- French Associates Institute for Agriculture and Biotechnology of Drylands, Jacob Blaustein Institutes for Desert Research, Ben-Gurion University of the Negev, Midreshet Ben-Gurion, Israel
Stem cells are commonly defined by their developmental capabilities, namely, self-renewal and multitype differentiation, yet the biology of stem cells and their inherent features both in plants and animals are only beginning to be elucidated. In this review article we highlight the stem cell state in plants with reference to animals and the plastic nature of plant somatic cells often referred to as totipotency as well as the essence of cellular dedifferentiation. Based on recent published data, we illustrate the picture of stem cells with emphasis on their open chromatin conformation. We discuss the process of dedifferentiation and highlight its transient nature, its distinction from re-entry into the cell cycle and its activation following exposure to stress. We also discuss the potential hazard that can be brought about by stress-induced dedifferentiation and its major impact on the genome, which can undergo stochastic, abnormal reorganization leading to genetic variation by means of DNA transposition and/or DNA recombination.
Introduction: Totipotency in Plants and Animals
Totipotency (Box 1) conveys the idea that many plant mature cells are not terminally differentiated but rather retain developmental plasticity. Except for certain types of terminally differentiated cells (e.g., tracheary elements, sieve-tube cells, and highly lignified cells such as mature fibers and sclereids) plant cells are capable under certain conditions to dedifferentiate, re-enter the cell cycle, proliferate and regenerate tissues, organs and entire fertile plants. Given the importance of in vitro plant regeneration for a wide range of applications including basic research, micropropagation, germplasm conservation, and formation of genetically modified plants, there are numerous reports demonstrating the totipotency nature of plant cells (Vasil and Vasil, 1972; Thorpe, 2007). Recently, Sugimoto et al. (2011) cast doubt on the idea that many of the plant cells are totipotent and retain capacity for switching fate and regeneration. The authors suggested that parenchymatous cells surrounding the vasculature, that is, pericycle or “pericycle-like cells” function as adult stem cells and serve as the major origin of regenerative tissue. Conceivably, pericycle cells represent an example of totipotent cells, but no evidence exists to show that pericycle cells are genuine stem cells (having characteristic features of stem cells, see below) neither evidence that these are the major cells from which regeneration of plant tissues and organs initiated. In fact, this hypothesis stands in contrast with numerous reports demonstrating the totipotent nature of mesophyll cells and protoplasts and their ability to re-enter the cell cycle, proliferate and form callus from which shoots and roots can be formed to give rise to whole fertile plants (Takebe et al., 1971; Frearson et al., 1973; Vasil and Vasil, 1974; Shepard and Totten, 1977). Furthermore, in contrast to the idea that callus is formed via activation of stem cell-like pericycle cells rather than dedifferentiation (see Box 1) of somatic cells (Sugimoto et al., 2010, 2011), Iwase et al. (2011) showed an induction of callus-like cells from the epidermal cell layer of roots, hypocotyls and cotyledons of Arabidopsis thaliana.
Box 1. Totipotency – a term signifying that cells maintain whole developmental capacity and under appropriate conditions can multiply and give rise to all types of cells that make up a new organism. An apparent example of a totipotent cell is the zygote, which is formed during fertilization from the union of the male gamete (sperm) with the female gamete (ovum) eventually leading to embryo development. In plants and various vertebrates, somatic differentiated cells may regain totipotency via dedifferentiation.
Differentiation – differentiation is often referred to as the sum of developmental processes whereby apparently unspecialized cells attain their mature form and function (Merriam-Webster’s Medical Dictionary) or as a process by which cells acquire or possess a character or function different from that of the original cell type (American Heritage Dictionary). At the gene expression level differentiation refers to the acquisition or possession of a specific pattern of gene expression (an interplay between transcribed and untranscribed genes), which is different from that of the previous (often primordial) cell type that bring about specific form and function of the cell.
Dedifferentiation – this process has often been studied with respect to cell proliferation leading to the erroneous assumption that re-entry into the cell cycle represents the actual event of dedifferentiation and that callus cells are essentially dedifferentiating cells (Grafi, 2004). However, the prefix “de” indicates doing the opposite of, or going in reverse, and thus the term dedifferentiation was initially coined to describe the reversal of cells from a given differentiated state into a more primordial state (“an indifferent embryonic cell type”) as deduced from changes in cell shape and morphology (Champy, 1913 as cited by Uhlenhuth, 1915). Apparently, dedifferentiation and re-entry into the cell cycle are two distinct processes and it is suggested that dedifferentiation represents a transient phase conferring competence to switch fate and thus preceding not only re-entry to the cell cycle but also re-differentiation/trans-differentiation and even a commitment for cell death (see Figure 1).
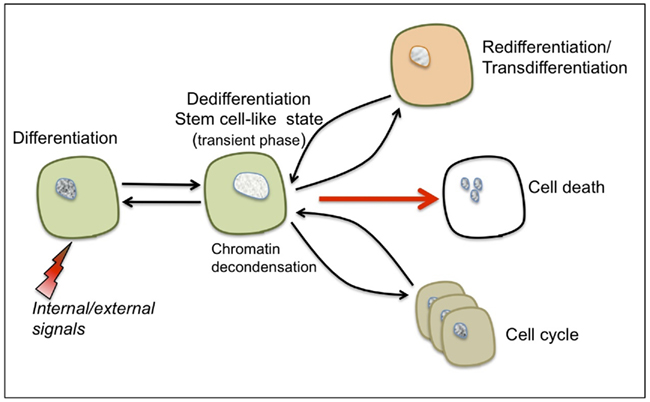
Figure 1. The characteristic features of cellular dedifferentiation. Somatic cells (such as parenchyma, collenchyma, or mesophyll cells) can be reprogrammed following exposure to various internal or external signals resulting in dedifferentiation and acquisition of a transient, stem cell-like state. This transient state is accompanied by global chromatin decondensation – a hallmark of stem cells. Depending on the type of stimulus, dedifferentiated cells can be induced to trans-differentiate/re-differentiate, re-enter the cell cycle or undergo a programmed cell death.
Re-differentiation/trans-differentiation – The term re-differentiation is often used to convey different meanings and as such the term may be confusing and misleading. Because “re” is a prefix indicating return to a previous condition, the term re-differentiation is often understood as “a process by which a group of once differentiated cells return to their original specialized form.” However, in plants, the term re-differentiation is commonly used not in the sense of returning to a previous differentiated state but rather to express the idea that differentiated plant cells do not lose their developmental capacity and are capable of repeated cycles of differentiation (re-differentiation). Thus when parenchyma cells are converted into tracheary elements (terminal differentiation) or into meristematic cells, in both cases, cells are said to have undergone re-differentiation although they have acquired completely different fates and certain cells may lose their capacity for repeated cycles of differentiation (e.g., tracheary elements). On the other hand, “trans” is a prefix meaning “across” or “beyond” and well conveys the idea that a given differentiated cell has acquired a different differentiated state “beyond” the previous one.
