- Sylvius Laboratory, Institute of Biology, Leiden University, Leiden, Netherlands
The promoter of the salicylic acid-inducible PR-1a gene of Nicotiana tabacum contains binding sites for transcription factor NtWRKY12 (WK-box at position −564) and TGA factors (as-1-like element at position −592). Transactivation experiments in Arabidopsis protoplasts derived from wild type, npr1-1, tga256, and tga2356 mutant plants revealed that NtWRKY12 alone was able to induce a PR-1a::β-glucuronidase (GUS) reporter gene to high levels, independent of co-expressed tobacco NtNPR1, TGA2.1, TGA2.2, or endogenous Arabidopsis NPR1, TGA2/3/5/6. By in vitro pull-down assays with GST and Strep fusion proteins and by Fluorescence Resonance Energy Transfer assays with protein–CFP and protein–YFP fusions in transfected protoplasts, it was shown that NtWRKY12 and TGA2.2 could interact in vitro and in vivo. Interaction of NtWRKY12 with TGA1a or TGA2.1 was not detectable by these techniques. A possible mechanism for the role of NtWRKY12 and TGA2.2 in PR-1a gene expression is discussed.
Introduction
Upon pathogen attack plants mobilize inducible defense systems. A classic example is the systemic acquired resistance (SAR) effective against a broad range of pathogens. The signal transduction route leading to SAR involves the induced synthesis of the endogenous signal molecule salicylic acid (SA). SAR is accompanied by the de novo synthesis of pathogenesis-related (PR) proteins of which many directly affect pathogen growth and disease proliferation. Although their exact function is still not fully characterized, the plant-wide conserved PR-1 proteins are generally considered as marker proteins for SAR (Ross, 1961; Durrant and Dong, 2004). In most plant species expression of the PR-1 genes is under transcriptional control.
Early work by the group of Chua in tobacco (Nicotiana tabacum) has indicated that gene expression controlled by the 35S promoter from Cauliflower mosaic virus is enhanced by SA and that this effect depends on the presence of activation sequence-1 (as-1), a DNA element in the 90-bp core promoter consisting of a TGACG tandem repeat (Qin et al., 1994). Mutation of the as-1 element in the 35S promoter resulted in reduced promoter activity and loss of DNA binding of the ASF-1 complex and of the basic leucine zipper (bZIP) transcription factor TGA1a, indicating that the ASF-1 complex and TGA1a have the same binding specificity for the as-1 element (Katagiri et al., 1989; Lam et al., 1989). More recently, the structurally related TGA2.2 was identified as the major DNA-binding component of ASF-1, while homolog TGA2.1 was present at lower amounts (Niggeweg et al., 2000a). While TGA2.2 was found to be of major importance for the expression of SA-inducible genes that contain as-1-like elements in their promoter, TGA2.1 was dispensable for this activation (Thurow et al., 2005).
Also promoters of several PR genes, such as Arabidopsis thaliana PR-1 and tobacco PR-1a contain as-1-(like) elements in promoter regions important for SA-induced expression. In tobacco the as-1-like element in the PR-1a promoter consists of a set of inverted TGACG motifs which were found to bind TGA transcription factors, while mutation of the element in a PR-1a promoter::GUS reporter gene affected SA-induced GUS expression (Strompen et al., 1998; Grüner et al., 2003). Likewise, a linker scanning analysis of the region of the Arabidopsis PR-1 promoter responsible for induced expression by the SA analog INA revealed the presence of an as-1 element with two direct TGACG motifs of which one is a positive regulatory element (LS7), while the other (LS5) mediates negative regulation of PR-1 expression (Lebel et al., 1998). Recently it was found that concurrent mutation of the LS5 and LS7 elements did not have a major effect on PR-1 promoter activity, and that the promoter in this situation is activated through a mechanism that requires the nearby upstream LS4 element, containing a consensus WRKY transcription factor binding site (Pape et al., 2010). Through knock-out analyses it was shown that the Arabidopsis bZIP transcription factors TGA2, TGA3, TGA5, and TGA6 act as redundant but essential activators of PR-1 expression and SAR (Zhang et al., 2003; Kesarwani et al., 2007). Recently it was shown that in a tga256 mutant background, 8 h after SA application the activation of PR-1 was impaired. However this was not observed for the 24-h time point, suggesting the involvement of other regulators in the SA-mediated activation of PR-1 (Blanco et al., 2009).
The ankyrin repeat protein NPR1 plays a central role in defense responses and is required for induction of PR gene expression and the establishment of SAR (Delaney et al., 1995; Cao et al., 1997; Wang et al., 2006). Upon pathogen-induced accumulation of SA, the redox state of the cell changes, resulting in release of reduced NPR1 monomers from cytoplasmic complexes and subsequent translocation to the nucleus where it interacts with TGA transcription factors to activate gene expression (Zhang et al., 1999; Després et al., 2000; Kinkema et al., 2000; Zhou et al., 2000; Mou et al., 2003). Recently it was shown that coactivation by NPR1 occurs in a pulse-wise manner and is regulated by degradation of NPR1 via the proteasome (Spoel et al., 2009).
In addition to TGAs, WRKY transcription factors are important for transcriptional programs induced in response to environmental signals (Eulgem and Somssich, 2007; Pandey and Somssich, 2009). Unlike the TGA transcription factors that are present at steady state levels (Johnson et al., 2003), many of the WRKY genes are transcriptionally activated upon biotic and abiotic stress. Of the 73 WRKY genes in Arabidopsis, 49 were differentially expressed upon Pseudomonas syringae infection or treatment with SA (Dong et al., 2003). In this respect it is interesting to note that SA biosynthesis itself may be under positive feedback regulation by WRKY transcription factors as we recently found that Arabidopsis WRKY28 and WRKY46 activate the SA biosynthesis genes ICS1 and PBS3, respectively (van Verk et al., 2011a,b). Many WRKY proteins bind to the W-box, a DNA motif with the core sequence TTGAC(T/C) and the overrepresentation of this motif in several WRKY genes suggests their expression is regulated by WRKY transcription factors. However, for several WRKY genes, SA-induced expression is dependent on NPR1 and TGAs, suggesting a similar activation strategy as was originally suggested for PR-1 (Dong et al., 2003; Wang et al., 2006).
In the same linker scanning study that identified the two as-1-like regulatory elements in the Arabidopsis PR-1 promoter, a consensus W-box motif with a strong negative effect was identified (Lebel et al., 1998), while recently, Pape et al. (2010) identified a W-box that conferred high-level inducibility to PR-1. These results suggest that WRKY factors are important for SA-mediated PR-1 gene expression (Lebel et al., 1998). The tobacco PR-1a promoter does not harbor a consensus W-box, however, NtWRKY12, a WRKY protein with a variant DNA-binding domain (BD), was found to bind to WK-boxes (TTTTCCAC) in the PR-1a promoter. Mutations in the WK-box at position −564 of the PR-1a promoter reduced SA-mediated PR-1a::GUS expression in transgenic tobacco or bacterial elicitor-mediated expression in agroinfiltrated leaves by 50–60%. In these assays, mutations in the as-1-like element at position −592 to −577 of the PR-1a promoter had little or no effect on PR-1a::GUS expression. However, combined mutation of the WK and as-1-like elements completely abolished inducible expression, indicating the importance of both NtWRKY12 and TGA transcription factors in regulation of PR-1a promoter activity (van Verk et al., 2008).
