- Department of Ophthalmology, The School of Medicine, University of Colorado, Aurora, CO, USA
The American Cancer Society estimates that there will be more than 1.5 million new cases of cancer in 2011, underscoring the need for identification of new therapeutic targets and development of novel cancer therapies. Previous studies have implicated the human aldo-ketoreductases AKR1B1 and AKR1B10 in cancer, and therefore we examined AKR1B1 and AKR1B10 expression across all major human cancer types using the Oncomine cancer gene expression database (Compendia Biosciences, www.oncomine.com). Using this database, we found that expression of AKR1B1 and AKR1B10 varies greatly by cancer type and tissue of origin, including agreement with previous reports that AKR1B10 is significantly over-expressed in cancers of the lungs and liver. AKR1B1 is more broadly over-expressed in human cancers than AKR1B10, albeit at a generally lower magnitude. AKR1B1 over-expression was found to be associated with shortened patient survival in acute myelogenous leukemias and multiple myelomas. High AKR1B10 expression tends to predict less aggressive clinical course generally, notably within lung cancers, where it tends to be highly over-expressed compared to normal tissue. These findings suggest that AKR1B1 inhibitors in particular hold great potential as novel cancer therapeutics.
Introduction
Cancer is the second leading cause of death in the U.S. behind heart disease, and the American Cancer Society estimates that there will be more than 1.5 million new cases of cancer in 2011. While improvements in detection, treatment, and prevention have led to decreases in cancer deaths and incidence for many cancer types in the U.S., the incidence rate for some cancers such as hepatocellular carcinoma is still rising. However, as the U.S. population ages, cancer incidence may reach a plateau or even rebound. Therefore, identification of new therapeutic targets and development of novel cancer therapies is still a pressing need. Previous studies have shown that the human aldo-ketoreductase AKR1B10 is over-expressed in cancers of the liver and lungs (Fukumoto et al., 2005; Woenckhaus et al., 2006; Heringlake et al., 2010; Kang et al., 2011; Schmitz et al., 2011) while AKR1B10 and the related enzyme AKR1B1 are both linked to drug resistance in cancer-derived cell lines (Dan et al., 2003; Plebuch et al., 2007; Matsunaga et al., 2011; Zhong et al., 2011). Several published studies have also pointed to role for AKR1B1 in colon carcinogenesis (Tammali et al., 2009, 2011a,b; Ramana et al., 2010). As well-characterized inhibitors for these enzymes are already in use or development, they would seem to be attractive targets for cancer therapeutic development. However, these studies have been conducted largely in model systems, and thus there is very little known about the involvement of AKR1B10 in cancer outside of the lungs and liver, and while AKR1B1 expression has been reported to be elevated in human cancers, this study was limited by a small number of available patient samples (Saraswat et al., 2006). Since AKR1B1 has been shown to be involved in many cellular processes relevant to cancer such as EMT (Zablocki et al., 2011), inflammation (Yadav et al., 2007, 2009, 2011), and angiogenesis (Tammali et al., 2011b,c), and AKR1B10 is known to have relevance to human cancers, we examined AKR1B1 and AKR1B10 expression across all major human cancer types using the Oncomine cancer gene expression database (Compendia Biosciences, www.oncomine.com).
Using this database, we found that expression of AKR1B1 and AKR1B10 varies greatly by cancer type and tissue of origin, including agreement with previous reports that AKR1B10 is significantly over-expressed in cancers of the lungs and liver (Fukumoto et al., 2005; Woenckhaus et al., 2006; Heringlake et al., 2010; Kang et al., 2011; Schmitz et al., 2011). While under-expression of AKRs in human cancers is less common than over-expression, AKR1B1 appears to be generally under-expressed in prostate cancers compared to normal tissue while AKR1B10 expression is reduced in colon tumors. AKR1B1 over-expression was associated with shortened patient survival in acute myelogenous leukemias and multiple myelomas. High AKR1B10 expression tends to predict less aggressive clinical course generally, notably within lung cancers, where it tends to be highly over-expressed compared to normal tissue. Neither AKR1B1 nor AKR1B10 appears to have notable associations with disease recurrence, and their associations with the presence of metastases are inconsistent.
These findings suggest that AKR1B1 in particular may be a promising drug target, due to its broad over-expression in solid tumors and leukemias. Previous drug development attempts centered on AKR1B1 inhibition in non-cancer disease states were halted due to unacceptable toxicity; however, the reported toxicities were milder than other chemotherapeutics currently in use. Newer AKR1B1 inhibitors such as those derived from natural products (Suryanarayana et al., 2004, 2007; Saraswat et al., 2008) may have lower toxicity than earlier compounds, and therefore hold great potential as novel therapeutics for cancers where AKR1B1 tends to be over-expressed.
