- 1 Integrated Toxicology Division, Department of Biochemistry and Cell Biology, United States Army Medical Research Institute of Infectious Diseases, Fort Detrick, MD, USA
- 2 Synaptic Research LLC, Baltimore, MD, USA
- 3 Office of the Chief Scientist, Fort Detrick, MD, USA
Botulinum neurotoxins are most potent of all toxins. Their N-terminal light chain domain (Lc) translocates into peripheral cholinergic neurons to exert its endoproteolytic action leading to muscle paralysis. Therapeutic development against these toxins is a major challenge due to their in vitro and in vivo structural differences. Although three-dimensional structures and reaction mechanisms are very similar, the seven serotypes designated A through G vastly vary in their intracellular catalytic stability. To investigate if protein phosphorylation could account for this difference, we employed Src-catalyzed tyrosine phosphorylation of the Lc of six serotypes namely LcA, LcB, LcC1, LcD, LcE, and LcG. Very little phosphorylation was observed with LcD and LcE but LcA, LcB, and LcG were maximally phosphorylated by Src. Phosphorylation of LcA, LcB, and LcG did not affect their secondary and tertiary structures and thermostability significantly. Phosphorylation of Y250 and Y251 made LcA resistant to autocatalysis and drastically reduced its kcat/Km for catalysis. A tyrosine residue present near the essential cysteine at the C-terminal tail of LcA, LcB, and LcG was readily phosphorylated in vitro. Inclusion of a competitive inhibitor protected Y426 of LcA from phosphorylation, shedding light on the role of the C-terminus in the enzyme’s substrate or product binding.
Introduction
Botulinum neurotoxins (BoNTs)1 are among the most lethal of all toxins and are also potential biowarfare agents (Cochrane, 1947; Gill, 1982; Montecucco and Schiavo, 1995; Arnon et al., 2001). Enormous efforts are devoted toward therapeutic development against these targets (see examples in the references (Schmidt and Stafford, 2005; Hines et al., 2008; Zuniga et al., 2008; Agarwal et al., 2009; Burnett et al., 2009; Capkova et al., 2009; Ludivico et al., 2009; Pang et al., 2009; Silhar et al., 2010; Hale et al., 2011). A thorough understanding of their structure under cellular environment is therefore essential.
These 150 kDa exotoxins are produced by strains of Clostridium botulinum as seven distinct serotypes, designated BoNT/A-G. After finding their way through oral, respiratory, or wound routes to the animal body, BoNTs travel to peripheral cholinergic neuronal cells and are internalized by endocytosis followed by translocational delivery of its 50 kDa light chain (Lc) from endosome into the cytosol (Simpson, 2004). The free Lc, a zinc-endopeptidase (Schiavo et al., 1992a), exerts its proteolytic activity at specific sites on one of the three synaptosomal proteins, SNAP-25, VAMP, or syntaxin. For example, the Lc of BoNT/A (LcA) and BoNT/E (LcE) cleave at specific but different sites on SNAP-25 (Schiavo et al., 1993), while Lc of BoNT/B (LcB) and BoNT/D (LcD) cleaves at distinct but different sites on VAMP (Schiavo et al., 1992b). The process prevents fusion of synaptosomes with the cell membrane, blocking acetylcholine release into the neuromuscular junction. The resultant effect is muscle paralysis and eventual death if the intoxication is severe and not treated.
The overall polypeptide fold and three-dimensional structures of all BoNT serotypes are almost identical (Lacy et al., 1998; Eswaramoorthy et al., 2002; Agarwal et al., 2004, 2005; Arndt et al., 2005, 2006; Jin et al., 2007; Kumaran et al., 2008a), and they appear to follow the same reaction mechanism using the conserved active site residues (Agarwal et al., 2004; Swaminathan et al., 2004; Kumaran et al., 2008a). In spite of their in vitro structural and mechanistic identity, duration of BoNT serotype-dependent paralysis in animal cells varies widely. For example, in humans BoNT/A-induced paralysis can persist for more than a year (Souayah et al., 2006). BoNT protease activity and toxicity in rat cerebellar and mouse spinal cord neurons lasts from 3 months for BoNT/A to less than a day for BoNT/E (Keller et al., 1999; Foran et al., 2003). Thus, the structure of the Lc domain inside neurons can be expected to vary widely to account for these differences. One possible source of structural variations may be through post-translocational modifications that include phosphorylation, palmitoylation, and ubiquitination, among others (Walsh, 2006a).
Post-translational modification of proteins in eukaryotic cells can generate 10 to 100-fold more variants than the 30,000 protein products of the human genome, imparting the same protein with various regulatory, secretory, catalytic, and structural functions (Walsh, 2006a). For example, SNAP-25 (Nagy et al., 2004), VAMP (Nielander et al., 1995), and syntaxin (Foster et al., 1998) among hundreds of synaptosomal proteins in neuronal cells undergo phosphorylation at specific sites (Munton et al., 2007) in performing and regulating various functions. Because the prokaryotic BoNT Lc exerts its catalytic action on synaptosomal proteins and survives within the eukaryotic neurons for extended times (Keller et al., 1999; Souayah et al., 2006), it might be subjected to phosphorylation. Indeed, in an attractive demonstration, Ferrer-Montiel et al. (1996) reported that BoNT/A Lc undergoes tyrosine phosphorylation inside PC12 cells and in vitro (Encinar et al., 1998; Blanes-Mira et al., 2001; Ibanez et al., 2004). The non-receptor tyrosine kinase, Src and PYK2 that are abundant in these cells was efficient and specific in this phosphorylation reaction. Src-catalyzed phosphorylation of LcA displayed elevated proteolytic activity and thermal stability. Because both Src and PYK2 are highly abundant in brain and neuroendocrine cells, it was postulated that tyrosine phosphorylation may modulate Lc activity within neurons (Ferrer-Montiel et al., 1996). This ex vivo demonstration of Lc phosphorylation has not been verified independently.
In this paper, we report our in vitro investigation of tyrosine phosphorylation of eight versions of Lc from the six serotypes of BoNT/A, BoNT/B, BoNT/C1, BoNT/D, BoNT/E, and BoNT/G. Our results showed while LcA, LcB, LcC1, and LcG were readily phosphorylated, LcD and LcE were poorly phosphorylated. One tyrosine residue near the interchain disulfide-forming cysteine at the C-terminus was phosphorylated in LcA, LcB, and LcG. In addition, two C-terminally truncated LcA forms, LcA420, and LcA424, were also phosphorylated. Phosphorylation of LcA was accompanied by loss of catalytic activity, without changes in secondary and tertiary structures and thermal denaturation but made it highly resistant to autocatalytic degradation due to phosphorylation of two tyrosine residues at the autocatalytic site. Phosphorylation of LcB marginally increased thermal stability and catalytic activity.
Experimental Procedures
Materials
Recombinant BoNT protease light chains of serotype A (LcA) and serotype B (LcB) were purified as described (Ahmed et al., 2003; Jensen et al., 2003; Gilsdorf et al., 2006), and similar purification of serotypes D (LcD), and G will be published elsewhere; LcE was purchased from BBtech (MA). Truncated versions of LcA containing the first 420 (LcA420; Segelke et al., 2004) and 424 residues (LcA424; Kumaran et al., 2008a), and of LcC1 containing the first 430 residues (Rawat et al., 2008) were purified as described. Although the full-length LcA used here contains an extra valine after the initial methionine at the N-terminus, residue numbering used in this manuscript are according to GenBank ID LcA1: AAQ06331; LcB1:BAE48264; LcC1: CAA44263; LcD1: AAB24244; LcE1: BAB86845; LcF1:ADA79551; LcG: CAA52275. Before the phosphorylation reaction, each light chain (0.5–1 mg/ml), with the exception of LcA, was brought to room temperature and then incubated with 0.25 mM ZnCl2 on ice for 30 min, followed by gel-filtration on a PD-10 column in 10 mM Na-phosphate, pH 7.4. LcA was directly applied to a PD-10 column without pre-treatment with ZnCl2. Src (1255 units/mg, 0.1 mg/ml), monoclonal anti-Src (GD11 and 4G10) antibody kits, and Western blot reagents were from Millipore, Billerica, MA, USA (previously Upstate Biotechnology Inc. Temecula, CA, USA). Sequence-derived substrates from SNAP-25 for LcA (SNKTRIDEANQ-RATKML), and from VAMP for LcB (LSELDDRADALQAGASQ-FETSAAKLKRKYWWKNLK; Foran et al., 1994), and another VAMP sequence-derived substrate peptide for LcD (LQQTQAQVDEVVDIMRVNVDKVLERDQK-LSELDD; Rowe et al., 2010), all having N-terminal acetylated and C-terminal amidated, were custom-synthesized and purified to >95% by Quality Controlled Biochemicals (Hampton, MS, USA).