In this study we used pull-down assays and fluorescence resonance energy transfer (FRET) analysis to identify protein–protein interactions between NtWRKY12 and TGA factors in vitro and in vivo, respectively. In addition, we performed transactivation experiments in Arabidopsis protoplasts to study the effect of combinations of NtWRKY12, TGAs, and NtNPR1 on PR-1a gene expression. Our findings revealed that NtWRKY12 alone was able to induce PR-1a expression to high levels. Although we established that NtWRKY12 specifically interacts with tobacco TGA2.2, their co-expression in Arabidopsis protoplasts did not have measurable effect on PR-1a::GUS expression. Furthermore, we found that NtWRKY12-induced PR-1a::GUS expression was independent of Arabidopsis NPR1 or TGAs.
Results
Protein–Protein Interactions between NtWRKY12, TGAs, and NPR1
Our previous work pointed to cooperation between NtWRKY12 and TGA transcription factors in the activation of the PR-1a promoter. To analyze a possible protein–protein interaction between NtWRKY12 and tobacco TGA factors in vivo and in vitro, we used FRET analysis and in vitro pull-down assays, respectively.
To elaborate the cellular localization of NtWRKY12, TGA1a, TGA2.1, TGA2.2, and NtNPR1 for the FRET analyses we transfected Arabidopsis protoplasts with plasmids in which the corresponding cDNAs were cloned upstream of the YFP or CFP coding sequence. Examples of imaging of the fusion proteins in living protoplasts by confocal laser scanning microscopy are shown in Figure 1. Whereas fluorescence of unfused CFP and YFP was dispersed throughout the cytoplasm and nucleus (Figures 1A,B), NtWRKY12:CFP, TGA2.1:YFP, and TGA2.2:YFP (Figures 1C–E) fluorescence localized mainly in the nucleus. The same results were obtained when the proteins were fused to the other chromophore (data not shown). Interestingly, the signals of both NtNPR1:CFP and NtNPR1:YFP were always concentrated in small nuclear spots (Figure 1F). Furthermore, it is noteworthy that we never detected fluorescence in protoplasts transformed with constructs containing TGA1a fused to either CFP or YFP. These results show that tobacco TGAs 2.1 and 2.2 localize to the nucleus, as was previously found in tobacco and for the Arabidopsis homologs (Pontier et al., 2002; Johnson et al., 2003; Thurow et al., 2005). Due to the extreme brightness of the uneven distributed small nuclear spots of the NtNPR1 chromophore fusions, these could not be used for FRET analysis.
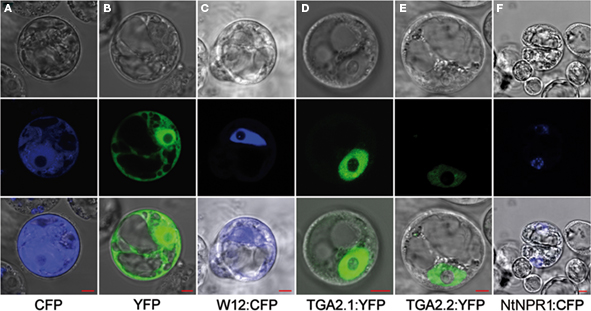
Figure 1. Nuclear localization of NtWRKY12 and TGAs. The panels on the middle row show confocal laser scanning microscopy images obtained of Arabidopsis protoplasts transfected with expression plasmids encoding unfused CFP (A) and YFP (B), and fusion proteins NtWRKY12:CFP [(C) W12:CFP], TGA2.1:YFP (D), TGA2.2:YFP (E), and NtNPR1:CFP (F). Panels on the bottom row show overlays with bright-field images (top row) of the same protoplasts. Localizations were visualized with a 63× objective. The red rulers indicate 5 μm.
Fluorescence resonance energy transfer analysis is based on overlapping emission/excitation spectra of donor fluorophore CFP and acceptor fluorophore YFP. Emitted fluorescence from CFP can only excite YFP when both fluorophores are in close (less than 10 nm apart) spatial proximity (Wu and Brand, 1994). Thus, a close association of two proteins with fusions to the respective fluorophores would result in an increase of acceptor fluorescence and quenching of the donor fluorescence. As a positive control for FRET, Arabidopsis protoplasts were transfected with an expression plasmid encoding a YFP:CFP tandem fusion, while cotransfection with uncoupled CFP- and YFP-encoding plasmids was used as negative control. The protoplasts were incubated for 24 h, after which FRET measurements were performed. The result is shown in Figure 2A. For the negative control, protoplasts were selected that showed both CFP (475 nm) and YFP (527 nm) emission after excitation of the respective fluorophores to confirm transfection with both CFP and YFP plasmids. Excitation of CFP with 457 nm UV light in these protoplasts resulted in an emission spectrum with a maximum at 475 nm and a certain level of bleeding at 527 nm. CFP excitation of the YFP:CFP fusion protein in the positive control protoplasts resulted in quenched emission at 475 nm, as part of the emission energy was used to excite the YFP fluorophore of the fusion protein, which was subsequently emitted at 527 nm. Thus, the slope of the line connecting the normalized emission intensities at 475 and 527 nm is a measure of the amount of FRET. Similarly, FRET assays were performed on protoplasts cotransfected with combinations of plasmids encoding NtWRKY12 and TGA chromophore fusion proteins.
