- Division for Therapies against Intractable Diseases, Institute for Comprehensive Medical Science, Fujita Health University, Toyoake, Japan
Skeletal muscle comprises approximately 40% of body weight, and is important for locomotion, as well as for metabolic homeostasis. Adult skeletal muscle mass is maintained by a fine balance between muscle protein synthesis and degradation. In response to cytokines, nutrients, and mechanical stimuli, skeletal muscle mass is increased (hypertrophy), whereas skeletal muscle mass is decreased (atrophy) in a variety of conditions, including cancer cachexia, starvation, immobilization, aging, and neuromuscular disorders. Recent studies have determined two important signaling pathways involved in skeletal muscle mass. The insulin-like growth factor-1 (IGF-1)/Akt pathway increases skeletal muscle mass via stimulation of protein synthesis and inhibition of protein degradation. By contrast, myostatin signaling negatively regulates skeletal muscle mass by reducing protein synthesis. In addition, the discovery of microRNAs as novel regulators of gene expression has provided new insights into a multitude of biological processes, especially in skeletal muscle physiology. We summarize here the current knowledge of microRNAs in the regulation of skeletal muscle hypertrophy, focusing on the IGF-1/Akt pathway and myostatin signaling.
Introduction
Skeletal muscle is the most abundant tissue in our body and is important in locomotion and metabolic adaptation. Adult skeletal muscle mass is mainly determined by a balance between muscle protein synthesis and degradation. Exercise, nutrients, and exogenous stimuli increase the rate of muscle protein synthesis and increase skeletal muscle mass (hypertrophy), whereas starvation, immobilization, aging, and diseases increase the rate of protein degradation and markedly reduce skeletal muscle mass (atrophy). In addition, some muscular disorders produce a larger but much weaker muscle (pseudo-hypertrophy) through degeneration and regeneration of myofibers (Tyler, 2003). Several signaling pathways are shown to be involved in the regulation of skeletal muscle mass. The insulin-like growth factor-1 (IGF-1)/Akt pathway and β-adrenergic pathway are positive regulators of skeletal muscle growth. In contrast, myostatin signaling, NF-κβ signaling, and glucocorticoid signaling negatively regulate skeletal muscle mass (Sandri, 2008).
MicroRNAs (miRNAs) are small non-coding RNAs that are highly conserved in eukaryotes. Currently, the important role of miRNAs is evident in diverse biological processes, including development, differentiation, homeostasis, and disease in vertebrate species (Sayed and Abdellatif, 2011). miRNAs function to fine tune gene expression by accelerating degradation of mRNA and/or by inhibiting translation (Bartel, 2009). Identification of muscle-specific miRNAs called myomiR (miR-1, miR-133a/b, miR-206, miR-208b, miR-499, and miR-486) has extended our knowledge of the molecular network in skeletal muscle. The myomiRs form a feedback regulatory loop with myogenic regulatory factors, such as MyoD and myogenin, to precisely regulate skeletal muscle plasticity. For example, miR-1, mir-27, miR-206, and miR-486 promote myoblast differentiation through the repression of Pax7 and Pax3 expression (Crist et al., 2009; Chen et al., 2010; Dey et al., 2011). Meanwhile, the expression of myomiRs is strictly controlled by MyoD, myogenin, MEF2, and Pax7 (Rao et al., 2006; Rosenberg et al., 2006; Liu et al., 2007; Dey et al., 2011).
This mini-review highlights the recent findings of miRNAs in skeletal muscle hypertrophy, and focuses on the IGF-1/Akt pathway and myostatin signaling, because the fine balance between these two signaling pathways is essential for maintaining normal skeletal muscle mass (Schiaffino et al., 2013). The role of miRNAs on the IGF-1/Akt pathway and myostatin signaling is summarized in Figure 1. miRNAs also function in skeletal muscle atrophy and their role has been described in detail in recent reviews (Ma et al., 2012; Wang, 2013).
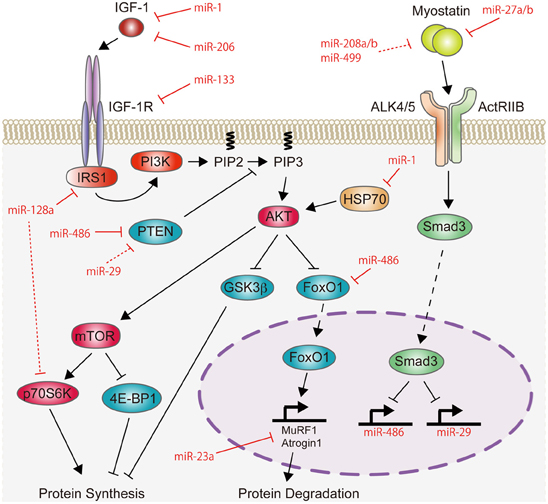
Figure 1. Schematic representation of miRNAs involved in the regulation of the IGF-1/Akt pathway and myostatin signaling. IGF-1 increases the activity of Akt protein. This activation of Akt is attenuated by PTEN. The activated state of Akt stimulates protein synthesis through mTOR. Akt also inhibits protein degradation by suppressing the activity of GSK3β and FoxO1, which induces protein degradation by activating the expression of MuRF1 and Atrogin1. Therefore, the IGF-1/Akt pathway induces skeletal muscle hypertrophy. Recent studies have shown that miR-1, miR-133, miR-206, and miR-128a negatively regulate the IGF-1/Akt pathway by targeting positive regulators of the IGF-1/Akt pathway (IGF-1, IGF-1R, IRS1, HSP70, or p70S6K), while miR-29, miR-486, and miR-23a positively regulate this pathway by targeting negative regulators (PTEN, FoxO1, MuRF1, or Atrogin1). In contrast to the IGF-1/Akt pathway, myostatin signaling is a negative regulator of skeletal muscle mass. Myostatin activates Smad3 protein and may inhibit protein synthesis by repressing the transcription of miR-486 and miR-29, which target PTEN and FoxO1 proteins. Conversely, myostatin expression is regulated by miR-27a/b, miR-208a/b, and miR-499. Red lines indicate the inhibitory function of miRNAs identified in skeletal muscle, while red-dashed lines represent the inhibitory function of miRNAs identified in cardiac muscle and cultured cells.