Phosphorylation Reaction
Phosphorylation reaction was performed according to the manufacturer’s protocol. Briefly, LcA (0.55 mg/ml), LcA424 (0.28 mg/ml), LcA420 (0.18 mg/ml), LcB (0.17 mg/ml, LcC (0.49 mg/ml), LcD (0.29 mg/ml), LcE (0.02 mg/ml), or LcG (0.27 mg/ml) was incubated on ice with 20 mM HEPES pH 7.4, 2 mM DTT (final concentrations) followed by addition of 1 mM EGTA, 20 mM MgCl2, 2 mM ATP, and three units of Src (0.025 mg/ml) in a total reaction volume of 0.1 ml. Concentration of the reagents, and the ratio of Lc (mg) to Src (units) were same as in the literature except that 2 mM used here was higher than 0.5 mM (Ferrer-Montiel et al., 1998; Blanes-Mira et al., 2001; Ibanez et al., 2004). The final reaction mixtures were incubated at 30°C (20°C for LcA) for 24 h. For mass spectrometric analysis, 6 μl of the reaction mixture was removed at various times and 6 μl of 0.5% trifluoroacetic acid was added to each aliquot. For SDS-PAGE and Western blot analyses, 12 μl of 2× SDS-load buffer was added to 6 μl aliquot of the stopped reaction mixture. Duplicate SDS-PAGE gels were run simultaneously, one for protein visualization by Coomassie stain, the other for Western blot using horseradish peroxide-anti-phosphotyrosine antibody. For catalytic activity measurements, the reaction mixture was diluted with ice-cold 50 mM HEPES pH 7.4 for immediate activity determination by UPLC. Alternately, in large-scale preparations, phosphorylation reaction was stopped by removing the Src with sepharose beads containing monoclonal anti-Src antibody. Unless otherwise stated, Western blot bands were visualized using electrochemical luminescent stain.
Preparative phosphorylation utilized LcA (0.58 mg/ml), LcB (0.9 mg/ml), or LcG (1.5 mg/ml) in volumes of 1.5–3.0 ml containing all other components. Aliquots were tested for completion of phosphorylation at intervals from 15 min to 48 h for Western blot and mass spectrometric analyses. If the phosphorylation reaction was incomplete, incubation was continued for up to 5 days with additional Src and ATP. The reactions were stopped by addition of monoclonal anti-Src-agarose followed by storage at 4°C for 24 h. Phosphorylated product in the soluble fraction was passed through a PD-10 column and collected in 50 mM Na-phosphate buffer, pH 6.8. The dilute protein fractions were combined, concentrated on Centricon-10 microconcentrators, washed, and collected in the same buffer for further analyses.
The standard tyrosine phosphorylation reaction component of the Src supplier included 20 mM MgCl2 and 20 mM Tris-HCl, pH 7.4. We replaced Tris with HEPES in the assays because it gave optimum Lc catalytic activity while the manufacturer suggested Tris-HCl is inhibitory to Lc enzyme activity (Ahmed and Smith, 2000). After 3–24 h incubation with tyrosine kinase Src, Coomassie staining of the proteins after SDS-PAGE showed (Figure 1) all BoNT serotypes remained stable in retaining their linear structures in the Src reaction mixture (0.1 ml). In large-scale phosphorylation reactions using LcB and LcG (∼3 mg, 2 ml), the proteins remained soluble after 48 h of incubation at 30°C. Large-scale (2 mg, 2 ml) reactions with LcA using standard conditions resulted in precipitation of most of the protein within 24 h. Close observation revealed the appearance of fine LcA precipitate soon after start of the incubation, similar to the one observed by gentle stirring at a lower temperature and at various protein concentrations (Toth et al., 2009). Most of the remaining soluble protein showed autocatalyzed fragmentation products, most likely due to presence of the divalent Mg++ ion (Ahmed et al., 2004) and a prolonged incubation temperature of 30°C (Ahmed et al., 2001, 2003, 2004). We therefore excluded ZnCl2 pre-treatment of LcA and varied the temperature of Src reaction incubation. We found that incubation at 20°C for up to 120 h prevented LcA precipitation and autocatalytic fragmentation. Therefore, a preparative amount of LcA-phos was prepared by incubating 3 ml (1.6 mg) LcA with 0.02 mg Src at 20°C for 48 h. Because the enzymatic tyrosine phosphorylation activity of Src varied from batch to batch, its concentration in the reaction mixture was adjusted between experiments to yield saturating phosphorylation as monitored by ESI-MS at various time points (see later).
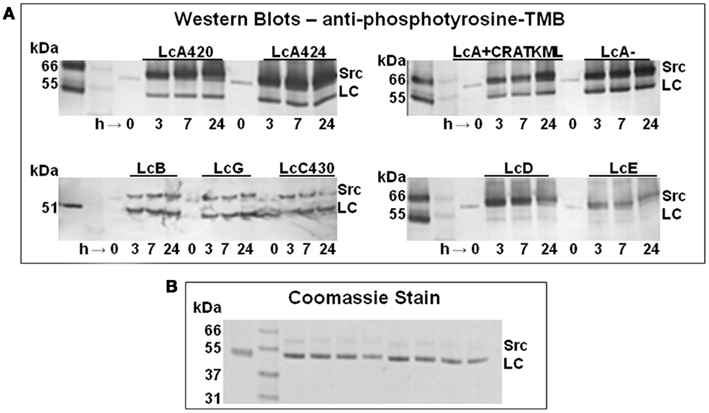
Figure 1. (A) Anti-phosphotyrosine Western blot of the time course of tyrosine phosphorylation of a truncated LcA having residues 1–420 (LcA420), another truncated version of LcA having residues 1–424 (LcA424), full-length LcA alone (LcA) or in presence of 200 μM of an inhibitor (LcA + CRATKML), LcB, LcG, a truncated version of LcC1 having residues 1–430, LcD, and LcE. Each sample was incubated with three units of Src for 3, 7, and 24 h at 30°C. Unlabeled lanes on the far left left of each panel contain molecular mass marker proteins. (B) Coomassie staining of LcA420, LcA424, LcA−, LcA+, LcB, LcG, LcC, and LcD after 24 h incubation are shown in this order on the right of molecular mass marker lane 2; LcA control is on the left, at lane 1.
Molecular mass of Lc target proteins, ∼51 kDa, was close to that of the Src of ∼60 kDa, and Src underwent self-phosphorylation. These two factors may lead to an incorrect interpretation of the Western blot results. We therefore continued electrophoresis (120 V, 90 mAmp) for a minimum of 1.5 h to clearly separate the two proteins (Figure 1A).
UV-Visible Absorption, Circular Dichroism, and Fluorescence Measurements
To determine protein concentration and to assess purity, UV-visible absorption spectra were recorded at 22°C with a Hewlett-Packard 8452 diode array spectrophotometer. Lc concentration was determined using A0.1% (1 cm path length) value of 1.0 at 278 nm (Ahmed et al., 2001) or by BCA assay (Pierce) with BSA as standard. Both methods gave the same result.
Circular dichroism spectra were recorded at 20°C with a Jasco 718 spectropolarimeter with quartz cuvettes of 2 mm path length. An average of five scans was recorded to increase signal-to-noise ratio at a scan speed of 20 nm/min with a response time of 8 s. In all measurements a buffer blank was recorded separately and subtracted from sample recordings.
