- 1 Department of Stem Cell Biology, Centre for Organismal Studies Heidelberg, Heidelberg University, Heidelberg, Germany
- 2 Max Planck Institute for Developmental Biology, Tübingen, Germany
As sessile organisms, plants are exposed to extreme variations in environmental conditions over the course of their lives. Since plants grow and initiate new organs continuously, they have to modulate the underlying developmental program accordingly to cope with this challenge. At the heart of this extraordinary developmental plasticity are pluripotent stem cells, which are maintained during the entire life-cycle of the plant and that are embedded within dynamic stem cell niches. While the complex regulatory principles of plant stem cell control under artificial constant growth conditions begin to emerge, virtually nothing is known about how this circuit adapts to variations in the environment. In addition to the local feedback system constituted by the homeodomain transcription factor WUSCHEL (WUS) and the CLAVATA signaling cascade in the center of the shoot apical meristem (SAM), the bZIP transcription factor PERIANTHIA (PAN) not only has a broader expression domain in SAM and flowers, but also carries out more diverse functions in meristem maintenance: pan mutants show alterations in environmental response, shoot meristem size, floral organ number, and exhibit severe defects in termination of floral stem cells in an environment dependent fashion. Genetic and genomic analyses indicate that PAN interacts with a plethora of developmental pathways including light, plant hormone, and meristem control systems, suggesting that PAN is as an important regulatory node in the network of plant stem cell control.
Introduction
In contrast to most animals, plants continue to form new organs throughout their lives. This remarkable capacity is dependent on the continuous presence of undifferentiated and self-renewing stem cells over long periods of time. These stem cells reside at the growing points of a plant, the tips of roots and shoots, and are embedded into specialized structures called meristems (Barton, 2010).
Several genes affecting meristem and stem cell function have been identified by mutant screens in Arabidopsis thaliana. Most notably WUSCHEL (WUS) and SHOOTMERISTEMLESS (STM) are required for the maintenance of the shoot meristem (Barton and Poethig, 1993; Laux et al., 1996; Long et al., 1996; Mayer et al., 1998). Their inactivation causes premature differentiation and the eventual exhaustion of the stem cell pool, leading to the termination of the shoot meristem. Another group of genes, the CLAVATA (CLV) genes, have an opposite effect on meristems and if defective, shoot meristems overproliferate and expand inappropriately (Clark et al., 1993, 1995; Kayes and Clark, 1998).
With the exception of CLV2, all genes mentioned above are expressed in small domains in the shoot apical meristem (SAM). Elegant genetic studies have shown that WUS and CLV3 are connected by a negative feedback loop to control the size of the stem cell pool. WUS, which is expressed in the organizing center, induces the expression of CLV3 in the overlying true stem cells, which in turn signals back to the organizing center to keep WUS expression in check (Brand et al., 2000; Schoof et al., 2000). In addition to these local regulatory interactions, meristem function is affected by global hormone signaling pathways, including auxin and cytokinin circuitries. While STM mediates cytokinin biosynthesis (Jasinski et al., 2005; Yanai et al., 2005) to allow cell proliferation in the meristem, its expression is repressed by auxin (Furutani et al., 2004), which in turn allows organ initiation on the flanks of the SAM. In contrast, WUS does not interfere with cytokinin biosynthesis, but directly regulates A-type ARABIDOPSIS RESPONSE REGULATORS (ARRs; Leibfried et al., 2005; Busch et al., 2010) that act in the negative feedback regulation of cytokinin response (To et al., 2004). This feedback system of cytokinin signal transduction is also connected to auxin signaling and ARR7 and ARR15 are directly repressed by the AUXIN RESPONSE FACTOR5/MONOPTEROS transcription factor (Zhao et al., 2010). A-type ARRs execute important meristematic functions (Leibfried et al., 2005; Buechel et al., 2010; Zhao et al., 2010) by so far undiscovered mechanisms (Leibfried et al., 2005; Zhao et al., 2010).
Cells that leave the shoot meristem during the initial, vegetative phase of the life-cycle give rise to leaves and meristems of axillary shoots. After the transition to the reproductive phase, meristems that newly arise at the flanks of the SAM will develop into flowers instead. This is due to the redundant activity of meristem identity genes such as LEAFY (LFY) and APETALA1 (AP1). In contrast to the shoot apex, which is indeterminate, flowers are determinate and stem cell activity ceases after a fixed number of organs have been formed. In plants that lack LFY activity, flowers are converted into partially indeterminate shoot-like structures (Weigel et al., 1992).
