- Department of Biotechnology, The University of Tokyo, Tokyo, Japan
It has long been regarded that the primary function of fungal peroxisomes is limited to the β-oxidation of fatty acids, as mutants lacking peroxisomal function fail to grow in minimal medium containing fatty acids as the sole carbon source. However, studies in filamentous fungi have revealed that peroxisomes have diverse functional repertoires. This review describes the essential roles of peroxisomes in the growth and survival processes of filamentous fungi. One such survival mechanism involves the Woronin body, a Pezizomycotina-specific organelle that plugs the septal pore upon hyphal lysis to prevent excessive cytoplasmic loss. A number of reports have demonstrated that Woronin bodies are derived from peroxisomes. Specifically, the Woronin body protein Hex1 is targeted to peroxisomes by peroxisomal targeting sequence 1 (PTS1) and forms a self-assembled structure that buds from peroxisomes to form the Woronin body. Peroxisomal deficiency reduces the ability of filamentous fungi to prevent excessive cytoplasmic loss upon hyphal lysis, indicating that peroxisomes contribute to the survival of these multicellular organisms. Peroxisomes were also recently found to play a vital role in the biosynthesis of biotin, which is an essential cofactor for various carboxylation and decarboxylation reactions. In biotin-prototrophic fungi, peroxisome-deficient mutants exhibit growth defects when grown on glucose as a carbon source due to biotin auxotrophy. The biotin biosynthetic enzyme BioF (7-keto-8-aminopelargonic acid synthase) contains a PTS1 motif that is required for both peroxisomal targeting and biotin biosynthesis. In plants, the BioF protein contains a conserved PTS1 motif and is also localized in peroxisomes. These findings indicate that the involvement of peroxisomes in biotin biosynthesis is evolutionarily conserved between fungi and plants, and that peroxisomes play a key role in fungal growth.
Introduction
Peroxisomes are ubiquitous organelles in eukaryotic cells and typically contain enzymes involved in the β-oxidation of fatty acids and detoxification of reactive oxygen species. Additionally, peroxisomes are known to have various physiological functions based on their roles in diverse metabolic activities. For example, mammalian peroxisomes participate in the lipid biosynthesis such as ether phospholipids, and in the oxidation of amino acids and polyamines (Wanders and Waterham, 2006). In plants, peroxisomes are involved in the glyoxylate cycle (Mano and Nishimura, 2005), photorespiration (Reumann and Weber, 2006), male-female gametophyte recognition (Boisson-Dernier et al., 2008) and biosynthesis of the hormones jasmonic acid and auxin (Weber, 2002; Woodward and Bartel, 2005). Peroxisomes also play important roles in higher eukaryotes, with defects in peroxisome biogenesis resulting in severe human disease, such as Zellweger syndrome, neonatal adrenoleukodystrophy, and Refsums disease (Waterham and Ebberink, 2012). In plants, loss of peroxisomal function causes embryo lethality, suggesting that peroxisomes have an essential role in growth and development (Hu et al., 2002; Schumann et al., 2003; Sparkes et al., 2003; Tzafrir et al., 2004; Fan et al., 2005).
The primary role of fungal peroxisomes is the β-oxidation of fatty acids, as fungal mutants lacking peroxisomes fail to grow in minimal medium containing fatty acids as the sole carbon source (Erdmann et al., 1989; Hynes et al., 2008). Peroxisomes are also required for methanol metabolism in methylotrophic yeasts, including Pichia pastoris (van der Klei et al., 2006). In filamentous fungi, peroxisomes are also involved in secondary metabolism including the biosynthesis of penicillin, AK (Alternaria kikuchiana) toxin, and paxilline (Saikia and Scott, 2009; Imazaki et al., 2010; Bartoszewska et al., 2011), plant pathogenicity (Kimura et al., 2001; Asakura et al., 2006), and sexual development (Bonnet et al., 2006; Peraza-Reyes et al., 2008). While fungal peroxisomes are known to proliferate massively on oleate and acetate, inducing substrates for this organelle (van der Klei and Veenhuis, 2006), it is apparent that many peroxisomes constitutively exist in the cell of filamentous fungi under the normal growth condition e.g., on glucose (Tanabe et al., 2011). The delayed growth and aberrant organelle morphologies observed in peroxisome-deficient mutants (Bonnet et al., 2006; Idnurm et al., 2007; Hynes et al., 2008) suggest that peroxisomes have fundamental roles for the growth of filamentous fungi. However, the molecular mechanisms underlying these severe growth effects remain unknown. In this review, evidence for the roles of peroxisomes in fungal growth, particularly the involvement of the Woronin body, a peroxisome-derived organelle with wound-healing function, and the recently identified function of peroxisomes in vitamin biosynthesis are presented.
The Woronin Body, An Organelle Specific to Pezizomycotina Species, Differentiates from Peroxisomes
Species of Pezizomycotina (filamentous ascomycetes) grow via elongation of the hyphal tip to form straight primary hyphae with branches. The hyphae are divided into distinct cells by the formation of septa, and thus filamentous fungi are characterized by multicellularity. The septum is proposed to have several possible functions, including increasing the mechanical integrity of hyphae and division of mycelium into sections that undergo distinct developmental processes. However, septa do not completely separate adjacent cells in the hyphae due to the presence of a septal pore, which allows the passage of cytoplasm and organelles between adjacent cells (Markham, 1994; Freitag et al., 2004; Lew, 2005; Tey et al., 2005; Ng et al., 2009). This intercellular communication resembles that found in higher eukaryotes, such as gap junctions in animal cells and plasmodesmata in plant cells, and suggests that filamentous fungi possess a cell-to-cell channel that modulates responses to environmental changes and development processes necessary for multicellularity.
Cytoplasmic continuity between adjacent cells through the septal pore is associated with catastrophic risk of cytoplasmic loss by adjacent cells due to hyphal lysis. This risk was clearly demonstrated by the exposure of the filamentous fungus Aspergillus oryzae grown on agar medium to hypotonic shock, which caused most of the hyphal tips to burst and lose the cytoplasmic constituents (Figure 1A) (Maruyama et al., 2005). However, as evidenced by differential interference contrast (DIC) and fluorescence microscopy, ~80% of the immediately adjacent cells retained their cytoplasmic constituents (Figure 1B) (Maruyama et al., 2005), allowing these cells to initiate regrowth by creating a new hyphal tip (Maruyama et al., 2006; Maruyama and Kitamoto, 2007). This process represents a type of defense system that aims to promote the survival of these multicellular organisms by preventing the excessive loss of cytoplasm upon hyphal lysis.
