- 1Science and Technology Department, BGI-Shenzhen, Beishan Industrial Zone, Shenzhen, China
- 2Key Lab for Zoonosis Research, Ministry of Education, Institute of Zoonosis, Jilin University, Changchun, China
- 3Jiangsu Co-innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, Yangzhou, China
The recent discovery of DNA methylation in the nematode T.spiralis may raise the possibility of using it as a potential model organism for epigenetic studies instead of C. elegans, which is deficient in this important epigenetic modification. In contrast to the free-living nematode C. elegans, T. spiralis is a parasitic worm that possesses a complicated life cycle and undergoes a complex developmental regulation of genes. We emphasize that the differential methylomes in the different life-history stages of T. spiralis can provide insight on how DNA methylation is triggered and regulated. In particular, we have demonstrated that DNA methylation is involved in the regulation of its parasitism-related genes. Further computational analyses indicated that the regulatory machinery for DNA methylation can also be found in the T. spiralis genome. By a logical extension of this point, we speculate that comprehensively addressing the epigenetic machinery of T. spiralis may help to understand epigenetics in invertebrates. Furthermore, considering the implication of epigenetics in metazoan parasitism, using T. spiralis as an epigenetic model organism may further contribute to drug development against metazoan parasites.
Current Model Organisms in Epigenetic Studies
Research activities using model organisms have provided critical breakthroughs in understanding fundamental questions in biology. The most popular animal models are the nematode Caenorhabditis elegans, the fruit fly Drosophila melanogaster, the zebrafish Danio rerio, the African clawed frog Xenopus laevis, the chicken Gallus gallus domesticus, the mouse Mus musculus and the rat Rattus norvegicus (Cogburn et al., 2007; Jenner and Wills, 2007; Kennedy, 2008). Benefiting from technological advancements, the complete genomes of multiple model organisms have been obtained. However, our knowledge about how the information encoded in their genomes is regulated or interpreted is still limited. Thus, understanding epigenetic phenomena has become a major focus of research activity in the current post-genomics era.
The term epigenetics refers to the study of any potentially stable and ideally heritable change that alters gene expression without altering DNA sequences in an organism (Goldberg et al., 2007). Among a variety of mechanisms involved, DNA methylation and post-translationally modified histones within their N termini can result in changes in the accessibility of chromatin to chromatin-remodeling complexes, transcriptional complexes and polymerases. Epigenetic studies also necessarily require good experimental models. Indeed, various model organisms have been applied to date, through which a wealth of knowledge has been acquired from several landmark epigenetic discoveries (David Allis and Danny Reinberg, 2007). Different model organisms offer different advantages, and all are important for learning about the processes and mechanisms involved in epigenetic regulation. For example, multiple species of yeast have been used as model systems to study chromatin structure (David Allis and Danny Reinberg, 2007). Because DNA methylation is not observed in yeast, the presence of DNA methylation in the fungus Neurospora crassa made it a model organism for DNA methylation studies and contributed to the discovery of repeat-induced point mutation (RIP), which is a genome defense mechanism (David Allis and Danny Reinberg, 2007). Histone variants, the first histone acetyltransferase, histone lysine methylation, histone phosphorylation and one RNAi pathway were discovered in the protozoan Tetrahymena thermophila (David Allis and Danny Reinberg, 2007). The studies on the fruit fly D. melanogaster have led to the discovery of chromatin remodeling proteins and histone modifying proteins (Lecuyer et al., 2007). Plants, such as Arabidopsis thaliana, have epigenetic mechanisms as sophisticated as those of mammals, including RNAi pathways, DNA methylation, histone modification, and chromosome remodeling complexes (Henderson and Jacobsen, 2007). Finally, mice, as mammals, are more similar to humans than any of these model systems and are used as models for epigenetic research, particularly in embryology, stem cell research and environmental studies, including the effects of behavior and nutrition on epigenetic states.