The IGF-1/Akt Pathway and microRNAs
IGF-1 binds to its own tyrosine kinase receptor IGF-1R and activates phosphatidylinositol-3-kinase (PI3K) through IRS1. Activated PI3K produces phosphatidylinositol-3,4,5,triphosphates (PIP3) and then induces activation of the Akt protein. Subsequently, Akt activates mammalian target of rapamycin (mTOR) and regulates down stream effector p70 ribosomal protein S6 kinase (p70S6K) and 4E-BP1, both of which control protein synthesis (Rommel et al., 2001). Akt also inactivates glycogen synthase kinase 3β (GSK3β), which blocks protein translation (Cross et al., 1995). In addition, Akt inhibits the nuclear translocation of FoxO family of transcription factors. Because FoxOs play a key role in the regulation of muscle atrophy-related genes, MuRF1, and Atrogin1 (Sandri et al., 2004), inactivation of FoxOs prevents muscle protein degradation. In this manner, the IGF-1/Akt pathway plays a central role in skeletal muscle hypertrophy. Indeed, ectopic Akt expression is sufficient for increasing the size of myofibers in adult mice (Bodine et al., 2001; Takahashi et al., 2002; Lai et al., 2004). Although the effect of IGF-1 on skeletal muscle mass mainly depends on development, IGF-1 can lead to increase skeletal muscle mass even in adulthood (Adams and McCue, 1998; Barton-Davis et al., 1998; Alzghoul et al., 2004; Schertzer et al., 2006).
Recent studies have identified several miRNAs that can modulate the muscle IGF-1/Akt pathway. In this section, we review the recently discovered miRNAs that target the IGF-1/Atk pathway in skeletal muscle.
MicroRNA-1, -133, and -206
The expression levels of miR-1 and miR-133 are greatly increased during myogenesis (Ge and Chen, 2011), whereas the levels of their expression are reduced during skeletal muscle hypertrophy. Functional overload in mice decreases the levels of miR-1 and miR-133 expression with an increase in muscle weight (McCarthy and Esser, 2007). Resistance training with amino acid ingestion also decreases miR-1 expression in young men (Drummond et al., 2008). Overload and acute resistance exercise are shown to increase the levels of IGF-1 peptide in rats (Adams et al., 1999) and the activity of the Akt/mTOR in humans, respectively (Mayhew et al., 2009). Therefore, reduced miR-1 and miR-133 expression could contribute to the activation of the IGF-1/Akt pathway. Indeed, miR-1 and miR-133 inhibit the IGF-1/Akt pathway in C2C12 cells by targeting IGF-1, IGF-1R, and HSP70 (Elia et al., 2009; Huang et al., 2011; Kukreti et al., 2013), however, their targets in adult skeletal muscle remain to be defined.
Notably, inhibition of miR-206 in tilapia skeletal muscle promotes body growth with an increase in IGF-1 expression (Yan et al., 2013). However, although inhibition of miR-206 robustly increases C2C12 myotube width, gain and loss of function of miR-206 do not affect skeletal muscle size in vivo (Winbanks et al., 2013).
MicroRNA-128a
To the best of our knowledge, miR-128a is the only miRNA that has the potential to increase skeletal muscle mass. In mice, miR-128a is highly expressed in brain and skeletal muscle (Lee et al., 2008), and its expression is increased during myoblast differentiation (Chen et al., 2006, 2010; Sun et al., 2010). Motohashi et al. (2013) identified IRS1 as a target of miR-128a in mice. They showed that overexpression of miR-128a repressed myoblast proliferation with a decrease in IRS1 protein and Akt activity. Interestingly, inhibition of miR-128a increased the size of C2C12 myotubes, with an increase in IRS1 protein and Akt activity. Notably, 4 weeks' administration of antisense miR-128 significantly induced skeletal muscle hypertrophy in mice. Given that miR-128 inhibits glioma tumor growth by targeting p70S6K (Shi et al., 2012), miR-128a may act as a negative regulator against multiple components of the IGF-1/Akt anabolic pathway.
MicroRNA-486
An alternative promoter, located upstream of exon 39a of the Ank1 gene, produces cardiac and skeletal muscle-specific Ank1.5 protein (also referred to as sAnk1), which connects sarcomeres to the sarcoplasmic reticulum (Zhou et al., 1997; Porter et al., 2005). miR-486 is transcribed from the final intron of the Ank1.5 gene, and is co-expressed with Ank1.5 mRNA in cardiac and skeletal muscles (Small et al., 2010).
miR-486 was initially identified as a downstream target of myocardin-related transcription factor-A in cardiomyocytes (Small et al., 2010). MyoD also directly stimulates the promoter activity of miR-486/Ank1.5 and increases their expression during myoblast differentiation (Dey et al., 2011). miR-486 promotes the differentiation of satellite cells, adult skeletal muscle stem cells, by targeting Pax7 mRNA (Dey et al., 2011), whereas muscle-specific transgenic overexpression of miR-486 shows delayed regeneration when skeletal muscle is injured (Alexander et al., 2011).
In addition, miR-486 is involved in the regulation of Akt activity by directly targeting the 3′ UTR of phosphatase and tensin homolog (PTEN) and FoxO1. Because PTEN inactivates the Akt protein (Stambolic et al., 1998), miR-486 increases the phosphorylation status of Akt with a decrease in PTEN and FoxO1 protein levels in mouse primary myotubes (Xu et al., 2012). miR-486 also decreases protein levels of PTEN, FoxO1, PDGFRβ, SFSR1, and SFSR3 in human myoblasts and myotubes (Alexander et al., 2011). We recently showed that miR-486 is necessary to maintain Akt activity in C2C12 myotubes. Furthermore, inhibition of miR-486 activity in vivo induced myofiber atrophy (Hitachi et al., 2014). Interestingly, ectopic miR-486 expression blocked the suppression of Akt activity caused by chronic kidney disease (CKD) and was sufficient for preventing skeletal muscle atrophy induced by CKD in mice (Xu et al., 2012). Therefore, miR-486 contributes to maintain skeletal muscle mass through the regulation of Akt activity.