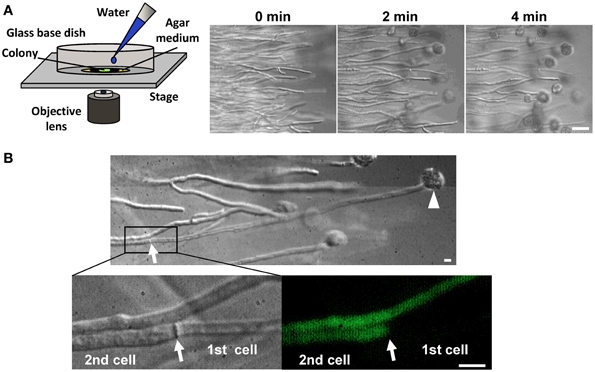
Figure 1. Hyphal tip bursting upon hypotonic shock in the filamentous fungus A. oryzae. (A) Time-lapse observation of hyphal tip bursting upon hypotonic shock. Hyphal tips at the edge of a colony grown on agar medium were observed by DIC microscopy before and after flooding hyphae with water. Bar: 50μm. (B) Excessive loss of cytoplasmic constituents is prevented upon hyphal tip bursting induced by hypotonic shock. The cytoplasm was labeled by EGFP. An arrowhead and arrow indicate a burst hyphal tip and the adjacent septum, respectively. Note that the cell (2nd) adjacent to the lysed cell (1st) retains its cytoplasmic constituents, as determined by DIC and fluorescence microcopy. Bar: 10μm.
The Woronin body is a unique organelle present in Pezizomycotina species that plugs the septal pore upon hyphal lysis and prevents excessive cytoplasmic loss from the cell adjacent to the lysed cell (Figure 2A) (Markham and Collinge, 1987). This organelle has two morphologically distinct subclasses; it is generally observed by transmission electron microscopy as a spherical electron-dense structure in the vicinity of the septum (Figure 2B), although a limited number of species, such as Neurospora crassa, form hexagonal crystalline Woronin bodies that are occasionally visible by light microscopy (Markham, 1994).
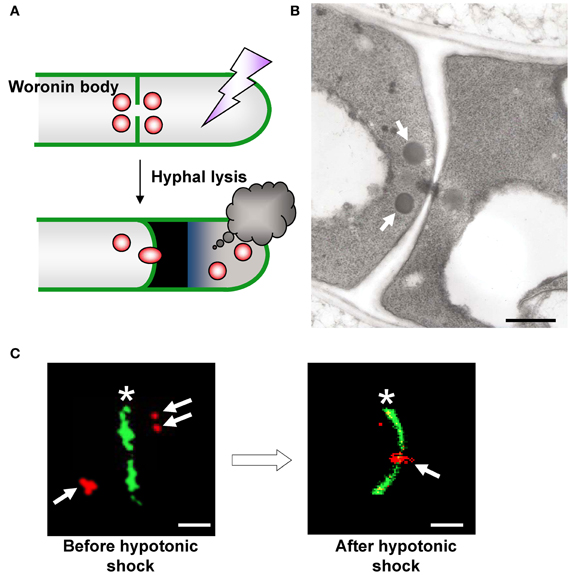
Figure 2. Morphology and function of the Woronin body. (A) Schematic model of Woronin body function. (B) Transmission electron microscopic observation of Woronin bodies (arrows) in A. oryzae (Maruyama et al., 2005). Bar: 500 nm. (C) Confocal images of Woronin bodies (red, arrows) and septa (green, asterisks) before (left) and after (right) hyphal tip bursting induced by hypotonic shock. Woronin bodies and septa were fluorescently labeled by expressing DsRed2–AoHex1 and RNase T1–EGFP fusion proteins, respectively (Maruyama et al., 2005). Bar: 2μm.
Jedd and Chua (2000) first identified Hex1 as a major protein of the Woronin body in N. crassa. Genes encoding the Hex1 protein are conserved in Pezizomycotina species (Jedd and Chua, 2000; Asiegbu et al., 2004; Curach et al., 2004; Soundararajan et al., 2004; Maruyama et al., 2005; Beck and Ebel, 2013). Self-assembly of Hex1 provides the Woronin body with a mechanically solid core that provides resistance to the protoplasmic streaming pressure arising from hyphal lysis (Jedd and Chua, 2000; Yuan et al., 2003). Phosphorylation of Hex1 has a role in the formation of the multimeric core of the Woronin body (Tenney et al., 2000; Juvvadi et al., 2007). Deletion of the hex1 gene results in the disappearance of Woronin bodies and is associated with severe cytoplasmic bleeding upon hyphal lysis (Jedd and Chua, 2000; Tenney et al., 2000; Maruyama et al., 2005). In the case of A. oryzae, hex1 deletion (Δhex1) significantly reduces the ability of this strain to prevent excessive cytoplasmic loss (Maruyama et al., 2005, 2010; Escaño et al., 2009). Using fluorescence microscopy, Woronin bodies were demonstrated to plug the septal pore adjacent to a lysed cell upon hyphal lysis in the A. oryzae wild-type strain (Figure 2C) (Maruyama et al., 2005). Recently, Bleichrodt et al. (2012) reported that the Woronin body reversibly closes the septal pore during normal growth of A. oryzae, a function that contrasts the behavior of this organelle conventionally observed during hyphal lysis. In addition, although wild-type A. oryzae has heterogeneous distribution of hyphae and gene expression activity, the absence of Woronin bodies results in uniform activity distribution of different cells (Bleichrodt et al., 2012). Collectively, Woronin bodies impede cytoplasmic continuity between adjacent cells during normal growth and help maintain hyphal heterogeneity in mycelia. This function of Woronin bodies may represent the most primitive way to regulate cell-to-cell channels in multicellularity by a simple plugging behavior similar to that upon hyphal lysis. Additionally, the roles of Woronin bodies in conidiation (asexual spore formation), survival under nitrogen starvation and efficient plant pathogenesis were reported (Yuan et al., 2003; Soundararajan et al., 2004).