Materials and Methods
Meta-analysis of AKR1B1 and AKR1B10 gene expression in human cancers and normal tissues as well as related statistical analysis were conducted using the Oncomine gene expression database (www.oncomine.com, Compendia biosciences, Ann Arbor, MI, USA). Where appropriate, raw data was downloaded from Oncomine and scrutinized to ensure consistent comparisons and definitions such as “high grade” were used across different studies. For example, in prostate tumors “high gleason score” is defined by the highest grade tumors within the study being considered, which for some studies is GS7 and in others GS10. P-values were determined by Student’s t-test and those less than 0.05 were considered significant. Gene rank represents the ordered numerical rank of that gene’s p-value against all other genes for that comparison – i.e., a gene with a rank of 5 has a more significant difference in expression level for the two conditions examined than for all but four other genes. In data presented considering over-expression versus under-expression in a given cancer type, the most significant p-value and gene rank are presented even in cases where neither were significant as defined by p < 0.05. Only studies based on human clinical samples were included in our analyses. Where an “overall p-value” is listed, the p-value generated by simultaneously considering all available data within Oncomine for the given comparison is displayed, i.e., the overall p-value for the cancer versus normal comparisons listed in Table 1 includes all studies for which gene expression for cancerous and corresponding normal tissue was available within Oncomine. Median gene ranks displayed are similarly inclusive of all available data in Oncomine. All graphics displayed in figures are Oncomine.svg file output modified by Adobe Illustrator.
Results
To determine whether the aldo-ketoreductases AKR1B1 and AKR1B10 were differentially expressed between cancerous and normal tissues, we broadly examined microarray data from patient samples contained within the Oncomine database. Results from cancer types where a statistically significant difference in AKR expression between the cancerous and corresponding normal tissue exists are summarized in Table 1. The cancers where gene expression for AKRs was compared to the corresponding normal tissue, but no overall significant difference was found were certain brain tumors (oligodendrogliomas, mixed gliomas), ductal and lobular breast cancers, acute myeloid leukemias, myelomas, and ovarian cancers (not shown). Data for cancers where only one study was available for analysis is also not shown. AKR1B1 expression is significantly elevated compared to the corresponding normal tissue in bladder, brain (astrocytomas and glioblastomas), cervical, esophageal, head and neck, kidney, leukemias (T-cell acute, B-cell acute, and chronic), lymphomas, and melanomas (Table 1; Figure 1A). The fold change in gene expression versus the normal tissue is summarized by study in Table 1, with AKR1B1 expression ranging from ∼1.2- to 5-fold the normal tissue in the majority of cancers where it is significantly over-expressed. The most significant differences between AKR1B1 expression in cancerous and normal tissue are seen in leukemias (Table 1; Figure 1A). AKR1B1 expression is significantly lower than the corresponding normal tissue in prostate cancers (Table 1). As previously reported, AKR1B10 is over-expressed in liver and lung tumors (Table 1; Fukumoto et al., 2005; Woenckhaus et al., 2006; Heringlake et al., 2010; Kang et al., 2011; Schmitz et al., 2011), with fold change relative to normal tissue ranging from 12- to 67-fold in liver cancers; 2- to 75-fold in squamous cell lung cancers; and 1.5- to 5.5-fold in lung adenocarcinomas (Table 1). AKR1B10 is also significantly over-expressed in leukemias (T-cell acute, B-cell acute, and chronic) and pancreatic cancers (Table 1; Figure 1A). AKR1B10 over-expression thus appears to be less common than AKR1B1 over-expression in cancer, and AKR1B10 is under-expressed in colon, gastric, and head and neck cancers (Table 1). It should be noted that these associations are those that hold true across the studies contained within Oncomine, and multiple studies may have individually held a significant association of AKR expression with either the cancerous or normal state, but not in the broader comparison. Our methods also necessarily exclude studies not contained within the Oncomine database.