Although totipotency is an apparent feature of many plant cells, animal cells retain, at least to some extent, totipotency as well. This is exemplified in somatic cell nuclear transfer whereby transplantation of a somatic cell nucleus into an enucleated oocyte induces nuclear reprogramming eventually giving rise to the formation of a zygote-like cell from which viable offspring can be generated (reviewed in Gurdon and Wilmut, 2011). Likewise, totipotency is retained in vertebrates such as fish and salamanders, which are capable of regenerating amputated appendages such as limbs, fins, and tails. This regeneration capacity is accomplished via formation of a blastema – a mass of proliferating cells at the amputation site from which missing body parts are reformed (Stocum, 1968; Kragl et al., 2009; Knopf et al., 2011). Although blastema cells can be formed, at least partly, from adult stem cells (e.g., resident muscle stem cells/satellite cells, Slack et al., 2004) there are many reports demonstrating that blastema cells are formed from adult somatic cells via dedifferentiation and that dedifferentiation of mature cells is a prerequisite for proliferation and blastema formation (Hay, 1959; Echeverri et al., 2001; Echeverri and Tanaka, 2002; Slack, 2006; Brockes and Kumar, 2008; Kragl et al., 2009; Knopf et al., 2011). Recent data showed that the blastema is composed of a heterogenous cell population with restricted developmental capacity inasmuch as most proliferating cell types appear to keep a memory of their tissue of origin (Kragl et al., 2009; Knopf et al., 2011). This restricted potential has been interpreted as a sign that regeneration is achieved without complete transition into a pluripotent state (Kragl et al., 2009; Knopf et al., 2011). Yet, we cannot exclude the possibility that blastema cells are in fact pluripotent, but their developmental potential is restricted due to persistence of signal(s) (i.e., markers of differentiation) of their tissue of origin, which favors their differentiation into cell types related to the donor cell. Indeed, it has recently been shown that induced pluripotent stem cells (iPSCs) produced from different cell types retain, at least at early passages, a transient epigenetic memory of their somatic cells of origin demonstrated by their differential gene expression and differentiation capacity; at late passages these differences are attenuated (Kim et al., 2010a; Polo et al., 2010).
Stem Cells in Plants
It is commonly accepted that stem cells, in plants, are localized at distinct regions called meristems. Meristems are of several types and include the apical meristems, which are commonly located at the apices of roots and shoots, and are responsible for primary growth leading to elongation of the shoot and the root axes. Many woody resprouter plants have adventitious apical meristems hidden underneath the bark (epicormic buds) or located at lignotuber – a starchy swelling of the root crown, which sprout following exposure to stress, particularly fire (James, 1984; Waters et al., 2010). Other meristems responsible for secondary growth and thickening of the shoot and root axes are known as lateral meristems and include the cambium that gives rise to secondary vascular tissues (xylem and phloem) and the phellogen (cork cambium) responsible for the development of the periderm (Fahn, 1990). Another putative meristem common to grasses is the intercalary meristem often located at internodes or at the bases of leaves and responsible for rapid re-growth of damaged leaves. Since the meristem is composed of several distinguishable domains, namely, the central zone, the rib meristem, and the peripheral zone it is not clear whether stem cells are restricted to a specific domain (central zone) or are populating all domains within the meristem. The latter possibility may prevail inasmuch as cells capable of mediating complete organ regeneration are dispersed in the plant meristem and are not restricted to the “so-called” stem cell niche; even young leaves/leaf primordia (about 200 μm in length) appeared to contain (stem) cells capable to re-pattern complex tissues (Sena et al., 2009). Thus, it appears that the stem cell state may persist in plant tissues and organs during initial stages of their development and that cells within the different domains of the meristem have stem cell properties (Laux, 2003). This may be supported by a recent transcriptome analysis of the shoot apical meristem (SAM) domains (Yadav et al., 2009). Scatter plot analysis of the available microarray dataset compiled by Yadav et al. (2009) showed that the gene expression profile of the central zone (CZ CLV3) is highly similar to those of the rib meristem (RM WUS) and the peripheral zone (PZ FIL; data not shown). Considering that most plant cells are fixed in place, except for those displaying significant intrusive growth (Lev-Yadun, 2001), and cannot migrate from one site to another as animal cells can do, the biological importance of having adult stem cells embedded in somatic plant tissues may be insignificant. Instead, cellular plasticity has evolved in plants as an adaptive trait providing many somatic cells with developmental flexibilities and with the capacity for dedifferentiation and acquisition of pluripotent state.
What are Stem Cells?
A notable problem in stem cell biology is their definition by developmental means, namely, self-renewal and differentiation into multiple cell types, rather than by their intrinsic features (Potten and Loeffler, 1990; McKay, 2000). This led to the incorrect assumption that re-entry into the cell cycle for the purpose of “self-renewal” represents an inherent feature of stem cells and that stem cell features can be fully maintained under culture conditions (Grafi and Avivi, 2004). Consequently, the attempts to uncover the “stem cell signature” in humans via transcriptome analyses of different stem cell culture lines failed as these experiments yielded different “signatures” and non-overlapping “stemness genes” (Ivanova et al., 2002; Ramalho-Santos et al., 2002; Fortunel et al., 2003). It appears that contrary to the idea that stem cells represent a unique entity that is characterized by the expression of specific genes, stem cells appear to represent a unique transient state characterized by promiscuous expression of differentiation-specific genes (Zipori, 2004). In addition, a high number of reports demonstrated a fundamental feature of stem cells, that is, they acquire open, decondensed chromatin architecture, which is essential though not sufficient for initiating large scale gene transcription (Gaspar-Maia et al., 2011; Grafi et al., 2011b).
The open chromatin conformation characteristic of animal stem cells was initially revealed by electron microscopy examination of erythropoietic cells. These studies showed that stem cells are characterized by large nuclei and homogenous euchromatin, while maturation is accompanied by an increase in nuclear condensation (MacRae and Meetz, 1970; Murphy et al., 1971; Miura et al., 1974). These observations were supported by micrococcal nuclease assays showing that the nucleosome repeat length (NRL) increases from 190 to 212 base pairs during erythropoiesis in the chick (Weintraub, 1978), which is required for the formation of the 30-nm DNA fiber-induced chromatin compaction (Routh et al., 2008). Furthermore, concomitantly with increasing NRL, a significant increase was observed in the concentration of histone H5 (Weintraub, 1978) – a linker histone necessary for stabilization of higher order chromatin structure (Robinson and Rhodes, 2006). Similarly to animal stem cells, ultrastructural observations of the nuclei in the shoot apex of the plant Tradescantia paludosa showed that a large proportion of the chromatin is organized as less condensed, diffused euchromatin fibrils (Booker and Dwivedi, 1973).
Switching Cell Fate in Plants
The withdrawal of somatic cells from their differentiated state is likely to occur following internal (developmental) and external signals (stress) that force the cell to stop performing its current function and undergo reprogramming (dedifferentiation) to become competent for switching fate (e.g., re-entry to the cell cycle, trans-/re-differentiation, and cell death). Dedifferentiation is uncovered by multiple phenomena in plants including cell culturing, or abnormal growths often referred to as galls, tumors, or neoplasms. These growth abnormalities can be induced by abiotic factors (e.g., irradiation), pests, or by pathogenic organisms such as bacteria, fungi, and viruses (Morrow and Tibbitts, 1988, and references therein).
Switching fate can occur during the normal developmental program of plants. This is exemplified in the development of secondary meristems such as the interfascicular cambium, which arises from parenchyma cells located between vascular bundles, or the cork cambium, which arises from parenchyma or collenchyma cells beneath the epidermis (Fahn, 1990).
Stress is probably a major factor leading somatic cells to switch fate via dedifferentiation (Grafi et al., 2011a). A well-known example is the formation of regenerative xylem from cortical parenchyma cells following wounding, which is often referred to as a process of re-differentiation (Sachs, 1981). Accordingly, wounding of Cucurbita stems resulted in a curved pattern of xylem regeneration immediately around the wound, which was derived from interfascicular parenchyma, to restore continuity of the severed longitudinal vascular bundle (Aloni and Barnett, 1996). In addition, wounding induced the development of phloem anastomoses – naturally occurring regenerative sieve tubes, which differentiate from interfascicular parenchyma cells between the longitudinal vascular bundles (Aloni and Barnett, 1996). Similarly, xylem regeneration from parenchyma cells was reported in inflorescence stems of A. thaliana following wounding (Flaishman et al., 2003). Furthermore, treatment of differentiated leaf cells with cell wall degrading enzymes results in production of protoplast cells displaying characteristic features of stem cells including open chromatin conformation and the capacity to acquire different fates depending of the type of stimulus supplied (Zhao et al., 2001; Williams et al., 2003; Grafi, 2004). Senescence is another example for stress-induced dedifferentiation associated with chromatin remodeling (Damri et al., 2009). Arabidopsis leaves induced to senesce by exposure to dark exhibited transcriptome profile with high similarity to dedifferentiating protoplast cells (Lin and Wu, 2004; Damri et al., 2009). Further study of dark-induced premature senescence of tobacco leaves revealed features of stem cells including open chromatin configuration, disrupted nucleolus and condensation of rRNA gene clusters (Damri et al., 2009). Indeed, molecular, biochemical, and bioinformatic analyses suggest that Arabidopsis plants responding to various stress conditions feature dedifferentiation and acquisition of stem cell-like state prior to a switch in cell fate (Grafi et al., 2011a, and references therein). Likewise, it has recently been proposed that cell dedifferentiation may occur in mammalian somatic cells as an adaptation for extreme stress conditions (Shoshani and Zipori, 2011).