A relationship between peroxisomes and the Woronin body is suggested from the fact that Hex1 contains peroxisomal targeting signal sequence 1 (PTS1) at the C-terminus (Jedd and Chua, 2000). Time-lapse imaging demonstrated that Woronin bodies bud from peroxisomes in N. crassa (Tey et al., 2005) and that Woronin body biogenesis requires the presence of peroxins that mediate peroxisomal protein import (Ramos-Pamplona and Naqvi, 2006; Managadze et al., 2007; Liu et al., 2008). The peripheral membrane peroxisomal protein Pex11 is implicated in peroxisomal proliferation and division (Erdmann and Blobel, 1995; Marshall et al., 1995), and in the absence of Pex11, filamentous fungi only contain few enlarged peroxisomes (Figure 3A, EGFP-PTS1) (Hynes et al., 2008; Escaño et al., 2009; Opaliński et al., 2012). It was also demonstrated that ability of Pex11-deficient strain of A. oryzae to prevent the excessive loss of cytoplasm is reduced by ~30% compared to wild type (Escaño et al., 2009), indicating that Pex11 is involved in Woronin body function. Under fluorescence microscopy, Woronin bodies are typically observed as small dots independent of peroxisomes (Figure 3A, mDsRed-AoHex1). In the absence of Pex11, however, the Woronin body protein Hex1 forms a structure that attaches to the matrix side of the peroxisomal membrane, but the mature Woronin body fails to differentiate from peroxisomes (Figure 3A) (Escaño et al., 2009). The Pezizomycotina-specific protein WSC (Woronin body sorting complex) recruits the Hex1 assembly to the matrix side of the peroxisomal membrane and facilitates the budding of the Woronin body (Liu et al., 2008). It has been suggested that Pex11 elongates the peroxisomal membrane to facilitate the division of peroxisomes by dynamin-related proteins (Koch et al., 2003, 2004; Schrader, 2006). Heterologous expression of Hex1 in the yeast Saccharomyces cerevisiae suggested that dynamin-related proteins participate in the budding of Woronin bodies from peroxisomes (Würtz et al., 2008). ApsB, a component of the microtubule-organizing center (MTOC), has been shown to interact with Hex1 and to localize to peroxisomes via peroxisomal targeting signal sequence 2 (PTS2) (Zekert et al., 2010). Hex1 physically associates with the essential matrix import peroxin Pex26 and promotes the enrichment of Pex26 in the membranes of differentiated peroxisomes (Liu et al., 2011). After Woronin bodies differentiate from peroxisomes, evidence suggests that the Pezizomycotina-specific protein Leashin (LAH) tethers the Woronin bodies to the vicinity of the septum (Ng et al., 2009). A schematic model of Woronin body differentiation from peroxisomes is presented in Figure 3B. Although a number of proteins functionally/spatially related to the Woronin body have been identified (Engh et al., 2007; Fleissner and Glass, 2007; Kim et al., 2009; Maruyama et al., 2010; Lai et al., 2012; Yu et al., 2012), the molecular mechanism for Woronin body biogenesis remains to be completely resolved.
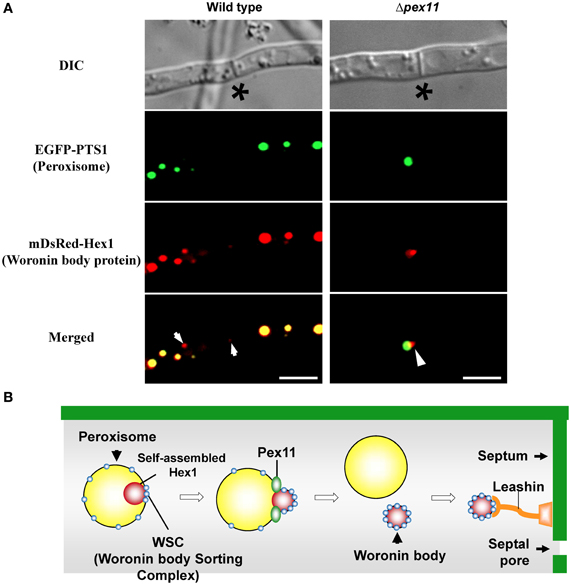
Figure 3. Differentiation of the Woronin body from peroxisomes. (A) Fluorescence microscopic analysis of wild-type and Δpex11 strains of A. oryzae expressing EGFP-PTS1 and mDsRed-AoHex1 fusion proteins for visualization of peroxisomes and the major Woronin body protein, respectively (Escaño et al., 2009). Asterisks denote septa and arrows indicate Woronin bodies (red) independent of peroxisomes (green). Arrowheads represent assembly of Hex1 attached to the matrix side of the peroxisome. Bars: 5 μm. (B) Schematic model of Woronin body differentiation from peroxisomes.
Involvement of Peroxisomes in Biotin Biosynthesis in Fungi
Biotin is an essential cofactor involved in a number of carboxylation and decarboxylation reactions (Knowles, 1989). In eukaryotes, plants and numerous fungal species are capable of synthesizing biotin. The studies of plants and fungi have revealed that the final four reactions in the biosynthetic process, which convert pimeloyl-CoA to biotin, are conserved (Figure 4A) (Streit and Entcheva, 2003). In plants, the enzymes BioF, Bio1, Bio3, and Bio2 mediate the final four steps of biotin biosynthesis. It was previously reported that BioF, a 7-keto-8-aminopelargonic acid (KAPA) synthase catalyzing the conversion of pimeloyl-CoA to KAPA, is localized to the cytoplasm (Pinon et al., 2005). The final three reactions converting KAPA to biotin occur in mitochondria. The BIO3 and BIO1 genes are unidirectionally aligned and expressed as a chimeric transcript, resulting in the production of Bio3-Bio1 as a bifunctional protein catalyzing desthiobiotin (DTB) synthase and 7, 8-diaminopelargonic acid (DAPA) synthase reactions (Muralla et al., 2008). Bio3-Bio1 contains a mitochondrial targeting sequence (MTS) and localizes in mitochondria (Muralla et al., 2008; Cobessi et al., 2012). The Bio2 protein, a biotin synthase catalyzing the conversion of DTB to biotin, also contains a MTS and must be mitochondrially localized for biotin prototrophy (Baldet et al., 1997; Picciocchi et al., 2001; Arnal et al., 2006). It was therefore suggested that plant biotin biosynthesis occurs in both the cytoplasm and mitochondria (Rébeillé et al., 2007).
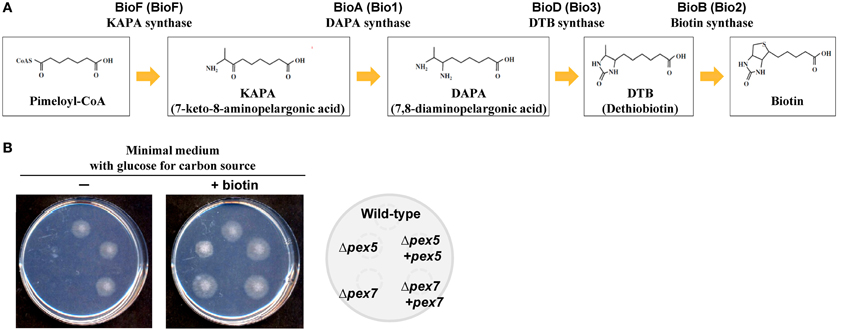
Figure 4. Biotin biosynthetic pathway and biotin auxotrophy in peroxisome-deficient strains. (A) The final four reactions of the biotin biosynthetic pathway and involved enzymes from fungi and plants (in parentheses). (B) Growth impairment of strains defective in peroxisomal targeting signal receptors (Δpex5 and Δpex7) grown on minimal medium containing glucose as the sole carbon source. Growth of the wild-type, Δpex5, and Δpex7 strains and the complemented strains (indicated to the right) on medium with and without biotin.