Biology and Genome of T. spiralis
In contrast to the free-living nematode C. elegans, Trichinella spiralis (T. spiralis) is a parasitic worm that possesses a complicated life cycle and undergoes complex developmental regulation of its genes (Figure 1) (Mitreva and Jasmer, 2006). T. spiralis begins its life cycle as muscle larvae contained in infected meat. With the aid of host gastric juices, the muscle larvae are then released into the host's stomach and mature into sexually active adults in the host's intestines. After the second generation larvae are born, they migrate throughout the entire body of the host and invade the skeletal muscles. The second generation larvae can survive in host for years (up to 40 years in humans), and this hypobiotic stage is maintained until being ingested by a new host, thereby the new generation begin (Mitreva and Jasmer, 2006).
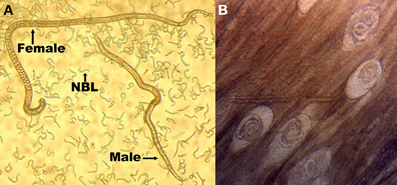
Figure 1. Three main life-stage forms of T. spiralis. (A) Adults and new born larvae (NBL) and (B) infective muscle larvae. Female adults, which develop in the small intestine, will produce new-born larvae, which then disseminate through the bloodstream, invade skeletal muscles, and encyst in a collagen capsule to form a new generation of muscle larvae.
T. spiralis exhibits a wide, global distribution, and this species is the most important etiological agent to cause disease in humans (Mitreva and Jasmer, 2006). Trichinellosis affects approximately ten million people globally (Tantrawatpan et al., 2013), with the common signs and symptoms in humans including fever, diarrhea, periorbital oedema and myalgia. Humans acquire this disease by ingesting raw or insufficiently cooked meat containing the infective larvae of T. spiralis (Wang et al., 2013). This disease not only is a public-health hazard but also represents an economic problem in porcine animal production and food safety (Mitreva and Jasmer, 2010). However, until now the mechanism by which T. spiralis infective larvae distinguish, attack, creep within the intestinal epithelium, and create their intramuscular niches have been inadequately explained, which has made the therapy against trichinellosis much more difficult (Basyoni and El-Sabaa, 2013).
In the evolution of the Nematoda, T. spiralis is a member of a clade that diverged early. It differs substantially in its biological and molecular characteristics from other crown groups. The completion of the T. spiralis draft genome enables further investigations on this interesting parasitic nematode, particularly with respect to either its evolutional position or comparative and functional genomics (Mitreva et al., 2011).
Preliminary Discovery in Epigenetics of T. spiralis
Extensive epigenetic studies have been performed on the genome of the well-annotated model nematode C. elegans, addressing its functional genomic elements including histone modifications (Gerstein et al., 2010; Liu et al., 2011). However, the most studied epigenetic mechanism, DNA methylation, is absent in C. elegans (Simpson et al., 1986). By extension, conventional thinking has held that DNA methylation is missing throughout the nematode phylum. However, recent progress has been made with the discovery, for the first time, of the presence of DNA methylation in T. spiralis (Gao et al., 2012). Not only have genes that are homologous to canonical dnmt1 and dnmt3 in the T. spiralis genome been identified, but also the presence of DNA methylation was probed directly by several methods. As T. spiralis belongs to a basal clade of Nematoda, having diverged from C. elegans several hundred million years ago in the late Precambrian (Mitreva et al., 2011), this species likely retained DNA methylation machinery that may have been present in the ancestral nematode. Furthermore, potential homologs of canonical histone-modifying enzymes, chromatin remodeling factors and other relevant proteins were also revealed in the released draft genome sequence (Mitreva et al., 2011) and in our own computational analyses (data not shown), suggesting the presence of a fully developed epigenetic system in T. spiralis. These results revealed a new and unique characterization of this species that differs from other nematode species.
T. spiralis May Provide a New System for Probing Epigenetic Regulation
Model organisms not only have some characteristics in common, such as easy culture, non-specialist living requirements and amenability in experimental manipulation but also have one or more special features that qualify them to be outstanding subjects in research. For example, in C. elegans, the fate of each constituent cell can be exactly traced, and in D. melanogaster, mutations are easily induced with the resulting phenotypes being easily observed.