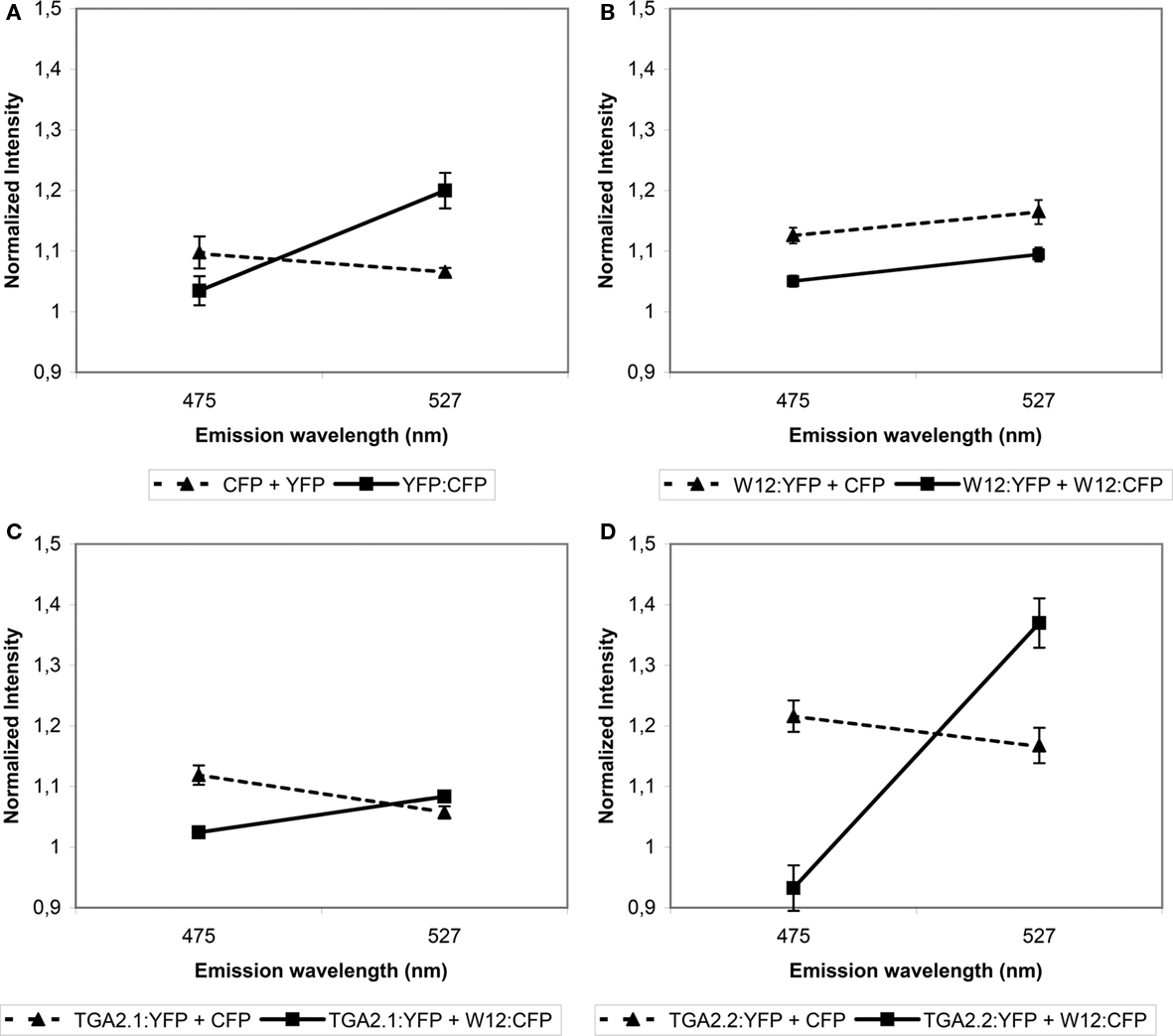
Figure 2. Fluorescence resonance energy transfer analysis of NtWRKY12 interacting with NtWRKY12, TGA2.1, and TGA2.2. Arabidopsis protoplasts were cotransfected with the indicated expression constructs. (A) Protoplasts transfected with a plasmid encoding a YFP:CFP tandem fusion (solid line) and protoplasts transfected with plasmids expressing unfused CFP and YFP (stippled line) were used as positive and negative FRET controls, respectively. (B–D) FRET data from protoplasts transfected with a combination of NtWRKY12:YFP (B), TGA2.1:YFP (C) or TGA2.2:YFP (D), and NtWRKY12:CFP (solid lines), compared to unfused CFP (stippled lines). After excitation at 457 nm, emission energies were measured in a total of 30, 5 nm wide intervals between 468 and 587 nm using confocal microscopy. Data from five protoplasts were averaged and normalized. FRET is presented by the slopes of lines connecting emission intensities at 475 nm (CFP quenching) and 527 nm (YFP emission). Error bars represent the SEM.
The control experiments with combinations of NtWRKY12:YFP, TGA2.1:YFP, or TGA2.2:YFP with unfused CFP did not result in increased 527 nm emission (dashed lines in Figures 2B–D, respectively), showing that neither NtWRKY12 nor the TGAs interacted with the CFP chromophore, which would preclude the use of FRET for analyzing interactions between these proteins. The angles of the solid lines in Figures 2B–D indicate the amount of FRET obtained between the various YFP and CFP fusion proteins. In addition to providing the control that the YFP chromophore does not interact with NtWRKY12, the lack of raised 527 nm emission with the combination of NtWRKY12:YFP/NtWRKY12:CFP indicates that NtWRKY12 is not able to form homodimers (Figure 2B). Similarly, although 475 nm emission in the protoplasts transfected with the NtWRKY12:CFP/TGA2.1:YFP plasmids was quenched, 527 nm emission was not significantly higher than in the control protoplasts, showing that no strong interaction occurred between TGA2.1 and NtWRKY12 (Figure 2C). On the other hand, the large amount of FRET in the protoplasts expressing the combination NtWRKY12:CFP/TGA2.2:YFP demonstrates that NtWRKY12 strongly interacted with TGA2.2 (Figure 2D). Although we could not detect the TGA1a:chromophore fusion proteins in our localization experiments (see above), we did perform a cotransfection of protoplasts with TGA1a:YFP and NtWRKY12:CFP. While it was not surprising to find no YFP signal in these protoplasts, we had not expected the reproducible total absence of protoplasts showing CFP emission.
To confirm the interaction between NtWRKY12 and TGA2.2, in vitro pull-down assays were performed with Escherichia coli-expressed GST and Strep/HIS fusion proteins purified using affinity chromatography. In addition to the interaction between NtWRKY12 and TGA2.2, also interactions with TGA1a, TGA2.1, and NtNPR1 were assayed. Figure 3 shows the results of these pull-down assays; the data obtained in Figures 3A–C are summarized in Figure 3D. Figure 3A shows the interactions between different TGA proteins and NtNPR1. GST:NtNPR1 was incubated with various Strep:TGA:HIS fusion proteins and with a Strep:NtNPR1:HIS fusion, after which the complexes were pulled down using Streptactin beads. The pulled-down proteins were analyzed on Western blots using anti-GST antibody conjugate. Strong NtNPR1–TGA2.2 and NtNPR1–NtNPR1 interactions were observed (Figure 3A, lanes 3 and 5), whereas no interactions between NtNPR1 and TGA2.1 or TGA1a were detectable (Figure 3A, lanes 2 and 4). Figure 3A, lanes 6 to 10 show the controls with single fusion proteins. The low background signal obtained with GST:NtNPR1 (Figure 3A, lane 6) was also visible in Figure 3A, lane 4. Homodimer formation as seen with the tobacco NtNPR1 has been reported for Arabidopsis NPR1, where it relies on interaction of NPR1’s BTB/POZ domain, which is independent of SA (Boyle et al., 2009).
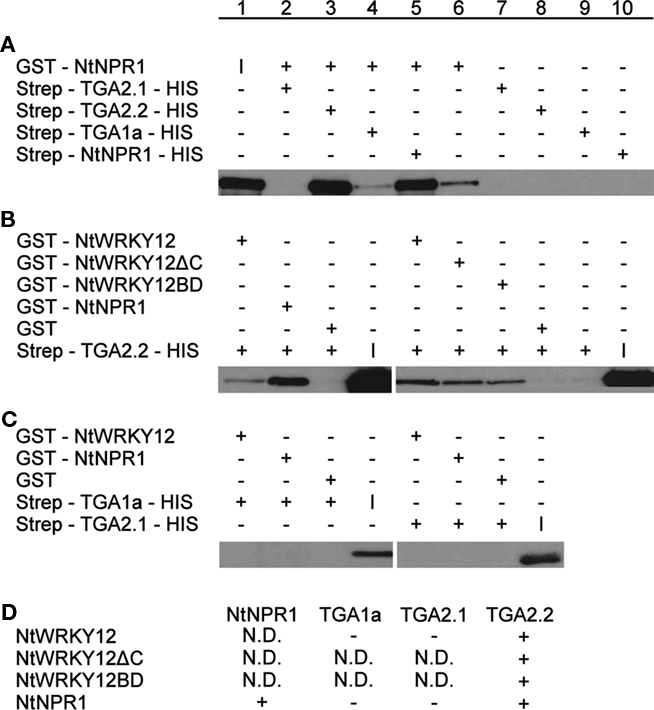
Figure 3. Pull-down assays of NtWRKY12, TGA1a, TGA2.1, TGA2.2, and NtNPR1. GST-proteins were incubated with Strep/HIS purified fusion proteins and complexes were pulled down with Streptactin–Sepharose beads (A) or Glutathione–Sepharose 4B beads (B,C). After SDS-PAGE and Western blotting fusion proteins were detected with anti-GST antibodies (A) or anti-HIS antibodies (B,C). Plus and minus signs denote the presence or absence in the incubation mixtures of the proteins indicated at the left. The input protein was loaded separately on gel and is indicated by (I). The table in (D) summarizes the results of the pull-down assays. Plus-sign, interaction, minus-sign, no interaction, N.D., not determined.