In Aspergillus species, mutants of the pex5 and pex7 genes are defective in protein import into the peroxisomal matrix due to the lack of PTS1 and PTS2 receptors, respectively (Hynes et al., 2008; Tanabe et al., 2011). These mutants fail to grow in minimal medium containing oleic acid as the sole carbon source due to defective peroxisomal β-oxidation; however, unlike the corresponding yeast mutants, the mutants of Aspergillus species also exhibit growth defects when grown on glucose (Hynes et al., 2008; Tanabe et al., 2011). Surprisingly, the growth defects are restored by the addition of biotin (Figure 4B). In fungi, biotin is synthesized through the sequential activities of three Bio proteins: BioF, a KAPA synthase; BioD/A, a chimeric protein composed of DTB and DAPA synthases; and BioB, a biotin synthase (Figure 4A) (Magliano et al., 2011a,b; Tanabe et al., 2011). The BioD/A protein localizes in mitochondria, suggesting that this is where KAPA is converted to biotin (Tanabe et al., 2011). Protein sequence analysis of fungal BioF proteins revealed that the C-terminal PTS1 sequences are conserved in ascomycete and basidiomycete species (Figure 5A). Consistent with these findings, BioF protein localizes in the peroxisomes via PTS1 (Figure 5B), and the peroxisomal targeting of this KAPA synthase is required for biotin biosynthesis (Magliano et al., 2011a; Tanabe et al., 2011). Yeast species appear to have lost the gene encoding BioF, as evidenced by their biotin auxotrophy, although several species have reacquired biotin prototrophy by horizontal gene transfer and gene duplication followed by neofunctionalization (Hall and Dietrich, 2007).
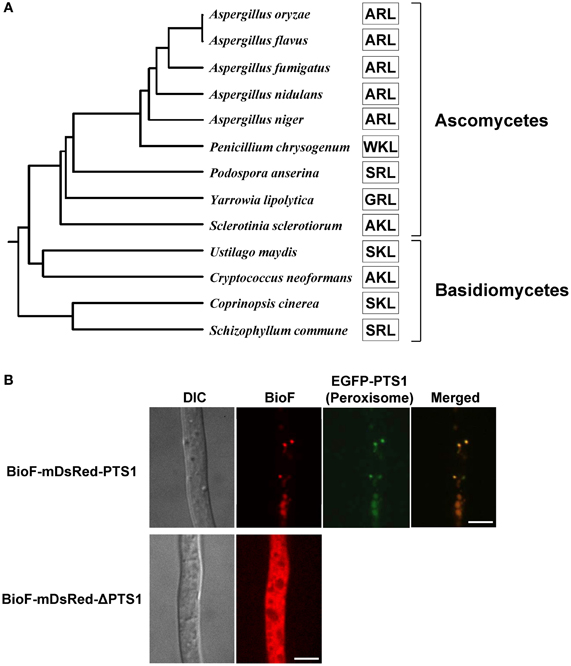
Figure 5. Phylogenetic relationship and peroxisomal localization of fungal BioF proteins. (A) Phylogenetic analysis of fungal BioF proteins. The amino acid residues of the C-terminal peroxisomal targeting signals (PTS1) are indicated by open boxes. The full-length amino acid sequences of the fungal BioF proteins were aligned using the Clustal W program (version 2.1), and then the phylogenetic tree was constructed. The Genbank accession numbers for the sequences used in the analysis are as follows: Aspergillus oryzae, XP_001817022.1; Aspergillus flavus, XP_002383037.1; Aspergillus fumigatus, XP_747713.1; Aspergillus nidulans, ACR44939.1; Aspergillus niger, XP_001396737.1; Penicillium chrysogenum, XP_002563821.1; Podospora anserina, XP_001903515.1; Yarrowia lipolytica, XP_504066.1; Sclerotinia sclerotiorum, XP_001590700.1; Ustilago maydis, XP_757344.1; Cryptococcus neoformans, XP_566616.1; Coprinopsis cinerea, XP_001836705.2; and Schizophyllum commune, XP_003028193.1. (B) Peroxisomal localization of fungal BioF protein. Note that BioF (BioF-mDsRed-PTS1) co-localizes with peroxisomes (EGFP-PTS1), but disperses in the absence of PTS1 (BioF-mDsRed-ΔPTS1) (Tanabe et al., 2011). Bars: 5 μm.
A new model for biotin biosynthesis in fungi is proposed in Figure 6. In this biotin biosynthesis pathway, the production of pimeloyl-CoA may involve proteins containing PTS1 and PTS2 (Tanabe et al., 2011), while peroxisomal β-oxidation is also involved (Magliano et al., 2011a). Ohsugi et al. (1988) reported that pimelic acid, a putative pimeloyl-CoA precursor, were produced from long chain fatty acids such as oleic acid in yeasts, which may support a relevance of β-oxidation to supplying a precursor substrate for biotin biosynthesis. In peroxisomes, KAPA is first synthesized from pimeloyl-CoA by BioF protein and is then likely transported from peroxisomes to mitochondria, where it serves as a substrate for the final series of biosynthesis reactions that convert KAPA to biotin by the action of the BioD/A and BioB proteins. Thus, functionally coupling between peroxisomes and mitochondria appears to be required for biotin biosynthesis.
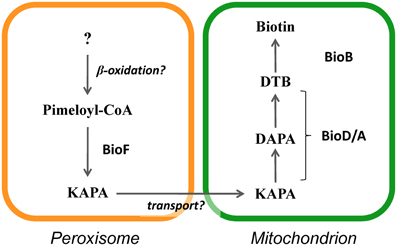
Figure 6. Model of subcellular compartmentalization of the biotin biosynthetic pathway in eukaryotes.
Conserved Peroxisomal Localization of Biof Protein and its Possible Relevance to Fungal Growth/Developmental Processes
As described above, plant BioF protein functions as a KAPA synthase and was shown to be cytosolic by GFP fusion at the C-terminus (Pinon et al., 2005). Phylogenetic analysis revealed that BioF proteins from various plant species possess PTS1 at the C-terminus (Figure 7A) (Tanabe et al., 2011; Maruyama et al., 2012), suggesting that the peroxisomal localization of BioF proteins is conserved throughout the plant kingdom. An N-terminal GFP-BioF fusion protein co-localizes with peroxisomes, and deletion of PTS1 causes cytosolic localization, suggesting that BioF is localized to peroxisomes via the PTS1 sequence (Figure 7B) (Tanabe et al., 2011).