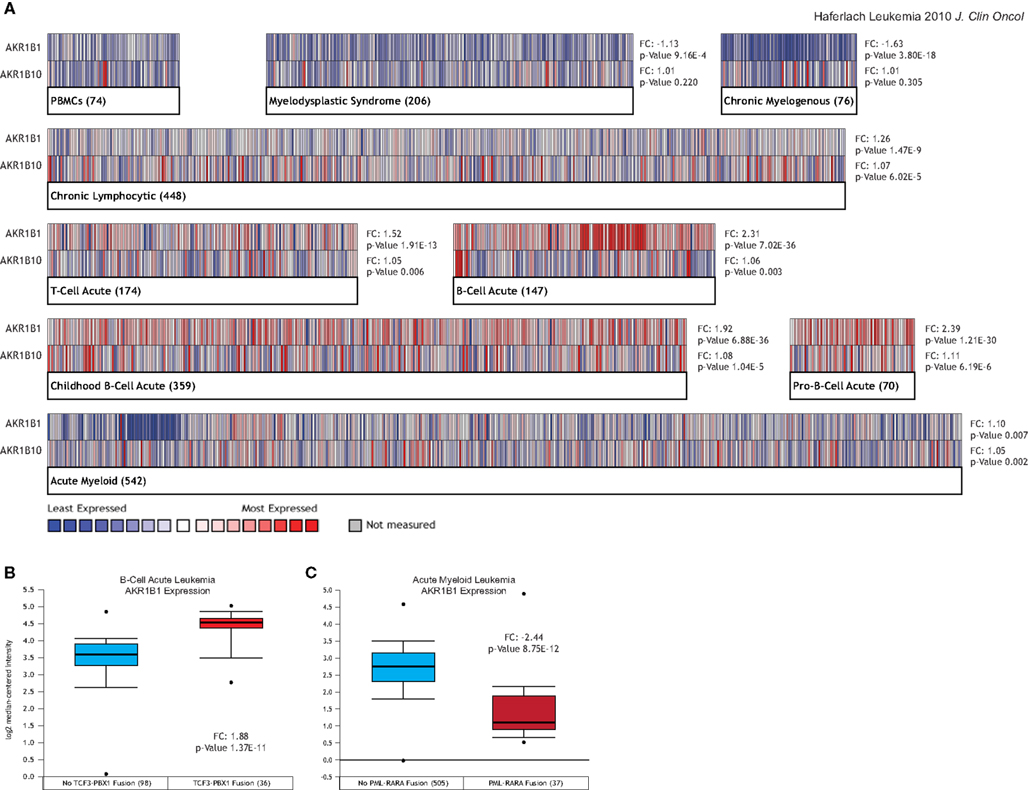
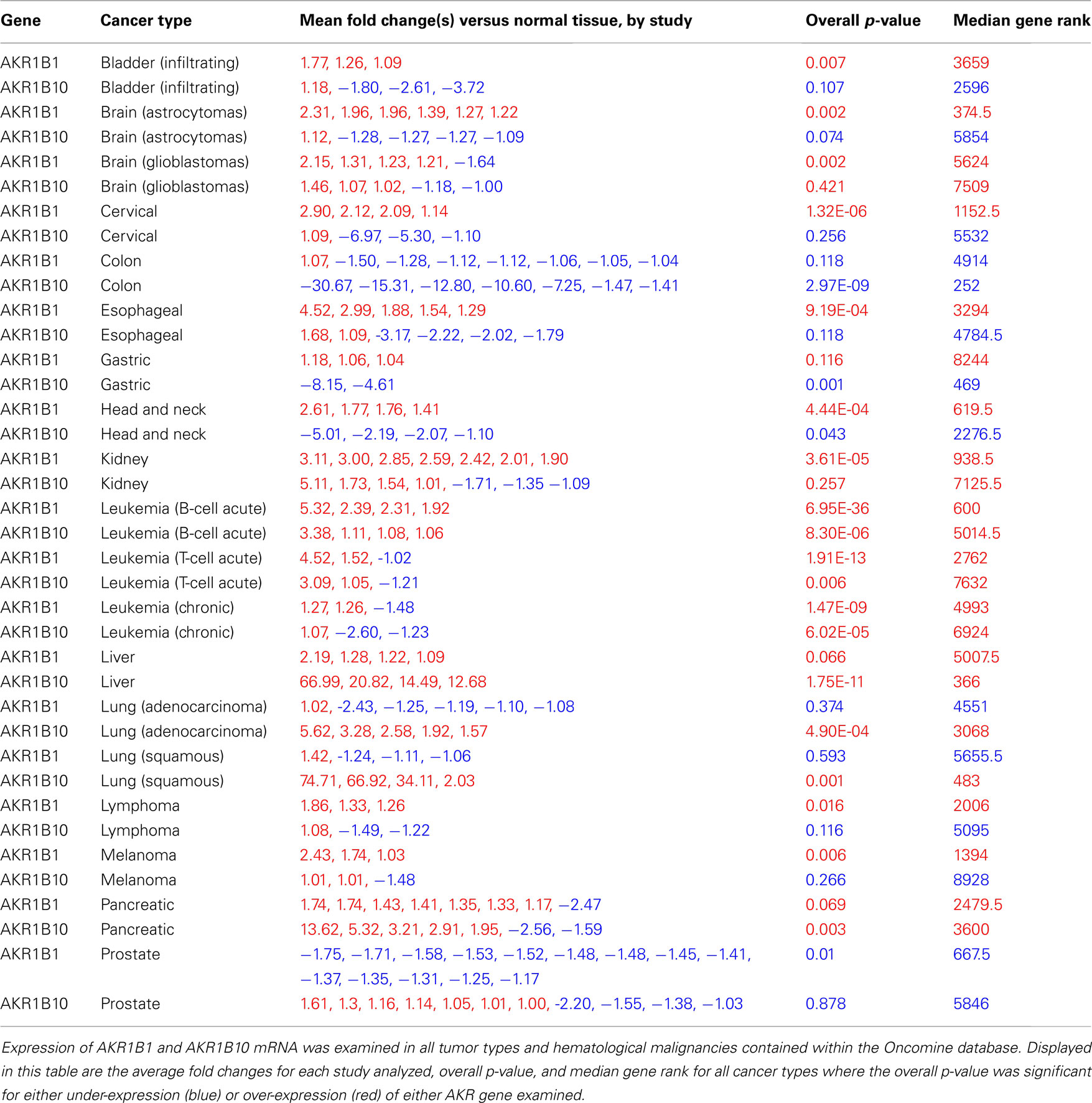
Figure 1. AKR expression in leukemia patients. Expression of AKR1B1 and AKR1B10 mRNA was examined in 2093 human leukemia patients from the Haferlach et al. (2010) study in the Oncomine database. (A) Heatmap display of AKR1B1 (top rows) and AKR1B10 (bottom rows) expression within the indicated leukemia types. Fold changes relative to peripheral blood mononuclear cells (PBMCs) and p-value for that comparison within each leukemia type are listed next to their respective heatmaps. Numbers in parentheses next to the label of each heatmap represent the number of patients contained within that group in this study. (B) AKR1B1 mRNA expression in B-cell acute leukemia patients without (blue box) and with (red box) the presence of the TCF3-PBX1 fusion gene. (C) AKR1B1 mRNA expression in acute myeloid leukemia patients without (blue box) and with (red box) the presence of the PML-RARA fusion gene.
As shown in Figure 1, even for leukemia types in which AKRs are over-expressed compared to normal tissue at a high level of statistical significance, there is considerable heterogeneity amongst patients in terms of AKR1B1 and AKR1B10 expression (Figure 1A). This led us to ask whether AKR expression could identify certain types of patients within these leukemias, and we found that high levels of AKR1B1 expression within B-cell leukemia patients was strongly associated with the presence of the TCF3-PBX1 gene fusion (Figure 1B), while under-expression of AKR1B1 in chronic myelogenous leukemias was associated with the presence of the PML-RARA gene fusion (Figure 1C). Across all translocations and gene fusions in all leukemia types, AKR1B1 