Tryptophan fluorescence emission spectra were recorded at 20°C in a PTI QuantaMaster Spectrofluorimeter, Model RTC 2000 equipped with a Peltier controlled thermostat and Felix software package. Emission and excitation slit widths were set at 1 nm and excitation wavelength at 295 nm. Each spectrum was an average of five scans.
Mass Spectrometry
Tryptic peptide data were acquired using a Finnigan LCQ quadrupole ion trap mass spectrometer (Thermo Electron Corporation, San Jose, CA, USA) equipped with a New Objective nanoelectrospray source (Woburn, MA, USA) and an Agilent 1100 nanoHPLC system (San Jose, CA, USA). Eight microliters of each tryptic digest were injected onto a New Objective capillary LC column (10 cm × 75 μm) packed with Biobasic C18 (5 μm particle size, 300 Å pore size). Peptides were eluted using a flow rate of 500 nL/min with flow splitting and the following linear gradient: 0–80% B in 60 min. Solvents A and B consisted of 0.1% formic acid and 90% acetonitrile in 0.1% formic acid, respectively. The electrospray voltage was 2.0 kV. The mass spectrometer was operated using data-dependent MS/MS acquisition.
Phosphorylation sites were identified using Bioworks 3.2 software (Thermo Electron Corporation, San Jose, CA, USA) and the NCBInr database (13Oct06). Identified peptides were further evaluated using a C. botulinum taxonomy filter and charge state versus cross-correlation scores (XCorr). The XCorr criteria for positive identification of tryptic peptides were values >1.9 for singly charged ions, values >2.2 for doubly charged ions, and values >2.9 for triply charged peptides.
Enzymatic Activity Assays
Activity assays were based on UPLC separation and measurement of the cleaved products from a 17-residue SNAP-25 peptide for LcA, 35-residue VAMP peptide for LcB, and 34-residue VAMP peptide for LcD (Rowe et al., 2010). A master reaction mixture lacking the Lc was prepared and aliquots were stored at –20°C. Stocks of 0.05–0.07 mg/ml Lc in 50 mM Na-HEPES, pH 7.4 containing 0.05% Tween-20 were stored at -20°C. Before assay, a Lc stock was thawed and diluted further in 50 mM HEPES, pH 7.4 containing bovine serum albumin (BSA). At the time of assay, 5 μl of the diluted LC was added to 25 μl of the thawed master mix to initiate the enzymatic reaction. Components and final concentration in this 30 μl reaction mixture were 0.9 mM substrate peptide, 0.2 mg/ml BSA, 0.0026 mg/ml LC, 0.25 mM ZnCl2, 5 mM dithiothreitol, and 50 mM Na-HEPES, pH 7.4. After 5–10 min at 37°C, the reactions were stopped by adding 90 μl of 1% trifluoroacetic acid.
The amounts of uncleaved substrate and the products were measured after separation by a Waters Acquity UPLC system equipped with Empower Pro software employing a reverse-phase C18 column (2.1 mm × 50 mm, 1.7-μm particle size) with 0.1% trifluoroacetic acid as solvent A and 70% acetonitrile/0.1% trifluoroacetic acid as solvent B at a flow rate of 0.5 ml/min (Rowe et al., 2010). LcA and LcC1 substrate and products were resolved by UPLC with a 0–42% gradient of the solvent B over 2 min, followed by column regeneration for 0.7 min. LcB substrate and products were resolved by UPLC with a 0–100% gradient of the solvents over 2 min, held at 100% B for 0.5 min, followed by column regeneration for 0.5 min (Rowe et al., 2010). LcD substrate and products were resolved by UPLC with a 10–25% B over 1 min, 25–55% B for 0.5 min, held at 55% B for 10 s, 100% B for 1.1 min, followed by column regeneration for 0.7 min (Rowe et al., 2010).
Results
Tyrosine Phosphorylation of BoNT Lcs
To understand structural and environmental factors affecting the stability of BoNT Lcs (Ahmed et al., 2001, 2003, 2004; Toth et al., 2009), we investigated in vitro tyrosine phosphorylation of LcA (including LcA424 and LcA420), LcB, LcC, LcD, LcE, and LcG by Src. Because purified LcF was not available to us, we could not investigate this light chain. Phosphorylation was monitored by Western blot probed with anti-phosphotyrosine antibody (Figure 1A). LcA, LcB, LcC, and LcG were rapidly phosphorylated within 3 h. LcB and LcG phosphorylation reached a maximum within first 3 h of incubation, but intensity of the phosphorylated LcA band continued to increase with incubation time, probably because it had more than one phosphorylation site (see later). Bands of reacted LcD and LcE, on the other hand, were very faint to undetectable, suggesting these Lc were not good substrates for phosphorylation by Src-kinase. Incubations of Lcs for up to 24 h did not appear to degrade the proteins by autocatalysis (Ahmed et al., 2003), as indicated by similar intensity of Coomassie-stained bands before and after incubation (Figure 1B). Long incubation times were chosen to ensure completion of the reactions of very low concentrations (3∼30 μM) of the Lc substrates employed in these experiments which could be much lower than their Km for Src; a few cases where protein substrate Km for Src was determined by others were in the millimolar range. Moreover, quantitative kcat value (0.7/s) available in literature for a Src places it as one of the poorest enzyme catalysts.
Two C-terminally truncated versions of LcA, both lacking a particular tyrosine residue, Y426, were included in our experiments. Like the full-length LcA the truncated versions, LcA420 and LcA424, were phosphorylated within 3 h. To find out if an active site ligand would affect phosphorylation, we also included a competitive peptide inhibitor, CRATKML (Schmidt and Stafford, 2002) with the full-length LcA. The Western blot results (Figure 1A) could not differentiate the rate of phosphorylation from control by the inhibitor, although MS analyses (see later) clearly identified a reduction in phosphorylation.
ESI-MS analysis of time course samples indicated a progressive decrease in the molecular ion peak of unmodified LcB with concomitant increase in a molecular mass 80 Da higher representing a single tyrosine residue phosphorylation. Similarly ESI-MS analysis of LcG and LcA reactions showed increasing molecular ion peaks 160 and 240 Da greater (not shown) than unmodified Lc, characteristic for two and three tyrosine residue phosphorylations, respectively; this difference is most likely due to 1, 2, and multiple sites of phosphorylation in these three light chains respectively (see Table 2, later). LcC1 showed an increase of 80 Da for monophosphorylation, and a minor component for diphosphorylation. A representative spectrum of LcB-phos in Figure 2A, inset shows a gain of 2 amu in the three peaks (1590/1592, 1641/1643, and 1696/1698) from their unphosphorylated forms. The plot of the extent of phosphorylation versus time showed more than 90% LcB was phosphorylated within 8 h. Phosphorylation reaction of LcB when followed by Western blot also showed a similar time-dependent increase in intensity (Figure 2B). Similar results were also observed for LcA (see later.)