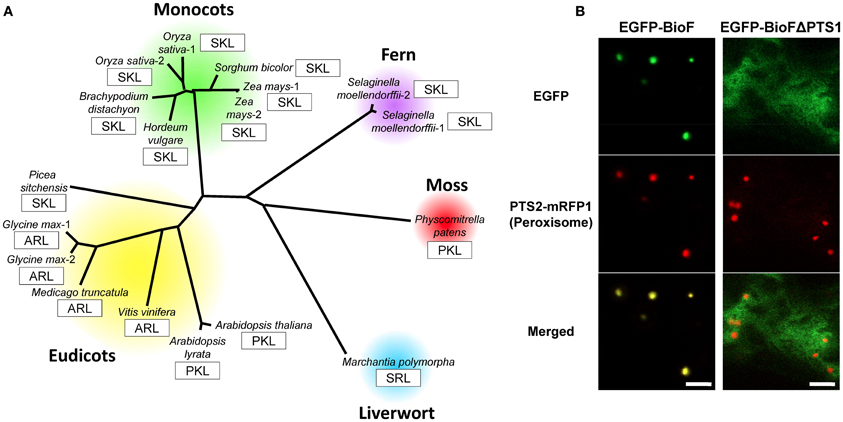
Figure 7. Phylogenetic relationship and peroxisomal localization of plant BioF proteins. (A) Phylogenetic analysis of plant BioF proteins (Maruyama et al., 2012). The amino acid residues of C-terminal peroxisomal targeting signals (PTS1) are indicated by open boxes. The full-length amino acid sequences of the plant BioF proteins were aligned using the method described in Figure 5. The Genbank accession numbers for the sequences used in the analysis are as follows: Arabidopsis thaliana, NP_974731.1; Arabidopsis lyrata, XP_002871105.1; Oryza sativa-1, BAD87813.1; Oryza sativa-2, NP_001065381.1; Hordeum vulgare, BAK03504.1; Brachypodium distachyon, XP_003574335.1; Sorgham bicolor, XP_002467492.1; Zea mays-1, ACG35792.1; Zea mays-2, ACG35881.1; Selaginella moellendorffii-1, XP_002969752.1; Selaginella moellendorffii-2, XP_002981364.1; Physcomitrella patens, XP_001769874.1; Picea sitchensis, ABR18106.1; Vinis vinifera, XP_002268950.1; Medicago truncatula, XP_003598166.1; Glycine max-1, XP_003527547.1; and Glycine max-2, XP_003522881.1. The amino acid sequence of the BioF protein of Marchantia polymorpha was confirmed by PCR amplification and cDNA sequencing based on information obtained from the Marchantia expression sequence tag database (Maruyama et al., 2012). (B) Peroxisomal localization of plant BioF protein. Note that BioF (EGFP-BioF-PTS1) co-localizes with the peroxisomes (PTS2-mRFP1), but disperses in the absence of PTS1 (EGFP-BioF-ΔPTS1) (Tanabe et al., 2011). Bars: 5 μm.
Plant biotin-auxotrophic mutants exhibit embryo lethality, indicating that biotin biosynthesis is vital for plant growth and development (Schneider et al., 1989; Shellhammer and Meinke, 1990; Patton et al., 1998; Tzafrir et al., 2004; Arnal et al., 2006). Embryo development also requires peroxisomal functions (Hu et al., 2002; Schumann et al., 2003; Sparkes et al., 2003; Tzafrir et al., 2004; Fan et al., 2005). Tanabe et al. (2011) suggested that fungi and plants use an evolutionarily conserved pathway for biotin biosynthesis that involves both peroxisomes and mitochondria. These findings suggest that biotin biosynthesis might be one of the reasons why peroxisomal deficiency results in embryo lethality. The Aspergillus peroxisome-deficient strains showing biotin auxotrophy exhibit abnormal polar growth (Tanabe et al., 2011), and impairment of sexual development by peroxisomal malfunction was reported in filamentous fungi (Bonnet et al., 2006). These similarities in the fungal and plant phenotypes indicate that growth and developmental defects due to peroxisomal deficiency may be partially or entirely attributed to biotin auxotrophy. More extensive studies will provide insight into the importance of biotin biosynthesis and peroxisomal function during growth and development of fungi and plants.
Conclusion
The primary function of fungal peroxisomes was long thought to be limited to the β-oxidation of fatty acids. During the two past decades, an increasing number of studies have unmasked the functional diversity of fungal peroxisomes (Pieuchot and Jedd, 2012), including the very recent findings that peroxisomes contain siderophore biosynthetic enzymes and are involved in iron acquisition (Gründlinger et al., 2013), and that several glycolysis enzymes possess cryptic PTS1 motifs that are activated by alternative splicing and stop codon read-through (Freitag et al., 2012). The present review has described the findings that demonstrate the fundamental involvement of fungal peroxisomes in the regulation of multicellular growth and the biosynthesis of the essential vitamin biotin. Further investigations, including proteomic/metabolomic approaches and genomic bioinformatics, will lead to a comprehensive understanding of the newly emerged functions of fungal peroxisomes.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Jun-ichi Maruyama was supported by a Grant-in-Aid for Young Scientists from the Japan Society for the Promotion of Science.
References
Arnal, N., Alban, C., Quadrado, M., Grandjean, O., and Mireau, H. (2006). The Arabidopsis Bio2 protein requires mitochondrial targeting for activity. Plant Mol. Biol. 62, 471–479. doi: 10.1007/s11103-006-9034-x
Asakura, M., Okuno, T., and Takano, Y. (2006). Multiple contributions of peroxisomal metabolic function to fungal pathogenicity in Colletotrichum lagenarium. Appl. Environ. Microbiol. 72, 6345–6354. doi: 10.1128/AEM.00988-06
Asiegbu, F. O., Choi, W., Jeong, J. S., and Dean, R. A. (2004). Cloning, sequencing and functional analysis of Magnaporthe grisea MVP1 gene, a hex-1 homolog encoding a putative “woronin body” protein. FEMS Microbiol. Lett. 230, 85–90. doi: 10.1016/S0378-1097(03)00858-9
Baldet, P., Alban, C., and Douce, R. (1997). Biotin synthesis in higher plants: purification of bioB gene product equivalent from Arabidopsis thaliana overexpressed in Escherichia coli and its subcellular localization in pea leaf cells. FEBS Lett. 419, 206–210. doi: 10.1016/S0014-5793(97)01458-0
Bartoszewska, M., Opaliński, L., Veenhuis, M., and van der Klei, I. J. (2011). The significance of peroxisomes in secondary metabolite biosynthesis in filamentous fungi. Biotechnol. Lett. 33, 1921–1931. doi: 10.1007/s10529-011-0664-y
Beck, J., and Ebel, F. (2013). Characterization of the major Woronin body protein HexA of the human pathogenic mold Aspergillus fumigatus. Int. J. Med. Microbiol. 303, 90–97. doi: 10.1016/j.ijmm.2012.11.005
Bleichrodt, R. J., van Veluw, G. J., Recter, B., Maruyama, J., Kitamoto, K., and Wösten, H. A. (2012). Hyphal heterogeneity in Aspergillus oryzae is the result of dynamic closure of septa by Woronin bodies. Mol. Microbiol. 86, 1334–1344. doi: 10.1111/mmi.12077
Boisson-Dernier, A., Frietsch, S., Kim, T. H., Dizon, M. B., and Schroeder, J. I. (2008). The peroxin loss-of-function mutation abstinence by mutual consent disrupts male-female gametophyte recognition. Curr. Biol. 18, 63–68. doi: 10.1016/j.cub.2007.11.067
Bonnet, C., Espagne, E., Zickler, D., Boisnard, S., Bourdais, A., and Berteaux-Lecellier, V. (2006). The peroxisomal import proteins PEX2, PEX5 and PEX7 are differently involved in Podospora anserina sexual cycle. Mol. Microbiol. 62, 157–169. doi: 10.1111/j.1365-2958.2006.05353.x
Cobessi, D., Dumas, R., Pautre, V., Meinguet, C., Ferrer, J. L., and Alban, C. (2012). Biochemical and structural characterization of the Arabidopsis bifunctional enzyme dethiobiotin synthetase-diaminopelargonic acid aminotransferase: evidence for substrate channeling in biotin synthesis. Plant Cell 24, 1608–1625. doi: 10.1105/tpc.112.097675
Curach, N. C., Te'o, V. S., Gibbs, M. D., Bergquist, P. L., and Nevalainen, K. M. (2004). Isolation, characterization and expression of the hex1 gene from Trichoderma reesei. Gene 331, 133–140. doi: 10.1016/j.gene.2004.02.007
Engh, I., Würtz, C., Witzel-Schlömp, K., Zhang, H. Y., Hoff, B., Nowrousian, M., and et al. (2007). The WW domain protein PRO40 is required for fungal fertility and associates with Woronin bodies. Eukaryot. Cell 6, 831–843. doi: 10.1128/EC.00269-06
Erdmann, R., and Blobel, G. (1995). Giant peroxisomes in oleic acid induced Saccharomyces cerevisiae lacking the peroxisomal membrane protein Pmp27p. J. Cell Biol. 128, 509–523. doi: 10.1083/jcb.128.4.509
Erdmann, R., Veenhuis, M., Mertens, D., and Kunau, W. H. (1989). Isolation of peroxisome-deficient mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 86, 5419–5423. doi: 10.1073/pnas.86.14.5419
Escaño, C. S., Juvvadi, P. R., Jin, F. J., Takahashi, T., Koyama, Y., Yamashita, S., and et al. (2009). Disruption of the Aopex11-1 gene involved in peroxisome proliferation leads to impaired Woronin body formation in Aspergillus oryzae. Eukaryot. Cell 8, 296–305.
