al. (34) studied FLT3 and NPM1 exon 12 mutations in a cohort of 44 adult patients with normal karyotype AML. HRM was used as a screening tool for confirmatory sequencing. In the pediatric samples we tested, HRM demonstrate 100% accuracy when compared to the gold standard Sanger sequencing in the randomly selected samples. This was consistent with the previous reports using HRM to detect mutations in different cancer types (33, 35–38). Compared to Sanger sequencing, the cost of HRM was lower and turn-around time was shorter (33). It was shown that FLT3-ITD with large base pair insertion could be analyzed with agarose gel electrophoresis (39). However, gel electrophoresis will not be able to differentiate mutant from wild-type when inserted fragments are shorter, such as the NPM1 mutations in our study, in particular, most of which have only 4 bp insertions.
High-resolution melting analysis also had its limitations. HRM can only serve as a screening method and it will not give the exact mutational status. The allelic ratio of FLT3-ITD also could not be identified using HRM. Furthermore, as we experienced, this test was not sensitive enough to be used as MRD monitoring. However, HRM is time- and cost-saving, with the major advantage of preventing sample contamination due to the closed tube system. HRM could be a suitable initial clinical screening tool for resource underprivileged countries with large patient populations. Once a mutation is detected, Sanger sequencing could be used as confirmation if necessary. Overall, it can be considered cost-effective in these regions.
Our analysis included 99 children with AML. The frequencies of FLT-ITD, FLT3-TKD, and NPM1 identified in this study are similar to previous reports in the pediatric series (4–8, 11, 12). In our whole cohort of patients, FLT3-ITD mutations were significantly associated with worse EFS. In patients with FLT3-TKD, no significance in clinical outcome was found. These results were also in agreement with other reports (4–8). Similarly, the favorable outcome among patients with the NPM1 mutation in our patient cohort was consistent with previously published data (8, 11, 12).
Regarding the prognostic significance of gene mutations in the CN-AML group, we and others found that FLT3-ITD and NPM1 were still the most common among the mutated genes examined (4–8, 11, 12). However, we failed to find a significant difference in the outcome between CN-AML patients with and without FLT3-ITD mutations, while the survival advantage in patients with NPM1 mutations remained significant. The discrepancy in the outcome of CN-AML patients with FLT3-ITD mutations in our study when compared to published data could be due to some of the patients possibly not having a high FLT3-ITD allelic ratio, or small sample size. In addition, some of the CN-AML patients may not be truly “cytogenetic normal” because FISH was not performed in most of the patients due to the high cost of the test ($350 USD per test).
Our study also established an RQ-PCR analysis for MLL-PTD and identified that 4% of patients harbored MLL-PTD mutations, similar to the published results (13–15). The survival results of this group were extremely poor. All of the patients with MLL-PTD mutations died of recurrent or refractory disease. However, the number is too small to make any meaningful conclusions, and further studies are warranted.
In conclusion, HRM is a fast, cost-effective screening diagnostic method for detection of the gene mutations FLT3-ITD, FLT3-TKD, and NPM1, especially in resource underprivileged countries. The frequencies and outcome prediction values of FLT-ITD, FLT3-TKD, and NPM1 gene mutations identified in this study were similar to previous reports in other pediatric studies. Further studies are planned using Sanger sequencing to validate the HRM analysis in all patient samples in current pediatric AML trials.
Author Contributions
Yin Liu wrote the first draft of this paper. Yin Liu, Jingyan Tang, Shuhong Shen, Peter Gaynon, and Weili Sun were responsible for the design and conception of this manuscript. Peter Wakamatsu contributed for statistical analysis. Jingyan Tang, Huiliang Xue, and Jing Chen were responsible for the provision of patients and data acquisition. Yin Liu and Shuhong Shen performed experiments. All authors provided critical review of the manuscript and approval final version submitted.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The Review Editor Matthew James Oberley declares that, despite being affiliated to the same institution as authors Peter Wakamatsu, Paul S. Gaynon and Weili Sun the review process was handled objectively and no conflict of interest exists.