One set of genes that is directly controlled by the LFY transcription factor includes homeotic genes that specify the fate of the different floral organs (Parcy et al., 1998; Busch et al., 1999). We have previously shown that LFY acts together with WUS, which also encodes a transcription factor, to contribute to the transcriptional activation of the homeotic gene AGAMOUS (AG) in the center of young flowers. AG in turn, not only specifies the fate of the floral reproductive organs, but also terminates stem cell maintenance by negative feedback on WUS expression (Lohmann et al., 2001). The bZIP transcription factor PERIANTHIA (PAN) is expressed in the SAM, as well as in developing flowers, where it overlaps with STM, WUS, the CLV transcripts, and AG, respectively (Chuang et al., 1999). Loss-of PAN function leads to an increase in the number of perianth organs, the sepals and petals, while on a gross morphological level the SAM seems unaffected (Running and Meyerowitz, 1996). In flowers, PAN genetically interacts with ABC homeotic genes, however these interactions appear mostly additive (Running and Meyerowitz, 1996). PAN protein expression was shown to be independent of the meristematic regulators CLV1 and CLV3 as well as of floral meristem identity genes, such as LFY or AP1, demonstrating that PAN also acts in parallel to these factors (Chuang et al., 1999). It has been shown that PAN interacts with the NPR1-like proteins BLADE ON PETIOLE 1 (BOP1) and BOP2 in yeast and that bop mutants share some of pan mutant features (Hepworth et al., 2005). However, their expression domains only overlap marginally, suggesting that PAN primarily acts together with other co-factors. It was shown that PAN plays important roles in the activation of AG (Das et al., 2009; Maier et al., 2009), which are strikingly modified in various day-length settings. While PAN brings about the termination of floral stem cell fate by the direct transcriptional activation of AG, its function in the SAM, where it is also strongly and specifically expressed, remains poorly understood.
Results and Discussion
Since we had noted before that the floral functions of PAN are strongly dependent on the environment (Maier et al., 2009), we carefully analyzed vegetative phenotypes of wild-type Columbia and pan mutant plants under various growth conditions and found that day-length had a substantial impact on the penetrance of pan related defects. In contrast to the reproductive phase, where pan mutants showed the most dramatic aberrations under short-day conditions, pan plants at the early vegetative stage were largely undistinguishable from wild-type in short days (SD; Figures 1A,D). Conversely, pan mutants exhibited pleiotropic phenotypes when exposed to long days (LD), including elongated petioles, curled leaves, and a twisted rosette (Figures 1B,E). Under continuous light (CL), Col and pan phenotypes were less distinct, but pan plants continued to show more extreme leaf-curling and rosette twisting. In addition to the morphological traits, we observed that pan mutants flowered slightly early and on average formed 1.5 or 2.5 rosette leaves less than wild-type under LD or CL, respectively (Figure 3A; n = 50). Furthermore, we realized that pan mutants are extremely sensitive to variations in diverse environmental conditions, including water and nutrient availability, as well as biotic and abiotic stress (data not shown). Taken together, these phenotypes indicated that PAN might act to stabilize the developmental program of the shoot apex and thus buffers the impact of diverse environmental inputs.

Figure 1. Vegetative phenotypes in response to environmental conditions (A–F). Phenotype of wild-type (A–C) and pan mutant (D–F) plants grown under short-day [SD (A,D)], long-day [LD (B,E)], and continuous light [CL (C,F)] conditions for 21 days. Note leaf-curling, elongated petioles and twisted leaf rosettes under LD and CL conditions.
Since the activity of the SAM is mainly determined by the WUS–CLV feedback system, which acts on the stem cell population, as well as the repression of differentiation throughout the meristem provided by STM, we investigated their regulatory and genetic interaction with PAN. Using in situ hybridization on serial histological sections, we first analyzed in detail the mRNA-expression patterns of PAN in the inflorescence meristem and found that, consistent with a buffering function, PAN mRNA is most highly expressed in a ring-shaped domain surrounding the stem cells (Figures 2A–D). We detected weaker signals throughout the center of the SAM, suggesting that PAN might execute slightly different functions depending on expression levels. Similar to the situation identified for WUS, which was shown to bind to distinct cis-regulatory motifs with different affinity (Busch et al., 2010), these functions could be mediated by distinct sets of PAN downstream targets. However, in situ detection of PAN protein on sections of the SAM did not show the ring domain, but rather suggested that PAN is found throughout the meristem (Chuang et al., 1999). Unfortunately, we were unable to resolve whether these differences were of technical nature, or reflected relevant biology. Hybridizations on cross sections demonstrated that PAN mRNA is strongly reduced even in early organ primordia (Figures 2E–H). We next investigated how the SAM regulatory system is affected by the loss-of PAN function. First, we noticed that the SAM was significantly increased in size (Figures 2I,M) and that the WUS expression domain is substantially wider compared to the wild-type situation (Figures 2J,N). Interestingly, the stem cell domain marked by CLV3 expression remained largely unaffected despite the expanded stem cell niche (Figures 2K,O), suggesting that the regulatory interaction between WUS and CLV3 is partially uncoupled in pan mutants. In line with the enlarged meristem, we found expanded STM expression in pan apices (Figures 2L,P) and the absence of STM transcripts from emerging organ primordia was less pronounced in pan when compared to wild-type. Taken together, these results demonstrate that PAN function is required for normal SAM development, which might be mediated by its effects on the expression of the canonical meristem regulators. To address how PAN is integrated into the regulatory network of the SAM, we analyzed its expression in wus and clv3 mutants, which represent the extremes in meristem dis-regulation. Since wus mutants rarely form inflorescence meristems, we focused our analysis on the seedling stage and found accumulation of PAN mRNA mostly in the center of the SAM in wild-type. In addition, we detected weaker signals on the periphery of the meristem and at the adaxial sides of young leaves (Figure 2Q). Consistent with the loss-of a fully developed SAM in wus, we were unable to detect PAN transcripts in central tissue of this mutant, however, strong expression was found in leaf-primordia and young leaves (Figure 2R). While Chuang et al. (1999) had reported that PAN protein expression is mostly independent of CLV3, we observed that PAN transcripts accumulated throughout the SAM, with a ring of strong expression toward the base with weaker signals toward the top of the expanded clv3 meristem (Figures 2S,T).