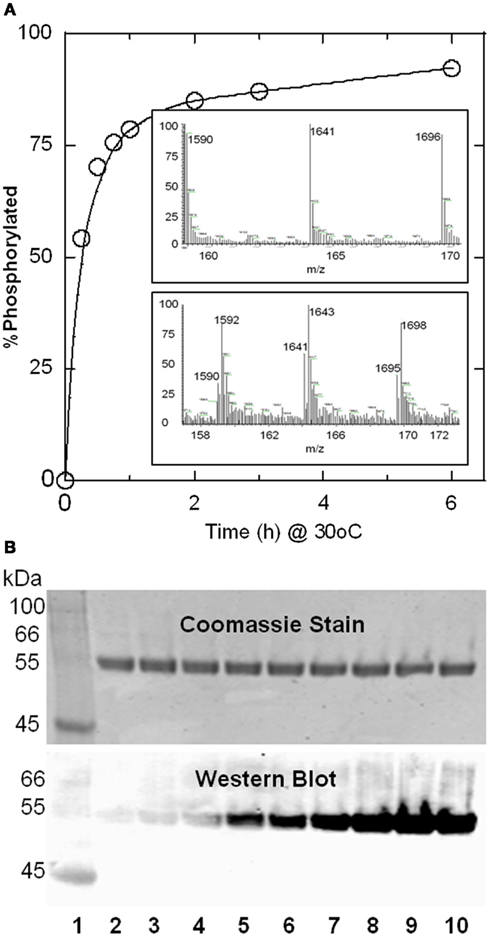
Figure 2. Progress of Src-catalyzed phosphorylation of LcB as followed by mass spectrometry (A) anti-phosphotyrosine antibody Western blot (B). (A) Relative abundance of the m/z species representing phosphorylated LcB was plotted as a% of total LcB (phosphorylated + unphosphorylated) as a function of time. Upper and lower panels in the inset show representative LC-ESI-MS spectra for the LcB phosphorylation reaction product at 0 and 4 h, respectively, showing the +32, +31, and +30 ion peaks of LcB (1590, 1641, 1696), and +32, +31, and +30 ion peaks of LcB-phos corresponds to 1592, 1643, 1698, respectively. LC-ESI-MS was an effective tool to evaluate the degree of phosphorylation. Additionally, deconvolution of the ion peaks gave molecular weights for LcB as 50844.9 Da and for LcB-phos as 50922.0 indicating a single phosphorylated site. (B) Lanes 2-10 represent samples 0, 0.25, 0.5, 0.75, 1, 2, 3, 6, and 8 h after start of the phosphorylation reaction catalyzed by 0.5 units of Src. Lane 1 has molecular mass markers. Absence of the phosphorylated Src band (as seen in Figure 1A is due to much lower amount of Src (0.5 units versus 3 units) used in this experiment. Because there were no detectable bands, the gel picture in (B) was truncated after 31 kDa for better presentation.
Secondary and Tertiary Structures and Stability
To determine if phosphorylation affected secondary and tertiary structures of the LC, we collected CD and tryptophan fluorescence spectra, respectively. Because salts are known to affect protein structure and stability, we used two different buffers: 10 mM Na-phosphate, pH 7.4 and PBS, pH 7.4 containing physiological concentration (0.85%) of NaCl. We did not investigate LcD, LcE, LcA424, LcA420, and LcC430 because the first two did not show significant phosphorylation (Figure 1A), and the last three were truncated proteins. Far-UV CD spectra of LcA-phos, LcB-phos (Figure 3), and LcG-phos (not shown) remained essentially identical to the unphosphorylated forms between 200 and 260 nm. Spectra were not affected by either 10 mM phosphate buffer pH 7.4 or PBS (Figure 3). Spectra below 200 nm were more affected by the buffer composition than by phosphorylation. Similarly, the tryptophan fluorescence spectrum of LcA was not noticeably affected by phosphorylation (Figure 4). Tryptophan fluorescence spectrum of LcB-phos (not shown) was not different than its unphosphorylated form although fluorescence intensity was somewhat lower than that of LcB. These results suggested that both secondary and tertiary structures of LcA and LcB were not appreciably affected by phosphorylation. In contrast to our results, from FTIR DSC, and catalytic activity analyses, Ferrer-Montiel et al. (Encinar et al., 1998; Ibanez et al., 2004) reported 2–3°C increased thermostability of LcA-phos due to an increase in the helical content.
Figure 3. Far-UV circular dichroism spectra of LcA-phos (A) and LcB-phos (B) compared with their unphosphorylated counterparts. Average of 5 scans at 20°C of each protein (0.2 mg/ml) in a 2 mm cuvette was recorded and reported here after subtraction of a corresponding buffer blank. Identity of each spectrum shown in the insets represent LcA-phos and LcB-phos in PB, closed circle; LcA-phos and LcB-phos in PBS, open circle; LcA and LcB in PB, open triangle; LcA and LcB in PBS, closed triangle.
Figure 4. Tryptophan fluorescence spectra of LcA-phos compared with its unpohsphorylated form. Average of five scans (λex = 295 nm) at 20°C of each protein (0.02 mg/ml) in a 2 mm cuvette was recorded and reported here after subtraction of a corresponding buffer blank. Identity of each spectrum shown in the inset represents LcA, closed circle and LcA-phos, open circle.
Post-translational modifications are known to affect protein stability (Li et al., 2002; Walsh, 2006b). We followed CD at 220 nm as a measure of thermostability when the temperature of the cuvette was continuously increased. Thermal denaturation profiles of LcA or LcB in 10 mM phosphate buffer pH 7.4 and of LcA-Phos or LcB-Phos in PBS underwent sharp, typical unfolding transitions (Figure 5) resulting in irreversible aggregate precipitate formation. Conversely, thermal denaturation profiles of LcA-Phos or LcB-Phos in 10 mM phosphate buffer were significantly different and suggested resistance to unfolding. In addition, thermal denaturation in 100 mM phosphate buffer pH 7.4 (data not shown) exhibited the typical sharp transition forming irreversible aggregate precipitate. Results indicate cooperative irreversible aggregate formation in high buffer and salt concentrations that rapidly precipitates from solution. Thus, behavior of the phosphorylated proteins in low ionic strength buffer probably is an exception. The midpoint of thermal denaturation, Tm of LcA-phos remained unchanged but those of LcB-phos and LcG-phos were 1–2°C higher than those of their unphosphorylated forms in PBS (Table 1).
Figure 5. Thermal denaturation patterns of LcA-phos (A) and LcB-phos (B) are compared with those of their unphosphorylated counterparts in two different buffers. Circular dichroism signals at 222 nm of 0.18–0.2 mg/ml protein samples were monitored in at a temperature gradient of 1°C/min from 20 to 90°C. Symbols: LcA-phos and LcB-phos in PBS, X (crosx); LcA-phos and LcB-phos in PB, – (dashed line); LcA and LcB in PB, O (open circle); LcA and LcB in PBS, Δ (open triangle).
LcA Autocatalysis
LcA is very prone to autocatalysis especially in the presence of divalent metal ions such as zinc, and increases with temperature leading to reduced catalytic activity (Ahmed et al., 2001, 2003, 2004). We observed autocatalysis when the phosphorylation reaction mixture (that contained MgCl2) was incubated at 30–37°C (see Experimental Procedures). LcA catalytic activity was also greatly reduced by phosphorylation (see below). Because autocatalysis and catalysis of LcA occurs at the same active site (Ahmed et al., 2003), we were interested to know if the autocatalysis reaction would be affected by phosphorylation. When incubated at room temperature for 5 days in the presence of ZnCl2, most LcA fragmented but LcA-phos remained largely intact (Figure 6). Thus phosphorylation made the LcA resistant to autocatalysis.
Figure 6. Phosphorylation of LcA makes it highly resistant to autocatalysis. Purified LcA before (upper panels) and after (lower panels) the phosphorylation reaction was incubated without (right panels) or with (left panels) 0.25 mM ZnCl2 at 4°C for 0–5 days (d). Aliquots were analyzed by SDS-PAGE and Coomassie staining. The LcA control at time zero represents the sample that was sitting at 4°C during the phosphorylation reaction, and until the start of the autocatalysis experiment. Thus, it shows substantial autocatalysis at time 0 h (h). Lanes labeled 1d and 5d represent samples incubated for 1 and 5 days, respectively. Lane 1 in both panels have molecular mass markers (Mr).
Phosphorylation Sites
We identified the sites of phosphorylation by mass spectrometric analyses of tryptic peptides. Three tyrosine residues in LcA, Y250, Y251, and Y426, were readily phosphorylated by Src (Tables 2 and 3). Tyrosines equivalent to Y250 and Y251 of LcA were not present in any other serotype.

Figure 7. Multiple sequence alignment (MSA) of conserved and non-conserved tyrosine containing peptides that were tyrosine-phosphorylated by Src. Clustal multiple sequence alignment of all of ∼450 residues of subtype 1 of BoNT Lc sequences were constructed. Only those stretches of the sequence alignment are shown here that contained a tyrosine residue being phosphorylated (bold) in any serotype sequence (underlined) by Src in our experiments. Because of high degeneracy, the sequence number 9 was also manually adjusted by keeping the identical cysteine fixed and the phosphorylated tyrosine aligned. GenBank ID: LcA1: AAQ06331; LcB1:BAE48264; LcC1: CAA44263; LcD1: AAB24244; LcE1: BAB86845; LcF1:ADA79551; LcG: CAA52275.