over-expression is associated with the TCF3-PBX1 gene fusion and 11q23 MLL rearrangements, while under-expression is associated with the PML-RARA and ETV6-RUNX1 gene fusions (Figure 2). Other gene fusions, translocations, and point mutations examined in leukemias did not have a statistically significant, consistent pattern (Figure 2 and data not shown).
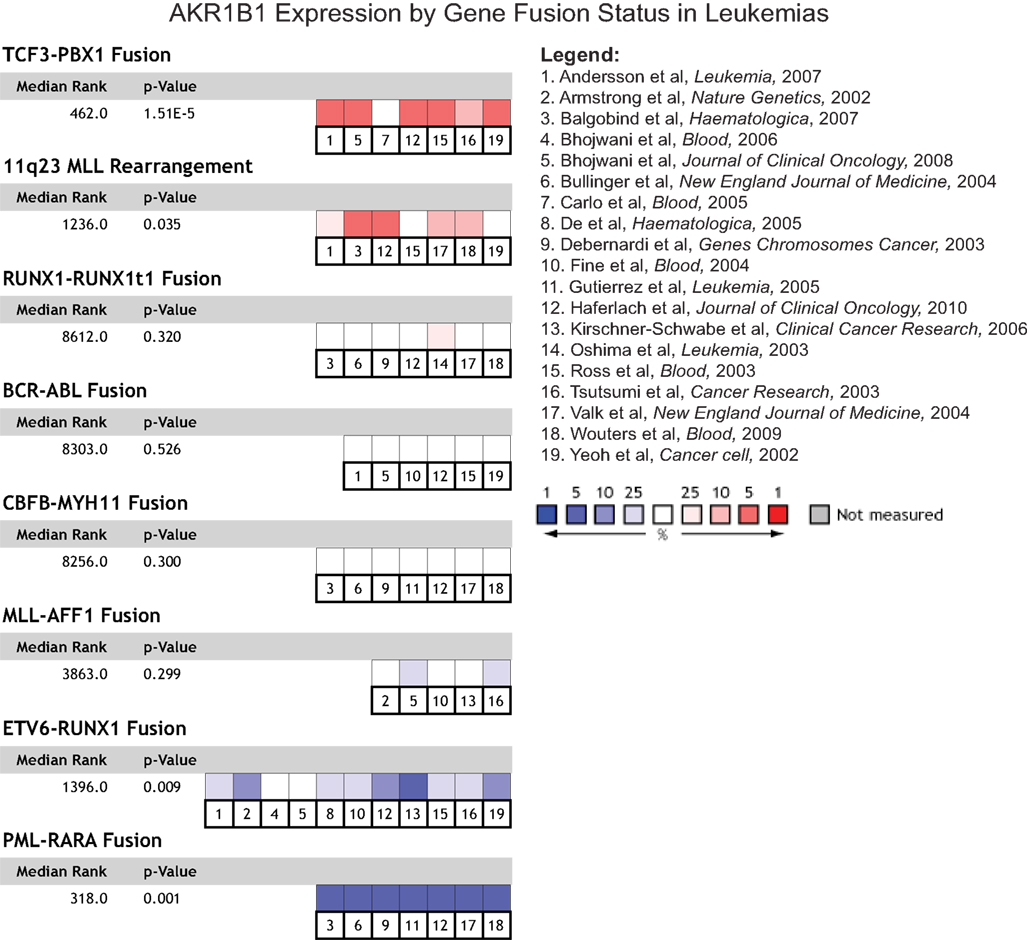
Figure 2. AKR1B1 expression by gene fusion status in leukemia patients. AKR1B1 mRNA expression in leukemia patients with specific gene fusions and chromosomal rearrangements was compared to corresponding leukemia patients without the fusion across all leukemia types and for all such events where Oncomine contained multiple studies with such data. The heatmaps represent the relative expression in patients with the indicated fusions compared to those without, with red indicating over-expression in patients bearing the fusion and blue under-expression. Median ranks and p-values consider all indicated studies simultaneously.
We next asked whether expression of AKRs might be able to predict clinical outcome, specifically in terms of patient survival, disease recurrence, tumor grade, and metastasis. We found no significant associations with expression of AKR1B1 and the presence of metastasis, tumor grade, or with disease recurrence in any cancer type, though some individual studies sometimes contained a significant relationship that did not hold up when all available data for that cancer type was considered (data not shown). AKR1B1 over-expression was associated with decreased patient survival at 1 year post-prognosis in acute myeloid leukemias (Figure 3A), as well as decreased patient survival at 1 year