Fan, J., Quan, S., Orth, T., Awai, C., Chory, J., and Hu, J. (2005). The Arabidopsis PEX12 gene is required for peroxisome biogenesis and is essential for development. Plant Physiol. 139, 231–239. doi: 10.1104/pp.105.066811
Fleissner, A., and Glass, N. L. (2007). SO, a protein involved in hyphal fusion in Neurospora crassa, localizes to septal plugs. Eukaryot. Cell 6, 84–94. doi: 10.1128/EC.00268-06
Freitag, J., Ast, J., and Bölker, M. (2012). Cryptic peroxisomal targeting via alternative splicing and stop codon read-through in fungi. Nature 485, 522–525. doi: 10.1038/nature11051
Freitag, M., Hickey, P. C., Raju, N. B., Selker, E. U., and Read, N. D. (2004). GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet. Biol. 41, 897–910. doi: 10.1016/j.fgb.2004.06.008
Gründlinger, M., Yasmin, S., Lechner, B. E., Geley, S., Schrettl, M., Hynes, M., and et al. (2013). Fungal siderophore biosynthesis is partially localized in peroxisomes. Mol. Microbiol. 88, 862–875. doi: 10.1111/mmi.12225
Hall, C., and Dietrich, F. S. (2007). The reacquisition of biotin prototrophy in Saccharomyces cerevisiae involved horizontal gene transfer, gene duplication and gene clustering. Genetics 177, 2293–2307. doi: 10.1534/genetics.107.074963
Hu, J., Aguirre, M., Peto, C., Alonso, J., Ecker, J., and Chory, J. (2002). A role for peroxisomes in photomorphogenesis and development of Arabidopsis. Science 297, 405–409. doi: 10.1126/science.1073633
Hynes, M. J., Murray, S. L., Khew, G. S., and Davis, M. A. (2008). Genetic analysis of the role of peroxisomes in the utilisation of acetate and fatty acids in Aspergillus nidulans. Genetics 178, 1355–1369. doi: 10.1534/genetics.107.085795
Idnurm, A., Giles, S. S., Perfect, J. R., and Heitman, J. (2007). Peroxisome function regulates growth on glucose in the basidiomycete fungus Cryptococcus neoformans. Eukaryot. Cell 6, 60–72. doi: 10.1128/EC.00214-06
Imazaki, A., Tanaka, A., Harimoto, Y., Yamamoto, M., Akimitsu, K., Park, P., and et al. (2010). Contribution of peroxisomes to secondary metabolism and pathogenicity in the fungal plant pathogen Alternaria alternata. Eukaryot. Cell 9, 682–694. doi: 10.1128/EC.00369-09
Jedd, G., and Chua, N. H. (2000). A new self-assembled peroxisomal vesicle required for efficient resealing of the plasma membrane. Nat. Cell Biol. 2, 226–231. doi: 10.1038/35008652
Juvvadi, P. R., Maruyama, J., and Kitamoto, K. (2007). Phosphorylation of the Aspergillus oryzae Woronin body protein, AoHex1, by protein kinase C: evidence for its role in the multimerization and proper localization of the Woronin body protein. Biochem. J. 405, 533–540. doi: 10.1042/BJ20061678
Kim, K. H., Willger, S. D., Park, S. W., Puttikamonkul, S., Grahl, N., Cho, Y., and et al. (2009). TmpL, a transmembrane protein required for intracellular redox homeostasis and virulence in a plant and an animal fungal pathogen. PLoS Pathog. 5:e1000653. doi: 10.1371/journal.ppat.1000653
Kimura, A., Takano, Y., Furusawa, I., and Okuno, T. (2001). Peroxisomal metabolic function is required for appressorium-mediated plant infection by Colletotrichum lagenarium. Plant Cell 13, 1945–1958.