The next predominant phosphorylated Y426 near the C-terminus (of the 449-residue LcA) is conserved in LcB, LcC1, and LcG, and was readily phosphorylated in LcB and LcG. The LcC1 protein used in this experiment was a truncated form (residues 1–430) lacking the C-terminus; this tyrosine (Y430) located at the end of the chain was not recognized by Src for phosphorylation.
The most predominant phosphorylation in the earliest time point samples of LcA was diphosphorylation at Y250–Y251, followed by monophosphorylation of Y251 and monophosphorylation of Y426 (Table 3). Because monophosphorylation at Y251 but not at Y250 was observed, it is clear that reaction at the former led to phosphorylation of the latter residue. Thus the order of ease of phosphorylation was Y251 > Y250 > Y426. During some phosphorylation reactions, LcA degraded into two major fragments of approximately 23 kDa containing Y251 and approximately 28 kDa containing Y250. Only Y251 in the smaller fragment was phosphorylated. Y250–Y251 residues are found at the end of a flexible loop bordering the S3′ substrate-binding pocket of LcA (Kumaran et al., 2008a) which also forms the most susceptible autocatalytic cleavable peptide bond (Ahmed et al., 2003).
Because the conserved Y72 residue in Figure 7 was reported to be the only Src-catalyzed phosphorylation site in BoNT Lcs (Encinar et al., 1998; Blanes-Mira et al., 2001; Ibanez et al., 2004), we repeated our experiment at least seven times with LcA under a variety of reaction conditions to determine if this residue was phosphorylated. Instead, we found several other minor phosphorylation sites: Y99, Y180, Y349, and Y366 (Tables 2 and 3). Of these residues, only Y366 is fully conserved (Figure 7) but it was phosphorylated only in LcA although there are five more (Y72, Y73, Y77, Y321, Y366, and Y384, LcA numbering) conserved residues. Thus, some conserved tyrosine residues although were phosphorylated in one (see later) serotypes, remained unaffected in other serotypes. Our results provide a generalized picture of preferential tyrosine phosphorylation across the BoNT serotypes but did not show a pattern of consistency for in vitro phosphorylation across different serotypes.
In another experiment where LcA was incubated at 20°C for 24 h, 85% of Y250–Y251 was diphosphorylated, 15% of Y250, 21% of Y426, and 15% of Y387 was phosphorylated. Please note that Y387 was not phosphorylated at 30°C (Table 2). Addition of 0.05% Tween-20 to this reaction mixture almost completely abolished Y250–Y251 diphosphorylation but increased Y251, and Y426 monophosphorylations to 70 and 38%, respectively, while decreased Y387 phosphorylation to 7%. Thus, it appears that temperature and detergents like Tween-20 can change the LcA conformation in exposing new tyrosine residues and their availability for phosphorylation by Src. We did not investigate the effects of temperature and detergent with other Lc serotypes.
Interestingly, only one residue Y432 in LcB (corresponding to Y426 in LcA) (Figure 7) was rapidly phosphorylated. Its equivalent in LcG, Y431, was also readily phosphorylated. Another non-conserved residue in LcG, Y71, also reacted but measuring the corresponding mass peaks at all time points, showed that its phosphorylated form was always much lower than the corresponding Y431; after 24 h of incubation, 78% of Y430, 18% of Y71, and 25% of both were phosphorylated.
From the preceding results, tyrosine residue 3–6 positions upstream from the essential and conserved disulfide-forming cysteine (C430 in LcA, C437 in LcB, and C436 in LcG) appears to be the only common residues in most of the BoNT serotypes that are phosphorylated by Src. Although present in LcC1 native sequence as Y431, it was absent in our LcC1 construct (residues 1–430). We suspect this residue in full-length LcC1 would be as susceptible to Src-catalyzed phosphorylation as in LcA, LcB, and LcG. This tyrosine residue readily gets phosphorylated in LcB and LcG but much slower in LcA.
Role of C-terminus of LcA on Catalytic Activity
The C-terminus of LcA undergoes autocatalytic processing (Ahmed et al., 2001), and C-terminally truncated LcA has reduced catalytic activity (Baldwin et al., 2004), suggesting that the C-terminus approaches the active site in solution (Ahmed et al., 2003). We therefore included two C-terminally truncated LcA (both lacking Y426) and a competitive inhibitor with full-length LcA in the Src reactions to determine the effect on phosphorylation of other tyrosines (Table 3). Three important results were obtained. First, the competitive inhibitor CRATKML (Schmidt and Stafford, 2002) inhibited phosphorylation of Y426 at the C-terminus and of Y180. Second, no measurable phosphorylations of Y199, Y180, Y349, and Y366 were detected in the C-terminally truncated Lcs (Table 3). Third, addition of the inhibitor with full-length LcA and truncation of C-terminal residues beyond 420 exposed a new Y233 to phosphorylation. These results strongly support the earlier suggestion that the C-terminus interacts with the active site (Ahmed et al., 2003; Baldwin et al., 2004). In addition, interactions of the C-terminus with other parts of the protein must induce subtle changes in the protein’s conformation so that some tyrosine residues become exposed to Src-catalyzed phosphorylation. Like Y366, Y233 is also a conserved residue but this too was not phosphorylated in any other serotype.
Catalytic Activity of Phosphorylated LcA and LcB
We assayed LcA and LcB catalytic activities as a function of phosphorylation. Aliquots of the phosphorylation reaction mixtures were removed at set times for catalytic activity measurements and for assessing the extent of phosphorylation. LcA catalytic activity was significantly decreased but LcB activity appeared to be slightly stimulated by increased phosphorylation (Figure 8). The LcB catalytic activity was stable for at least 4 weeks when stored at 4°C. Loss of LcA activity in the phosphorylation reaction mixture (Figure 8) was verified in a purified preparation. At the end of 48 h incubation at 20oC, un-reacted components of the reaction mixture were first removed by anti-Src affinity precipitation followed by repeatedly washing the LcA-phos on a Centricon-10 microconcentrator. As seen before (Table 1), ESI-MS of the tryptic digests of the purified sample identified Y250–Y251 as the major phosphorylation site along with Y426 as another prominent phosphorylation site.
Figure 8. Catalytic activities (closed circle) of LcA (A) and LcB (B) are compared to the extent of phosphorylation (open circle) as a function of phosphorylation incubation time. Aliquots of the reaction mixtures were immediately diluted with substrate for activity measurements, SDS-PAGE loading buffer for Western blot, and with TFA for mass spectrometric analyses. Phosphorylation of LcA was computed by densitometric scanning of the Western blot images of Src reaction mixture (20°C) aliquots at various time intervals. Phosphorylation of LcB (30°C) was computed from the ratio of phosphorylated to non-phosphorylated ionic mass peaks. A low temperature of 20°C necessitated a longer incubation of LcA for completion of the phosphorylation reaction.
Loss of activity of the LcA-phos was measured in the purified sample as due to effects of phosphorylation on both Km and Kcat (Figure 9). Because phosphorylation had little impact on the LcB catalytic activity (Figure 8B), we did not attempt to determine its kinetic parameters. Although we prepared a large amount of LcG (see above), we could not determine its catalytic activity, due to the lack of a dependable assay method.