T. spiralis is also an easy organism to raise in the lab, with high fertility, relatively short generation time and easy accessibility of all life-history stages, which ensure that it is an accessible model to study. More importantly, it has special features that are advantageous to being an epigenetics model organism compared with other existing non-mammalian model organisms. First, as a nematode, T. spiralis appears to have developed a relatively complete epigenetic system. In addition to evolutionally conserved histone modifications, similar characteristics of the T. spiralis methylome compared with vertebrate genomes were found, although methylated cytosines are relatively sparse. The methylation of transposable elements was previously viewed as a unique feature of vertebrate genomes and was thought to be mostly absent from invertebrates (Suzuki and Bird, 2008). However, in the T. spiralis genome, transposable elements appear to be more heavily methylated than the genomic background. Furthermore, the DNA methylation levels of gene upstream regions have a negative correlation with gene expression levels, and non-expressed genes in particular had different patterns of DNA methylation with the methylation levels in their upstream regulatory regions being higher than in the coding sequences. This phenomenon fits with the model of vertebrate animals but is different from some non-vertebrate organisms, such as silkworm (Xiang et al., 2010). Second, by screening and sequencing methylated DNA present at the new-born, larval, and adult stages, virtually no methylation during the new-born stage was found. In the larval stage, when the worms move from the intestine to the muscle, however, methylation begins to occur, with adult worms carrying the most methylated DNA. The methylation machinery appears to be suddenly switched to the “on” state in the muscle larvae stage from the “off” state in the NBL stage. This finding is in stark contrast to DNA methylation patterns in other animals, including humans, which generally stay consistent throughout the organism's lifetime. Thus, T. spiralis may provide an excellent model for studying the pathways leading to the initiation, establishment, maintenance and plasticity of methylation states. Understanding such pathways will be extremely useful in dissecting the functions of DNA methylation in organismal development or differentiation and in disease occurrence.
T. spiralis May Provide a New System for Studying the Epigenetic Mechanism of Metazoan Parasitism
Parasites, either protozoan or metazoan, must be able to rapidly transit between complex life cycles and use long-term antigenic variation to avoid immune attacks in their hosts. To achieve this outcome, the parasite must initiate complex differentiation in response to environmental cues, which requires drastic and rapid alterations of their gene expression profiles. The transcriptional regulation of gene expression is therefore employed.
As an essential means of transcriptional regulation, epigenetics has emerged as a crucial aspect of parasite biology over the past decade. A major focus of investigation has been the characterization of histone modifications and the identification of the responsible enzymes. Currently, intensive studies in the protozoan parasites Plasmodium falciparum, Toxoplasma gondii and Trypanosoma brucei have been performed, which was reviewed nicely by Croken et al. (2012). The existence and functional roles of histone modifications in the epigenetic control of platyhelminth schistosome transcription have also been established [reviewed in Geyer and Hoffmann (2012)]. Through these studies, the relative conservation of histone-modifying enzymes across different species, including both protozoan and metazoan parasites, has been associated with evolutionarily conserved functions.
Observations of the most highly studied epigenetic mechanism of DNA methylation in mammals appeared variable in parasites. Among the protozoan parasites, T. brucei and Entamoeba histolytica have a DNA methyltransferase and methylated DNA detectable by mass spectrometry (Fisher et al., 2004; Militello et al., 2008). The genomes of T. gondii and Plasmodium and Cryptosporidium species do encode candidate methyltransferases, although no detectable methylated cytosines were discovered in previous studies (Gissot et al., 2008). The detection of DNA methylation is highly dependent on the sensitivity of the applied technology, and thus DNA methylation may be restricted to a small number of loci that were not detected by the methodology used. In the metazoan parasites, a previous study convincingly demonstrated that the platyhelminth Schistosoma mansoni contained a methylated genome and that methylation was found to be differentially present across the parasite's lifecycle (Geyer et al., 2011). For other nematodes in addition to C. elegans and T. spiralis, although a canonical DNA methyltransferase (dnmt3) homolog among 11 nematodes has not been found, we should not exclude the possibility of the existence of cytosine methylation in their genomes. Exceptionally, the silkworm uses DNA methylation, but its genome does not contain dnmt3 (Xiang et al., 2010). Moreover, the model protozoan ciliate does in fact utilize DNA methylation during a certain part of its life cycle (Bracht et al., 2012) even though no homologs of dnmts were found in its genome. Taken together, DNA methylation might also be an ancient epigenetic mechanism occurring in parasites and may play essential roles in gene regulation. Recent innovations in high-throughput sequencing may enable researchers to infer methylation at a single-base resolution [10], thereby raising the possibility of a thorough investigation of the conservation and divergence of methylation patterns in different parasites. As a parasitic nematode, T. spiralis shares many characteristics with other parasites regarding parasitism. Parasitism-related genes were also found to be regulated by DNA methylation between the life cycle stages of T. spiralis, suggesting the potential biological implications of epigenetics in parasitism. Although further studies are required to reveal the implication of other epigenetic mechanisms; based on its fully developed epigenetic system, T. spiralis may serve as a good model for studying the implication of epigenetics in metazoan parasitism.