Knowles, J. R. (1989). The mechanism of biotin-dependent enzymes. Annu. Rev. Biochem. 58, 195–221. doi: 10.1146/annurev.bi.58.070189.001211
Koch, A., Schneider, G., Luers, G. H., and Schrader, M. (2004). Peroxisome elongation and constriction but not fission can occur independently of dynamin-like protein 1. J. Cell Sci. 117, 3995–4006. doi: 10.1242/jcs.01268
Koch, A., Thiemann, M., Grabenbauer, M., Yoon, Y., McNiven, M. A., and Schrader, M. (2003). Dynamin-like protein 1 is involved in peroxisomal fission. J. Biol. Chem. 278, 8597–8605. doi: 10.1074/jbc.M211761200
Lai, J., Koh, C. H., Tjota, M., Pieuchot, L., Raman, V., Chandrababu, K. B., and et al. (2012). Intrinsically disordered proteins aggregate at fungal cell-to-cell channels and regulate intercellular connectivity. Proc. Natl. Acad. Sci. U.S.A. 109, 15781–15786. doi: 10.1073/pnas.1207467109
Lew, R. R. (2005). Mass flow and pressure-driven hyphal extension in Neurospra crassa. Microbiology 151, 2685–2692. doi: 10.1099/mic.0.27947-0
Liu, F., Lu, Y., Pieuchot, L., Dhavale, T., and Jedd, G. (2011). Import oligomers induce positive feedback to promote peroxisome differentiation and control organelle abundance. Dev. Cell 21, 457–468. doi: 10.1016/j.devcel.2011.08.004
Liu, F., Ng, S. K., Lu, Y., Low, W., Lai, J., and Jedd, G. (2008). Making two organelles from one: woronin body biogenesis by peroxisomal protein sorting. J. Cell Biol. 180, 325–339. doi: 10.1083/jcb.200705049
Magliano, P., Flipphi, M., Arpat, B. A., Delessert, S., and Poirier, Y. (2011a). Contributions of the peroxisome and β-oxidation cycle to biotin synthesis in fungi. J. Biol. Chem. 286, 42133–42140. doi: 10.1074/jbc.M111.279687
Magliano, P., Flipphi, M., Sanglard, D., and Poirier, Y. (2011b). Characterization of the Aspergillus nidulans biotin biosynthetic gene cluster and use of the bioDA gene as a new transformation marker. Fungal Genet. Biol. 48, 208–215. doi: 10.1016/j.fgb.2010.08.004
Managadze, D., Würtz, C., Sichting, M., Niehaus, G., Veenhuis, M., and Rottensteiner, H. (2007). The peroxin PEX14 of Neurospora crassa is essential for the biogenesis of both glyoxysomes and Woronin bodies. Traffic 8, 687–701. doi: 10.1111/j.1600-0854.2007.00560.x
Mano, S., and Nishimura, M. (2005). Plant peroxisomes. Vitam. Horm. 72, 111–154. doi: 10.1016/S0083-6729(05)72004-5
Markham, P. (1994). Occlusions of septal pores in filamentous fungi. Mycol. Res. 98, 1089–1106. doi: 10.1016/S0953-7562(09)80195-0
Markham, P., and Collinge, A. J. (1987). Woronin bodies of filamentous fungi. FEMS Microbiol. Rev. 46, 1–11. doi: 10.1111/j.1574-6968.1987.tb02448.x
Marshall, P. A., Krimkevich, Y. I., Lark, R. H., Dyer, J. M., Veenhuis, M., and Goodman, J. M. (1995). Pmp27 promotes peroxisomal proliferation. J. Cell Biol. 129, 345–355. doi: 10.1083/jcb.129.2.345
Maruyama, J., Escaño, C. S., and Kitamoto, K. (2010). AoSO protein accumulates at the septal pore in response to various stresses in the filamentous fungus Aspergillus oryzae. Biochem. Biophys. Res. Commun. 391, 868–873.
Maruyama, J., Juvvadi, P. R., Ishi, K., and Kitamoto, K. (2005). Three-dimensional image analysis of plugging at the septal pore by Woronin body during hypotonic shock inducing hyphal tip bursting in the filamentous fungus Aspergillus oryzae. Biochem. Biophys. Res. Commun. 331, 1081–1088. doi: 10.1016/j.bbrc.2005.03.233
Maruyama, J., Kikuchi, S., and Kitamoto, K. (2006). Differential distribution of the endoplasmic reticulum network as visualized by the BipA-EGFP fusion protein in hyphal compartments across the septum of the filamentous fungus, Aspergillus oryzae. Fungal Genet. Biol. 43, 642–654. doi: 10.1016/j.fgb.2005.11.007
Maruyama, J., and Kitamoto, K. (2007). Differential distribution of the endoplasmic reticulum network in filamentous fungi. FEMS Microbiol. Lett. 272, 1–7. doi: 10.1111/j.1574-6968.2007.00758.x
Maruyama, J., Yamaoka, S., Matsuo, I., Tsutsumi, N., and Kitamoto, K. (2012). A newly discovered function of peroxisomes: involvement in biotin biosynthesis. Plant Signal. Behav. 7, 1589–1593. doi: 10.4161/psb.22405
Muralla, R., Chen, E., Sweeney, C., Gray, J. A., Dickerman, A., Nikolau, B. J., and et al. (2008). A bifunctional locus (BIO3-BIO1) required for biotin biosynthesis in Arabidopsis. Plant Physiol. 146, 60–73. doi: 10.1104/pp.107.107409
Ng, S. K., Liu, F., Lai, J., Low, W., and Jedd, G. (2009). A tether for Woronin body inheritance is associated with evolutionary variation in organelle positioning. PLoS Genet. 5:e1000521. doi: 10.1371/journal.pgen.1000521
Ohsugi, M., Miyauchi, K., Tachibana, K., and Nakao, S. (1988). Formation of a biotin precursor, pimelic acid, in yeasts from C18 fatty acids. J. Nutr. Sci. Vitaminol. 34, 343–352. doi: 10.3177/jnsv.34.343
Opaliński, £., Bartoszewska, M., Fekken, S., Liu, H., de Boer, R., van der Klei, I., and et al. (2012). De novo peroxisome biogenesis in Penicillium chrysogenum is not dependent on the Pex11 family members or Pex16. PLoS ONE 7:e35490. doi: 10.1371/journal.pone.0035490
Patton, D. A., Schetter, A. L., Franzmann, L. H., Nelson, K., Ward, E. R., and Meinke, D. W. (1998). An embryo-defective mutant of Arabidopsis disrupted in the final step of biotin synthesis. Plant Physiol. 116, 935–946. doi: 10.1104/pp.116.3.935
Peraza-Reyes, L., Zickler, D., and Berteaux-Lecellier, V. (2008). The peroxisome RING-finger complex is required for meiocyte formation in the fungus Podospora anserina. Traffic 9, 1998–2009. doi: 10.1111/j.1600-0854.2008.00812.x
Picciocchi, A., Douce, R., and Alban, C. (2001). Biochemical characterization of the Arabidopsis biotin synthase reaction. The importance of mitochondria in biotin synthesis. Plant Physiol. 127, 1224–1233. doi: 10.1104/pp.010346
Pieuchot, L., and Jedd, G. (2012). Peroxisome assembly and functional diversity in eukaryotic microorganisms. Annu. Rev. Microbiol. 66, 237–263. doi: 10.1146/annurev-micro-092611-150126
Pinon, V., Ravanel, S., Douce, R., and Alban, C. (2005). Biotin synthesis in plants. The first committed step of the pathway is catalyzed by a cytosolic 7-keto-8-aminopelargonic acid synthase. Plant Physiol. 139, 1666–1676. doi: 10.1104/pp.105.070144
Ramos-Pamplona, M., and Naqvi, N. I. (2006). Host invasion during rice-blast disease requires carnitine-dependent transport of peroxisomal acetyl-CoA. Mol. Microbiol. 61, 61–75. doi: 10.1111/j.1365-2958.2006.05194.x
Rébeillé, F., Alban, C., Bourguignon, J., Ravanel, S., and Douce, R. (2007). The role of plant mitochondria in the biosynthesis of coenzymes. Photosynth. Res. 92, 149–162. doi: 10.1007/s11120-007-9167-z
Reumann, S., and Weber, A. P. (2006). Plant peroxisomes respire in the light: some gaps of the photorespiratory C2 cycle have become filled–others remain. Biochim. Biophys. Acta 1763, 1496–1510. doi: 10.1016/j.bbamcr.2006.09.008
Saikia, S., and Scott, B. (2009). Functional analysis and subcellular localization of two geranylgeranyl diphosphate synthases from Penicillium paxilli. Mol. Genet. Genomics 282, 257–271. doi: 10.1007/s00438-009-0463-5
Schneider, T., Dinkins, R., Robinson, K., Shellhammer, J., and Meinke, D. W. (1989). An embryo-lethal mutant of Arabidopsis thaliana is a biotin auxotroph. Dev. Biol. 131, 161–167. doi: 10.1016/S0012-1606(89)80047-8
Schrader, M. (2006). Shared components of mitochondrial and peroxisomal division. Biochim. Biophys. Acta 1763, 531–541. doi: 10.1016/j.bbamcr.2006.01.004
Schumann, U., Wanner, G., Veenhuis, M., Schmid, M., and Gietl, C. (2003). AthPEX10, a nuclear gene essential for peroxisome and storage organelle formation during Arabidopsis embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 100, 9626–9631. doi: 10.1073/pnas.1633697100
Shellhammer, J., and Meinke, D. W. (1990). Arrested embryos from the bio1 auxotroph of Arabidopsis contain reduced levels of biotin. Plant Physiol. 93, 1162–1167. doi: 10.1104/pp.93.3.1162
Soundararajan, S., Jedd, G., Li, X., Ramos-Pamploña, M., Chua, N. H., and Naqvi, N. I. (2004). Woronin body function in Magnaporthe grisea is essential for efficient pathogenesis and for survival during nitrogen starvation stress. Plant Cell 6, 1564–1574.