post-prognosis in multiple myeloma (Figure 3B). AKR1B1 over-expression was also associated with decreased survival in pancreatic cancer, however, only one small study (27 patients) within Oncomine contained patient survival data (data not shown). While no significant associations of patient survival with AKR1B10 expression were observed, it is noteworthy that in the solid tumors where AKR1B10 is most highly over-expressed, namely liver cancer and squamous cell lung carcinoma, there is a strong trend for AKR1B10 over-expression predicting longer patient survival (Figure 3C).
Figure 3. AKR gene expression and clinical outcome. The relationship between AKR1B1 and AKR1B10 mRNA expression and clinical outcomes were examined using the Oncomine database. (A) Comparison of AKR1B1 expression in acute myeloid leukemia patients who were alive (blue boxes) and dead (red boxes) at 1 year post diagnosis. Individual p-values are indicated within each box plot and the p-value and median gene rank for all three studies is at the right of the panel. (B) Comparison of AKR1B1 expression in multiple myeloma patients who were alive (blue boxes) and dead (red boxes) at 1 year post diagnosis. Individual p-values are indicated within each box plot and the p-value and median gene rank for all three studies is at the right of the panel. (C) AKR1B10 expression relative to clinical outcome in liver cancer and squamous cell lung cancer. Colored boxes are a heatmap-style representation of AKR1B10 expression in patients dead relative to those alive at 3 years post diagnosis, with blue indicating under-expression and red over-expression. Median rank and p-value for this panel considers all indicated studies simultaneously.