More importantly, understanding epigenetic regulation using the T. spiralis model opens new methods for the diagnosis and treatment of metazoan parasites, especially with respect to the development of new drugs affecting the epigenetic machinery. To develop new compounds that interfere with pathogenic states, the targeted epigenetic regulators should be specific compared with their host orthologs. Previous analyses on class I HDACs (SmHDAC8) and sirtuins (SmSirt1 and 2) in metazoan schistosomes have shown such potential regarding schistosomes (Pierce et al., 2011). Our analyses on dnmts and mbd-like proteins of T. spiralis also show notable differences in their catalytic domains compared with their mammalian orthologs (Figure 2). Therefore, given the importance of epigenetics in metazoan parasite biology, drug discovery by targeting epigenetic regulators should be promising as a paradigm for drug development against metazoan parasites.
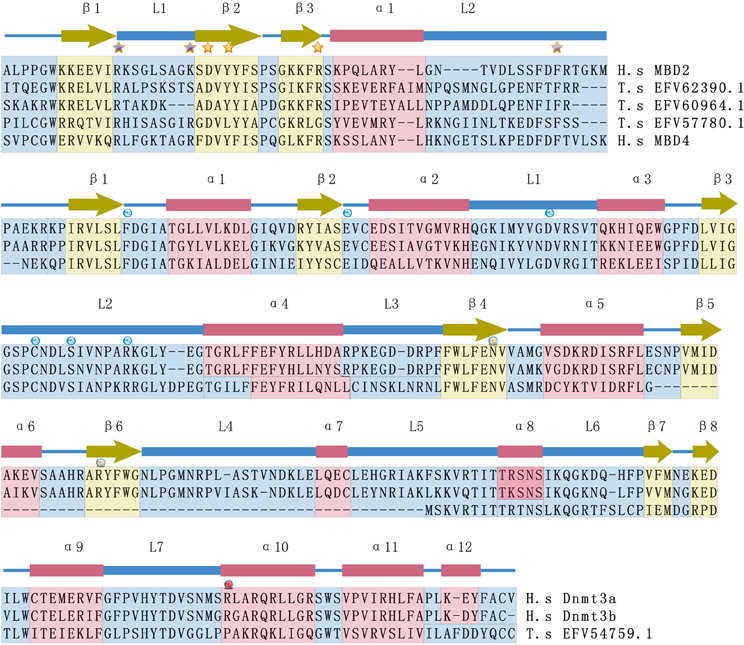
Figure 2. Comparison of the secondary structures of MBD domains and Dnmt3 catalytic domains between T. spiralis and H. sapiens. The secondary structures of human MBD2 and Dnmt3a are indicated as blocks, including alpha helices (α), beta sheets (β), and loops (L), that are marked with different colors (blue = alpha helices, yellow = beta sheets, and red = loops). Important residues responsible for methyl-CpG binding are marked with asterisks. The functional sites of the Dnmt3 catalytic domain are marked with circles, including reported S-Adenosyl methionine (SAM) binding sites and active sites. H.s and T.s are short for H. sapiens and T. spiralis, respectively. T.s EFV 62390.1, T.s EFV 60964.1 and T.s EFV57780 are proteins in T. spiralis that show high sequence similarity with human Mbd proteins. T.s EFV54759.1 is a protein of T.spiralis that has sequence similarity with human Dnmt3 protein.