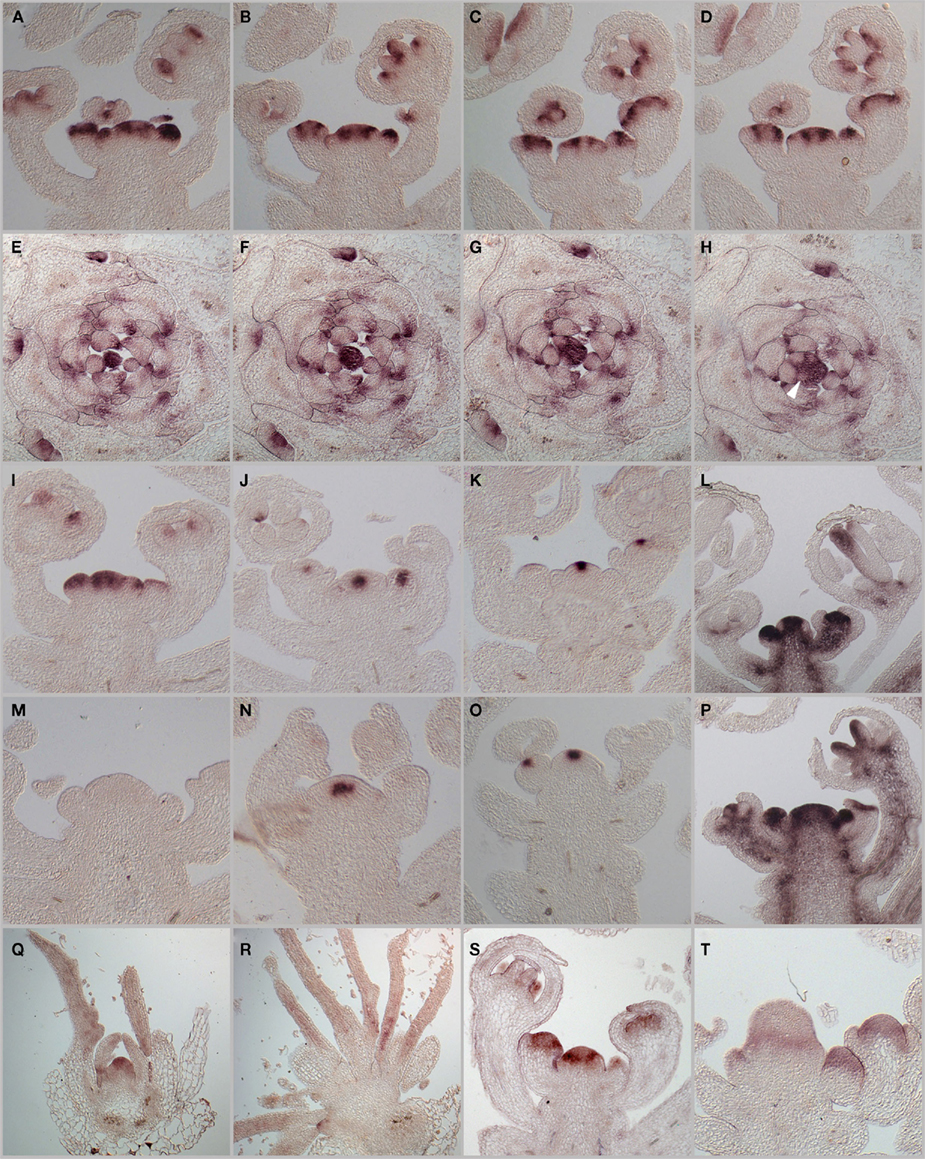
Figure 2. mRNA-expression patterns of PAN and SAM regulators WUS, CLV3, STM. In situ hybridizations were used to analyze PAN mRNA-expression patterns. (A–D) Serial longitudinal sections of wild-type inflorescence apices after 25 days of growth LD. (E–L) Serial cross sections of a vegetative apex grown in 23 days in SD. PAN mRNA shows varying expression with a local maximum in a ring domain around the central zone. PAN expression is reduced in newly arising organ primordia [P3-P0, see arrowhead in (H)]. Expression patterns of PAN, WUS, CLV3, and STM in inflorescence apices of wild-type (I–L) and pan mutant plants (M–P). PAN(I,M), WUS (J,N), CLV3 (K,O), and STM (L,P). PAN mRNA-expression in vegetative tissues of wild-type (Q) and wus mutants (R). PAN expression in wild-type (S) and ring-like expression in enlarged floral tissues on clv3 mutant (T).