Sparkes, I. A., Brandizzi, F., Slocombe, S. P., El-Shami, M., Hawes, C., and Baker, A. (2003). An Arabidopsis pex10 null mutant is embryo lethal, implicating peroxisomes in an essential role during plant embryogenesis. Plant Physiol. 133, 1809–1819. doi: 10.1104/pp.103.031252
Streit, W. R., and Entcheva, P. (2003). Biotin in microbes, the genes involved in its biosynthesis, its biochemical role and perspectives for biotechnological production. Appl. Microbiol. Biotechnol. 61, 21–31.
Tanabe, Y., Maruyama, J., Yamaoka, S., Yahagi, D., Matsuo, I., Tsutsumi, N., and et al. (2011). Peroxisomes are involved in biotin biosynthesis in Aspergillus and Arabidopsis. J. Biol. Chem. 286, 30455–30461. doi: 10.1074/jbc.M111.247338
Tenney, K., Hunt, I., Sweigard, J., Pounder, J. I., McClain, C., Bowman, E. J., and et al. (2000). Hex-1, a gene unique to filamentous fungi, encodes the major protein of the Woronin body and functions as a plug for septal pores. Fungal Genet. Biol. 31, 205–217. doi: 10.1006/fgbi.2000.1230
Tey, W. K., North, A. J., Reyes, J. L., Lu, Y. F., and Jedd, G. (2005). Polarized gene expression determines Woronin body formation at the leading edge of the fungal colony. Mol. Biol. Cell 16, 2651–2659. doi: 10.1091/mbc.E04-10-0937
Tzafrir, I., Pena-Muralla, R., Dickerman, A., Berg, M., Rogers, R., Hutchens, S., and et al. (2004). Identification of genes required for embryo development in Arabidopsis. Plant Physiol. 135, 1206–1220. doi: 10.1104/pp.104.045179
van der Klei, I. J., and Veenhuis, M. (2006). Yeast and filamentous fungi as model organisms in microbody research. Biochim. Biophys. Acta 1763, 1364–1373. doi: 10.1016/j.bbamcr.2006.09.014
van der Klei, I. J., Yurimoto, H., Sakai, Y., and Veenhuis, M. (2006). The significance of peroxisomes in methanol metabolism in methylotrophic yeast. Biochim. Biophys. Acta 1763, 1453–1462. doi: 10.1016/j.bbamcr.2006.07.016
Waterham, H. R., and Ebberink, M. S. (2012). Genetics and molecular basis of human peroxisome biogenesis disorders. Biochim. Biophys. Acta 1822, 1430–1441. doi: 10.1016/j.bbadis.2012.04.006
Wanders, R. J., and Waterham, H. R. (2006). Biochemistry of mammalian peroxisomes revisited. Annu. Rev. Biochem. 75, 295–332. doi: 10.1146/annurev.biochem.74.082803.133329
Weber, H. (2002). Fatty acid-derived signals in plants. Trends Plant Sci. 7, 217–224. doi: 10.1016/S1360-1385(02)02250-1
Woodward, A. W., and Bartel, B. (2005). Auxin: regulation, action, and interaction. Ann. Bot. 95, 707–735. doi: 10.1093/aob/mci083
Würtz, C., Schliebs, W., Erdmann, R., and Rottensteiner, H. (2008). Dynamin-like protein-dependent formation of Woronin bodies in Saccharomyces cerevisiae upon heterologous expression of a single protein. FEBS J. 275, 2932–2941. doi: 10.1111/j.1742-4658.2008.06430.x
Yu, Y., Jiang, D., Xie, J., Cheng, J., Li, G., Yi, X., and et al. (2012). Ss-Sl2, a novel cell wall protein with PAN modules, is essential for sclerotial development and cellular integrity of Sclerotinia sclerotiorum. PLoS ONE 7:e34962. doi: 10.1371/journal.pone.0034962
Yuan, P., Jedd, G., Kumaran, D., Swaminathan, S., Shio, H., Hewitt, D., and et al. (2003). A HEX-1 crystal lattice required for Woronin body function in Neurospora crassa. Nat. Struct. Biol. 4, 264–270. doi: 10.1038/nsb910
Keywords: peroxisome, fungi, Woronin body, biotin, mitochondria
Citation: Maruyama J and Kitamoto K (2013) Expanding functional repertoires of fungal peroxisomes: contribution to growth and survival processes. Front. Physiol. 4:177. doi: 10.3389/fphys.2013.00177
Received: 27 May 2013; Paper pending published: 11 June 2013;
Accepted: 23 June 2013; Published online: 17 July 2013.
Edited by:
Vladimir I. Titorenko, Concordia University, CanadaReviewed by:
Vladimir I. Titorenko, Concordia University, CanadaMarc Fransen, Katholieke Universiteit Leuven, Belgium
Véronique Berteaux-Lecellier, French National Research Center, France
Copyright © 2013 Maruyama and Kitamoto. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Jun-ichi Maruyama, Department of Biotechnology, The University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo 113-8657, Japan e-mail: amarujun@mail.ecc.u-tokyo.ac.jp