Discussion
In this report we show that AKR1B1 and AKR1B10 are over-expressed, and less frequently under-expressed, in a cancer-type-specific manner. AKR1B10 is most prominently up-regulated in cancers of the liver and lungs, consistent with previous reports (Fukumoto et al., 2005; Woenckhaus et al., 2006; Heringlake et al., 2010; Kang et al., 2011; Schmitz et al., 2011). AKR1B1 over-expression is more common amongst different tumor types than AKR1B10 over-expression, but at a generally lower magnitude (Table 1). Under-expression is less common for either AKR, with AKR1B1 under-expressed in prostate tumors and AKR1B10 under-expressed in colon and head and neck cancers (Table 1). Increased AKR1B1 expression is also associated with the TCF3-PBX1 gene fusion and 11q23 MLL rearrangement in acute leukemias, while decreased expression is associated with the PML-RARA and ETV6-RUNX1 gene fusions (Figure 2). Only AKR1B1 expression has a significant association with clinical outcome, being associated with reduced survival in acute myeloid leukemias and multiple myeloma (Figure 3). Recent reports have implicated AKRs in cellular responses to various stresses, including promotion of hypoxia-driven HIF1a signaling, inflammation, and resistance to chemotherapeutics (Dan et al., 2003; Plebuch et al., 2007; Yadav et al., 2007, 2009, 2011; Matsunaga et al., 2011; Zhong et al., 2011). AKR1B1 over-expression has also been associated with an EMT-like phenotype, is implicated in colon carcinogenesis, and notably, increased AKR1B1 protein expression and enzymatic activity has been reported in several cancer types (Saraswat et al., 2006; Tammali et al., 2009, 2011a,b; Ramana et al., 2010; Zablocki et al., 2011), further suggesting that AKRs play a functional role in tumor growth. Given the broad over-expression of AKRs, particularly AKR1B1, in human cancers and the critical processes that they appear to regulate, AKRs have potential to be useful therapeutic targets. AKR inhibitors have been in development for complications related to diabetes for many years, as AKR1B1 and the polyol pathway have been implicated in the pathogenesis of diabetic retinopathy, nephropathy, and cataract (Makiishi et al., 2003; Suryanarayana et al., 2004, 2007; Wolford et al., 2006; Reddy et al., 2008, 2011; Zablocki et al., 2011). While many of these AKR inhibitor drug development efforts have been halted due to toxicity, they exhibit much lower toxicity than many current cancer therapies.
AKR1B1 expression is increased by high blood glucose via NF-κB (Yang et al., 2008), providing a potential mechanism by which diabetes and elevated risk of developing certain cancers may be linked. AKR1B1 and the polyol pathway also contribute to hyperglycemic pseudohypoxia, which one could imagine linking the Warburg effect to tumor angiogenesis through HIF1a and perhaps bolstering neovascularization at oxygen tensions that would not normally promote it. Consistent with this, VEGF has been linked to diabetic retinopathy and nephropathy (Aiello et al., 1994; Cha et al., 2000; Ozaki et al., 2000), perhaps downstream of AKR1B1-driven pseudohypoxic effects. Intriguingly, patients with Von Hippel–Lindau disease often develop retinal angiomas and kidney tumors, suggesting that VHL-associated malignancies and diabetic complications may differ primarily by the degree of HIF1a and/or VEGF-dysregulation present. It is possible that diabetics are effectively primed to promote tumorigenesis by virtue of an already abnormally high level of hypoxia/HIF1a signaling. In light of all the signs pointing to the involvement of AKRs in human cancers, we hypothesize that AKRs are functionally linked to cancer progression, if not initiation as well. We also propose that AKR inhibitors would have value as cancer therapeutics in cancers that typically feature AKR over-expression, especially in the case of AKR1B1.
References for Studies Analyzed Within the Oncomine Database
We apologize to our colleagues whose papers used in the meta-analyses are not cited here due the cumbersome nature of including these hundreds of references. For a list of studies used in the analyses for a given tumor type, please contact the authors.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge the assistance of Dr. Dean Tang with meta-analysis techniques, and the members of the Petrash lab for helpful discussions and critical reading of this manuscript. This work was funded in part by NIH grants EY005856 and EY021498.
References
Aiello, L. P., Avery, R. L., Arrigg, P. G., Keyt, B. A., Jampel, H. D., Shah, S. T., Pasquale, L. R., Thieme, H., Iwamoto, M. A., Park, J. E., Nguyen, H. V., Aiello, L. M., Ferrara, N., and King, G. L. (1994). Vascular endothelial growth factor in ocular fluid of patients with diabetic retinopathy and other retinal disorders. N. Engl. J. Med. 331, 1480–1487.
Cha, D. R., Kim, N. H., Yoon, J. W., Jo, S. K., Cho, W. Y., Kim, H. K., and Won, N. H. (2000). Role of vascular endothelial growth factor in diabetic nephropathy. Kidney Int. Suppl. 77, S104–S112.
Dan, S., Shirakawa, M., Mukai, Y., Yoshida, Y., Yamazaki, K., Kawaguchi, T., Matsuura, M., Nakamura, Y., and Yamori, T. (2003). Identification of candidate predictive markers of anticancer drug sensitivity using a panel of human cancer cell lines. Cancer Sci. 94, 1074–1082.