Having shown that PAN is more tightly connected to the regulatory system of the SAM than previously anticipated, we extended our analysis to test the functional interaction of PAN with CLV3, WUS, and STM using genetics. Plants that carry mutations in CLV3 are characterized by an enlarged SAM, an increase in the number of lateral organs developing from the SAM and over-proliferation of floral meristems. When we combined the clv3–7 loss-of-function allele with pan, we observed a substantial enhancement of the clv3 phenotype (Figure 3A). Compared to clv3 single mutants, SAMs of pan clv3 double mutants were even further enlarged (arrowheads in Figures 3C,D) and developed even more lateral organs (Figures 3C,D). Consistent with an enhancement of meristem phenotypes by the pan mutation, we observed a drastic reduction SAM function when we combined wus and pan (Figure 3F). In contrast to wus mutants, which develop a bushy stature because of the stop and go phenotype of the meristem (Laux et al., 1996), stem cell activity in wus pan double mutants ceased after the formation of leaves and elongated shoots were never formed. Since CLV3 and WUS act in the same pathway and both showed synergistic genetic interactions with PAN, we next wondered how PAN would interact with STM, whose activity is independent of the WUS–CLV system. To our surprise we found that the stm phenotype was partially suppressed in pan stm double mutants, which developed a substantially larger number of lateral organs and shoots compared to stm plants (Figure 3G). In some cases we even observed flowers with a regular arrangement of floral organs, however these flowers remained sterile. Thus, while in the case of WUS and CLV3 PAN behaved as a molecular buffer, which is able to stabilize SAM function in the absence of other meristem regulators, this function was not observed when pan was combined with stm, suggesting that they have antagonistic activities.

Figure 3. Genetic interactions of PAN with CLV3, WUS, and STM. (A) From left to right the following genotypes are shown: wild-type, clv3, pan clv3, and pan. Top views of inflorescence apices of wild-type (B), clv3(C), pan clv3 (D), and pan (E) inflorescences. (F) Two wus mutant plants (left) are shown in comparison to two pan wus double mutants (right). Note the inhibition of shoot outgrowth in the double mutant. At later developmental stages a reduced number of shoots grows at a slow rate. (G) Two stm mutants (left) and two pan stm double mutant plants (right). Note the elevated number of shoots and branches, as well as floral buds in the pan stm double mutant.
To elucidate some of the mechanisms that could underlie these complex meristematic functions of PAN, we recorded the molecular phenotype of pan single mutants by transcript profiling. Wild-type and pan mutants were grown in LD for 25 days before we sampled two independent pools of 50 inflorescence meristems of each genotype by removing developing flowers older than stage 8. After Affymetrix Ath1 profiling we applied GC-RMA to normalize the data and derive expression values (Wu et al., 2004) followed by Rank Products to identify differentially expressed genes at a false discovery rate of 0.05 (Breitling et al., 2004). One hundred sixty transcripts showed increased abundance (Table 1), while 120 mRNAs were found to be significantly reduced in inflorescence apices of pan mutants compared to wild-type (Table 2). To obtain a first insight into the potential function of PAN downstream genes we used Gene Ontology (GO) analysis on the level of the annotation of biological function, as well as using molecular function as a readout. Interestingly, we found the “response to stimulus” category as highly enriched among the genes with increased as well as reduced expression. Among the increased mRNAs we found diverse functional sub-categories indicating that PAN plays a role in stress and environmental response (Figure 4). A prominent example was GIGANTEA (GI), whose expression is controlled by the circadian clock and whose activity is necessary for normal clock function and promotion of flowering under LD (Fowler et al., 1999; Park et al., 1999). To test if GI plays a relevant role as PAN downstream gene, we created pan gi double mutants and compared them to the respective parental genotypes. Strikingly, we found that loss-of PAN function was able to fully suppress the late flowering phenotype of gi mutants in LD (Figure 5), demonstrating that GI and PAN act in the same pathway.