Myostatin Signaling and microRNAs
Myostatin is a cytokine belonging to the transforming growth factor-β (TGF-β) superfamily. In contrast to the IGF-1/Akt pathway, myostatin signaling acts as a negative regulator of skeletal muscle mass. The importance of myostatin in skeletal muscle mass was first shown by McPherron et al. (1997). They generated mice with deletion of the myostatin gene, which resulted in doubled skeletal muscle weight due to an increase in muscle fiber size (hypertrophy) and increased muscle fiber number (hyperplasia). This groundbreaking research led to better understanding of the mechanism for excessive skeletal muscle growth observed in cattle, sheep, and dogs, which have naturally occurring mutations in the myostatin gene (Grobet et al., 1997; McPherron and Lee, 1997; Clop et al., 2006; Mosher et al., 2007). Importantly, a naturally occurring mutation in the myostatin gene was found in human with increased skeletal muscle mass (Schuelke et al., 2004). The importance of myostatin in muscle size was further supported by the finding that systemically administrated myostatin induces skeletal muscle atrophy in mice (Zimmers et al., 2002). Notably elevated myostatin expression is correlated with muscle wasting in cancer cachexia, heart failure, HIV, CKD, chronic obstructive pulmonary disease, and aging (Gonzalez-Cadavid et al., 1998; Yarasheski et al., 2002; Shyu et al., 2006; Costelli et al., 2008; Plant et al., 2010; Verzola et al., 2011). Currently, blockade of myostatin function is believed to be a promising therapy against muscle wasting caused by neuromuscular disorders, cancer, and aging (Han et al., 2013).
Myostatin signaling is transmitted intracellularly through serine/threonine kinase receptors, including activin receptor type IIB (ActRIIB) and activin receptor-like kinases (ALK4 or ALK5) (Rebbapragada et al., 2003). After binding of myostatin to its receptors, ALK4/ALK5 activates Smad2 and Smad3 by phosphorylation of the C-terminal domain (Langley et al., 2002; Rebbapragada et al., 2003). Phosphorylated Smad2/3 can form a complex together with Smad4, and it then moves to the nucleus to mediate transcriptional regulation of target genes. In early studies, satellite cells were once considered to be a major source of myostatin deficiency-induced muscular hypertrophy because myostatin inhibits satellite cell activation and self-renewal (McCroskery et al., 2003). Subsequently, several groups have shown a direct effect of myostatin on myofibrillar protein synthesis (Taylor et al., 2001; Welle et al., 2006, 2009, 2011). Recent studies have shown that satellite cells minimally contribute to skeletal muscle hypertrophy by inhibition of myostatin (Amthor et al., 2009; Lee et al., 2012; Wang and McPherron, 2012).
In 2006, a relationship between myostatin and miRNA was first shown in sheep. Clop et al. found a G to C single nucleotide polymorphism in the 3′ UTR of the myostatin gene in Texel sheep, which show remarkable skeletal muscle hypertrophy (Clop et al., 2006). This single nucleotide polymorphism changes the 3′ UTR of myostatin into the target of miR-206. Accordingly, the expression level of myostatin is low in Texel sheep. After this discovery, several miRNAs have been shown to be involved in myostatin signaling. In this section, we summarize the current knowledge of miRNAs related to myostatin signaling.
MicroRNA-27
miR-27 consists of miR-27a and miR-27b, which differ by one nucleotide at position 19. During mice embryogenesis, miR-27b is expressed in the limb bud, tail bud, neural tube, heart, and somites (Crist et al., 2009). miR-27b expression is also upregulated during satellite cell differentiation in adult mice (Crist et al., 2009). Gain and loss of function of miR-27b initially indicated that miR-27b promotes myogenic differentiation by downregulating Pax3 expression (Crist et al., 2009). Consistent with this finding, miR-27b and Pax3 mRNA are exclusively expressed in somites. miR-27b is expressed in myotomes and sclerotomes, while Pax3 mRNA is expressed in dermomyotomes (Crist et al., 2009).
Skeletal muscle mainly consists of three types of fibers, which are slow oxidative, fast oxidative glycolytic, and fast glycolytic fibers. Myostatin mRNA is highly expressed in fast-twitch muscle, while its expression is low in slow-twitch muscle (Allen et al., 2008). Surprisingly, the expression level of myostatin pre-mRNA is indistinguishable between these two types of muscle. Allen and Loh resolved this paradox to discover post-transcriptional control of myostatin mRNA by miR-27. They showed that miR-27a/b is highly expressed in slow-twitch muscle and reduced the stability of myostatin mRNA by direct binding to the 3′ UTR (Allen and Loh, 2011). Therefore, low myostatin levels in slow-twitch muscle are maintained by miR-27 in a post-transcriptional mechanism. In mouse embryos at day 10.5, myostatin mRNA is expressed in the myotome compartment (McPherron et al., 1997) where miR-27b is expressed (Crist et al., 2009). Accordingly, the regulatory mechanism of myostatin mRNA may differ between adult and embryonic muscle.
Piedmontese cattle show the double-muscled phenotype, which is inherited, and is caused by a point mutation in the myostatin gene. Missense mutation of the myostatin gene G938A is translated to C313Y, and this alters the activity of myostatin protein (Berry et al., 2002), and induces muscle hypertrophy and hyperplasia (Kambadur et al., 1997; McPherron and Lee, 1997). Although 96% of Piedmontese cattle have a homozygosis mutation of the myostatin gene, different phenotypes of muscularity are observed in this breed. Greatly increased miR-27b expression with a decrease in myostatin expression have been found in Piedmontese cattle (Miretti et al., 2013). However, the cause of differential expression of miR-27b in this breed is unclear.
In humans, ingestion of leucine-enriched essential amino acids quickly stimulates muscle protein synthesis (Fujita et al., 2007) with a decrease in myostatin mRNA expression (Drummond et al., 2009). Expression levels of miR-27 have not yet been determined in this situation. However, considering the fact that treatment of 1 mM leucine decreases myostatin expression with an increase in miR-27a expression in C2C12 cells (Chen et al., 2013), miR-27 might contribute to reduce myostatin expression even in human skeletal muscle.