Chromatin Structure and the Stem Cell State
The nucleosome represents the basic structural unit of chromatin and is made from DNA wrapped around a core of histone proteins (two of each) H2A, H2B, H3, and H4. All core histone proteins share a common structural motif called the histone fold which is necessary for the interactions between core histone proteins and for the interaction between histone octamer and duplex DNA (Arents and Moudrianakis, 1995). The histone amino-terminal tail is unstructured and protruding outside the nucleosomal disk (Luger et al., 1997) where it can contact with neighboring nucleosomes or with proteins or protein complexes that affect chromatin structure and function. The structure of chromatin is flexible due to multiple types of reversible chemical modifications that occur on the DNA (cytosine methylation) or on the DNA interacting core histone proteins. Most modifications of histone proteins occur on the N-terminal tails and include acetylation, methylation, phosphorylation, and ubiquitination. Reversibility of DNA and histone modifications as well as chromatin remodeling often occur following exposure to various biotic and abiotic stress conditions – a topic covered by multiple recent reviews (Alvarez et al., 2010; Kim et al., 2010b; Luo et al., 2011; Vaahtera and Brosché, 2011; Yaish et al., 2011; Zhu et al., 2011). Changes in DNA and histone chemical modifications are made possible due to the activities of several groups of chromatin modifier enzymes that add or remove specific chemical group (e.g., methyl, acetyl) from the DNA or from the histone proteins. DNA methylation is catalyzed by various types of DNA methyltransferases, which transfer a methyl group from S-adenosyl-L-methionine to position 5 of the pyrimidine ring of cytosine (for recent review see Vanyushin and Ashapkin, 2011). Active DNA demethylation in plants is generally carried out by a base excision repair (BER) pathway that can be induced following the activity of the DME/ROS1 family of DNA glycosylases or by the coupled activities of 5-methylcytosine deaminase that converts 5-methyl-C to thymidine, and G/T mismatch DNA glycosylase that corrects the G/T mismatch (reviewed in Zhu, 2009). An acetyl group is added by histone acetyltransferases (HATs) and are removed by histone deacetylases (HDACs), while a methyl group is added by a large group of histone methyltransferases (SET containing proteins) and removed by the activities of lysine-specific demethylase 1 (LSD1) and the jumonji classes of histone demethylases (for recent reviews on chromatin modifier enzymes see Chen et al., 2010; Chen et al., 2011; Luo et al., 2011; Thorstensen et al., 2011).
Gene promoters can be found in three fundamental states determined by their histone modification marks, namely, restrictive/inactive (e.g., trimethylated H3K9/K27), permissive/active (e.g., trimethylated H3K4, acetylated H3K9), or both restrictive and permissive (e.g., trimethylated H3K27 and trimethylated H3K4) also known as “bivalent” state. SET domain-containing histone methyltransferase proteins, such as Clr4 (Cryptic locus regulator) in Schizosaccharomyces pombe, SUV39H1 and SUV39H2 in humans and Kryptonite/SUVH4 in Arabidopsis are enzymes that methylate histone H3 specifically at lysine 9 (Rea et al., 2000; Jackson et al., 2002; Malagnac et al., 2002). This methylation generates a binding site for heterochromatin protein 1 (HP1; Bannister et al., 2001; Lachner et al., 2001; Jackson et al., 2002; Zemach et al., 2006), which may lead to the formation of a condensed, repressive chromatin. Conversely, histone acetylation and/or methylation of histone H3 at lysine 4, 36, and 79 are often associated with “open” chromatin configuration and gene transcription (Eberharter and Becker, 2002; Shilatifard, 2006; Lee and Workman, 2007; Eissenberg and Shilatifard, 2010). More recently it has been shown that some non-expressed genes or genes expressed at low levels in human embryonic stem cells carry both permissive (trimethylated H3K4) and restrictive (trimethylated H3K27) chromatin marks (Azuara et al., 2006; Bernstein et al., 2006). This unique “bivalent” chromatin state suggests a model in which many tissue-specific regulatory genes are “primed” but their transcription is delayed until entry into a specific differentiation pathway that dictates either activation (e.g., recruitment of H3K27 demethylases) or silencing (e.g., recruitment of H3K4 demethylases) of the gene locus (Lan et al., 2008). Notably, bivalent domains were found to occur also in the genomes of somatic cells of Arabidopsis and rice where a large proportion of genes possessing the repressive mark H3K27me1 or H3K27me3 also contained permissive marks such as H3K4me3, acetylated H3K9, H3K36me3, and ubiquitinated H2B (Deal and Henikoff, 2010; He et al., 2010; Roudier et al., 2011). The prevalence of methylated H3K27 on poised gene promoters may point to the importance of polycomb proteins and PRC2 complex in regulating the stem cell maintenance in plants (Mosquna et al., 2009; Lafos et al., 2011). Thus, as mentioned above, a unique mechanism has been evolved in plants to cope with the fixed nature of their cells, namely, plasticity and the capacity for dedifferentiation. This is demonstrated by plant somatic cells displaying some attributes of animal embryonic stem cells whereby gene promoters may be primed for transcription but held in check until specific signals determine the fate of genes either for expression or repression.
Open chromatin configuration is the fingerprint of pluripotent stem cells and might serve as a major feature for defining the stem cell state. It might confer stem cells with the capacity for differentiation into multiple cell types and with the flexibility for rapid switching into the appropriate transcriptional program upon induction of differentiation. The open chromatin conformation can be maintained by chromatin modifier genes (CMGs) in a quantitative or qualitative manner (Grafi et al., 2011b). The quantitative way suggests that open chromatin configuration is achieved by overrepresentation of CMGs, an approach that is well demonstrated in Arabidopsis SAM stem cells (Yadav et al., 2009). Further analysis of the available microarray dataset of the SAM stem cells revealed that among the 445 CMGs, which were represented on the ATH1 array, stem cells expressed 297 genes whose expression signal is higher than 256 (>28). The qualitative way proposes that open chromatin state is maintained by selective expression and repression of genes that promote the formation of permissive and restrictive chromatin, respectively. Accordingly, transcriptome profiling of dedifferentiating protoplast cells (Damri et al., 2009) showed that among the 465 CMGs on these ATH1 arrays only 95 genes displayed an expression signal higher than 256 (>28). However, close examination of these CMGs showed that many of the genes that were down-regulated in dedifferentiating protoplasts, such as histone deacetylase encoding genes SRT2, HDA14, and HDT4, as well as linker histone genes Hon1 and Hon2 are implicated in chromatin compaction. On the other hand, many of the CMGs that were upregulated, such as histone acetyltransferase encoding genes HAF1, HAC5, and HAG3 as well as histone demethylase genes JMJ21 and JMJ13 are implicated in chromatin decondensation. Thus, the expression profile of CMGs in dedifferentiating protoplast cells favors the acquisition of an open chromatin conformation.