Fukumoto, S., Yamauchi, N., Moriguchi, H., Hippo, Y., Watanabe, A., Shibahara, J., Taniguchi, H., Ishikawa, S., Ito, H., Yamamoto, S., Iwanari, H., Hironaka, M., Ishikawa, Y., Niki, T., Sohara, Y., Kodama, T., Nishimura, M., Fukayama, M., Dosaka-Akita, H., and Aburatani, H. (2005). Overexpression of the aldo-ketoreductase family protein AKR1B10 is highly correlated with smokers’ non-small cell lung carcinomas. Clin. Cancer Res. 11, 1776–1785.
Haferlach, T., Kohlmann, A., Wieczorek, L., Basso, G., Kronnie, G. T., Béné, M. C., De Vos, J., Hernández, J. M., Hofmann, W. K., Mills, K. I., Gilkes, A., Chiaretti, S., Shurtleff, S. A., Kipps, T. J., Rassenti, L. Z., Yeoh, A. E., Papenhausen, P. R., Liu, W. M., Williams, P. M., and Foà, R. (2010). Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the International Microarray Innovations in Leukemia Study Group. J. Clin. Oncol. 28, 2529–2537.
Heringlake, S., Hofdmann, M., Fiebeler, A., Manns, M. P., Schmiegel, W., and Tannapfel, A. (2010). Identification and expression analysis of the aldo-ketoreductase1-B10 gene in primary malignant liver tumours. J. Hepatol. 52, 220–227.
Kang, M. W., Lee, E. S., Yoon, S. Y., Jo, J., Lee, J., Kim, H. K., Choi, Y. S., Kim, K., Shim, Y. M., Kim, J., and Kim, H. (2011). AKR1B10 is associated with smoking and smoking-related non-small-cell lung cancer. J. Int. Med. Res. 39, 78–85.
Makiishi, T., Araki, S., Koya, D., Maeda, S., Kashiwagi, A., and Haneda, M. (2003). C-106T polymorphism of AKR1B1 is associated with diabetic nephropathy and erythrocyte aldose reductase content in Japanese subjects with type 2 diabetes mellitus. Am. J. Kidney Dis. 42, 943–951.
Matsunaga, T., Yamane, Y., Iida, K., Endo, S., Banno, Y., El-Kabbani, O., and Hara, A. (2011). Involvement of the aldo-ketoreductase, AKR1B10, in mitomycin-c resistance through reactive oxygen species-dependent mechanisms. Anticancer Drugs 22, 402–408.
Ozaki, H., Seo, M. S., Ozaki, K., Yamada, H., Yamada, E., Okamoto, N., Hofmann, F., Wood, J. M., and Campochiaro, P. A. (2000). Blockade of vascular endothelial cell growth factor receptor signaling is sufficient to completely prevent retinal neovascularization. Am. J. Pathol. 156, 697–707.
Plebuch, M., Soldan, M., Hungerer, C., Koch, L., and Maser, E. (2007). Increased resistance of tumor cells to daunorubicin after transfection of cDNAs coding for anthracycline inactivating enzymes. Cancer Lett. 255, 49–56.
Ramana, K. V., Tammali, R., and Srivastava, S. K. (2010). Inhibition of aldose reductase prevents growth factor-induced G1-S phase transition through the AKT/phosphoinositide 3-kinase/E2F-1 pathway in human colon cancer cells. Mol. Cancer Ther. 9, 813–824.
Reddy, A. B., Tammali, R., Mishra, R., Srivastava, S., Srivastava, S. K., and Ramana, K. V. (2011). Aldose reductase deficiency protects sugar-induced lens opacification in rats. Chem. Biol. Interact. 191, 346–350.
Reddy, G. B., Satyanarayana, A., Balakrishna, N., Ayyagari, R., Padma, M., Viswanath, K., and Petrash, J. M. (2008). Erythrocyte aldose reductase activity and sorbitol levels in diabetic retinopathy. Mol. Vis. 14, 593–601.
Saraswat, M., Mrudula, T., Kumar, P. U., Suneetha, A., RaoRao, T. S., Srinivasulu, M., and Reddy, B. (2006). Overexpression of aldose reductase in human cancer tissues. Med. Sci. Monit. 12, CR525–CR529.
Saraswat, M., Muthenna, P., Suryanarayana, P., Petrash, J. M., and Reddy, G. B. (2008). Dietary sources of aldose reductase inhibitors: prospects for alleviating diabetic complications. Asia Pac. J. Clin. Nutr. 17, 558–565.
Schmitz, K. J., Sotiropoulos, G. C., Baba, H. A., Schmid, K. W., Müller, D., Paul, A., Auer, T., Gamerith, G., and Loeffler-Ragg, J. (2011). AKR1B10 expression is associated with less aggressive hepatocellular carcinoma: a clinicopathological study of 168 cases. Liver Int. 31, 810–816.
Suryanarayana, P., Kumar, P. A., Saraswat, M., Petrash, J. M., and Reddy, G. B. (2004). Inhibition of aldose reductase by tannoid principles of Emblica officinalis: implications for the prevention of sugar cataract. Mol. Vis. 10, 148–154.