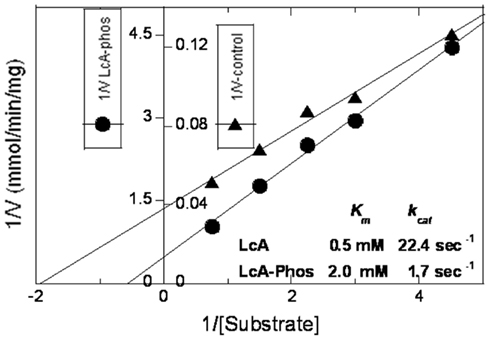
Figure 9. Lineweaver–Burke plots of reaction velocity versus substrate concentration of reactions catalyzed by phosphorylated (closed circles) versus unphosphorylated (closed triangles) LcA. The 30-μl assay reaction mixtures contained 5 mM dithiothreitol, 0.25 mM ZnCl2, 0.2 mg/ml BSA, 0.028 mg/ml LcA, or 0.45 mg/ml LcA-phos and variable (0.22, 0.33, 0.44, 0.67, and 1.13 mM) substrate (SNKTRIDEANQ-RATKML) concentrations in 50 mM Na-HEPES pH 7.4. The y-axis has two scales, one for LcA (inner scale, closed triangle), and the other for LcA-phos (outer scale, closed circle). Each data point represents an average of five assays.
Discussion
A major challenge in the development of therapeutics as medical intervention to botulinum BoNT intoxication is the persistence of clinical symptoms for certain serotypes, particularly BoNT/A,/B, and/C compared to the short-lived/E (Adler et al., 2001; Foran et al., 2003). For example, half-life of BoNT/A action in rat cerebellar neurons was 31 days, for BoNT/B was 10 days, and for BoNT/E it was less than 1 day (Foran et al., 2003). Besides, after 15 days of start of intoxication, only 12% recovery was achieved from BoNT/A-induced rat muscle paralysis compared to 94% recovery of BoNT/E-induced paralysis (Adler et al., 2001). Because the reaction mechanism and overall three-dimensional structure of all BoNT catalytic domains are essentially identical (Lacy et al., 1998; Eswaramoorthy et al., 2002; Agarwal et al., 2004, 2005; Swaminathan et al., 2004; Arndt et al., 2005, 2006; Jin et al., 2007; Kumaran et al., 2008a), it was expected that the proteins might undergo sequence-dependent covalent modification inside animal host cells.
Of all the post-translational protein modifications, phosphorylation is probably the most common and also most extensively studied (Walsh, 2006b). In addition to playing a central role in protein-based signaling pathways, phosphorylation can impart a proteins’ stabilization (Li et al., 2002) and stimulation of activity (Patwardhan and Miller, 2007). We investigated if (a) phosphorylation can occur in the catalytic domains of various serotypes of BoNTs, and (b) if differences in their phosphorylation patterns might correlate with differences in their persistence of their catalytic activity inside neurons.
In this study, we employed tyrosine kinase Src as the phosphorylating enzyme. Ferrer-Montiel et al. (1996) had reported that only tyrosine kinase Src and no other serine-threonine kinases were effective for BoNT/A phosphorylation. We showed that, with the exception of LcD and LcE, tyrosine residues in other BoNT Lcs were rapidly phosphorylated by Src (Figure 1A; Table 2). Although we did not detect any phosphorylation of LcE, the results remain inconclusive because the concentration of LcE was very low due to its poor solubility. Our results differ with previous reports concerning phosphorylation of the invariant Y71 of LcA (our numbering Y72) and Y67 of LcE (Encinar et al., 1998; Blanes-Mira et al., 2001; Ibanez et al., 2004). Because this residue was reported as the only Src-catalyzed BoNT Lc phosphorylation site based on mutational studies with LcA and LcE (Encinar et al., 1998; Blanes-Mira et al., 2001; Ibanez et al., 2004), we repeated our experiments several times with LcA and LcB but could not detect Y71 (our numbering Y72) phosphorylation using trypsin digestion of the phosphorylated Lc and mass spectrometry. In our studies, both conserved and non-conserved (Figure 7) tyrosines of LcA were phosphorylated (Tables 2 and 3) but no conserved tyrosine was phosphorylated across the serotypes.
From the available data, phosphorylation of Y426 of LcA, Y432 of LcB, and of Y431 of LcG may have some functional significance because they are located very near the fully conserved C430 (of LcA) that is essential for translocation (Simpson et al., 2004). For example, wherever conserved, this residue was readily phosphorylated: Y432 of LcB and Y431 of LcG. Additional studies are needed to ascertain the significance of phosphorylation of this residue.
Consequence of Tyrosine Phosphorylation
The in vitro phosphorylation results described here do not support any detectable change in the protein’s secondary or tertiary structure, nor any significant impact on their thermal stability. Slightly elevated (1∼2°C) midpoint of thermal transition (Figure 5; Table 1) and activity of LcB (Figure 8B) and dramatic reduction in LcA activity (Figures 7, 8) by our in vitro tyrosine phosphorylation experiments (Figures 1, 2; Tables 2, 3) are in contradiction to the well-established, increased catalytic stability of BoNT/A over BoNT/B in neuronal cells (Keller et al., 1999; Foran et al., 2003). From a structural point, the increased Km and decreased kcat of LcA-phos (Figure 9) can be easily explained by the fact that phosphorylation of Y250 and Y251 introduces two bulky groups on the 250-loop that normally closes onto the active site during catalysis (Kumaran et al., 2008b). This may limit the access of the substrate to the active site. Because no ex vivo quantitative specific activity data are available, one cannot be sure if LcA activity is low inside neurons that would support our in vitro data (Figure 8), even though LcA activity persists there for a very long time (Keller et al., 1999; Foran et al., 2003). Because LcB does not have a site near the active site that can be phosphorylated, the LcB activity remained little affected by phosphorylation. This consideration of steric hindrance by phosphate will equally explain the stability of LcA-phos from autocatalytic fragmentation (Figure 6) that occurs at the LcA active site (Ahmed et al., 2003, 2008).
Conclusion
In vitro reaction of LcA, LcB, LcC1, LcD, LcE, and LcG with Tyrosine kinase Src resulted in phosphorylation of several tyrosine residues. One of these residues is fully conserved but not all the conserved tyrosine residues were phosphorylated in each serotype. Phosphorylation of Y250 and Y251 in LcA took place most readily and made the protein highly resistant to autocatalysis but drastically reduced its catalytic efficiency. Phosphorylation of the tyrosine residue located near to the essential, conserved cysteine residue (C430 of LcA) might have a functional significance for LcA, LcB, LcC1, and LcG.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mr. Matthew Ludivico for conducting the UPLCTM analyses, kinetic and circular dichroism experiments, and Dr. Mizanur Rahman for careful reading of the manuscript. This project received support from the Defense Threat Reduction Agency-Joint Science and Technology Office for Chemical and Biological Defense (Grant #s CBS.MEDBIO.01.10.RD.002 and JSTOCBD3.10012_06_RD_B to S. Ashraf Ahmed). Opinions, interpretations, conclusions, and recommendations are those of the author and are not necessarily endorsed by the U.S. Army.
Abbreviations
BoNT, botulinum neurotoxin; BoNT/A-G, BoNT serotypes A, B, C, D, E, F, G; BSA, bovine serum albumin; CD: circular dichroism; CFP, cyan fluorescent protein; DSC: differential scanning calorimetry; DTT, dithiothreitol; DPBS, Dulbecco’s phosphate-buffered saline; HEPES, (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid); EGTA, ethylene glycol tetra acetic acid; FTIR: Fourier transform infra red; HRP, horseradish peroxidase; LC, light chain LcA, light chain of serotype A; LcA-phos, phosphorylated LcA; LcB-Phos, phosphorylated LcB; LcB, light chain of serotype B; LcC1, light chain of serotype C1; LcD, light chain of serotype D; LcE, light chain of serotype E; LcG, light chain of serotype G; LcG-Phos, phosphorylated LcG; SBP: streptavidin binding protein; SNAP-25, synaptosome-associated protein of 25 kDa; TFA, trifluoroacetic acid; Tm: melting temperature; TMB, 3,3′,5,5′-tetramethylbenzidine; UPLC, ultra performance liquid chromatography; VAMP, vesicle-associated membrane protein.
References
Adler, M., Keller, J. E., Sheridan, R. E., and Deshpande, S. S. (2001). Persistence of botulinum neurotoxin A demonstrated by sequential administration of serotypes A and E in rat EDL muscle. Toxicon 39, 233–243.