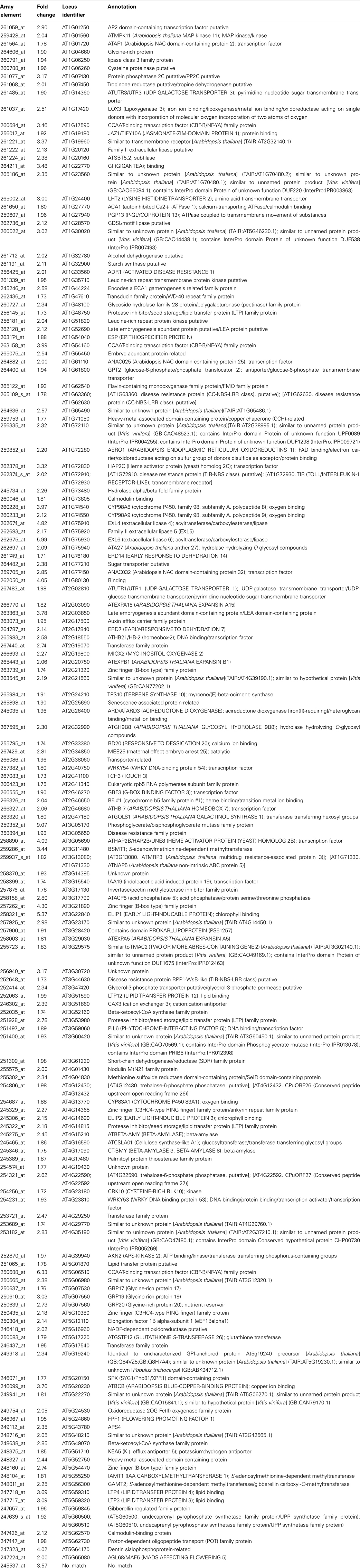
Table 1. Genes with significantly increased expression in inflorescence apices of pan mutants (Rank Products FDR 0.05).
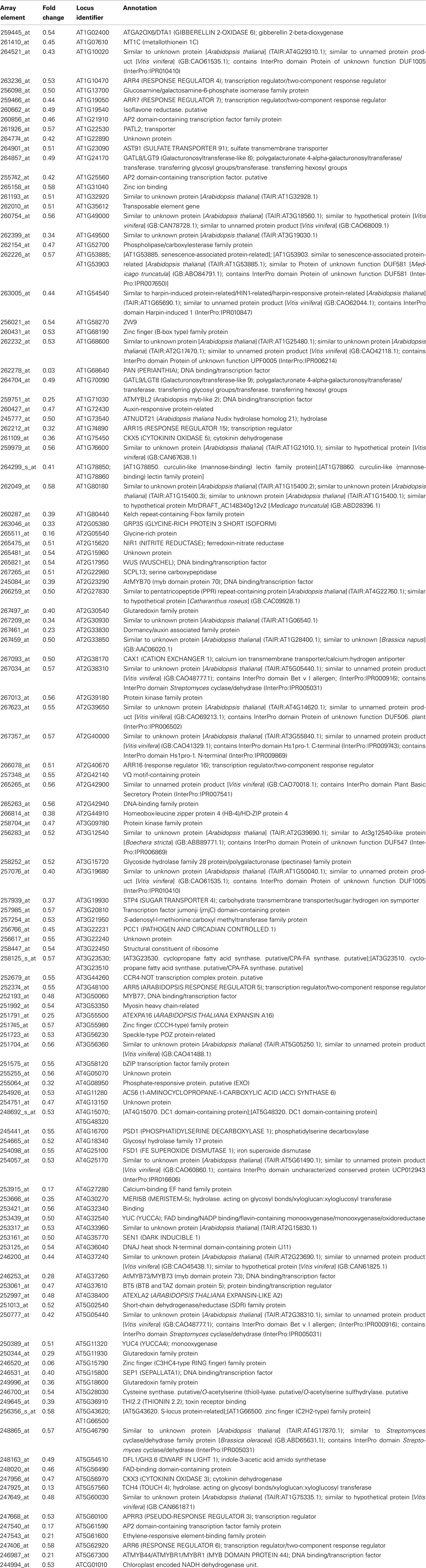
Table 2. Genes with significantly reduced expression in inflorescence apices of pan mutants (Rank Products FDR 0.05).