MicroRNA-208a, -208b, and -499
Three related myomiRs, miR-208a, miR-208b, and miR-499, are encoded by introns of the myosin genes (miR-208a/Myh6, miR-208b/Myh7, and miR-499/Myh7b). Expression of miR-208a is heart specific, while miR-208b and miR-499 are expressed in cardiac and slow-twitch skeletal muscle (van Rooij et al., 2009). In skeletal muscle, miR-208b and miR-499 promote the formation of slow-twitch fibers, because these miRNAs increase the expression of slow muscle genes by repressing Sox6, Pur-β, and Sp3 (van Rooij et al., 2009). Accordingly, double knockout of miR-208b and miR-499 in mice leads to a loss of slow fibers in the soleus (van Rooij et al., 2009).
Recent studies have shown the direct association of miR-208a, miR-208b, and miR-499 with the 3′ UTR of myostatin by luciferase assays (Callis et al., 2009; Bell et al., 2010). Although miR-208a induces cardiac hypertrophy with decreased myostatin expression (Callis et al., 2009), whether miR-208b and miR-499 have potential to induce skeletal muscle hypertrophy has not been described. It is noteworthy that nutrient ingestion increases miR-208b and miR-499 expressions with a decrease in myostatin expression in human vastus lateralis muscle (Drummond et al., 2009). Therefore, future studies are required to determine the function of miR-208b and miR-499 in the regulation of skeletal muscle mass.
MicroRNAs Under the Control of Myostatin Signaling
Thus far, we have described miRNAs that affect myostatin expression. Various studies have also investigated the opposite situation of whether myostatin-related signaling alters the expression of miRNAs. Davis et al. (2008) found that, in response to TGF-β and BMP, Smad proteins (Smad3 and Smad1/5) accelerated the processing of miR-21 in human smooth muscle cells. In this situation, Smad proteins interacted with RNA helicase p68, a component of the Drosha complex. Smad proteins were later shown to directly bind to the RNA-Smad binding element of primary transcripts of miRNAs and facilitate the interaction between miRNAs and the Drosha complex (Davis et al., 2010). BMP-2 is also likely to regulate the biogenesis of miR-206 using Smad proteins in C2C12 cells (Sato et al., 2009). Although altered expression of miR-1, miR-133a/b, and miR-206 is found in myostatin knockout mice (Rachagani et al., 2010), there is still no evidence that myostatin directly regulates miRNA processing.
miR-29 was initially shown to accelerate muscle differentiation by suppressing YY1 (Wang et al., 2008) and Akt3 (Wei et al., 2013). miR-29 also attenuates the inhibitory effect of TGF-β in muscle differentiation, and its expression is under regulation of TGF-β and Smad3 (Winbanks et al., 2011; Zhou et al., 2012). Interestingly, miR-29 activates the Akt pathway by targeting PTEN in cultured cells (Kong et al., 2011; Tumaneng et al., 2012). Goodman et al. (2013) recently showed that in vivo transfection of Smad3 reduced the Akt/mTOR activity and induced myofiber atrophy. In this situation, increased PTEN mRNA translation with a decrease in miR-29 promoter activity was observed. However, whether myostatin is involved in the regulation of miR-29 expression and the function of miR-29 in the regulation of skeletal muscle mass remain to be elucidated.
Most recently, we found greatly increased miR-486 expression in myostatin knockout mice. We showed that myostatin signaling repressed miR-486 expression at the transcriptional level using Smad3 protein (Hitachi et al., 2014). As described above, miR-486 increases the Akt activity by targeting PTEN, therefore, several miRNAs including miR-486 would contribute to induce skeletal muscle hypertrophy from inhibition of myostatin by activating the Akt pathway.
Conclusion
Although miRNAs have been implicated in skeletal muscle development, regeneration, and function, our understanding of the molecular mechanisms underlying the regulation of skeletal muscle mass by miRNAs is still limited. Exercise, nutrition, and disease affect skeletal muscle mass postnatally, and they were also recently shown to alter the levels of skeletal muscle miRNA expression (Pasiakos and McClung, 2013; Zacharewicz et al., 2013). Therefore, to understand the exact role of miRNAs in skeletal muscle hypertrophy and atrophy, it is necessary to specifically investigate the molecular function of miRNAs in adult skeletal muscle without affecting development of skeletal muscle. Indeed, using electric pulse-mediated gene transfer in adult mice, Wada et al. (2011) clearly showed that ectopic expression of miR-23a was sufficient to prevent dexamethasone-induced skeletal muscle atrophy. Future studies using in vivo techniques, such as conditional transgenic and knockout technology, viral and electrophoretic transfer of DNA and RNA, or tail vein injection of nucleic acids with a modified backbone, will further define the exact role of miRNAs in the regulation of skeletal muscle mass.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported in part by JSPS KAKENHI (23790347, 24590363, 25126726, and 25860151), an Intramural Research Grant (23-5) for Neurological and Psychiatric Disorders from NCNP, a Grant-in-Aid from the YOKOYAMA Foundation for Clinical Pharmacology, and a Grant-in-Aid from the NAKATOMI Foundation.
References
Adams, G. R., Haddad, F., and Baldwin, K. M. (1999). Time course of changes in markers of myogenesis in overloaded rat skeletal muscles. J. Appl. Physiol. 87, 1705–1712.
Adams, G. R., and McCue, S. A. (1998). Localized infusion of IGF-I results in skeletal muscle hypertrophy in rats. J. Appl. Physiol. 84, 1716–1722.