Acknowledgments
We like to thank Ms. Betty Liu and Ms. Amelia MacRae from Children’s Hospital Los Angeles for reviewing the manuscript. This study was supported by grants from the National Science Foundation of China (Grant number: 30670883) and the Science and Technology Commission of Shanghai (Grant number: 08411953800 and 074119610).
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Journal/10.3389/fped.2014.00096/abstract
Abbreviations
FLT3, FMS-like tyrosine kinase 3; HRM, high-resolution melting curve; ITD, internal tandem duplication; MLL-PTD, partial tandem duplication within the mixed linage leukemia; NPM1, nucleophosmin 1; TKD, tyrosine kinase domain.
References
1. Gamis AS, Alonzo TA, Perentesis JP, Meshinchi S, COG Acute Myeloid Leukemia Committee. Children’s oncology group’s 2013 blueprint for research: acute myeloid leukemia. Pediatr Blood Cancer (2013) 60:964–71. doi:10.1002/pbc.24432
2. Radhi M, Meshinchi S, Gamis A. Prognostic factors in pediatric acute myeloid leukemia. Curr Hematol Malig Rep (2010) 5:200–6. doi:10.1007/s11899-010-0060-z
3. Manola KN. Cytogenetics of pediatric acute myeloid leukemia. Eur J Haematol (2009) 83:391–405. doi:10.1111/j.1600-0609.2009.01308.x
4. Meshinchi S, Alonzo TA, Stirewalt DL, Zwaan M, Zimmerman M, Reinhardt D, et al. Clinical implications of FLT3 mutations in pediatric AML. Blood (2006) 108:3654–61. doi:10.1182/blood-2006-03-009233
5. Meshinchi S, Woods WG, Stirewalt DL, Sweetser DA, Buckley JD, Tjoa TK, et al. Prevalence and prognostic significance of Flt3 internal tandem duplication in pediatric acute myeloid leukemia. Blood (2001) 97:89–94. doi:10.1182/blood.V97.1.89
6. Kottaridis PD, Gale RE, Frew ME, Harrison G, Langabeer SE, Belton AA, et al. The presence of a FLT3 internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: analysis of 854 patients from the United Kingdom Medical Research Council AML 10 and 12 trials. Blood (2001) 98:1752–9. doi:10.1182/blood.V98.6.1752
7. Zwaan CM, Meshinchi S, Radich JP, Veerman AJ, Huismans DR, Munske L, et al. FLT3 internal tandem duplication in 234 children with acute myeloid leukemia: prognostic significance and relation to cellular drug resistance. Blood (2003) 102:2387–94. doi:10.1182/blood-2002-12-3627
8. Brown P, McIntyre E, Rau R, Meshinchi S, Lacayo N, Dahl G, et al. The incidence and clinical significance of nucleophosmin mutations in childhood AML. Blood (2007) 110:979–85. doi:10.1182/blood-2007-02-076604
9. Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med (2005) 352:254–66. doi:10.1056/NEJMoa041974
10. Cazzaniga G, Dell’Oro MG, Mecucci C, Giarin E, Masetti R, Rossi V, et al. Nucleophosmin mutations in childhood acute myelogenous leukemia with normal karyotype. Blood (2005) 106:1419–22. doi:10.1182/blood-2005-03-0899
11. Thiede C, Creutzig E, Reinhardt D, Ehninger G, Creutzig U. Different types of NPM1 mutations in children and adults: evidence for an effect of patient age on the prevalence of the TCTG-tandem duplication in NPM1-exon 12. Leukemia (2007) 21:366–7. doi:10.1038/sj.leu.2404519
12. Braoudaki M, Papathanassiou C, Katsibardi K, Tourkadoni N, Karamolegou K, Tzortzatou-Stathopoulou F. The frequency of NPM1 mutations in childhood acute myeloid leukemia. J Hematol Oncol (2010) 3:41. doi:10.1186/1756-8722-3-41
13. Shimada A, Taki T, Tabuchi K, Taketani T, Hanada R, Tawa A, et al. Tandem duplications of MLL and FLT3 are correlated with poor prognoses in pediatric acute myeloid leukemia: a study of the Japanese childhood AML Cooperative Study Group. Pediatr Blood Cancer (2008) 50:264–9. doi:10.1002/pbc.21318