Stress-Induced Dedifferentiation and Genetic Variation
Barbra McClintock (1984) has recognized the dramatic effect that stress (such as tissue culturing and pathogen infection) might impose on the genome predicting that aberrant genome responses to stress are likely to be induced by mobilization of transposable elements. Genome restructuring was first reported in maize following infection with the barley-stripe-mosaic virus (BSMV) resulting in increased frequencies of deficiencies or distortion of marker genes (Sprague et al., 1963). Insertional mutation in the maize Adh1 gene following BSMV infection (Mottinger et al., 1984) resulted from activation of the Bs1 transposable element – a low copy number, copia-like transposon (Johns et al., 1985). Numerous reports have further demonstrated the effect of various biotic and abiotic stresses including tissue culturing, wounding, protoplasting, pathogen infection, UV light, salt, heat, and drought on genomic restructuring via DNA recombination and DNA transposition (Table 1, see also Wessler, 1996; Tanurdzic et al., 2008; Mirouze and Paszkowski, 2011; Rigal and Mathieu, 2011). We suggest that genetic variation occurs in cells in the course of stress-induced cellular dedifferentiation via DNA recombination and DNA transposition (Grafi et al., 2007; Grafi, 2009). Notably, a high range of nuclear processes can be affected by TE activation including mutation of genes when inserted into coding regions, chromosome breakage, genome rearrangement, as well as the expression of neighboring genes by altering patterns of splicing and polyadenylation or by acting as enhancers or promoters (Girard and Freeling, 1999; Slotkin and Martienssen, 2007; Lisch, 2009; Boyko and Kovalchuk, 2011; Lisch and Bennetzen, 2011).
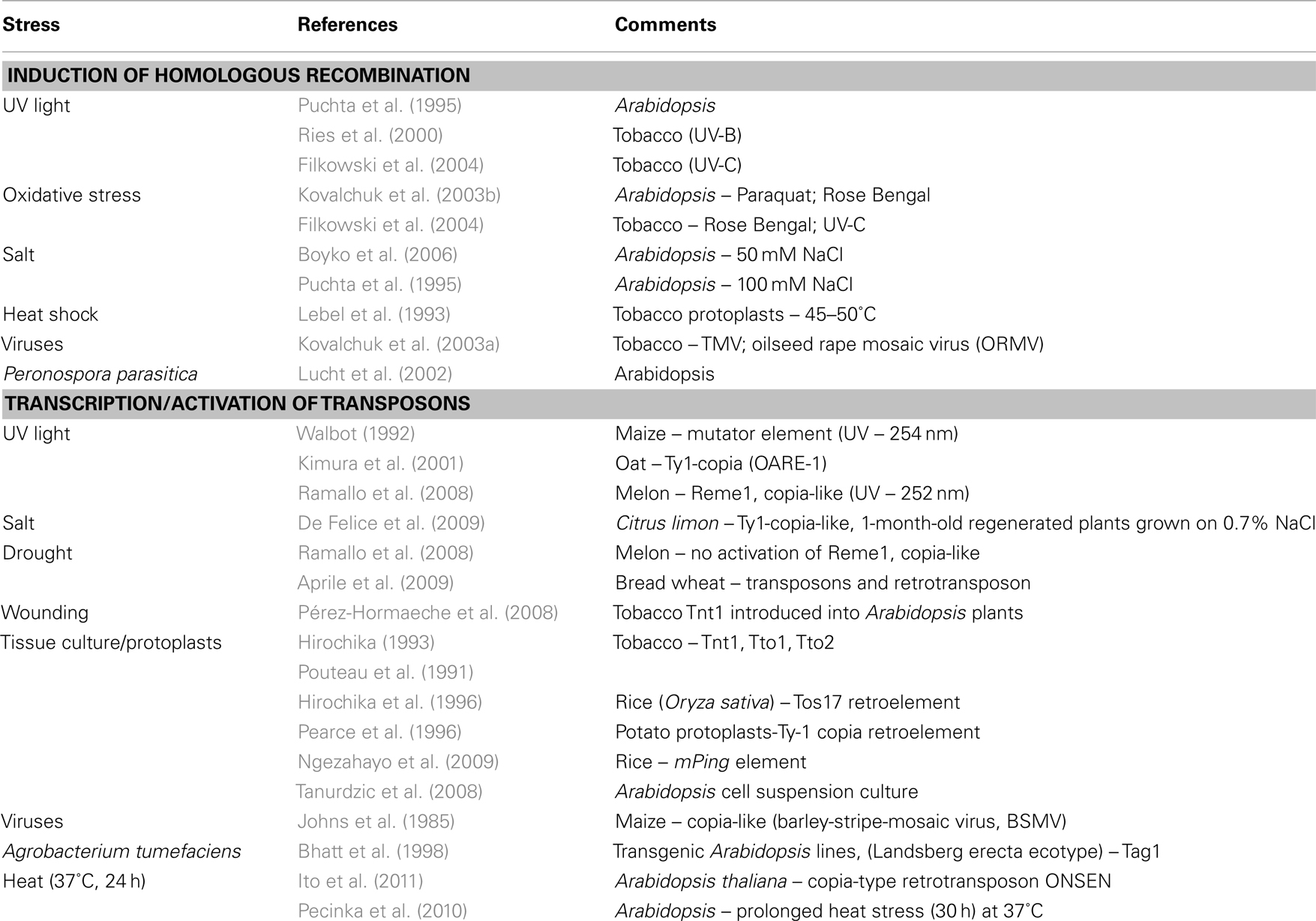
Table 1. Induction of homologous recombination and activation of transposons under various biotic and abiotic stress conditions.
Commonly, researchers are not aware that removal of cells or tissues from the plant body into tissue culture environment might impose an extreme stress over the cells resulting in reprogramming, dedifferentiation and acquisition of stem cell-like state prior to re-entry into the cell cycle (Figure 2). In this respect, it is worth mentioning McClintock’s view on cell culturing stating, “Some responses to stress are especially significant for illustrating how a genome may modify itself when confronted with unfamiliar conditions. Changes induced in genomes when cells are removed from their normal locations and placed in tissue culture surroundings are outstanding examples of this” (McClintock, 1984). It is thus conceivable that the genotype(s) of somatic cells entering the cell cycle may not be identical to the genotype of the original somatic cells and that a genotype(s) conferring increasing fitness for tissue culture conditions may prevail.
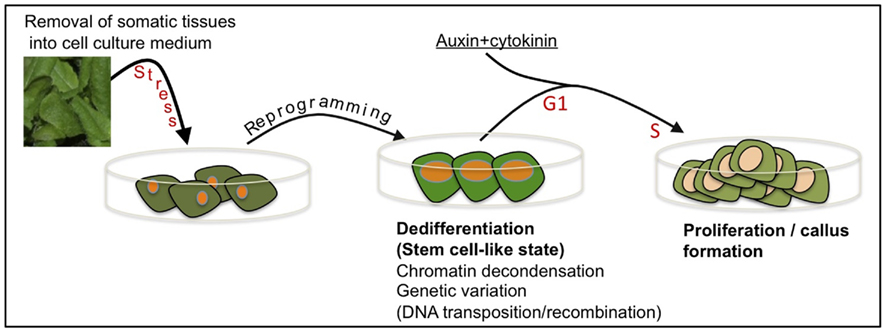
Figure 2. Cell culturing – a special case of stress-induced dedifferentiation and genetic variation. Removal of tissues from the plant body and placing them in cell culture environment is stressful leading to cellular reprogramming and acquisition of a transient, dedifferentiated, stem cell-like state. This transition is associated with widespread chromatin decondensation often accompanied by DNA transposition and/or recombination. Further signals induce cell cycle re-entry (G1 to S transition), proliferation, and callus formation. Notably, cells entering the cell cycle, each may possess a genotype, which is different from that of the original somatic cell.