Conclusion
We highlight that the current findings on the DNA methylation of T. spiralis may raise the possibility of using it as a potential model organism in epigenetic studies. We emphasize that the differential methylomes in the different life-history stages of T. spiralis can provide insight into how DNA methylation is triggered and regulated. In particular, T. spiralis is a parasitic worm that diverged early in the evolution of nematodes, and epigenetics is involved in regulation of its parasitism-related genes. By a logical extension of this point, we speculate that comprehensively addressing the epigenetic machinery of T. spiralis may help to deeply understand the implication of epigenetics in metazoan parasitism and further contribute to the drug development against metazoan parasites.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Yaping Huang for preparing Figure 2 in the manuscript.
References
Basyoni, M. M., and El-Sabaa, A. A. (2013). Therapeutic potential of myrrh and ivermectin against experimental Trichinella spiralis infection in mice. Korean J. Parasitol. 51, 297–304. doi: 10.3347/kjp.2013.51.3.297
Bracht, J. R., Perlman, D. H., and Landweber, L. F. (2012). Cytosine methylation and hydroxymethylation mark DNA for elimination in Oxytricha trifallax. Genome Biol. 13, R99. doi: 10.1186/gb-2012-13-10-r99
Cogburn, L. A., Porter, T. E., Duclos, M. J., Simon, J., Burgess, S. C., Zhu, J. J., et al. (2007). Functional genomics of the chicken–a model organism. Poult. Sci. 86, 2059–2094.
Croken, M. M., Nardelli, S. C., and Kim, K. (2012). Chromatin modifications, epigenetics, and how protozoan parasites regulate their lives. Trends Parasitol. 28, 202–213. doi: 10.1016/j.pt.2012.02.009
David Allis, C., and Danny Reinberg, T. J. (2007). “Overview and Concepts,” in Epigenetics, eds T. Jenuwein, C. David Allis, and D. Reinberg (New York, NY: Cold Spring Harbor Laboratory Press), 23–62.
Fisher, O., Siman-Tov, R., and Ankri, S. (2004). Characterization of cytosine methylated regions and 5-cytosine DNA methyltransferase (Ehmeth) in the protozoan parasite Entamoeba histolytica. Nucleic Acids Res. 32, 287–297. doi: 10.1093/nar/gkh161
Gao, F., Liu, X., Wu, X. P., Wang, X. L., Gong, D., Lu, H., et al. (2012). Differential DNA methylation in discrete developmental stages of the parasitic nematode Trichinella spiralis. Genome Biol. 13, R100. doi: 10.1186/gb-2012-13-10-r100
Gerstein, M. B., Lu, Z. J., Van Nostrand, E. L., Cheng, C., Arshinoff, B. I., Liu, T., et al. (2010). Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330, 1775–1787. doi: 10.1126/science.1196914
Geyer, K. K., and Hoffmann, K. F. (2012). Epigenetics: a key regulator of platyhelminth developmental biology? Int. J. Parasitol. 42, 221–224. doi: 10.1016/j.ijpara.2012.02.003
Geyer, K. K., Rodriguez Lopez, C. M., Chalmers, I. W., Munshi, S. E., Truscott, M., Heald, J., et al. (2011). Cytosine methylation regulates oviposition in the pathogenic blood fluke Schistosoma mansoni. Nat. Commun. 2, 424. doi: 10.1038/ncomms1433
Gissot, M., Choi, S. W., Thompson, R. F., Greally, J. M., and Kim, K. (2008). Toxoplasma gondii and Cryptosporidium parvum lack detectable DNA cytosine methylation. Eukaryotic Cell 7, 537–540. doi: 10.1128/EC.00448-07
Goldberg, A. D., Allis, C. D., and Bernstein, E. (2007). Epigenetics: a landscape takes shape. Cell 128, 635–638. doi: 10.1016/j.cell.2007.02.006
Henderson, I. R., and Jacobsen, S. E. (2007). Epigenetic inheritance in plants. Nature 447, 418–424. doi: 10.1038/nature05917