14. Steudel C, Wermke M, Schaich M, Schakel U, Illmer T, Ehninger G, et al. Comparative analysis of MLL partial tandem duplication and FLT3 internal tandem duplication mutations in 956 adult patients with acute myeloid leukemia. Genes Chromosomes Cancer (2003) 37:237–51. doi:10.1002/gcc.10219
15. Schnittger S, Kinkelin U, Schoch C, Heinecke A, Haase D, Haferlach T, et al. Screening for MLL tandem duplication in 387 unselected patients with AML identify a prognostically unfavorable subset of AML. Leukemia (2000) 14:796–804. doi:10.1038/sj.leu.2401773
16. Meshinchi S, Stirewalt DL, Alonzo TA, Boggon TJ, Gerbing RB, Rocnik JL, et al. Structural and numerical variation of FLT3-ITD in pediatric AML. Blood (2008) 111:4930–3. doi:10.1182/blood-2008-01-117770
17. Abu-Duhier FM, Goodeve AC, Wilson GA, Gari MA, Peake IR, Rees DC, et al. FLT3 internal tandem duplication mutations in adult acute myeloid leukaemia define a high-risk group. Br J Haematol (2000) 111:190–5. doi:10.1046/j.1365-2141.2000.02317.x
18. Kang HJ, Hong SH, Kim IH, Park BK, Han KS, Cho HI, et al. Prognostic significance of FLT3 mutations in pediatric non-promyelocytic acute myeloid leukemia. Leuk Res (2005) 29:617–23. doi:10.1016/j.leukres.2004.11.006
19. Mead AJ, Linch DC, Hills RK, Wheatley K, Burnett AK, Gale RE. FLT3 tyrosine kinase domain mutations are biologically distinct from and have a significantly more favorable prognosis than FLT3 internal tandem duplications in patients with acute myeloid leukemia. Blood (2007) 110:1262–70. doi:10.1182/blood-2006-04-015826
20. Liu Q, Wang B, Kong Y, Cheng KK. China’s primary health-care reform. Lancet (2011) 377:2064–6. doi:10.1016/S0140-6736(11)60167-0
21. Wagstaff A, Moreno-Serra R. Social health insurance and labor market outcomes: evidence from central and Eastern Europe, and Central Asia. Adv Health Econ Health Serv Res (2009) 21:83–106.
22. Cordell JL, Pulford KA, Bigerna B, Roncador G, Banham A, Colombo E, et al. Detection of normal and chimeric nucleophosmin in human cells. Blood (1999) 93:632–42.
23. Whitman SP, Liu S, Vukosavljevic T, Rush LJ, Yu L, Liu C, et al. The MLL partial tandem duplication: evidence for recessive gain-of-function in acute myeloid leukemia identifies a novel patient subgroup for molecular-targeted therapy. Blood (2005) 106:345–52. doi:10.1182/blood-2005-01-0204
24. Yamamoto Y, Kiyoi H, Nakano Y, Suzuki R, Kodera Y, Miyawaki S, et al. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood (2001) 97:2434–9. doi:10.1182/blood.V97.8.2434
25. Montgomery JL, Sanford LN, Wittwer CT. High-resolution DNA melting analysis in clinical research and diagnostics. Expert Rev Mol Diagn (2010) 10:219–40. doi:10.1586/erm.09.84
26. Taylor CF. Mutation scanning using high-resolution melting. Biochem Soc Trans (2009) 37:433–7. doi:10.1042/BST0370433
27. Erali M, Voelkerding KV, Wittwer CT. High resolution melting applications for clinical laboratory medicine. Exp Mol Pathol (2008) 85:50–8. doi:10.1016/j.yexmp.2008.03.012
28. Chen D, Wang YY, Chuai ZR, Huang JF, Wang YX, Liu K, et al. High-resolution melting analysis for accurate detection of BRAF mutations: a systematic review and meta-analysis. Sci Rep (2014) 4:4168. doi:10.1038/srep04168
29. Feldman E, Arlin Z, Ahmed T, Mittelman A, Puccio C, Chun H, et al. Homoharringtonine in combination with cytarabine for patients with acute myelogenous leukemia. Leukemia (1992) 6:1189–91.