Presently, it is not clear why the genome become vulnerable under certain stress conditions. Stochastic epigenetic modifications associated with chromatin decondensation may release constraints over transposable elements resulting in their activation and transposition into other chromosomal sites. Indeed, transposition of mPing – an endogenous miniature inverted-repeat transposable element (MITE) in the rice genome was activated in tissue culture. This activation was correlated with alteration in cytosine methylation pattern of the mPing element (Ngezahayo et al., 2009). Similarly, MITEs and long terminal repeat (LTR) retrotransposons were activated in certain rice cultivars under spaceflight stress conditions; transposition was correlated with cytosine demethylation within the elements (Long et al., 2009; Ou et al., 2010). Introduction of the tobacco Tnt1 retroelement into Arabidopsis plants resulted in Tnt1 silencing mediated by small interfering RNA-induced non-CG methylation of its LTR; silencing was partly reversed by stresses (Pérez-Hormaeche et al., 2008). Also, infection of Arabidopsis leaves with the pathogen Pseudomonas syringae resulted in hypomethylation of centromeric DNA accompanied by structural alteration of chromatin (Pavet et al., 2006). It should be noted, however, that activation of repeated elements following exposure to stress (e.g., heat shock) can occur without notable changes in DNA and histone methylation (Pecinka et al., 2010; Tittel-Elmer et al., 2010); a reduction in nucleosome occupancy was correlated with the activation of these repeats (Pecinka et al., 2010). Interestingly, exposure to heat stress resulted in widespread decondensation of centromeric repeats (Pecinka et al., 2010), which may be indicative for the acquisition of dedifferentiated, stem cell-like state.
Stochastic genome reorganization and chromatin decondensation could bring highly similar or identical DNA sequences into close proximity and under appropriate conditions, such as double strand DNA break (DSB), homologous recombination (HR) can be induced. DSBs can be generated in the course of transposition and/or by specific enzymes such as topoisomerases (reviewed in Wang, 2002) and single-stranded DNA endonucleases that target weakly hydrogen bonded regions (e.g., AT-rich regions) within double strand DNA (Grafi and Larkins, 1995). Interestingly, topoisomerases are upregulated in plants following exposure to various stress conditions (Mudgil et al., 2002; Hettiarachchi et al., 2005) and their activities might be necessary for proper function of the meristem (Graf et al., 2010). Apparently, these HR events are stress-induced (Table 1) and could irreversibly modify the genome often leading to somaclonal variation in plants and to diseases in humans (Kaeppler et al., 2000; Griffin and Thacker, 2004; Hurles, 2005).
Concluding Remarks
An immense body of published data supports the idea that plant somatic cells retain plasticity, that is, the capacity for dedifferentiation and switching fate. Dedifferentiation is a unique, transient state that represents a developmental decision point having features of stem cells from which cells can take different fates including return to a previous or assume a new differentiation state (re-differentiation and trans-differentiation, respectively), re-enter to the cell cycle and even commit for death. Accumulating data suggest that the stem cell niche in the SAM is much broader that thought; the stem cell state is not restricted to cells localized at the central zone but characterizes all cells within the SAM including the peripheral zone and leaf initials. Open chromatin configuration emerges as a fundamental feature of stem cell, which might confer stem cells with the capacity for rapid switching into the appropriate transcriptional program upon induction of differentiation and with the flexibility needed for differentiation into multiple cell types. Finally, in recent years there is growing body of evidence implicating dedifferentiation in response to stress both in plants and animals. Exposure of plants to stress (e.g., cell culturing, virus infection) might lead to epigenetic and genetic variations and consequently to phenotypic alterations. This idea has been categorically expressed by McClintock (1984) with respect to plant regeneration by means of tissue culture: “It may be safe to state that no two of the callus derived plants are exactly alike, and none is just like the plant that donated the cell or cells for the tissue culture.” In light of this, it is possible that many of the dedifferentiation-derived cells obtained through re-differentiation, trans-differentiation, or re-entry into the cell cycle, each possesses a genotype, which is not exactly like the genotype of the original differentiated cell(s).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Simcha Lev-Yadun for critical reading of the manuscript. This work was supported by The Israel Science Foundation (ISF) grant No. 476/09 to Gideon Grafi.
References
Aloni, R., and Barnett, J. R. (1996). The development of phloem anastomoses between vascular bundles and their role in xylem regeneration after wounding in Cucurbita and Dahlia. Planta 198, 595–603.
Alvarez, M. E., Nota, F., and Cambiagno, D. A. (2010). Epigenetic control of plant immunity. Mol. Plant Pathol. 11, 563–576.
Aprile, A., Mastrangelo, A. M., De Leonardis, A. M., Galiba, G., Roncaglia, E., Ferrari, F., De Bellis, L., Turchi, L., Giuliano, G., and Cattivelli, L. (2009). Transcriptional profiling in response to terminal drought stress reveals differential responses along the wheat genome. BMC Genomics 10, 279. doi: 10.1186/1471-2164-10-279
Arents, G., and Moudrianakis, E. N. (1995). The histone fold: a ubiquitous architectural motif utilized in DNA compaction and protein dimerization. Proc. Natl. Acad. Sci. U.S.A. 92, 11170–11174.
Azuara, V., Perry, P., Sauer, S., Spivakov, M., Jørgensen, H. F., John, R. M., Gouti, M., Casanova, M., Warnes, G., Merkenschlager, M., and Fisher, A. G. (2006). Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 8, 532–538.
Bannister, A. J., Zegerman, P., Partridge, J. F., Miska, E. A., Thomas, J. O., Allshire, R. C., and Kouzarides, T. (2001). Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124.
Bernstein, B. E., Mikkelsen, T. S., Xie, X., Kamal, M., Huebert, D. J., Cuff, J., Fry, B., Meissner, A., Wernig, M., Plath, K., Jaenisch, R., Wagschal, A., Feil, R., Schreiber, S. L., and Lander, E. S. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326.
Bhatt, A. M., Lister, C., Crawford, N., and Dean, C. (1998). The transposition frequency of Tag1 elements is increased in transgenic Arabidopsis lines. Plant Cell 10, 427–434.
Booker, C. E., and Dwivedi, R. S. (1973). Ultrastructure of meristematic cells of dormant and released buds in Tradescantia paludosa. Exp. Cell Res. 82, 255–261.
Boyko, A., Greer, M., and Kovalchuk, I. (2006). Acute exposure to UVB has a more profound effect on plant genome stability than chronic exposure. Mutat. Res. 602, 100–109.
Boyko, A., and Kovalchuk, I. (2011). Genome instability and epigenetic modification–heritable responses to environmental stress? Curr. Opin. Plant Biol. 14, 260–266.
Brockes, J. P., and Kumar, A. (2008). Comparative aspects of animal regeneration. Annu. Rev. Cell Dev. Biol. 24, 525–549.
Champy, C. (1913). Dedifferentiation des tissus cultives en dehors de l’organisme. Bibliog. Anat. xxiii, :184.
Chen, M., Lv, S., and Meng, Y. (2010). Epigenetic performers in plants. Dev. Growth Differ. 52, 555–566.
Chen, X., Hu, Y., and Zhou, D. X. (2011). Epigenetic gene regulation by Plant Jumonji group of histone demethylase. Biochim. Biophys. Acta 1809, 421–426.
Damri, M., Granot, G., Ben-Meir, H., Avivi, Y., Plaschkes, I., Chalifa-Caspi, V., Wolfson, M., Fraifeld, V., and Grafi, G. (2009). Senescing cells share common features with dedifferentiating cells. Rejuvenation Res. 12, 435–443.
De Felice, B., Wilson, R. R., Argenziano, C., Kafantaris, I., and Conicella, C. (2009). A transcriptionally active copia-like retroelement in Citrus limon. Cell. Mol. Biol. Lett. 14, 289–304.
Deal, R. B., and Henikoff, S. (2010). A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev. Cell 18, 1030–1040.
Eberharter, A., and Becker, P. B. (2002). Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 3, 224–229.
Echeverri, K., Clarke, J. D., and Tanaka, E. M. (2001). In vivo imaging indicates muscle fiber dedifferentiation is a major contributor to the regenerating tail blastema. Dev. Biol. 236, 151–164.
Echeverri, K., and Tanaka, E. M. (2002). Ectoderm to mesoderm lineage switching during axolotl tail regeneration. Science 298, 1993–1996.