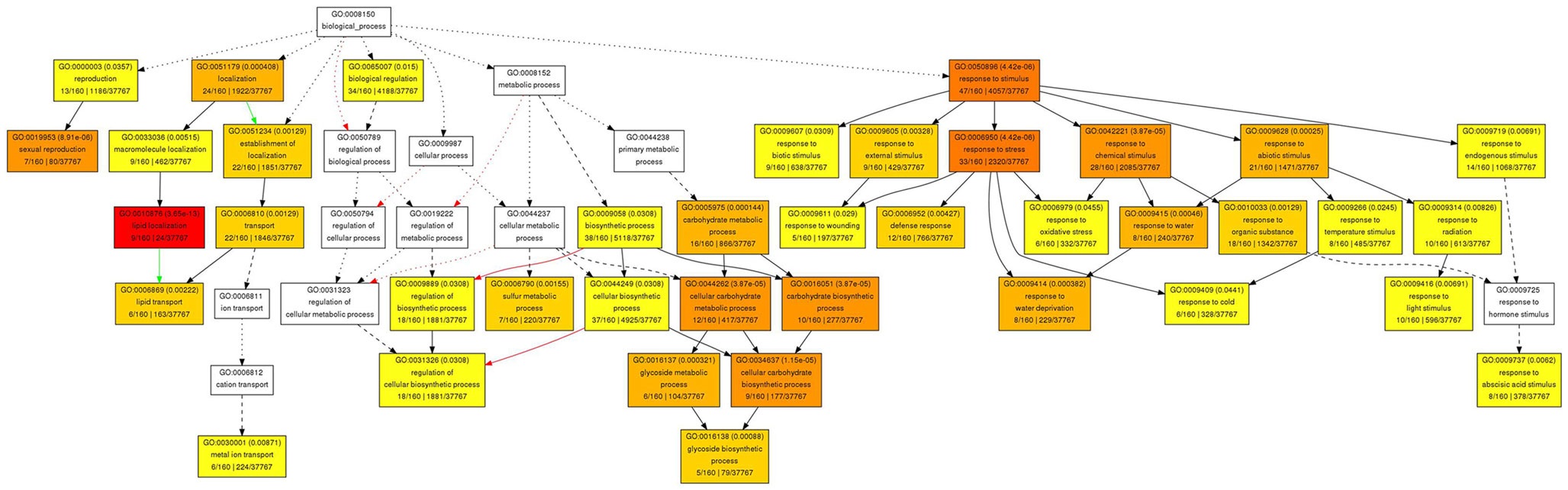
Figure 4. Gene Ontology analysis of biological functions for genes with increased expression in pan mutant inflorescence apices. Significantly enriched GO categories are shown in yellow, orange, and red.
In contrast to the rather diverse GO categories observed in the list of genes with increased expression, the reduced transcripts revealed a much more specific developmental signature. Among them we identified a substantial overrepresentation of genes with annotated functions in hormone signaling, specifically for gibberellin, ethylene, auxin and, most prominently, cytokinin response (Figure 6). This developmental signature was also apparent in the GO analysis for molecular functions with “transcription regulator activity” and “two-component response regulator activity” as the most overrepresented annotation terms (Figure 7). Two-component response regulators build the backbone of cytokinin signal transduction and response, with B-type ARRs acting as cytokinin dependent transcription factors directly upstream of A-type ARRs as immediate early cytokinin response genes with roles in negative feedback regulation (Werner and Schmülling, 2009). Strikingly, only the expression of A-type ARRs was affected in pan mutants and ARR4, ARR5, ARR6, ARR7, ARR15, and ARR16, were among the transcripts with significantly reduced abundance, a result which we independently confirmed using quantitative real-time RT-PCR (data not shown). In addition to cytokinin response genes, we identified two cytokinin oxidases, CKX3 and CKX5, as genes with reduced expression. Since CKX proteins irreversibly degrade cytokinin (Mok and Mok, 2001; Werner et al., 2003) and because A-type ARRs counteract cytokinin signaling (To et al., 2004), a reduction of their expression in pan mutants suggests that PAN acts to limit cytokinin activity in the SAM. This interpretation is consistent with the finding that SAM size is increased in pan mutants reminiscent of plants with increased cytokinin levels (Bartrina et al., 2011). In addition, we had previously identified ARR5, ARR6, ARR7, and ARR15 as direct transcriptional targets of WUS, connecting these cytokinin response genes to the core regulatory system of the SAM. While from the list of genes with reduced expression an antagonistic interaction of PAN and cytokinin could be deduced, it also suggested that PAN acts to stimulate auxin signaling, since it contained YUCCA1 and YUCCA4, two genes coding for important auxin biosynthesis enzymes (Zhao et al., 2001). Since auxin directly represses transcription of ARR7 and ARR15 via the Auxin Response Factor MONOPTEROS (Zhao et al., 2010) in the SAM, PAN could act on the expression of A-type ARRs in multiple independent pathways. Strikingly, WUS was identified among the transcriptional regulators with reduced expression, confirming that PAN is intimately connected to the SAM regulatory network.