Alexander, M. S., Casar, J. C., Motohashi, N., Myers, J. A., Eisenberg, I., Gonzalez, R. T., et al. (2011). Regulation of DMD pathology by an ankyrin-encoded miRNA. Skelet. Muscle 1, 27. doi: 10.1186/2044-5040-1-27
Allen, D. L., Cleary, A. S., Speaker, K. J., Lindsay, S. F., Uyenishi, J., Reed, J. M., et al. (2008). Myostatin, activin receptor IIb, and follistatin-like-3 gene expression are altered in adipose tissue and skeletal muscle of obese mice. Am. J. Physiol. Endocrinol. Metab. 294, E918–E927. doi: 10.1152/ajpendo.00798.2007
Allen, D. L., and Loh, A. S. (2011). Posttranscriptional mechanisms involving microRNA-27a and b contribute to fast-specific and glucocorticoid-mediated myostatin expression in skeletal muscle. Am. J. Physiol. Cell Physiol. 300, C124–C137. doi: 10.1152/ajpcell.00142.2010
Alzghoul, M. B., Gerrard, D., Watkins, B. A., and Hannon, K. (2004). Ectopic expression of IGF-I and Shh by skeletal muscle inhibits disuse-mediated skeletal muscle atrophy and bone osteopenia in vivo. FASEB J. 18, 221–223. doi: 10.1096/fj.03-0293fje
Amthor, H., Otto, A., Vulin, A., Rochat, A., Dumonceaux, J., Garcia, L., et al. (2009). Muscle hypertrophy driven by myostatin blockade does not require stem/precursor-cell activity. Proc. Natl. Acad. Sci. U.S.A. 106, 7479–7484. doi: 10.1073/pnas.0811129106
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Barton-Davis, E. R., Shoturma, D. I., Musaro, A., Rosenthal, N., and Sweeney, H. L. (1998). Viral mediated expression of insulin-like growth factor I blocks the aging-related loss of skeletal muscle function. Proc. Natl. Acad. Sci. U.S.A. 95, 15603–15607. doi: 10.1073/pnas.95.26.15603
Bell, M. L., Buvoli, M., and Leinwand, L. A. (2010). Uncoupling of expression of an intronic microRNA and its myosin host gene by exon skipping. Mol. Cell. Biol. 30, 1937–1945. doi: 10.1128/MCB.01370-09
Berry, C., Thomas, M., Langley, B., Sharma, M., and Kambadur, R. (2002). Single cysteine to tyrosine transition inactivates the growth inhibitory function of Piedmontese myostatin. Am. J. Physiol. Cell Physiol. 283, C135–C141. doi: 10.1152/ajpcell.00458.2001
Bodine, S. C., Stitt, T. N., Gonzalez, M., Kline, W. O., Stover, G. L., Bauerlein, R., et al. (2001). Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 3, 1014–1019. doi: 10.1038/ncb1101-1014
Callis, T. E., Pandya, K., Seok, H. Y., Tang, R.-H., Tatsuguchi, M., Huang, Z.-P., et al. (2009). MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J. Clin. Invest. 119, 2772–2786. doi: 10.1172/JCI36154
Chen, J.-F., Mandel, E. M., Thomson, J. M., Wu, Q., Callis, T. E., Hammond, S. M., et al. (2006). The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38, 228–233. doi: 10.1038/ng1725
Chen, J.-F., Tao, Y., Li, J., Deng, Z., Yan, Z., Xiao, X., et al. (2010). microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J. Cell Biol. 190, 867–879. doi: 10.1083/jcb.200911036
Chen, X., Huang, Z., Chen, D., Yang, T., and Liu, G. (2013). MicroRNA-27a is induced by leucine and contributes to leucine-induced proliferation promotion in C2C12 cells. Int. J. Mol. Sci. 14, 14076–14084. doi: 10.3390/ijms140714076
Clop, A., Marcq, F., Takeda, H., Pirottin, D., Tordoir, X., Bibé, B., et al. (2006). A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat. Genet. 38, 813–818. doi: 10.1038/ng1810
Costelli, P., Muscaritoli, M., Bonetto, A., Penna, F., Reffo, P., Bossola, M., et al. (2008). Muscle myostatin signalling is enhanced in experimental cancer cachexia. Eur. J. Clin. Invest. 38, 531–538. doi: 10.1111/j.1365-2362.2008.01970.x
Crist, C. G., Montarras, D., Pallafacchina, G., Rocancourt, D., Cumano, A., Conway, S. J., et al. (2009). Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. Proc. Natl. Acad. Sci. U.S.A. 106, 13383–13387. doi: 10.1073/pnas.0900210106
Cross, D. A., Alessi, D. R., Cohen, P., Andjelkovich, M., and Hemmings, B. A. (1995). Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 378, 785–789. doi: 10.1038/378785a0
Davis, B. N., Hilyard, A. C., Lagna, G., and Hata, A. (2008). SMAD proteins control DROSHA-mediated microRNA maturation. Nature 454, 56–61. doi: 10.1038/nature07086
Davis, B. N., Hilyard, A. C., Nguyen, P. H., Lagna, G., and Hata, A. (2010). Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol. Cell 39, 373–384. doi: 10.1016/j.molcel.2010.07.011
Dey, B. K., Gagan, J., and Dutta, A. (2011). miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol. Cell. Biol. 31, 203–214. doi: 10.1128/MCB.01009-10
Drummond, M. J., Glynn, E. L., Fry, C. S., Dhanani, S., Volpi, E., and Rasmussen, B. B. (2009). Essential amino acids increase microRNA-499, -208b, and -23a and downregulate myostatin and myocyte enhancer factor 2C mRNA expression in human skeletal muscle. J. Nutr. 139, 2279–2284. doi: 10.3945/jn.109.112797
Drummond, M. J., McCarthy, J. J., Fry, C. S., Esser, K. A., and Rasmussen, B. B. (2008). Aging differentially affects human skeletal muscle microRNA expression at rest and after an anabolic stimulus of resistance exercise and essential amino acids. Am. J. Physiol. Endocrinol. Metab. 295, E1333–E1340. doi: 10.1152/ajpendo.90562.2008
Elia, L., Contu, R., Quintavalle, M., Varrone, F., Chimenti, C., Russo, M. A., et al. (2009). Reciprocal regulation of microRNA-1 and insulin-like growth factor-1 signal transduction cascade in cardiac and skeletal muscle in physiological and pathological conditions. Circulation 120, 2377–2385. doi: 10.1161/CIRCULATIONAHA.109.879429
Fujita, S., Dreyer, H. C., Drummond, M. J., Glynn, E. L., Cadenas, J. G., Yoshizawa, F., et al. (2007). Nutrient signalling in the regulation of human muscle protein synthesis. J. Physiol (Lond). 582, 813–823. doi: 10.1113/jphysiol.2007.134593
Ge, Y., and Chen, J. (2011). MicroRNAs in skeletal myogenesis. Cell Cycle 10, 441–448. doi: 10.4161/cc.10.3.14710
Gonzalez-Cadavid, N. F., Taylor, W. E., Yarasheski, K., Sinha-Hikim, I., Ma, K., Ezzat, S., et al. (1998). Organization of the human myostatin gene and expression in healthy men and HIV-infected men with muscle wasting. Proc. Natl. Acad. Sci. U.S.A. 95, 14938–14943. doi: 10.1073/pnas.95.25.14938
Goodman, C. A., McNally, R. M., Hoffmann, F. M., and Hornberger, T. A. (2013). Smad3 induces atrogin-1, inhibits mTOR and protein synthesis, and promotes muscle atrophy in vivo. Mol. Endocrinol. 27, 1946–1957. doi: 10.1210/me.2013-1194
Grobet, L., Martin, L. J., Poncelet, D., Pirottin, D., Brouwers, B., Riquet, J., et al. (1997). A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat. Genet. 17, 71–74. doi: 10.1038/ng0997-71
Han, H. Q., Zhou, X., Mitch, W. E., and Goldberg, A. L. (2013). Myostatin/activin pathway antagonism: molecular basis and therapeutic potential. Int. J. Biochem. Cell Biol. 45, 2333–2347. doi: 10.1016/j.biocel.2013.05.019
Hitachi, K., Nakatani, M., and Tsuchida, K. (2014). Myostatin signaling regulates Akt activity via the regulation of miR-486 expression. Int. J. Biochem. Cell Biol. 47, 93–103. doi: 10.1016/j.biocel.2013.12.003
Huang, M.-B., Xu, H., Xie, S.-J., Zhou, H., and Qu, L.-H. (2011). Insulin-like growth factor-1 receptor is regulated by microRNA-133 during skeletal myogenesis. PLoS ONE 6:e29173. doi: 10.1371/journal.pone.0029173
Kambadur, R., Sharma, M., Smith, T. P., and Bass, J. J. (1997). Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res. 7, 910–916.