Eissenberg, J. C., and Shilatifard, A. (2010). Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev. Biol. 339, 240–249.
Filkowski, J., Yeoman, A., Kovalchuk, O., and Kovalchuk, I. (2004). Systemic plant signal triggers genome instability. Plant J. 38, 1–11.
Flaishman, M. A., Loginovsky, K., and Lev-Yadun, S. (2003). Regenerative xylem in inflorescence stems of Arabidopsis thaliana. J. Plant Growth Regul. 22, 253–258.
Fortunel, N. O., Out, H. H., Ng, H. H., Chen, J., Mu, X., Chevassut, T., Li, X., Joseph, M., Bailey, C., Hatzfeld, J. A., Hatzfeld, A., Usta, F., Vega, V. B., Long, P. M., Libermann, T. A., and Lim, B. (2003). Comment on “‘Stemness’: transcriptional profiling of embryonic and adult stem cells” and “a stem cell molecular signature.” Science 302, 393.
Frearson, E. M., Power, J. B., and Cocking, E. C. (1973). The isolation, culture and regeneration of Petunia leaf protoplasts. Dev. Biol. 33, 130–137.
Gaspar-Maia, A., Alajem, A., Meshorer, E., and Ramalho-Santos, M. (2011). Open chromatin in pluripotency and reprogramming. Nat. Rev. Mol. Cell Biol. 12, 36–47.
Girard, L., and Freeling, M. (1999). Regulatory changes as a consequence of transposon insertion. Dev. Genet. 25, 291–296.
Graf, P., Dolzblasz, A., Würschum, T., Lenhard, M., Pfreundt, U., and Laux, T. (2010). MGOUN1 encodes an Arabidopsis type IB DNA topoisomerase required in stem cell regulation and to maintain developmentally regulated gene silencing. Plant Cell 22, 716–728.
Grafi, G. (2009). The complexity of cellular dedifferentiation: implications for regenerative medicine. Trends Biotechnol. 27, 329–332.
Grafi, G., and Avivi, Y. (2004). Stem cells: a lesson from dedifferentiation. Trends Biotechnol. 22, 388–389.
Grafi, G., Ben-Meir, H., Avivi, Y., Moshe, M., Dahan, Y., and Zemach, A. (2007). Histone methylation controls telomerase-independent telomere lengthening in cells undergoing dedifferentiation. Dev. Biol. 306, 838–846.
Grafi, G., Chalifa-Caspi, V., Nagar, T., Plaschkes, I., Barak, S., and Ransbotyn, V. (2011a). Plant response to stress meets dedifferentiation. Planta 233, 433–438.
Grafi, G., Ofir, R., Chalifa-Caspi, V., and Plaschkes, I. (2011b). “Illuminating hidden features of stem cells,” in Embryonic Stem Cells: Basic Biology to Bioengineering, ed. M. K. Kallos (INTECH Open Access Publisher), 265–282.
Grafi, G., and Larkins, B. A. (1995). Activity of single-stranded DNA endonucleases in mung bean is associated with cell division. Plant Mol. Biol. 29, 703–710.
Griffin, C. S., and Thacker, J. (2004). The role of homologous recombination repair in the formation of chromosome aberrations. Cytogenet. Genome Res. 104, 21–27.
Gurdon, J. B., and Wilmut, I. (2011). Nuclear transfer to eggs and oocytes. Cold Spring Harb. Perspect. Biol. 3, a002659.
Hay, E. D. (1959). Electron microscopic observations of muscle dedifferentiation in regenerating Amblystoma limbs. Dev. Biol. 1, 555–585.
He, G., Zhu, X., Elling, A. A., Chen, L., Wang, X., Guo, L., Liang, M., He, H., Zhang, H., Chen, F., Qi, Y., Chen, R., and Deng, X. W. (2010). Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell 22, 17–33.
Hettiarachchi, G. H., Reddy, M. K., Sopory, S. K., and Chattopadhyay, S. (2005). Regulation of TOP2 by various abiotic stresses including cold and salinity in pea and transgenic tobacco plants. Plant Cell Physiol. 46, 1154–1160.
Hirochika, H. (1993). Activation of tobacco retrotransposons during tissue culture. EMBO J. 12, 2521–2528.
Hirochika, H., Otsuki, H., Yoshikawa, M., Otsuki, Y., Sugimoto, K., and Takeda, S. (1996). Autonomous transposition of the tobacco retrotransposon Tto1 in rice. Plant Cell 8, 725–734.
Hurles, M. (2005). How homologous recombination generates a mutable genome. Hum. Genomics 2, 179–186.
Ito, H., Gaubert, H., Bucher, E., Mirouze, M., Vaillant, I., and Paszkowski, J. (2011). An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472, 115–119.
Ivanova, N. B., Dimos, J. T., Schaniel, C., Hackney, J. A., Moore, K. A., and Lemischka, I. R. (2002). A stem cell molecular signature. Science 298, 601–604.
Iwase, A., Mitsuda, N., Koyama, T., Hiratsu, K., Kojima, M., Arai, T., Inoue, Y., Seki, M., Sakakibara, H., Sugimoto, K., and Ohme-Takagi, M. (2011). The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr. Biol. 21, 508–514.
Jackson, J. P., Lindroth, A. M., Cao, X., and Jacobsen, S. E. (2002). Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416, 556–560.
James, S. (1984). Lignotubers and burls: their structure, function and ecological significance in mediterranean ecosystems. Bot. Rev. 50, 225–266.
Johns, M. A., Mottinger, J., and Freeling, M. (1985). A low copy number, copia-like transposon in maize. EMBO J. 4, 1093–1101.
Kaeppler, S. M., Kaeppler, H. F., and Rhee, Y. (2000). Epigenetic aspects of somaclonal variation in plants. Plant Mol. Biol. 43, 179–188.
Kim, K., Doi, A., Wen, B., Ng, K., Zhao, R., Cahan, P., Kim, J., Aryee, M. J., Ji, H., Ehrlich, L. I., Yabuuchi, A., Takeuchi, A., Cunniff, K. C., Hongguang, H., McKinney-Freeman, S., Naveiras, O., Yoon, T. J., Irizarry, R. A., Jung, N., Seita, J., Hanna, J., Murakami, P., Jaenisch, R., Weissleder, R., Orkin, S. H., Weissman, I. L., Feinberg, A. P., and Daley, G. Q. (2010a). Epigenetic memory in induced pluripotent stem cells. Nature 467, 285–290.
Kim, J. M., To, T. K., Nishioka, T., and Seki, M. (2010b). Chromatin regulation functions in plant abiotic stress responses. Plant Cell Environ. 33, 604–611.
Kimura, Y., Tosa, Y., Shimada, S., Sogo, R., Kusaba, M., Sunaga, T., Betsuyaku, S., Eto, Y., Nakayashiki, H., and Mayama, S. (2001). OARE-1, a Ty1-copia retrotransposon in oat activated by abiotic and biotic stresses. Plant Cell Physiol. 42, 1345–1354.
Knopf, F., Hammond, C., Chekuru, A., Kurth, T., Hans, S., Weber, C. W., Mahatma, G., Fisher, S., Brand, M., Schulte-Merker, S., and Weidinger, G. (2011). Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev. Cell 20, 713–724.
Kovalchuk, I., Kovalchuk, O., Kalck, V., Boyko, V., Filkowski, J., Heinlein, M., and Hohn, B. (2003a). Pathogen-induced systemic plant signal triggers DNA rearrangements. Nature 423, 760–762.
Kovalchuk, I., Filkowski, J., Smith, K., and Kovalchuk, O. (2003b). Reactive oxygen species stimulate homologous recombination in plants. Plant Cell Environ. 26, 1531–1539.
Kragl, M., Knapp, D., Nacu, E., Khattak, S., Maden, M., Epperlein, H. H., and Tanaka, E. M. (2009). Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature 460, 60–65.