In the experiments shown in Figures 3B,C, GST fusions of NtWRKY12 and NtNPR1 were incubated with various Strep:TGA:HIS fusions, and protein complexes were bound to Glutathione–Sepharose 4B beads. The pulled-down proteins were analyzed on Western blots using anti-HIS antibodies. Interactions of NtWRKY12 were observed with TGA2.2 (Figure 3B, lane 1), but not with TGA1a or TGA2.1 (Figure 3C, lanes 1 and 5). Moreover, the conclusion from Figure 3A that NtNPR1 interacts with TGA2.2, but not with TGA1a or TGA2.1 was confirmed in this system (Figure 3B, lane 2; Figure 3C, lanes 2 and 6).
As a first step toward the characterization of the NtWRKY12 sequence involved in the interaction with TGA2.2, two NtWRKY12 deletion mutants were made. NtWRKY12ΔC lacks the C-terminal 87 aa of the 220 aa long protein; NtWRKY12BD lacks the N-terminal 113 aa. Both mutants were found to interact with TGA2.2 (Figure 3B, lanes 6 and 7). Either the overlap between the two mutant proteins (aa 114–133) is involved in the interaction of NtWRKY12 with TGA2.2, or NtWRKY12 contains two independent binding sites for TGA2.2, possibly involved in the interaction with a TGA dimer.
Functional Domains of NtWRKY12
Previously, yeast one-hybrid screening for tobacco proteins binding to the PR-1a promoter resulted in the isolation of a protein corresponding to the C-terminal 107 aa of NtWRKY12 fused to the GAL4 activation domain. This protein contained the conserved WRKY and Zn-finger domains and, apparently, a DNA-BD (van Verk et al., 2008). Moreover, it was shown that full-length (220 aa) NtWRKY12 was able to activate PR-1a::HIS gene expression in yeast, indicating that in addition to a BD, NtWRKY12 also contains an activating domain (AD). To further characterize functional domains of NtWRKY12, deletion mutants of NtWRKY12 were assayed in the one-hybrid system in three different ways. First, the mutants were fused to the GAL4 BD and assayed for their ability to activate GAL4 promoter::Ade reporter gene expression (Figure 4; results summarized in the column with the caption “BD”). Fusions, which activated the reporter gene, were concluded to contain the NtWRKY12 AD. Second, the mutants were fused to the GAL4 AD and assayed for their ability to activate PR-1a::HIS gene expression (Figure 4; results summarized in the column with the caption “AD”). Fusions which activated gene expression were concluded to contain the NtWRKY12 BD. Third, the mutants were expressed as non-fused proteins and assayed for their ability to activate PR-1a::HIS expression (Figure 4; results summarized in the column with the caption “–”). Mutants which activated gene expression were concluded to contain both the AD and BD domains of NtWRKY12.
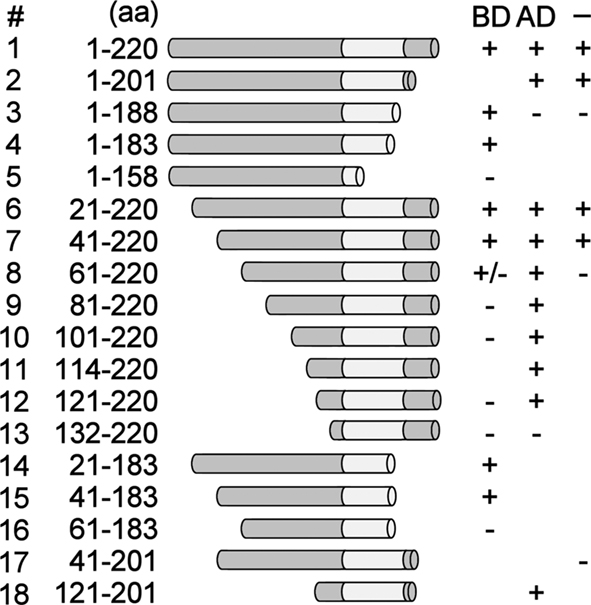
Figure 4. Mapping of the functional domains of NtWRKY12. NtWRKY12 peptides fused to the GAL4 DNA-binding domain were tested for their ability to activate GAL4::Ade gene expression in yeast (results with these constructs are indicated in column BD). NtWRKY12 peptides fused to the GAL4 activation domain were tested for their ability to bind to PR-1a promoter fragment controlling the HIS marker gene (results with these constructs are indicated in column AD). Unfused NtWRKY peptides were assayed for binding to the PR-1a promoter and activation of the HIS gene under its control (results with these constructs are indicated in column –). Column aa shows the ranges of the amino acid residues of the respective NtWRKY12 peptides. Plus signs in columns BD, AD, and – indicate growth of yeast; minus signs indicate absence of growth of yeast. The absence of plus or minus signs: not tested.
Figure 4 (column “BD”) shows that GAL4 BD fusions lacking the C-terminal 37 aa (construct 4) or N-terminal 40 aa (construct 7) of NtWRKY12, and a protein with both these deletions (construct 15) were able to activate GAL4::Ade expression. Apparently, the NtWRKY12 AD function is contained within the aa 41–183 region of the protein. The GAL4 AD fusion of the smallest NtWRKY12 deletion mutant that was able to activate PR-1a::HIS expression was construct 18 (Figure 4, column “AD”). Thus, the NtWRKY12 BD is localized in the sequence of aa 121–201. This region encompasses both the conserved WRKY and Zn-finger domains. Apparently, aa upstream of the WRKY domain are also necessary for DNA binding, as a deletion mutant with only seven aa in front of the WRKY domain (Figure 4, construct 13, aa 132–220) was not able to activate HIS gene expression. Construct 17 (aa 41–201) combined the minimal sequences with NtWRKY12 AD and BD activity (construct 15, aa 41–183, and construct 18, aa 121–201). However, the non-fused protein encoded by construct 17 was not able to activate PR-1a::HIS expression (Figure 4, column “–“). To permit both AD and BD activity, the sequence of construct 17 had to be extended by either the N-terminal 40 aa of NtWRKY12 (Figure 4, construct 2, aa 1–201) or the C-terminal 19 aa (Figure 4, construct 7, aa 41–220). Possibly, the lack of activity of the protein encoded by construct 17 (aa 41–201) was due to instability or misfolding of the polypeptide.
PR-1A::GUS Gene Expression in Arabidopsis Protoplasts Cotransfected with Plasmids Encoding NtWRKY12, NtNPR1, and TGAs
In our previous paper we showed that cotransfection of Arabidopsis protoplasts with 35S::NtWRKY12 and PR-1a::GUS constructs resulted in a strong increase in GUS expression (van Verk et al., 2008). NtWRKY12 activates PR-1a::GUS expression in Arabidopsis protoplasts by binding to the WK-box (TTTTCCAC) at position −564 in the PR-1a promoter (van Verk et al., 2008). The as-1-like element (CGTCA[N]6TGACG) at position −592 is the likely binding site for TGA transcription factors. This raises the possibility that NtWRKY12 and TGA2.2 bind to the PR-1a promoter in close proximity, and binding of NtWRKY12 and TGA2.2 might be stabilized by interactions between the two factors that were observed in vivo and in vitro (Figures 2 and 3). To further investigate the role of NtWRKY12, TGA and NPR1 on activation of PR-1a driven expression, additional transactivation assays were set up in protoplasts isolated from leaves of Arabidopsis seedlings grown on MS medium. To avoid interfering effects of NtWRKY12 binding to the far upstream WK binding site (−859), this WK site in the PR-1a::GUS reporter gene used in these experiments was mutated (TTTTCCAC into TCCCTTGC). Figure 5A shows the effects of overexpression of NtWRKY12, TGA2.1, TGA2.2, and NPR1 on PR-1a::GUS expression in wild-type Arabidopsis protoplasts. Obviously, NtWRKY12 greatly enhanced beta-glucuronidase expression from the PR-1a promoter (sixfold over background level). Overexpression of TGA2.1, TGA2.2 or NtNPR1, or combinations of the TGAs with NtNPR1 did not result in enhanced GUS expression. Neither did TGA2.2, alone or in combination with NtNPR1, affect the level of NtWRKY12 enhanced PR-1a::GUS expression, whereas overexpression of TGA2.1, alone or together with NtNPR1, notably reduced NtWRKY12 activated GUS expression.