30. Tang J, Liu Y, Chen J, Xue H, Pan C, Gu L. Homoharringtonine as a backbone drug for the treatment of newly diagnosed pediatric acute myeloid leukemia: a report from a single institution in China. Int J Hematol (2011) 93:610–7. doi:10.1007/s12185-011-0837-4
31. Picard C, Silvy M, Gabert J. Overview of real-time RT-PCR strategies for quantification of gene rearrangements in the myeloid malignancies. Methods Mol Med (2006) 125:27–68.
32. Eischeid AC. SYTO dyes EvaGreen outperform SYBR green in real-time PCR. BMC Res Notes (2011) 4:263. doi:10.1186/1756-0500-4-263
33. Ihle MA, Fassunke J, Konig K, Grunewald I, Schlaak M, Kreuzberg N, et al. Comparison of high resolution melting analysis, pyrosequencing, next generation sequencing and immunohistochemistry to conventional Sanger sequencing for the detection of p.V600E and non-p.V600E BRAF mutations. BMC Cancer (2014) 14:13. doi:10.1186/1471-2407-14-13
34. Tan AY, Westerman DA, Carney DA, Seymour JF, Juneja S, Dobrovic A. Detection of NPM1 exon 12 mutations and FLT3 – internal tandem duplications by high resolution melting analysis in normal karyotype acute myeloid leukemia. J Hematol Oncol (2008) 1:10. doi:10.1186/1756-8722-1-10
35. Kunstlinger H, Binot E, Merkelbach-Bruse S, Huss S, Wardelmann E, Buettner R, et al. High-resolution melting analysis is a sensitive diagnostic tool to detect imatinib-resistant and imatinib-sensitive PDGFRA exon 18 mutations in gastrointestinal stromal tumors. Hum Pathol (2014) 45:573–82. doi:10.1016/j.humpath.2013.10.025
36. Akiyoshi K, Yamada Y, Honma Y, Iwasa S, Kato K, Hamaguchi T, et al. KRAS mutations in patients with colorectal cancer as detected by high-resolution melting analysis and direct sequencing. Anticancer Res (2013) 33:2129–34.
37. Huet S, Jallades L, Charlot C, Chabane K, Nicolini FE, Michallet M, et al. New quantitative method to identify NPM1 mutations in acute myeloid leukaemia. Leuk Res Treatment (2013) 2013:756703. doi:10.1155/2013/756703
Keywords: FLT3, NPM1, MLL, high-resolution melting curve analysis, acute myeloid leukemia, childhood
Citation: Liu Y, Tang J, Wakamatsu P, Xue H, Chen J, Gaynon PS, Shen S and Sun W (2014) High-resolution melting curve analysis, a rapid and affordable method for mutation analysis in childhood acute myeloid leukemia. Front. Pediatr. 2:96. doi: 10.3389/fped.2014.00096
Received: 20 July 2014; Accepted: 24 August 2014;
Published online: 09 September 2014.
Edited by:
Mignon L. Loh, University of California, San Francisco, USAReviewed by:
Ulrike Bacher, MLL Munich Leukemia Laboratory, GermanyMatthew James Oberley, University of Southern California, USA
Copyright: © 2014 Liu, Tang, Wakamatsu, Xue, Chen, Gaynon, Shen and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuhong Shen, Department of Hematology/Oncology, Shanghai Children’s Medical Center, Shanghai Jiaotong University School of Medicine, 1678 Dongfang Road, Shanghai 200127, China e-mail: shenshuhong@scmc.com.cn
 Yin Liu
Yin Liu Jingyan Tang1
Jingyan Tang1
 Paul S. Gaynon
Paul S. Gaynon Shuhong Shen
Shuhong Shen Weili Sun
Weili Sun