Lachner, M., O’Carroll, D., Rea, S., Mechtler, K., and Jenuwein, T. (2001). Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120.
Lafos, M., Kroll, P., Hohenstatt, M. L., Thorpe, F. L., Clarenz, O., and Schubert, D. (2011). Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet. 7, e1002040. doi: 10.1371/journal.pgen.1002040
Lan, F., Nottke, A. C., and Shi, Y. (2008). Mechanisms involved in the regulation of histone lysine demethylases. Curr. Opin. Cell Biol. 20, 316–325.
Lebel, E. G., Masson, J., Bogucki, A., and Paszkowski, J. (1993). Stress-induced intrachromosomal recombination in plant somatic cells. Proc. Natl. Acad. Sci. U.S.A. 90, 422–426.
Lee, K. K., and Workman, J. L. (2007). Histone acetyltransferase complexes: one size doesn’t fit all. Nat. Rev. Mol. Cell Biol. 8, 284–295.
Lev-Yadun, S. (2001). Intrusive growth – the plant analog of dendrite and axon growth in animals. New Phytol. 150, 508–512.
Lin, J. F., and Wu, S. H. (2004). Molecular events in senescing Arabidopsis leaves. Plant J. 39, 612–628.
Lisch, D. (2009). Epigenetic regulation of transposable elements in plants. Annu. Rev. Plant Biol. 60, 43–66.
Lisch, D., and Bennetzen, J. L. (2011). Transposable element origins of epigenetic gene regulation. Curr. Opin. Plant Biol. 14, 156–161.
Long, L., Ou, X., Liu, J., Lin, X., Sheng, L., and Liu, B. (2009). The spaceflight environment can induce transpositional activation of multiple endogenous transposable elements in a genotype-dependent manner in rice. J. Plant Physiol. 166, 2035–2045.
Lucht, J. M., Mauch-Mani, B., Steiner, H. Y., Metraux, J. P., Ryals, J., and Hohn, B. (2002). Pathogen stress increases somatic recombination frequency in Arabidopsis. Nat. Genet. 30, 311–314.
Luger, K., Mäder, A. W., Richmond, R. K., Sargent, D. F., and Richmond, T. J. (1997). Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260.
Luo, M., Liu, X., Singh, P., Cui, Y., Zimmerli, L., and Wu, K. (2011). Chromatin modifications and remodeling in plant abiotic stress responses. Biochim. Biophys. Acta. doi: 10.1016/j.bbagrm.2011.06.008. [Epub ahead of print].
MacRae, E. K., and Meetz, G. D. (1970). Electron microscopy of the ammoniacal silver reaction for histones in the erythropoietic cells of the chick. J. Cell Biol. 45, 235–245.
Malagnac, F., Bartee, L., and Bender, J. (2002). An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J. 21, 6842–6852.
McClintock, B. (1984). The significance of responses of the genome to challenge. Science 226, 792–801.
Mirouze, M., and Paszkowski, J. (2011). Epigenetic contribution to stress adaptation in plants. Curr. Opin. Plant Biol. 14, 267–274.
Miura, A. B., Shibata, A., Akihama, T., Endo, Y., and Saito, Y. (1974). Ultrastructure of developing erythrocytes. Tohoku J. Exp. Med. 112, 299–313.
Morrow, R. C., and Tibbitts, T. W. (1988). Evidence for involvement of phytochrome in tumor development on plants. Plant Physiol. 88, 1110–1114.
Mosquna, A., Katz, A., Decker, E. L., Rensing, S. A., Reski, R., and Ohad, N. (2009). Regulation of stem cell maintenance by the Polycomb protein FIE has been conserved during land plant evolution. Development 136, 2433–2444.
Mottinger, J. P., Johns, M. A., and Freeling, M. (1984). Mutations of the Adh1 gene in maize following infection with barley stripe mosaic virus. Mol. Gen. Genet. 195, 367–369.
Mudgil, Y., Singh, B. N., Upadhyaya, K. C., Sopory, S. K., and Reddy, M. K. (2002). Cloning and characterization of a cell cycle-regulated gene encoding topoisomerase I from Nicotiana tabacum that is inducible by light, low temperature and abscisic acid. Mol. Genet. Genomics 267, 380–390.
Murphy, M. J. Jr., Bertles, J. F., and Gordon, A. S. (1971). Identifying characteristics of the hematopoietic precursor cell. J. Cell Sci. 9, 23–47.
Ngezahayo, F., Xu, C., Wang, H., Jiang, L., Pang, J., and Liu, B. (2009). Tissue culture-induced transpositional activity of mPing is correlated with cytosine methylation in rice. BMC Plant Biol. 9, 91. doi:10.1186/1471-2229-9-91
Ou, X., Long, L., Wu, Y., Yu, Y., Lin, X., Qi, X., and Liu, B. (2010). Spaceflight-induced genetic and epigenetic changes in the rice (Oryza sativa L.) genome are independent of each other. Genome 53, 524–532.
Pavet, V., Quintero, C., Cecchini, N. M., Rosa, A. L., and Alvarez, M. E. (2006). Arabidopsis displays centromeric DNA hypomethylation and cytological alterations of heterochromatin upon attack by Pseudomonas syringae. Mol. Plant Microbe Interact. 19, 577–587.
Pearce, S. R., Kumar, A., and Flavell, A. J. (1996). Activation of the Ty1-copia group retrotransposons of potato (Solanum tuberosum) during protoplast isolation. Plant Cell Rep. 15, 949–953.
Pecinka, A., Dinh, H. Q., Baubec, T., Rosa, M., Lettner, N., and Mittelsten Scheid, O. (2010). Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell 22, 3118–3129.
Pérez-Hormaeche, J., Potet, F., Beauclair, L., Le Masson, I., Courtial, B., Bouché, N., and Lucas, H. (2008). Invasion of the Arabidopsis genome by the tobacco retrotransposon Tnt1 is controlled by reversible transcriptional gene silencing. Plant Physiol. 147, 1264–1278.
Polo, J. M., Liu, S., Figueroa, M. E., Kulalert, W., Eminli, S., Tan, K. Y., Apostolou, E., Stadtfeld, M., Li, Y., Shioda, T., Natesan, S., Wagers, A. J., Melnick, A., Evans, T., and Hochedlinger, K. (2010). Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 28, 848–855.
Potten, C. S., and Loeffler, M. (1990). Stem cells: attributes, cycles, spirals, pitfalls and uncertainties. Lessons for and from the crypt. Development 110, 1001–1020.
Pouteau, S., Huttner, E., Grandbastien, M. A., and Caboche, M. (1991). Specific expression of the tobacco Tnt1 retrotransposon in protoplasts. EMBO J. 10, 1911–1918.
Puchta, H., Swoboda, P., Gal, S., Blot, M., and Hohn, B. (1995). Somatic intrachromosomal homologous recombination events in populations of plant siblings. Plant Mol. Biol. 28, 281–292.
Ramalho-Santos, M., Yoon, S., Matsuzaki, Y., Mulligan, R. C., and Melton, D. A. (2002). “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science 298, 597–600.
Ramallo, E., Kalendar, R., Schulman, A. H., and Martínez-Izquierdo, J. A. (2008). Reme1, a Copia retrotransposon in melon, is transcriptionally induced by UV light. Plant Mol. Biol. 66, 137–150.
Rea, S., Eisenhaber, F., O’Carroll, D., Strahl, B. D., Sun, Z. W., Schmid, M., Opravil, S., Mechtler, K., Ponting, C. P., Allis, C. D., and Jenuwein, T. (2000). Regulation of chromatin structure by site specific histone H3 methyltransferases. Nature 406, 593–599.
Ries, G., Buchholz, G., Frohnmeyer, H., and Hohn, B. (2000). UV-damage-mediated induction of homologous recombination in Arabidopsis is dependent on photosynthetically active radiation. Proc. Natl. Acad. Sci. U.S.A. 97, 13425–13429.