Agarwal, R., Binz, T., and Swaminathan, S. (2005). Structural analysis of botulinum neurotoxin serotype F light chain: implications on substrate binding and inhibitor design. Biochemistry 44, 11758–11765.
Agarwal, R., Eswaramoorthy, S., Kumaran, D., Binz, T., and Swaminathan, S. (2004). Structural analysis of botulinum neurotoxin type E catalytic domain and its mutant Glu212→Gln reveals the pivotal role of the Glu212 carboxylate in the catalytic pathway. Biochemistry 43, 6637–6644.
Agarwal, R., Schmidt, J. J., Stafford, R. G., and Swaminathan, S. (2009). Mode of VAMP substrate recognition and inhibition of Clostridium botulinum neurotoxin F. Nat. Struct. Mol. Biol. 16, 789–794.
Ahmed, S. A., Byrne, M. P., Jensen, M., Hines, H. B., Brueggemann, E., and Smith, L. A. (2001). Enzymatic autocatalysis of botulinum A neurotoxin light chain. J. Protein Chem. 20, 221–231.
Ahmed, S. A., Ludivico, M. L., and Smith, L. A. (2004). Factors affecting autocatalysis of botulinum A neurotoxin light chain. Protein J. 23, 445–451.
Ahmed, S. A., McPhie, P., and Smith, L. A. (2003). Autocatalytically fragmented light chain of botulinum a neurotoxin is enzymatically active. Biochemistry 42, 12539–12549.
Ahmed, S. A., Olson, M. A., Ludivico, M. L., Gilsdorf, J., and Smith, L. A. (2008). Identification of residues surrounding the active site of type A botulinum neurotoxin important for substrate recognition and catalytic activity. Protein J. 27, 151–162.
Ahmed, S. A., and Smith, L. A. (2000). Light chain of botulinum A neurotoxin expressed as an inclusion body from a synthetic gene is catalytically and functionally active. J. Protein Chem. 19, 475–487.
Arndt, J. W., Chai, Q., Christian, T., and Stevens, R. C. (2006). Structure of botulinum neurotoxin type D light chain at 1.65 A resolution: repercussions for VAMP-2 substrate specificity. Biochemistry 45, 3255–3262.
Arndt, J. W., Yu, W., Bi, F., and Stevens, R. C. (2005). Crystal structure of botulinum neurotoxin type G light chain: serotype divergence in substrate recognition. Biochemistry 44, 9574–9580.
Arnon, S. S., Schechter, R., Inglesby, T. V., Henderson, D. A., Bartlett, J. G., Ascher, M. S., Eitzen, E., Fine, A. D., Hauer, J., Layton, M., Lillibridge, S., Osterholm, M. T., O’Toole, T., Parker, G., Perl, T. M., Russell, P. K., Swerdlow, D. L., and Tonat, K. (2001). Botulinum toxin as a biological weapon: medical and public health management. JAMA 285, 1059–1070.
Baldwin, M. R., Bradshaw, M., Johnson, E. A., and Barbieri, J. T. (2004). The C-terminus of botulinum neurotoxin type A light chain contributes to solubility, catalysis, and stability. Protein Expr. Purif. 37, 187–195.
Blanes-Mira, C., Ibanez, C., Fernandez-Ballester, G., Planells-Cases, R., Perez-Paya, E., and Ferrer-Montiel, A. (2001). Thermal stabilization of the catalytic domain of botulinum neurotoxin E by phosphorylation of a single tyrosine residue. Biochemistry 40, 2234–2242.
Burnett, J. C., Wang, C., Nuss, J. E., Nguyen, T. L., Hermone, A. R., Schmidt, J. J., Gussio, R., Wipf, P., and Bavari, S. (2009). Pharmacophore-guided lead optimization: the rational design of a non-zinc coordinating, sub-micromolar inhibitor of the botulinum neurotoxin serotype a metalloprotease. Bioorg. Med. Chem. Lett. 19, 5811–5813.
Capkova, K., Salzameda, N. T., and Janda, K. D. (2009). Investigations into small molecule non-peptidic inhibitors of the botulinum neurotoxins. Toxicon 54, 575–582.
Cochrane, R. C. (1947). “Biological warfare research in the United States,” in History of the Chemical Warfare Service in World War II (01 July 1940 – 15 August 1945). Fort Detrick: Office of Chief, Chemical Corps.
Encinar, J. A., Fernandez, A., Ferragut, J. A., Gonzalez-Ros, J. M., DasGupta, B. R., Montal, M., and Ferrer-Montiel, A. (1998). Structural stabilization of botulinum neurotoxins by tyrosine phosphorylation. FEBS Lett. 429, 78–82.
Eswaramoorthy, S., Kumaran, D., and Swaminathan, S. (2002). A novel mechanism for Clostridium botulinum neurotoxin inhibition. Biochemistry 41, 9795–9802.
Ferrer-Montiel, A. V., Canaves, J. M., DasGupta, B. R., Wilson, M. C., and Montal, M. (1996). Tyrosine phosphorylation modulates the activity of clostridial neurotoxins. J. Biol. Chem. 271, 18322–18325.
Ferrer-Montiel, A. V., Gutierrez, L. M., Apland, J. P., Canaves, J. M., Gil, A., Viniegra, S., Biser, J. A., Adler, M., and Montal, M. (1998). The 26-mer peptide released from SNAP-25 cleavage by botulinum neurotoxin E inhibits vesicle docking. FEBS Lett. 435, 84–88.
Foran, P., Shone, C. C., and Dolly, J. O. (1994). Differences in the protease activities of tetanus and botulinum B toxins revealed by the cleavage of vesicle-associated membrane protein and various sized fragments. Biochemistry 33, 15365–15374.
Foran, P. G., Mohammed, N., Lisk, G. O., Nagwaney, S., Lawrence, G. W., Johnson, E., Smith, L., Aoki, K. R., and Dolly, J. O. (2003). Evaluation of the therapeutic usefulness of botulinum neurotoxin B, C1, E, and F compared with the long lasting type A. Basis for distinct durations of inhibition of exocytosis in central neurons. J. Biol. Chem. 278, 1363–1371.
Foster, L. J., Yeung, B., Mohtashami, M., Ross, K., Trimble, W. S., and Klip, A. (1998). Binary interactions of the SNARE proteins syntaxin-4, SNAP23, and VAMP-2 and their regulation by phosphorylation. Biochemistry 37, 11089–11096.
Gilsdorf, J., Gul, N., and Smith, L. A. (2006). Expression, purification, and characterization of Clostridium botulinum type B light chain. Protein Expr. Purif. 46, 256–267.
Hale, M., Oyler, G., Swaminathan, S., and Ahmed, S. A. (2011). Basic tetrapeptides as potent intracellular inhibitors of type A botulinum neurotoxin protease activity. J. Biol. Chem. 286, 1802–1811.
Hines, H. B., Kim, A. D., Stafford, R. G., Badie, S. S., Brueggeman, E. E., Newman, D. J., and Schmidt, J. J. (2008). Use of a recombinant fluorescent substrate with cleavage sites for all botulinum neurotoxins in high-throughput screening of natural product extracts for inhibitors of serotypes A, B, and E. Appl. Environ. Microbiol. 74, 653–659.
Ibanez, C., Blanes-Mira, C., Fernandez-Ballester, G., Planells-Cases, R., and Ferrer-Montiel, A. (2004). Modulation of botulinum neurotoxin A catalytic domain stability by tyrosine phosphorylation. FEBS Lett. 578, 121–127.
Jensen, M. J., Smith, T. J., Ahmed, S. A., and Smith, L. A. (2003). Expression, purification, and efficacy of the type A botulinum neurotoxin catalytic domain fused to two translocation domain variants. Toxicon 41, 691–701.
Jin, R., Sikorra, S., Stegmann, C. M., Pich, A., Binz, T., and Brunger, A. T. (2007). Structural and biochemical studies of botulinum neurotoxin serotype C1 light chain protease: implications for dual substrate specificity. Biochemistry 46, 10685–10693.
Keller, J. E., Neale, E. A., Oyler, G., and Adler, M. (1999). Persistence of botulinum neurotoxin action in cultured spinal cord cells. FEBS Lett. 456, 137–142.