Kong, G., Zhang, J., Zhang, S., Shan, C., Ye, L., and Zhang, X. (2011). Upregulated microRNA-29a by hepatitis B virus X protein enhances hepatoma cell migration by targeting PTEN in cell culture model. PLoS ONE 6:e19518. doi: 10.1371/journal.pone.0019518
Kukreti, H., Amuthavalli, K., Harikumar, A., Sathiyamoorthy, S., Feng, P. Z., Anantharaj, R., et al. (2013). Muscle-specific microRNA1 (miR1) targets heat shock protein 70 (HSP70) during dexamethasone-mediated atrophy. J. Biol. Chem. 288, 6663–6678. doi: 10.1074/jbc.M112.390369
Lai, K.-M. V., Gonzalez, M., Poueymirou, W. T., Kline, W. O., Na, E., Zlotchenko, E., et al. (2004). Conditional activation of akt in adult skeletal muscle induces rapid hypertrophy. Mol. Cell. Biol. 24, 9295–9304. doi: 10.1128/MCB.24.21.9295-9304.2004
Langley, B., Thomas, M., Bishop, A., Sharma, M., Gilmour, S., and Kambadur, R. (2002). Myostatin inhibits myoblast differentiation by down-regulating MyoD expression. J. Biol. Chem. 277, 49831–49840. doi: 10.1074/jbc.M204291200
Lee, E. J., Baek, M., Gusev, Y., Brackett, D. J., Nuovo, G. J., and Schmittgen, T. D. (2008). Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA 14, 35–42. doi: 10.1261/rna.804508
Lee, S.-J., Huynh, T. V., Lee, Y.-S., Sebald, S. M., Wilcox-Adelman, S. A., Iwamori, N., et al. (2012). Role of satellite cells versus myofibers in muscle hypertrophy induced by inhibition of the myostatin/activin signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 109, E2353–E2360. doi: 10.1073/pnas.1206410109
Liu, N., Williams, A. H., Kim, Y., McAnally, J., Bezprozvannaya, S., Sutherland, L. B., et al. (2007). An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc. Natl. Acad. Sci. U.S.A. 104, 20844–20849. doi: 10.1073/pnas.0710558105
Ma, J. F., Hall, D. T., and Gallouzi, I. E. (2012). The impact of mRNA turnover and translation on age-related muscle loss. Ageing Res. Rev. 11, 432–441. doi: 10.1016/j.arr.2012.05.004
Mayhew, D. L., Kim, J.-S., Cross, J. M., Ferrando, A. A., and Bamman, M. M. (2009). Translational signaling responses preceding resistance training-mediated myofiber hypertrophy in young and old humans. J. Appl. Physiol. 107, 1655–1662. doi: 10.1152/japplphysiol.91234.2008
McCarthy, J. J., and Esser, K. A. (2007). MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J. Appl. Physiol. 102, 306–313. doi: 10.1152/japplphysiol.00932.2006
McCroskery, S., Thomas, M., Maxwell, L., Sharma, M., and Kambadur, R. (2003). Myostatin negatively regulates satellite cell activation and self-renewal. J. Cell Biol. 162, 1135–1147. doi: 10.1083/jcb.200207056
McPherron, A. C., Lawler, A. M., and Lee, S.-J. (1997). Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 387, 83–90. doi: 10.1038/387083a0
McPherron, A. C., and Lee, S.-J. (1997). Double muscling in cattle due to mutations in the myostatin gene. Proc. Natl. Acad. Sci. U.S.A. 94, 12457–12461. doi: 10.1073/pnas.94.23.12457
Miretti, S., Martignani, E., Accornero, P., and Baratta, M. (2013). Functional effect of mir-27b on myostatin expression: a relationship in Piedmontese cattle with double-muscled phenotype. BMC Genomics 14:194. doi: 10.1186/1471-2164-14-194
Mosher, D. S., Quignon, P., Bustamante, C. D., Sutter, N. B., Mellersh, C. S., Parker, H. G., et al. (2007). A mutation in the myostatin gene increases muscle mass and enhances racing performance in heterozygote dogs. PLoS Genetics 3:e79. doi: 10.1371/journal.pgen.0030079
Motohashi, N., Alexander, M. S., Shimizu-Motohashi, Y., Myers, J. A., Kawahara, G., and Kunkel, L. M. (2013). Regulation of IRS1/Akt insulin signaling by microRNA-128a during myogenesis. J. Cell Sci. 126, 2678–2691. doi: 10.1242/jcs.119966
Pasiakos, S. M., and McClung, J. P. (2013). miRNA analysis for the assessment of exercise and amino acid effects on human skeletal muscle. Adv. Nutr. 4, 412–417. doi: 10.3945/an.113.003699
Plant, P. J., Brooks, D., Faughnan, M., Bayley, T., Bain, J., Singer, L., et al. (2010). Cellular markers of muscle atrophy in chronic obstructive pulmonary disease. Am. J. Respir. Cell Mol. Biol. 42, 461–471. doi: 10.1165/rcmb.2008-0382OC