Jenner, R. A., and Wills, M. A. (2007). The choice of model organisms in evo-devo. Nat. Rev. Genet. 8, 311–319. doi: 10.1038/nrg2062
Kennedy, B. K. (2008). The genetics of ageing: insight from genome-wide approaches in invertebrate model organisms. J. Intern. Med. 263, 142–152. doi: 10.1111/j.1365-2796.2007.01903.x
Lecuyer, E., Yoshida, H., Parthasarathy, N., Alm, C., Babak, T., Cerovina, T., et al. (2007). Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131, 174–187. doi: 10.1016/j.cell.2007.08.003
Liu, T., Rechtsteiner, A., Egelhofer, T. A., Vielle, A., Latorre, I., Cheung, M. S., et al. (2011). Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res. 21, 227–236. doi: 10.1101/gr.115519.110
Militello, K. T., Wang, P., Jayakar, S. K., Pietrasik, R. L., Dupont, C. D., Dodd, K., et al. (2008). African trypanosomes contain 5-methylcytosine in nuclear DNA. Eukaryotic Cell 7, 2012–2016. doi: 10.1128/EC.00198-08
Mitreva, M., and Jasmer, D. P. (2006). “Biology and genome of Trichinella spiralis,” in WormBook, ed The C. elegans Research Community, 1–21. doi: 10.1895/wormbook.1.124.1
Mitreva, M., and Jasmer, D. P. (2010). Trichinella spiralis: genomic application to control a zoonotic nematode. Infect. Disord. Drug Targets 10, 376–384. doi: 10.2174/187152610793180830
Mitreva, M., Jasmer, D. P., Zarlenga, D. S., Wang, Z., Abubucker, S., Martin, J., et al. (2011). The draft genome of the parasitic nematode Trichinella spiralis. Nat. Genet. 43, 228–235. doi: 10.1038/ng.769
Pierce, R. J., Dubois-Abdesselem, F., Caby, S., Trolet, J., Lancelot, J., Oger, F., et al. (2011). Chromatin regulation in schistosomes and histone modifying enzymes as drug targets. Mem. Inst. Oswaldo Cruz 106, 794–801. doi: 10.1590/S0074-02762011000700003
Simpson, V. J., Johnson, T. E., and Hammen, R. F. (1986). Caenorhabds elegans DNA does not contain 5-methylcytosine at any time during development or aging. Nucleic Acids Res. 14, 9. doi: 10.1093/nar/14.16.6711
Suzuki, M. M., and Bird, A. (2008). DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 9, 465–476. doi: 10.1038/nrg2341
Tantrawatpan, C., Intapan, P. M., Thanchomnang, T., Sanpool, O., Janwan, P., Boonmars, T., et al. (2013). Early detection of Trichinella spiralis in muscle of infected mice by real-time fluorescence resonance energy transfer PCR. Vector Borne Zoonotic Dis. 13, 674–681. doi: 10.1089/vbz.2012.1221
Wang, L., Wang, Z. Q., Hu, D. D., and Cui, J. (2013). Proteomic analysis of Trichinella spiralis muscle larval excretory-secretory proteins recognized by early infection sera. Biomed Res. Int. 2013, 139745. doi: 10.1155/2013/139745
Keywords: trichinella spiralis, epigenetics, model nematode, metazoan, parasitism
Citation: Gao F, Wang R and Liu M (2014) Trichinella spiralis, potential model nematode for epigenetics and its implication in metazoan parasitism. Front. Physiol. 4:410. doi: 10.3389/fphys.2013.00410
Received: 10 October 2013; Accepted: 26 December 2013;
Published online: 10 January 2014.
Edited by:
Mackenzie Gavery, University of Washington, USAReviewed by:
Maurice R. Elphick, Queen Mary University of London, UKAriel D. Chipman, The Hebrew University of Jerusalem, Israel
Copyright © 2014 Gao, Wang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fei Gao, Science and Technology Department, BGI-Shenzhen, No.11, Beishan Industrial Zone, Yantian District, Shenzhen, 518083, China e-mail: flys828@gmail.com;
Mingyuan Liu, Key Lab for Zoonosis Research, Ministry of Education, Institute of Zoonosis, Jilin University, 5333 Xi An Road, Changchun, 130062, China e-mail: liumy@jlu.edu.cn
 Fei Gao
Fei Gao Rui Wang
Rui Wang Mingyuan Liu
Mingyuan Liu