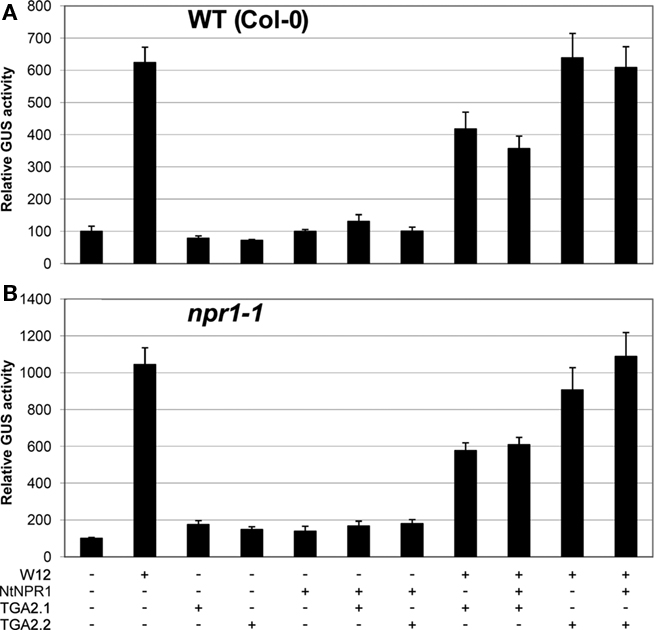
Figure 5. Activation of PR-1a::GUS in WT Col-0 and npr1-1mutant lines. Leaf protoplasts were cotransfected with PR-1a::GUS constructs together with expression plasmids containing 35S::NtWRKY12 (W12), 35S::NtNPR1 (NtNPR1), 35S::TGA2.1 (TGA2.1), 35S::TGA2.2 (TGA2.2), a combination, or with empty expression vector, as indicated by the plus and minus signs. (A) Expression in protoplasts isolated from seedlings of WT Col-0 Arabidopsis, (B) expression in npr1-1 protoplasts. The bars represent the percentage of GUS activity from triplicate experiments relative to that of the protoplasts cotransfected with the corresponding PR-1a::GUS construct and empty vector control. Error bars represent the SEM.
NtWRKY12 Activates PR-1A::GUS Gene Expression in Arabidopsis Protoplasts Independently of Endogenous NPR1 or TGA Factors
In Arabidopsis, PR gene expression is dependent on NPR1 and there is accumulating evidence that NPR1 orthologs similarly effect expression of PR genes in other plant species (Rayapuram and Baldwin, 2007; Anand et al., 2008; Le Henanff et al., 2009). We wondered whether the lack of effects of overexpressed NtNPR1 on PR-1a::GUS expression in the cotransfection experiments could be due to the presence of saturating levels of functionally equivalent Arabidopsis NPR1. However, the results of transactivation assays in protoplasts from npr1-1 mutant plants were virtually identical to those of the wild-type protoplasts (compare Figures 5A,B). This implies that PR-1a gene expression mediated by NtWRKY12 in Arabidopsis protoplasts is independent of NPR1.
On the basis of sequence homology, tobacco TGA2.2 belongs to the group II TGA proteins together with Arabidopsis TGAs 2, 5, and 6 (Xiang et al., 1997). To exclude the possibility that the absence of effects of overexpressed tobacco TGA on PR-1a::GUS expression in the Arabidopsis protoplasts was caused by functionally similar Arabidopsis TGAs, cotransfection experiments were performed in Arabidopsis protoplasts derived from tga2-1 tga5-1 tga6-1 (tga256) and tga2-1 tga3-1 tga5-1 tga6-1 (tga2356) triple and quadruple mutant plants (Figure 6). Also in these mutant backgrounds, overexpression of NtWRKY12 led to activation of PR-1a::GUS expression (ninefold over background level in the triple mutant (Figure 6A), although the enhancement in the quadruple mutant was greatly reduced (twofold, Figure 6B). Likely, this reduced GUS expression is the result of reduced production of NtWRKY12 from the transfected 35S::NtWRKY12 gene, similar to the reduced GUS activity from the 35S::GUS gene in tga2356 protoplasts compared to wild-type or tga256 protoplasts (Figure 6C).
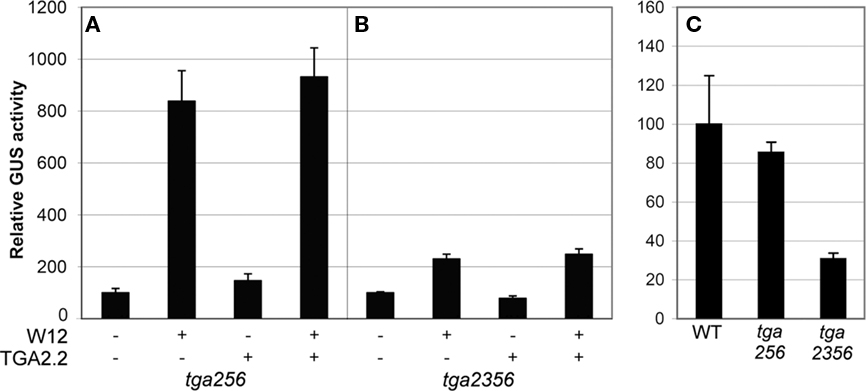
Figure 6. Activation of PR-1a::GUS in Arabidopsistga mutant lines. Leaf protoplasts were cotransfected with PR-1a::GUS constructs together with expression plasmids containing 35S::NtWRKY12 (W12), 35S::TGA2.2 (TGA2.2), a combination, or with empty expression vector, as indicated by the plus and minus signs in (A,B). Protoplasts in (C) were transfected with a 35S::GUS construct. Protoplasts were isolated from a TGA triple mutant (tga256), a TGA quadruple mutant (tga2356), or WT Col-0 (WT) as indicated below the panels. The bars represent the percentage of GUS activity from triplicate experiments relative to that of the protoplasts cotransfected with the corresponding promoter::GUS construct and empty vector control incubated overnight in W5 medium. Error bars represent the SEM.
Together, the results of the cotransfection assays in Arabidopsis leaf protoplasts suggest that NtWRKY12 is an important transcriptional activator of PR-1a::GUS expression and that TGA2.1, TGA2.2, or NtNPR1, alone or in combination, do not positively affect activation.