Porter, N. C., Resneck, W. G., O'Neill, A., van Rossum, D. B., Stone, M. R., and Bloch, R. J. (2005). Association of small ankyrin 1 with the sarcoplasmic reticulum. Mol. Membr. Biol. 22, 421–432. doi: 10.1080/09687860500244262
Rachagani, S., Cheng, Y., and Reecy, J. M. (2010). Myostatin genotype regulates muscle-specific miRNA expression in mouse pectoralis muscle. BMC Res. Notes 3:297. doi: 10.1186/1756-0500-3-297
Rao, P. K., Kumar, R. M., Farkhondeh, M., Baskerville, S., and Lodish, H. F. (2006). Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl. Acad. Sci. U.S.A. 103, 8721–8726. doi: 10.1073/pnas.0602831103
Rebbapragada, A., Benchabane, H., Wrana, J. L., Celeste, A. J., and Attisano, L. (2003). Myostatin signals through a transforming growth factor beta-like signaling pathway to block adipogenesis. Mol. Cell. Biol. 23, 7230–7242. doi: 10.1128/MCB.23.20.7230-7242.2003
Rommel, C., Bodine, S. C., Clarke, B. A., Rossman, R., Nunez, L., Stitt, T. N., et al. (2001). Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat. Cell Biol. 3, 1009–1013. doi: 10.1038/ncb1101-1009
Rosenberg, M. I., Georges, S. A., Asawachaicharn, A., Analau, E., and Tapscott, S. J. (2006). MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J. Cell Biol. 175, 77–85. doi: 10.1083/jcb.200603039
Sandri, M. (2008). Signaling in muscle atrophy and hypertrophy. Physiology (Bethesda) 23, 160–170. doi: 10.1152/physiol.00041.2007
Sandri, M., Sandri, C., Gilbert, A., Skurk, C., Calabria, E., Picard, A., et al. (2004). Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117, 399–412. doi: 10.1016/S0092-8674(04)00400-3
Sato, M. M., Nashimoto, M., Katagiri, T., Yawaka, Y., and Tamura, M. (2009). Bone morphogenetic protein-2 down-regulates miR-206 expression by blocking its maturation process. Biochem. Biophys. Res. Commun. 383, 125–129. doi: 10.1016/j.bbrc.2009.03.142
Sayed, D., and Abdellatif, M. (2011). MicroRNAs in development and disease. Physiol. Rev. 91, 827–887. doi: 10.1152/physrev.00006.2010
Schertzer, J. D., Plant, D. R., and Lynch, G. S. (2006). Optimizing plasmid-based gene transfer for investigating skeletal muscle structure and function. Mol. Ther. 13, 795–803. doi: 10.1016/j.ymthe.2005.09.019
Schiaffino, S., Dyar, K. A., Ciciliot, S., Blaauw, B., and Sandri, M. (2013). Mechanisms regulating skeletal muscle growth and atrophy. FEBS J. 280, 4294–4314. doi: 10.1111/febs.12253
Schuelke, M., Wagner, K. R., Stolz, L. E., Hübner, C., Riebel, T., Kömen, W., et al. (2004). Myostatin mutation associated with gross muscle hypertrophy in a child. N.Engl. J. Med. 350, 2682–2688. doi: 10.1056/NEJMoa040933
Shi, Z.-M., Wang, J., Yan, Z., You, Y.-P., Li, C.-Y., Qian, X., et al. (2012). MiR-128 inhibits tumor growth and angiogenesis by targeting p70S6K1. PLoS ONE 7:e32709. doi: 10.1371/journal.pone.0032709
Shyu, K. G., Lu, M. J., Wang, B. W., Sun, H. Y., and Chang, H. (2006). Myostatin expression in ventricular myocardium in a rat model of volume-overload heart failure. Eur. J. Clin. Invest. 36, 713–719. doi: 10.1111/j.1365-2362.2006.01718.x
Small, E. M., O'Rourke, J. R., Moresi, V., Sutherland, L. B., McAnally, J., Gerard, R. D., et al. (2010). Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc. Natl. Acad. Sci. U.S.A. 107, 4218–4223. doi: 10.1073/pnas.1000300107
Stambolic, V., Suzuki, A., de la Pompa, J. L., Brothers, G. M., Mirtsos, C., Sasaki, T., et al. (1998). Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell 95, 29–39. doi: 10.1016/S0092-8674(00)81780-8
Sun, Y., Ge, Y., Drnevich, J., Zhao, Y., Band, M., and Chen, J. (2010). Mammalian target of rapamycin regulates miRNA-1 and follistatin in skeletal myogenesis. J. Cell Biol. 189, 1157–1169. doi: 10.1083/jcb.200912093
Takahashi, A., Kureishi, Y., Yang, J., Luo, Z., Guo, K., Mukhopadhyay, D., et al. (2002). Myogenic Akt signaling regulates blood vessel recruitment during myofiber growth. Mol. Cell. Biol. 22, 4803–4814. doi: 10.1128/MCB.22.13.4803-4814.2002
Taylor, W. E., Bhasin, S., Artaza, J., Byhower, F., Azam, M., Willard, D. H., et al. (2001). Myostatin inhibits cell proliferation and protein synthesis in C2C12 muscle cells. Am. J. Physiol. Endocrinol. Metab. 280, E221–E228.