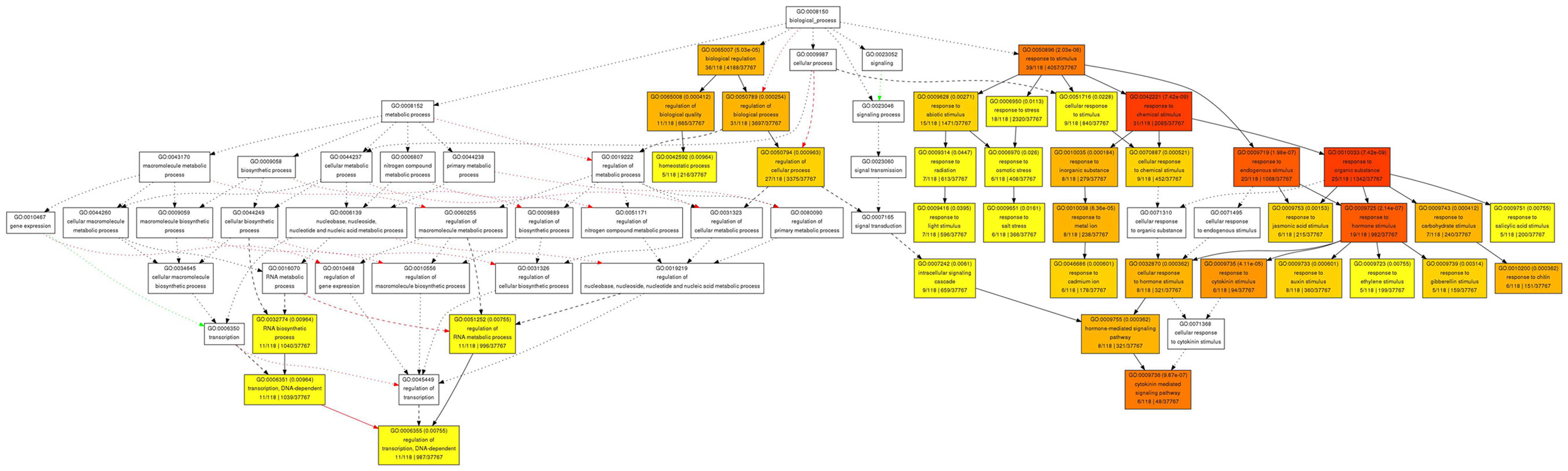
Figure 6. Gene Ontology analysis for biological function for genes with reduced expression in pan mutant inflorescence apices. Significantly enriched GO categories are shown in yellow, orange, and red.
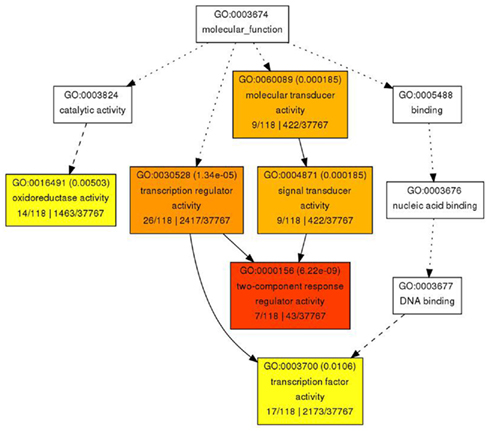
Figure 7. Gene Ontology analysis for molecular function for genes with reduced expression in pan mutant inflorescence apices. Significantly enriched GO categories are shown in yellow, orange, and red.
Having identified cytokinin and auxin signaling as major downstream effector pathways of PAN we next addressed the functional relevance of these regulatory interactions using genetics. We focused our analysis on ARR7 and ARR15, since both of them were shown to have important meristematic functions (Leibfried et al., 2005; Zhao et al., 2010), and combined these mutants (Figures 8D,E) with pan (Figure 8B) and clv3 (Figure 8C) in double and triple mutant combinations. While single A-type arr mutants have no phenotypes or very mild ones (Figures 8D,E; To et al., 2004), combination of arr7 and arr15 with pan lead to severe growth retardation (Figures 8G,H). Interestingly, while removing CLV3 function in the pan background lead to massive over-proliferation and meristem expansion beyond the regular clv3 defect (Figures 3B–E), this phenotype was completely suppressed in the pan clv3 arr7 combination (Figures 8F–I). However, the growth retardation was only transient and pan arr15 as well as pan arr15 clv3 plants recovered after about 2 weeks and developed plants with pentameric flowers, which closely resembled pan clv3 mutants. This capacity to overcome A-type ARR related developmental defects was also observed in plants carrying an over-activated form of ARR7 (Leibfried et al., 2005) and suggest that the cytokinin signaling system has a strong ability to adapt to perturbations. Mutation of multiple A-type ARRs, such as in an arr7 arr15 double mutant did not cause the phenotypes observed in the pan arr combinations (Figure 8J) underlining the important role of PAN in the SAM. Having observed a strong genetic interaction of PAN with components of the cytokinin response, we next tested its ability to modify auxin related defects. To this end we analyzed the interaction of PAN with PINFORMED-1 (PIN1), the major auxin efflux carrier responsible for generating local auxin maxima at the periphery of the SAM and thus organ initiation during shoot development (Gälweiler et al., 1998; Reinhardt et al., 2000). While pin1 mutants rarely developed flowers under our growth conditions (Figures 9A,C), pin1 pan double mutants exhibited a significantly increased number of flowers (Figures 9B,C), which were deformed and generally sterile. Again, as in the case of cytokinin signaling, these results demonstrated that PAN is able to modulate auxin dependent developmental functions, in line with the hypothesis that PAN might act as a multifunctional hub for diverse meristematic functions.