Discussion
Our previous studies pointed to NtWRKY12 as a regulator of PR-1a gene expression (van Verk et al., 2008). Mutations in the NtWRKY12 binding site (WK-box) in the PR-1a promoter reduced the SA-induced expression of PR-1a::GUS fusions in transgenic tobacco by 60%, whereas mutations in the as-1-like element resulted in a 30% reduction. Transient expression of PR-1a::GUS fusions, induced by bacterial elicitors in agroinfiltrated tobacco leaves, was not affected by mutations in the as-1-like element, but was reduced by 50% when mutations were made in the WK-box. Interestingly, when both the WK-box and as-1-like element were mutated, elicitor-induced expression was reduced by 95%. This result pointed to the importance of both NtWRKY12 and TGA transcription factors for activation the PR-1a promoter through the WK-box and the as-1-like element (van Verk et al., 2008). In the present work we further analyzed the role of NtWRKY12 and TGA transcription factors in PR-1a gene expression.
Fluorescence resonance energy transfer analysis of possible interactions between NtWRKY12 and the tobacco transcription factors TGA2.1 and TGA2.2 revealed a strong and specific interaction between NtWRKY12 and TGA2.2 in the nucleus of transfected Arabidopsis protoplasts (Figure 2). This interaction was confirmed by in vitro pull-down assays. In vitro, no interaction between NtWRKY12 and TGA1a, TGA2.1, or NtNPR1 was observed (Figure 3). Pull-down assays and studies with the yeast one-hybrid system permitted an initial localization of domains in NtWRKY12 involved in the interaction with TGA2.2, in DNA binding and in transcription activation (Figures 3 and 4). The role of NtWRKY12 and TGA2.2 in PR-1a gene expression was further investigated by transactivation studies in Arabidopsis protoplasts cotransfected with one vector expressing a transcription factor and another vector containing the PR-1a::GUS reporter construct. The major advantage of this system over SA-induced or elicitor-induced gene expression in whole plants is the possibility to analyze the response induced by well-defined single transcription factors.
Of the factors tested, TGA2.2 was the only factor that was found to interact with NtWRKY12 in FRET and in in vitro pull-down assays. However, it is possible that binding sites involved in protein–protein interactions are masked in the fusion proteins used in these assays. Further studies are required to reveal whether or not different TGA factors use different pathways in the activation of the PR-1a promoter.
In the transactivation experiments, PR-1a::GUS expression was activated by co-expression of NtWRKY12, but not by co-expression of NtNPR1, TGA2.1, or TGA2.2. This expression pattern was not affected in protoplasts from npr1-1 or tga Arabidopsis mutants (Figures 5 and 6). These results indicate that NtWRKY12 activates the PR-1a promoter independently of exogenously or endogenously expressed NPR1 or TGA factors. Recently we observed that in EMSA experiments a 47-bp fragment of the PR-1a promoter, harboring the as-1-like element at position −592, showed a specific band-shift with TGA2.2. Mutation of the as-1-like element abolished this band-shift (unpublished data). This indicates that TGA2.2 binds specifically to the as-1-like element at position −592 in the PR-1a promoter. Similar to TGA2.2, TGA1a has been shown to bind to the as-1-like element at position −592 in the PR-1a promoter (Strompen et al., 1998). Several studies have shown that TGAs are involved in PR-1a gene expression, either acting as positive or negative regulators (Strompen et al., 1998; Niggeweg et al., 2000b; Pontier et al., 2001).
A possible role of NtWRKY12 and TGA2.2 in PR-1a gene expression could be that in order to allow NtWRKY12 to effectively bind to the promoter at the WK-box and activate gene expression, it requires interaction with TGA2.2 as a binding enhancer or scaffold, perhaps to replace other tobacco DNA-binding proteins negatively acting on gene expression. This would explain why in tobacco, mutation of the as-1 element in the PR-1a promoter results in reduced GUS expression, even when sufficient endogenous NtWRKY12 is present (van Verk et al., 2008). Such repressors could be (other) TGAs binding at the as-1 element interfering with NtWRKY12’s binding at the nearby WK-box. In Arabidopsis protoplasts, lack of cognate as-1 elements for Arabidopsis TGAs in the tobacco promoter, would allow co-expressed NtWRKY12 to bind sufficiently efficient to the transfected PR-1a promoter to permit high GUS expression irrespective of the presence of co-expressed tobacco TGA2.2. This would also explain why co-expressed TGA2.1, which cannot interact with NtWRKY12, but which probably also binds to the as-1 element in the PR-1a promoter, has a negative effect on GUS expression in Arabidopsis protoplasts (Figure 5).
As was recently shown by Spoel et al. (2009), NPR1-mediated gene expression in Arabidopsis is largely dependent on its proteasome-mediated turnover. The authors found that SA-mediated induction of WRKY18, WRKY38, and WRKY62, target genes of NPR1 expressed early after induction (Kim et al., 2008), was reduced 50–60% after treatment with the proteasome inhibitor MG115. Surprisingly, the SA-induced expression of PR-1 was only affected 5–10% by MG115. This could indicate that NPR1, and consequently TGA transcription factors through which it acts as a co-activator, do not have a direct role in Arabidopsis PR-1 expression, which occurs much later after induction.
These observations fit well with the results of Pape et al. (2010), who found that NPR1 can activate the Arabidopsis PR-1 promoter through a functional LS5/LS7 unit. However, mutation of both LS5 and LS7 in transgenic Arabidopsis did not have much effect on promoter activity and postulated that in the absence of these sites NPR1 activates the PR-1 promoter via the LS4 element trough activation of WRKY factors that can bind there. Based on our findings in tobacco, such WRKY factors could be Arabidopsis homologs of NtWRKY12.
Using the npr1 background to screen for mutants that have regained the capacity to induce PR gene expression, sni1 (SUPPRESSOR OF NPR1) was identified (Li et al., 1999). SNI1 encodes an armadillo repeat nuclear protein that upon mutation restores the inducibility of the PR-1 promoter. Structural similarity searches with SNI1 suggest that it may form a scaffold for interaction with proteins that modulate transcription through local modification of chromatin structure (Mosher et al., 2006). Recently it was shown that SNI1 forms complexes with SSN2 and RAD51D that are involved in homologous DNA recombination, and that it is recruited to the PR-1 promoter through TGA7 and NPR1 (Martín et al., 2006; Durrant et al., 2007; Song et al., 2011). If in tobacco, like in Arabidopsis, local modification of chromatin structure requiring the action of NtNPR1 would be involved in regulation of PR-1a expression, such effects could be difficult to study using transient protoplast expression assays with transgenes likely lacking a normal chromatin structure. Also, we cannot exclude that the lack of effects of NtNPR1 in the transient assays is due to insufficient reduced monomer. Exogenous application of SA could not be used as it had major negative effects on protoplast fitness.
Materials and Methods
Bacterial Expression of Fusion Proteins
The open reading frames of NtWRKY12 and NtNPR1, and mutants encoding the 133 amino acids (aa) of the N-terminal half (NtWRKY12ΔC) or 107 aa of the C-terminal half (NtWRKY12BD) were cloned in frame behind the GST open reading frame of expression vector pGEX-KG (Guan and Dixon, 1991), expressed and purified according to van Verk et al. (2008).
The full-length coding sequence of N. tabacum TGA1a, TGA2.1, TGA2.2, and NPR1 were cloned in frame of expression vector pASK-IBA45 plus harboring a Strep and HIS tag (IBA). These plasmids were transformed into E. coli XL1. For induction of protein expression, cultures were grown to mid-log phase at 37°C, after which tetracycline was added to a final concentration of 0.2 μg mL−1 and incubation continued for 3.5 h at 29°C. The cells were harvested by centrifugation, resuspended in 1/25th volume lysis buffer [1× PBS containing 1% (v/v) NP40, 2 mM DTT, and 1/50th volume Complete (Roche) protease inhibitors] and lysed by sonication (Vibracell, Sonics en Materials Inc., USA). Soluble protein fraction was collected by centrifugation, and expressed fusion proteins were analyzed using 12% (w/v) SDS-PAGE.