Tumaneng, K., Schlegelmilch, K., Russell, R. C., Yimlamai, D., Basnet, H., Mahadevan, N., et al. (2012). YAP mediates crosstalk between the Hippo and PI(3)K–TOR pathways by suppressing PTEN via miR-29. Nat. Cell Biol. 14, 1322–1329. doi: 10.1038/ncb2615
Tyler, K. L. (2003). Origins and early descriptions of Duchenne muscular dystrophy. Muscle Nerve 28, 402–422. doi: 10.1002/mus.10435
van Rooij, E., Quiat, D., Johnson, B. A., Sutherland, L. B., Qi, X., Richardson, J. A., et al. (2009). A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev. Cell 17, 662–673. doi: 10.1016/j.devcel.2009.10.013
Verzola, D., Procopio, V., Sofia, A., Villaggio, B., Tarroni, A., Bonanni, A., et al. (2011). Apoptosis and myostatin mRNA are upregulated in the skeletal muscle of patients with chronic kidney disease. Kidney Int. 79, 773–782. doi: 10.1038/ki.2010.494
Wada, S., Kato, Y., Okutsu, M., Miyaki, S., Suzuki, K., Yan, Z., et al. (2011). Translational suppression of atrophic regulators by microRNA-23a integrates resistance to skeletal muscle atrophy. J. Biol. Chem. 286, 38456–38465. doi: 10.1074/jbc.M111.271270
Wang, H., Garzon, R., Sun, H., Ladner, K. J., Singh, R., Dahlman, J., et al. (2008). NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell 14, 369–381. doi: 10.1016/j.ccr.2008.10.006
Wang, Q., and McPherron, A. C. (2012). Myostatin inhibition induces muscle fibre hypertrophy prior to satellite cell activation. J. Physiol. (Lond) 590, 2151–2165. doi: 10.1113/jphysiol.2011.226001
Wang, X. H. (2013). MicroRNA in myogenesis and muscle atrophy. Curr. Opin. Clin. Nutr. Metab. Care 16, 258–266. doi: 10.1097/MCO.0b013e32835f81b9
Wei, W., He, H.-B., Zhang, W.-Y., Zhang, H.-X., Bai, J.-B., Liu, H.-Z., et al. (2013). miR-29 targets Akt3 to reduce proliferation and facilitate differentiation of myoblasts in skeletal muscle development. Cell Death Dis. 4, e668. doi: 10.1038/cddis.2013.184
Welle, S., Bhatt, K., and Pinkert, C. A. (2006). Myofibrillar protein synthesis in myostatin-deficient mice. Am. J. Physiol. Endocrinol. Metab. 290, E409–E415. doi: 10.1152/ajpendo.00433.2005
Welle, S., Burgess, K., and Mehta, S. (2009). Stimulation of skeletal muscle myofibrillar protein synthesis, p70 S6 kinase phosphorylation, and ribosomal protein S6 phosphorylation by inhibition of myostatin in mature mice. Am. J. Physiol. Endocrinol. Metab. 296, E567–E572. doi: 10.1152/ajpendo.90862.2008
Welle, S., Mehta, S., and Burgess, K. (2011). Effect of postdevelopmental myostatin depletion on myofibrillar protein metabolism. Am. J. Physiol. Endocrinol. Metab. 300, E993–E1001. doi: 10.1152/ajpendo.00509.2010
Winbanks, C. E., Beyer, C., Hagg, A., Qian, H., Sepulveda, P. V., and Gregorevic, P. (2013). miR-206 represses hypertrophy of myogenic cells but not muscle fibers via inhibition of HDAC4. PLoS ONE 8:e73589. doi: 10.1371/journal.pone.0073589
Winbanks, C. E., Wang, B., Beyer, C., Koh, P., White, L., Kantharidis, P., et al. (2011). TGF-beta regulates miR-206 and miR-29 to control myogenic differentiation through regulation of HDAC4. J. Biol. Chem. 286, 13805–13814. doi: 10.1074/jbc.M110.192625
Xu, J., Li, R., Workeneh, B., Dong, Y., Wang, X., and Hu, Z. (2012). Transcription factor FoxO1, the dominant mediator of muscle wasting in chronic kidney disease, is inhibited by microRNA-486. Kidney Int. 82, 401–411. doi: 10.1038/ki.2012.84
Yan, B., Zhu, C.-D., Guo, J.-T., Zhao, L.-H., and Zhao, J.-L. (2013). miR-206 regulates the growth of the teleost tilapia (Oreochromis niloticus) through the modulation of IGF-1 gene expression. J. Exp. Biol. 216, 1265–1269. doi: 10.1242/jeb.079590
Yarasheski, K. E., Bhasin, S., Sinha-Hikim, I., Pak-Loduca, J., and Gonzalez-Cadavid, N. F. (2002). Serum myostatin-immunoreactive protein is increased in 60-92 year old women and men with muscle wasting. J. Nutr. Health Aging 6, 343–348.
Zacharewicz, E., Lamon, S., and Russell, A. P. (2013). MicroRNAs in skeletal muscle and their regulation with exercise, ageing, and disease. Front. Physiol. 4:266. doi: 10.3389/fphys.2013.00266
Zhou, D., Birkenmeier, C. S., Williams, M. W., Sharp, J. J., Barker, J. E., and Bloch, R. J. (1997). Small, membrane-bound, alternatively spliced forms of ankyrin 1 associated with the sarcoplasmic reticulum of mammalian skeletal muscle. J. Cell Biol. 136, 621–631. doi: 10.1083/jcb.136.3.621
Zhou, L., Wang, L., Lu, L., Jiang, P., Sun, H., and Wang, H. (2012). Inhibition of miR-29 by TGF-beta-Smad3 signaling through dual mechanisms promotes transdifferentiation of mouse myoblasts into myofibroblasts. PLoS ONE 7:e33766. doi: 10.1371/journal.pone.0033766
Keywords: insulin-like growth factor-1, myostatin, protein kinase B (Akt), Smad3, skeletal muscle hypertrophy
Citation: Hitachi K and Tsuchida K (2014) Role of microRNAs in skeletal muscle hypertrophy. Front. Physiol. 4:408. doi: 10.3389/fphys.2013.00408
Received: 28 September 2013; Accepted: 25 December 2013;
Published online: 16 January 2014.
Edited by:
Wataru Aoi, Kyoto Prefectural University, JapanReviewed by:
Ravi C. Balijepalli, University of Wisconsin, USABruno Bastide, University of Lille Nord de France, France
Sue Bodine, University of California, Davis, USA
Gustavo Brum, Universidad de la República, Uruguay
Copyright © 2014 Hitachi and Tsuchida. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kunihiro Tsuchida, Division for Therapies against Intractable Diseases, Institute for Comprehensive Medical Science, Fujita Health University, 1-98 Dengakugakubo, Kutsukake-cho, Toyoake, Aichi 470-1192, Japan e-mail: tsuchida@fujita-hu.ac.jp
 Keisuke Hitachi
Keisuke Hitachi Kunihiro Tsuchida
Kunihiro Tsuchida