Kumaran, D., Rawat, R., Ludivico, M. L., Ahmed, S. A., and Swaminathan, S. (2008a). Structure- and substrate-based inhibitor design for Clostridium botulinum neurotoxin serotype A. J. Biol. Chem. 283, 18883–18891.
Kumaran, D., Rawat, R., Ahmed, S. A., and Swaminathan, S. (2008b). Substrate binding mode and its implication on drug design for botulinum neurotoxin A. PLoS Pathog. 4, e1000165. doi:10.1371/journal.ppat.1000165
Lacy, D. B., Tepp, W., Cohen, A. C., DasGupta, B. R., and Stevens, R. C. (1998). Crystal structure of botulinum neurotoxin type A and implications for toxicity. Nat. Struct. Biol. 5, 898–902.
Li, Y., Dowbenko, D., and Lasky, L. A. (2002). AKT/PKB phosphorylation of p21Cip/WAF1 enhances protein stability of p21Cip/WAF1 and promotes cell survival. J. Biol. Chem. 277, 11352–11361.
Ludivico, M., Smith, L. A., and Ahmed, S. A. (2009). Structure-based design of peptide inhibitors of botulinum neurotoxin serotypes A proteolytic activity. Botulinum J. 1, 297–308.
Montecucco, C., and Schiavo, G. (1995). Structure and function of tetanus and botulinum neurotoxins. Q. Rev. Biophys. 28, 423–472.
Munton, R. P., Tweedie-Cullen, R., Livingstone-Zatchej, M., Weinandy, F., Waidelich, M., Longo, D., Gehrig, P., Potthast, F., Rutishauser, D., Gerrits, B., Panse, C., Schlapbach, R., and Mansuy, I. M. (2007). Qualitative and quantitative analyses of protein phosphorylation in naive and stimulated mouse synaptosomal preparations. Mol. Cell Proteomics 6, 283–293.
Nagy, G., Reim, K., Matti, U., Brose, N., Binz, T., Rettig, J., Neher, E., and Sorensen, J. B. (2004). Regulation of releasable vesicle pool sizes by protein kinase A-dependent phosphorylation of SNAP-25. Neuron 41, 417–429.
Nielander, H. B., Onofri, F., Valtorta, F., Schiavo, G., Montecucco, C., Greengard, P., and Benfenati, F. (1995). Phosphorylation of VAMP/synaptobrevin in synaptic vesicles by endogenous protein kinases. J. Neurochem. 65, 1712–1720.
Pang, Y. P., Vummenthala, A., Mishra, R. K., Park, J. G., Wang, S., Davis, J., Millard, C. B., and Schmidt, J. J. (2009). Potent new small-molecule inhibitor of botulinum neurotoxin serotype A endopeptidase developed by synthesis-based computer-aided molecular design. PLoS ONE 4, e7730. doi:10.1371/journal.pone.0007730
Patwardhan, P., and Miller, W. T. (2007). Processive phosphorylation: mechanism and biological importance. Cell. Signal. 19, 2218–2226.
Rawat, R., Ahmed, S. A., and Swaminathan, S. (2008). High level expression of the light chain of botulinum neurotoxin serotype C1 and an efficient HPLC assay to monitor its proteolytic activity. Protein Expr. Purif. 60, 165–169.
Rowe, B., Schmidt, J. J., Smith, L. A., and Ahmed, S. A. (2010). Rapid product analysis and increased sensitivity for quantitative determinations of botulinum neurotoxin proteolytic activity. Anal. Biochem. 396, 188–193.
Schiavo, G., Rossetto, O., Catsicas, S., Polverino de Laureto, P., DasGupta, B. R., Benfenati, F., and Montecucco, C. (1993). Identification of the nerve terminal targets of botulinum neurotoxin serotypes A, D, and E. J. Biol. Chem. 268, 23784–23787.
Schiavo, G., Rossetto, O., Santucci, A., DasGupta, B. R., and Montecucco, C. (1992a). Botulinum neurotoxins are zinc proteins. J. Biol. Chem. 267, 23479–23483.
Schiavo, G., Benfenati, F., Poulain, B., Rossetto, O., Polverino de Laureto, P., DasGupta, B. R., and Montecucco, C. (1992b). Tetanus and botulinum-B neurotoxins block neurotransmitter release by proteolytic cleavage of synaptobrevin. Nature 359, 832–835.
Schmidt, J. J., and Stafford, R. G. (2002). A high-affinity competitive inhibitor of type A botulinum neurotoxin protease activity. FEBS Lett. 532, 423–426.
Schmidt, J. J., and Stafford, R. G. (2005). Botulinum neurotoxin serotype F: identification of substrate recognition requirements and development of inhibitors with low nanomolar affinity. Biochemistry 44, 4067–4073.
Segelke, B., Knapp, M., Kadkhodayan, S., Balhorn, R., and Rupp, B. (2004). Crystal structure of Clostridium botulinum neurotoxin protease in a product-bound state: evidence for noncanonical zinc protease activity. Proc. Natl. Acad. Sci. U.S.A. 101, 6888–6893.
Silhar, P., Capkova, K., Salzameda, N. T., Barbieri, J. T., Hixon, M. S., and Janda, K. D. (2010). Botulinum neurotoxin A protease: discovery of natural product exosite inhibitors. J. Am. Chem. Soc. 132, 2868–2869.
Simpson, L. L. (2004). Identification of the major steps in botulinum toxin action. Annu. Rev. Pharmacol. Toxicol. 44, 167–193.
Simpson, L. L., Maksymowych, A. B., Park, J. B., and Bora, R. S. (2004). The role of the interchain disulfide bond in governing the pharmacological actions of botulinum toxin. J. Pharmacol. Exp. Ther. 308, 857–864.
Souayah, N., Karim, H., Kamin, S. S., McArdle, J., and Marcus, S. (2006). Severe botulism after focal injection of botulinum toxin. Neurology 67, 1855–1856.
Swaminathan, S., Eswaramoorthy, S., and Kumaran, D. (2004). Structure and enzymatic activity of botulinum neurotoxins. Mov. Disord. 19(Suppl. 8), S17–S22.
Toth, S. I., Smith, L. A., and Ahmed, S. A. (2009). Extreme sensitivity of botulinum neurotoxin domains towards mild agitation. J. Pharm. Sci. 98, 3302–3311.
Walsh, C. T. (2006a). Posttranslational Modification of Proteins, 1st Edn. Englewood, CO: Roberts and Company Publishers.
Walsh, C. T. (2006b). Posttranslational Modification of Proteins: Expanding Nature’s Inventory, 1st Edn. Edgewood, CO: Roberts and Company Publishers.
Zuniga, J. E., Schmidt, J. J., Fenn, T., Burnett, J. C., Arac, D., Gussio, R., Stafford, R. G., Badie, S. S., Bavari, S., and Brunger, A. T. (2008). A potent peptidomimetic inhibitor of botulinum neurotoxin serotype A has a very different conformation than SNAP-25 substrate. Structure 16, 1588–1597.
Keywords: botulinum neurotoxin, tyrosine phosphorylation, zinc endoporotease, protease, clostridium botulinum, protein phosphorylation
Citation: Toth S, Brueggmann EE, Oyler GA, Smith LA, Hines HB and Ahmed SA (2012) Tyrosine phosphorylation of botulinum neurotoxin protease domains. Front. Pharmacol. 3:102. doi: 10.3389/fphar.2012.00102
Received: 13 February 2012; Accepted: 07 May 2012;
Published online: 04 June 2012.
Edited by:
Chiranjib Chakraborty, School of Bio-Sciences and Technology – VIT University, IndiaCopyright: © 2012 Toth, Brueggmann, Oyler, Smith, Hines and Ahmed. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: S. Ashraf Ahmed, USAMRIID, 1425 Porter Street, Fort Detrick, MD 21702, USA. e-mail: syed.ahmed@amedd.army.mil
†Present address: Stephen Toth, Mentor Biologics, Madison, WI 53711, USA.