In vitro pull-down
For the in vitro pull-down assay, GST-fusion proteins were mixed with Strep-fusion-HIS proteins in binding buffer [1× PBS, 1% (w/v) NP40, 2 mM DTT] and incubated on an orbital shaker for 1 h at room temperature. To this mixture Glutathione–Sepharose 4B beads (GE Healthcare) or Strep–Tactin Sepharose beads (IBA) in buffer W (100 mM Tris pH 8.0, 150 mM NaCl, 1 mM EDTA) were added, and incubation was continued for an additional hour. The beads were washed five times with PBS [with 1% (w/v) NP40 for Glutathione beads] after which the beads were collected, resuspended in Laemmli buffer, and heated at 95°C for 2 min.
The proteins bound to the beads were separated by SDS-PAGE and transferred onto Hybond P membrane (GE Healthcare). Membranes were incubated with the anti-GST antibody (GE Healthcare), or anti-HIS antibody (5 Prime) according to manufacturers instructions and exposed to X-ray film.
One-Hybrid Screening
A tetramer fragment of the tobacco PR-1a promoter corresponding to the region −605 to −513 relative to the transcription start site cloned in front of the HIS3 gene and integrated in the Saccharomyces cerevisiae genome of strain Y187 (van Verk et al., 2008) was used to screen for the DNA-BD of NtWRKY12 and presence of both an activation (AD) and BD. Deletion mutants of NtWRKY12 were cloned in pACT2 to screen for the presence of an BD or p415GPD-HA to screen for both the BD and AD. Mutants were screened for HIS-independent growth with addition of 3AT up to 20 mM. To locate the AD, deletion mutants were cloned into pAS2-1 and transformed in yeast strain PJ69-4A containing the Gal4 binding site in front of the Ade gene. Mutants were screened for adenine independent growth.
Protoplast Preparation and Transactivation Experiments
For microscopy protoplasts were prepared from Arabidopsis thaliana ecotype Col-0 cell suspensions according to van Verk et al. (2008).
The leaves from approximately 50 four-week-old seedlings (Col-0, npr1-1, tga256, tga2356) grown on sterile medium were cut in small pieces and protoplasts were prepared according to He et al. (2007). In total 1 × 105 protoplasts were transformed per transfection using polyethylene glycol [40% (w/v) PEG 4000, 0.2 M mannitol, 0.1 M CaCl2].
Protoplasts were cotransfected with 2 μg of plasmid carrying PR-1a promoter::GUS construct and 6 μg of 35S::effector plasmid pRT101 (Töpfer et al., 1987). As a control, cotransformation of PR-1a::GUS construct with the empty expression vector pRT101 was carried out. The protoplasts were harvested 16 hrs after transformation and GUS activity was determined. GUS activities from triplicate experiments were normalized against total protein level.
Microscopy and Förster Resonance Energy Transfer
Protoplasts were cotransfected with 10 μg of plasmid carrying protein::CFP and 10 μg of protein::YFP constructs. As controls 2.5 μg of plasmid containing unfused CFP/YFP or 10 μg YFP:CFP fusion was used. Protoplasts expressing the fusion proteins were analyzed with a Leica DM IRBE confocal laser scanning microscope with a 63× water objective, digital zoom, and 51% laser intensity. The fluorescence was visualized with an Argon laser for excitation at 457 nm with 471–481 nm emission filter for CFP and 514 nm excitation with a 522- to 532-nm filter for YFP. A transmitted light picture was used as reference. For FRET analysis Lambda scanning was performed by excitation at 457 nm and by measuring emission from 468 to 587 nm in a total of 30, 5 nm wide intervals using a RSP465 filter. Of every interval the intensity of the whole cell was quantified using ImageJ. The intensity of five protoplasts were averaged and normalized. The slopes between the 475- and 527-nm point were compared for differences in quenched donor emission and increased acceptor emission in comparison to the controls. Similar results were obtained for three independent transfections.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Dr. Xinnian Dong for kindly providing Arabidopsis tga256 and tga2356 mutant seed, Dr. Corné Pieterse for providing Arabidopsis npr1-1 mutant seed, Carlos Galvan-Ampudia for providing pART7CFP, pART7YFP, and FRET control plasmids, Roy Baas for assistance with several transactivation experiments, and Gerda Lamers and Ward de Winter for their help with microscopy and tissue culture, respectively.
References
Anand, A., Uppalapati, S. R., Ryu, C. M., Allen, S. N., Kang, L., Tang, Y., and Mysore, K. S. (2008). Salicylic acid and systemic acquired resistance play a role in attenuating crown gall disease caused by Agrobacterium tumefaciens. Plant Physiol. 146, 703–715.
Blanco, F., Salinas, P., Cecchini, N. M., Jordana, X., Van Hummelen, P., Alvarez, M. E., and Holuigue, L. (2009). Early genomic responses to salicylic acid in Arabidopsis. Plant Mol. Biol. 70, 79–102.
Boyle, P., Le Su, E., Rochon, A., Shearer, H. L., Murmu, J., Chu, J. Y., Fobert, P. R., and Després, C. (2009). The BTB/POZ domain of the Arabidopsis disease resistance protein NPR1 interacts with the repression domain of TGA2 to negate its function. Plant Cell 21, 3700–3713.
Cao, H., Glazebrook, J., Clarke, J. D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene ene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63.
Delaney, T. P., Friedrich, L., and Ryals, J. A. (1995). Arabidopsis signal transduction mutant defective efective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. U.S.A. 92, 6602–6606.
Després, C., De Long, C., Glaze, S., Liu, E., and Fobert, P. R. (2000). The Arabidopsis NPR1/NIM1 IM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12, 279–290.
Dong, J., Chen, C., and Chen, Z. (2003). Expression profile of the Arabidopsis WRKY gene superfamily uperfamily during plant defense response. Plant Mol. Biol. 51, 21–37.
Durrant, W. D., and Dong, X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209.
Durrant, W. E., Wang, S., and Dong, X. (2007). Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response. Proc. Natl. Acad. Sci. U.S.A. 104, 4223–4227.
Eulgem, T., and Somssich, I. E. (2007). Networks of WRKY transcription factors in defense signalingignaling. Curr. Opin. Plant Biol. 10, 366–371.
Grüner, R., Strompen, G., Pfitzner, A. J. P., and Pfitzner, U. M. (2003). Salicylic acid and the hypersensitive ypersensitive response initiate distinct signal transduction pathways in tobacco that converge on the as-1-like element of the PR-1a promoter. Eur. J. Biochem. 270, 4876–4886.
Guan, K. L., and Dixon, J. E. (1991). Eukaryotic proteins expressed in Escherichia coli: an improved mproved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192, 262–267.
He, P., Shan, L., and Sheen, J. (2007). The use of protoplasts to study innate immune responses. Methods Mol. Biol. 354, 1–9.
Johnson, C., Boden, E., and Arias, J. (2003). Salicylic acid and NPR1 induce the recruitment of f trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell 15, 1846–1858.
Katagiri, F., Lam, E., and Chua, N.-H. (1989). Two tobacco DNA-binding proteins with homology omology to the nuclear factor CREB. Nature 340, 727–730.
Kesarwani, M., Yoo, J., and Dong, X. (2007). Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol. 144, 336–346.