Rigal, M., and Mathieu, O. (2011). A “mille-feuille” of silencing: epigenetic control of transposable elements. Biochim. Biophys. Acta 1809, 452–458.
Robinson, P. J., and Rhodes, D. (2006). Structure of the ‘30 nm’ chromatin fibre: a key role for the linker histone. Curr. Opin. Struct. Biol. 16, 336–343.
Roudier, F., Ahmed, I., Bérard, C., Sarazin, A., Mary-Huard, T., Cortijo, S., Bouyer, D., Caillieux, E., Duvernois-Berthet, E., Al-Shikhley, L., Giraut, L., Després, B., Drevensek, S., Barneche, F., Dèrozier, S., Brunaud, V., Aubourg, S., Schnittger, A., Bowler, C., Martin-Magniette, M. L., Robin, S., Caboche, M., and Colot, V. (2011). Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 30, 1928–1938.
Routh, A., Sandin, S., and Rhodes, D. (2008). Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc. Natl. Acad. Sci. U.S.A. 105, 8872–8877.
Sachs, T. (1981). The control of the patterned differentiation of vascular tissues. Adv. Bot. Res. 9, 151–262.
Sena, G., Wang, X., Liu, H. Y., Hofhuis, H., and Birnbaum, K. D. (2009). Organ regeneration does not require a functional stem cell niche in plants. Nature 457, 1150–1153.
Shepard, J. F., and Totten, R. E. (1977). Mesophyll cell protoplasts of potato: isolation, proliferation, and plant regeneration. Plant Physiol. 60, 313–316.
Shilatifard, A. (2006). Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 75, 243–269.
Shoshani, O., and Zipori, D. (2011). Mammalian cell dedifferentiation as a possible outcome of stress. Stem Cell. Rev. 7, 488–493.
Slack, J. M. (2006). Amphibian muscle regeneration – dedifferentiation or satellite cells? Trends Cell Biol. 16, 273–275.
Slack, J. M., Beck, C. W., Gargioli, C., and Christen, B. (2004). Cellular and molecular mechanisms of regeneration in Xenopus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359, 745–751.
Slotkin, R. K., and Martienssen, R. (2007). Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8, 272–285.
Sprague, G. F., Mckinney, H. H., and Greeley, L. (1963). Virus as a mutagenic agent in maize. Science 141, 1052–1053.
Stocum, D. L. (1968). The urodele limb regeneration blastema: a self-organizing system. I. Morphogenesis and differentiation of autografted whole and fractional blastemas. Dev. Biol. 18, 457–480.
Sugimoto, K., Gordon, S. P., and Meyerowitz, E. M. (2011). Regeneration in plants and animals: dedifferentiation, transdifferentiation, or just differentiation? Trends Cell Biol. 21, 212–218.
Sugimoto, K., Jiao, Y., and Meyerowitz, E. M. (2010). Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev. Cell 18, 463–471.
Takebe, I., Labib, G., and Melchers, G. (1971). Regeneration of whole plants from isolated mesophyll protoplasts of tobacco. Naturwissenschaften 58, 318–320.
Tanurdzic, M., Vaughn, M. W., Jiang, H., Lee, T. J., Slotkin, R. K., Sosinski, B., Thompson, W. F., Doerge, R. W., and Martienssen, R. A. (2008). Epigenomic consequences of immortalized plant cell suspension culture. PLoS Biol. 6, e302. doi: 10.1371/journal.pbio.0060302
Thorstensen, T., Grini, P. E., and Aalen, R. B. (2011). SET domain proteins in plant development. Biochim. Biophys. Acta 1809, 407–420.
Tittel-Elmer, M., Bucher, E., Broger, L., Mathieu, O., Paszkowski, J., and Vaillant, I. (2010). Stress-induced activation of heterochromatic transcription. PLoS Genet. 6, e1001175. doi: 10.1371/journal.pgen.1001175
Uhlenhuth, E. (1915). The form of the epithelial cells in cultures of frog skin, and its relation to the consistency of the medium. J. Exp. Med. Xxii, :76.
Vaahtera, L., and Brosché, M. (2011). More than the sum of its parts – how to achieve a specific transcriptional response to abiotic stress. Plant Sci. 180, 421–430.
Vanyushin, B. F., and Ashapkin, V. V. (2011). DNA methylation in higher plants: past, present and future. Biochim. Biophys. Acta 1809, 360–368.
Vasil, I. K., and Vasil, V. (1972). Totipotency and embryogenesis in plant cell and tissue cultures. In vitro 8, 117–127.
Vasil, V., and Vasil, I. K. (1974). Regeneration of tobacco and petunia plants from protoplasts and culture of corn protoplasts. In vitro 10, 83–96.
Walbot, V. (1992). Reactivation of Mutator transposable elements of maize by ultra violet. Mol. Gen. Genet. 234, 353–360.
Wang, J. C. (2002). Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 3, 430–440.
Waters, D. A., Burrows, G. E., and Harper, J. D. (2010). Eucalyptus regnans (Myrtaceae): a fire-sensitive eucalypt with a resprouter epicormic structure. Am. J. Bot. 97, 545–556.
Weintraub, H. (1978). The nucleosome repeat length increases during erythropoiesis in the chick. Nucleic Acids Res. 5, 1179–1188.
Williams, L., Zhao, J., Morozova, N., Li, Y., Avivi, Y., and Grafi, G. (2003). Chromatin reorganization accompanying cellular dedifferentiation is associated with modifications of histone H3, redistribution of HP1, and activation of E2F-target genes. Dev. Dyn. 228, 113–120.
Yadav, R. K., Girke, T., Pasala, S., Xie, M., and Reddy, G. V. (2009). Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc. Natl. Acad. Sci. U.S.A. 106, 4941–4946.
Yaish, M. W., Colasanti, J., and Rothstein, S. J. (2011). The role of epigenetic processes in controlling flowering time in plants exposed to stress. J. Exp. Bot. 62, 3727–3735.
Zemach, A., Li, Y., Ben-Meir, H., Oliva, M., Mosquna, A., Kiss, V., Avivi, Y., Ohad, N., and Grafi, G. (2006). Different domains control the localization and mobility of like heterochromatin protein1 in Arabidopsis nuclei. Plant Cell 18, 133–145.
Zhao, J., Morozova, N., Williams, L., Libs, L., Avivi, Y., and Grafi, G. (2001). Two phases of chromatin decondensation during dedifferentiation of plant cells: distinction between competence for cell fate switch and a commitment for S phase. J. Biol. Chem. 276, 22772–22778.
Zhu, J. K. (2009). Active DNA demethylation mediated by DNA glycosylases. Annu. Rev. Genet. 43, 143–166.
Zhu, Y., Dong, A., and Shen, W. H. (2011). Histone variants and chromatin assembly in plant abiotic stress responses. Biochim. Biophys. Acta.. PMID:21839188. [Epub ahead of print].
Keywords: stem cell, dedifferentiation, totipotency, chromatin structure, chromatin modifiers, genetic variation, DNA transposition, stress
Citation: Grafi G, Florentin A, Ransbotyn V and Morgenstern Y (2011) The stem cell state in plant development and in response to stress. Front. Plant Sci. 2:53. doi: 10.3389/fpls.2011.00053
Received: 28 July 2011;
Accepted: 03 September 2011;
Published online: 04 October 2011.
Edited by:
Igor Kovalchuk, University of Lethbridge, CanadaReviewed by:
Fan Chen, Chinese Academy of Sciences, ChinaIgor Kovalchuk, University of Lethbridge, Canada
Copyright: © 2011 Grafi, Florentin, Ransbotyn and Morgenstern. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Gideon Grafi, French Associates Institute for Agriculture and Biotechnology of Drylands, Jacob Blaustein Institutes for Desert Research, Ben-Gurion University of the Negev, Sede Boqer Campus, Midreshet Ben-Gurion 84990, Israel. e-mail: ggrafi@bgu.ac.il