Suryanarayana, P., Saraswat, M., Petrash, J. M., and Reddy, G. B. (2007). Emblica officinalis and its enriched tannoids delay streptozotocin-induced diabetic cataract in rats. Mol. Vis. 13, 1291–1297.
Tammali, R., Reddy, A. B., Ramana, K. V., Petrash, J. M., and Srivastava, S. K. (2009). Aldose reductase deficiency in mice prevents azoxymethane-induced colonic preneoplastic aberrant crypt foci formation. Carcinogenesis 30, 799–807.
Tammali, R., Reddy, A. B., Saxena, A., Rychahou, P. G., Evers, B. M., Qiu, S., Awasthi, S., Ramana, K. V., and Srivastava, S. K. (2011a). Inhibition of aldose reductase prevents colon cancer metastasis. Carcinogenesis 32, 1259–1267.
Tammali, R., Saxena, A., Srivastava, S. K., and Ramana, K. V. (2011b). Aldose reductase inhibition prevents hypoxia-induced increase in hypoxia-inducible factor-1alpha (HIF-1alpha) and vascular endothelial growth factor (VEGF) by regulating 26 S proteasome-mediated protein degradation in human colon cancer cells. J. Biol. Chem. 286, 24089–24100.
Tammali, R., Reddy, A. B., Srivastava, S. K., and Ramana, K. V. (2011c). Inhibition of aldose reductase prevents angiogenesis in vitro and in vivo. Angiogenesis 14, 209–221.
Woenckhaus, M., Klein-Hitpass, L., Grepmeier, U., Merk, J., Pfeifer, M., Wild, P., Bettstetter, M., Wuensch, P., Blaszyk, H., Hartmann, A., Hofstaedter, F., and Dietmaier, W. (2006). Smoking and cancer-related gene expression in bronchial epithelium and non-small-cell lung cancers. J. Pathol. 210, 192–204.
Wolford, J. K., Yeatts, K. A., Red Eagle, A. R., Nelson, R. G., Knowler, W. C., and Hanson, R. L. (2006). Variants in the gene encoding aldose reductase (AKR1B1) and diabetic nephropathy in American Indians. Diabet. Med. 23, 367–376.
Yadav, U. C., Ramana, K. V., Aguilera-Aguirre, L., Boldogh, I., Boulares, H. A., and Srivastava, S. K. (2009). Inhibition of aldose reductase prevents experimental allergic airway inflammation in mice. PLoS ONE 4, e6535. doi:10.1371/journal.pone.0006535
Yadav, U. C., Shoeb, M., Srivastava, S. K., and Ramana, K. V. (2011). Aldose reductase deficiency protects from autoimmune- and endotoxin-induced uveitis in mice. Invest. Ophthalmol. Vis. Sci. 52, 8076–8085.
Yadav, U. C., Srivastava, S. K., and Ramana, K. V. (2007). Aldose reductase inhibition prevents endotoxin-induced uveitis in rats. Invest. Ophthalmol. Vis. Sci. 48, 4634–4642.
Yang, B., Hodgkinson, A., Oates, P. J., Millward, B. A., and Demaine, A. G. (2008). High glucose induction of DNA-binding activity of the transcription factor NFkappaB in patients with diabetic nephropathy. Biochim. Biophys. Acta 1782, 295–302.
Zablocki, G. J., Ruzycki, P. A., Overturf, M. A., Palla, S., Reddy, G. B., and Petrash, J. M. (2011). Aldose reductase-mediated induction of epithelium-to-mesenchymal transition (EMT) in lens. Chem. Biol. Interact. 191, 351–356.
Keywords: AKR1B1, AKR1B10, HSIR, aldose reductase, cancer, leukemia, meta-analysis
Citation: Laffin B and Petrash JM (2012) Expression of the Aldo-Ketoreductases AKR1B1 and AKR1B10 in Human Cancers. Front. Pharmacol. 3:104. doi: 10.3389/fphar.2012.00104
Received: 05 March 2012; Paper pending published: 09 April 2012;
Accepted: 13 May 2012; Published online: 06 June 2012.
Edited by:
Yi Jin, University of Pennsylvania, USAReviewed by:
Tea Lanisnik Rizner, University of Ljubljana, SloveniaKota V. Ramana, University of Texas Medical Branch, USA
Copyright: © 2012 Laffin and Petrash. This is an openaccess article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: J. Mark Petrash, Department of Ophthalmology, The School of Medicine, University of Colorado, 12800 East 19th Avenue, Mail Stop 8311RC1-North, 5100, Aurora, CO 80045, USA. e-mail: mark.petrash@ucdenver.edu