Kim, K.-C., Lai, Z., Fan, B., and Chen, Z. (2008). Arabidopsis WRKY38 and WRKY62 transcription ranscription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20, 2357–2371.
Kinkema, M., Fan, W., and Dong, X. (2000). Nuclear localization of NPR1 is required for activation ctivation of PR gene expression. Plant Cell 12, 2339–2350.
Lam, E., Benfey, P. N., Gilmartin, P. M., Fang, R.-X., and Chua, N.-H. (1989). Site-specific mutations alter in vitro factor binding and change promoter expression pattern in transgenic plants. Proc. Natl. Acad. Sci. U.S.A. 86, 7890–7894.
Le Henanff, G., Heitz, T., Mestre, P., Mutterer, J., Walter, B., and Chong, J. (2009). Characterization of Vitis vinifera NPR1 homologs involved in the regulation of pathogenesis-related gene expression. BMC Plant Biol. 9, 54. doi: 10.1186/1471-2229-9-54
Lebel, E., Heifetz, P., Thorne, L., Uknes, S., Ryals, J., and Ward, E. (1998). Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J. 16, 223–233.
Li, X., Zhang, Y., Clarke, J. D., Li, Y., and Dong, X. (1999). Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1–1. Cell 98, 329–339.
Martín, V., Chahwan, C., Gao, H., Blais, V., Wohlschlegel, J., Yates, J. R. III, McGowan, C. H., and Russell, P. (2006). Sws1 is a conserved regulator of homologous recombination in eukaryotic cells. EMBO J. 25, 2564–2574.
Mosher, R. A., Durrant, W. E., Wang, D., Song, J., and Dong, X. (2006). A comprehensive structure-function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity. Plant Cell 18, 1750–1765.
Mou, Z., Fan, W., and Dong, X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113, 935–944.
Niggeweg, R., Thurow, C., Kegler, C., and Gatz, C. (2000a). Tobacco transcription factor TGA2.2 is the main component of as-1-binding factor ASF-1 and is involved in salicylic acid- and auxin-inducible expression of as-1-containing target promoters. J. Biol. Chem. 275, 19897–19905.
Niggeweg, R., Thurow, C., Weigel, R., Pfitzner, U., and Gatz, C. (2000b). Tobacco TGA factors differ with respect to interaction with NPR1, activation potential and DNA-binding properties. Plant Mol. Biol. 42, 775–788.
Pandey, S. P., and Somssich, I. E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150, 1648–1655.
Pape, S., Thurow, C., and Gatz, C. (2010). The Arabidopsis thaliana PR-1 promoter contains multiple integration sites for the co-activator NPR1 and the repressor SNI1. Plant Physiol. 154, 1805–1818.
Pontier, D., Miao, Z. H., and Lam, E. (2001). Trans-dominant suppression of plant TGA factors reveals their negative and positive roles in plant defense responses. Plant J. 27, 529–538.
Pontier, D., Privat, I., Trifa, Y., Zhou, J. M., Klessig, D. F., and Lam, E. (2002). Differential regulation egulation of TGA transcription factors by post-transcriptional control. Plant J. 32, 641–653.
Qin, X.-F., Holuigue, L., Horvath, D. M., and Chua, N.-H. (1994). Immediate early transcription ranscription activation by salicylic acid via the cauliflower mosaic virus as-1 element. Plant Cell 6, 863–874.
Rayapuram, C., and Baldwin, I. T. (2007). Increased SA in NPR1-silenced plants antagonizes JA and JA-dependent direct and indirect defenses in herbivore-attacked Nicotiana attenuata in nature. Plant J. 52, 700–715.
Ross, A. F. (1961). Systemic acquired resistance induced by localized virus infections in plants. Virology 14, 340–358.
Song, J., Durrant, W. E., Wang, S., Yan, S., Tan, E. H., and Dong, X. (2011). DNA repair proteins are directly involved in regulation of gene expression during plant immune response. Cell 9, 115–124.
Spoel, S. H., Mou, Z., Tada, Y., Spivey, N. W., Genschik, P., and Dong, X. (2009). Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137, 860–872.
Strompen, G., Grüner, R., and Pfitzner, U. M. (1998). An as-1-like motif controls the level of expression xpression of the gene for the pathogenesis-related protein 1a from tobacco. Plant Mol. Biol. 37, 871–883.
Thurow, C., Schiermeyer, A., Krawczyk, S., Butterbrodt, T., Nickolov, K., and Gatz, C. (2005). Tobacco bZIP transcription factor TGA2.2 and related factor TGA2.1 have distinct roles in plant defense responses and plant development. Plant J. 44, 100–113.
Töpfer, R., Matzeit, V., Gronenborn, B., Schell, J., and Steinbiss, H. H. (1987). A set of plant expression xpression vectors for transcriptional and translational fusions. Nucleic Acids Res. 15, 5890.
van Verk, M. C., Bol, J. F., and Linthorst, H. J. M. (2011a). Prospecting for genes involved in transcriptional regulation of plant defenses, a bioinformatics approach. BMC Plant Biol. 11, 88. doi: 10.1186/1471-2229-11-88
van Verk, M. C., Bol, J. F., and Linthorst, H. J. M. (2011b). WRKY transcription factors involved in activation of SA biosynthesis genes. BMC Plant Biol. 11, 89. doi: 10.1186/1471-2229-11-89
van Verk, M. C., Pappaioannou, D., Neeleman, L., Bol, J. F., and Linthorst, H. J. M. (2008). A Novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol. 146, 1983–1995.
Wang, D., Amornsiripanitch, N., and Dong, X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2, e123. doi: 10.1371/journal.ppat.0020123
Wu, P., and Brand, L. (1994). Resonance energy transfer: methods and applications. Anal. Biochem. 218, 1–13.
Xiang, C., Miao, Z., and Lam, E. (1997). DNA-binding properties, genomic organization and expression xpression pattern of TGA6, a new member of the TGA family of bZIP transcription factors in Arabidopsis thaliana. Plant Mol. Biol. 34, 403–415.
Zhang, Y., Fan, W., Kinkema, M., Li, X., and Dong, X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. U.S.A. 96, 6523–6528.
Zhang, Y., Tessaro, M. J., Lassner, M., and Li, X. (2003). Knockout analysis of Arabidopsis transcription ranscription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15, 2647–2653.
Zhou, J.-M., Trifa, Y., Silva, H., Pontier, D., Lam, E., Shah, J., and Klessig, D. F. (2000). NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant Microbe Interact. 13, 191–202.
Keywords: NtWRKY12, TGA2.2, PR-1a, NPR1, protein–protein interaction, salicylic acid, plant defense, signal transduction
Citation: van Verk MC, Neeleman L, Bol JF and Linthorst HJ (2011) Tobacco transcription factor NtWRKY12 interacts with TGA2.2 in vitro and in vivo. Front. Plant Sci. 2:32. doi: 10.3389/fpls.2011.00032
Received: 20 April 2011;
Accepted: 07 July 2011;
Published online: 18 July 2011.
Edited by:
Corné M. J. Pieterse, Utrecht University, NetherlandsReviewed by:
Kemal Kazan, Commonwealth Scientific and Industrial Research Organization, AustraliaChristiane Gatz, Georg August Universität Göttingen, Germany
Copyright: © 2011 van Verk, Neeleman, Bol and Linthorst. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Huub J. M. Linthorst, Sylvius Laboratory, Leiden University, Sylviusweg 72, 2333 BE Leiden, Netherlands. e-mail: h.j.m.linthorst@biology.leidenuniv.nl