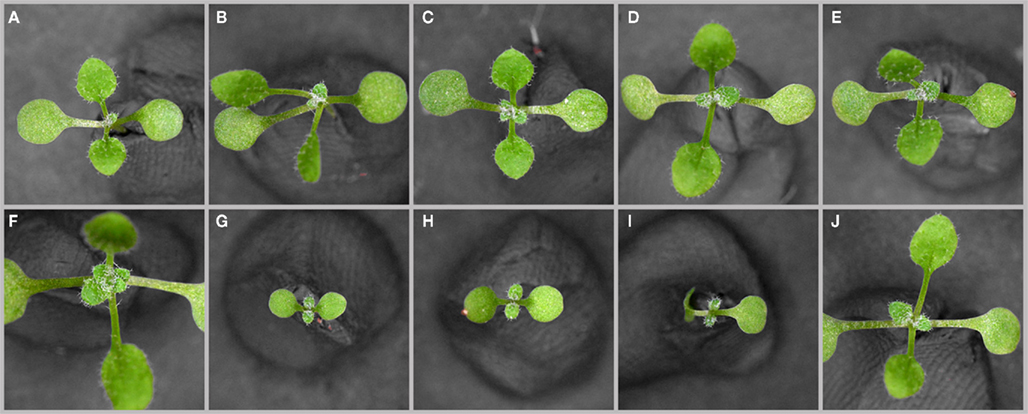
Figure 8. Genetic interaction of PAN with Cytokinin Signaling Components ARR7, ARR15, and CLV3. Ten-days-old soil grown seedlings of wild-type (A) and pan(B), clv3 (C), arr15 (D), arr7 (E) pan clv3–7 (F), pan arr7 (G), pan arr15 (H), pan arr15 clv3 (I), and arr7 arr15 (J) mutant plants. All plants were grown under LD and representative seedlings are shown.

Figure 9. Genetic interaction of PAN with PIN1. Primary shoot of pin1 mutant (A) and pan pin1 double mutant (B) and whole plant comparison (C) of pin1 (left) and pan pin1 (right) showing increased development of floral buds on the primary shoot of the pan pin1 double mutant.
Summary and Outlook
Taken together, we have shown here by molecular phenotyping and genetics that PAN is connected to a plethora of diverse input pathways and may act as an integrator to buffer shoot meristem activity. PAN inputs include pathways for environmental sensing, such as day-length and other abiotic factors, as well as hard-wired developmental circuitries, such as the WUS–CLV system. Strikingly, the same holds true for the PAN output network, which we found to include components of the circadian clock and stress response as examples for modulating environmental interactions. Furthermore, PAN downstream genes showed a strong developmental signature, which was most apparently represented by a number of plant hormone signaling systems. Based on our results we suggest that PAN might act as a node between cytokinin and auxin signaling pathways, with cytokinin outputs being repressed and auxin activity being induced by PAN. PAN is a member of the D-class of bZIP transcription factors (Jakoby et al., 2002) and thus groups with the TGA regulators, which are involved in mediating pathogen defense (Zander et al., 2010). The sequence similarity of PAN and TGA pathogen response regulators suggests that PAN function might have evolved from an environmental surveillance activity, which was enhanced to include developmental roles to give rise to an integrated buffering system.
Materials and Methods
Plant Material
Arabidopsis thaliana plants of the Columbia (Col-0) background were grown on soil at 23°C. Analyses were performed after growth under three different light conditions: CL, LD (16 h of light), or SD (8 h light) for 10 days for seedlings and 25 days for vegetative and reproductive tissues if not noted otherwise. The following mutant alleles used: arr7 (At1G19050): WiscDsLox485–488B15; arr15 (At1G74890): WiscDSLox334D02; clv3 (At2G27250): clv3–7; pan (AT1G68640): Salk N557190; wus (At2G17950): wus-4 in Columbia background (wus-mh; Leibfried et al., 2005); stm (At1G62360): GABI-Kat line 100F11; pin1 (At1G73590): GABI-Kat line 051A10; gi (At1G22770): gi-201. Phenotypic characterizations were carried out by growing mutants and controls at least three times independently and analyzing a total of at least 30 individuals for each genotype. Representative plants are shown.
In situ Hybridizations
Plant material was fixed and embedded using a Leica ASP300 and hybridized following standard protocols (Weigel and Glazebrook, 2002) adding 10% polyvinylalcohol (PVA) to the staining solution. Digoxigenin-labeled full-length RNA riboprobes were synthesized for CLV3 (At2G27250), PAN (AT1G68640), STM (At1G62360), and WUS (At2G17950) as described in Geier et al. (2008) and Maier et al. (2009) according to the manufacturer instruction (Roche).
Microarray Experiments
Pools of 50 microscopically dissected inflorescence apices of pan mutants and wild-type both carrying the KB14 AG::GUS reporter gene (Busch et al., 1999; Lohmann et al., 2001) were grown for 25 days in LD conditions and profiled in duplicate using the Affymetrix ATH1 platform. RNA extraction and microarray analyses were performed as described (Schmid et al., 2005; Buechel et al., 2010). Expression estimates were derived by GC-RMA (Wu et al., 2004) at standard settings implemented in R. We determined significant changes on a per-gene level by applying the Rank products algorithm (Breitling et al., 2004) using 100 permutations and a false discovery rate cut-off of 5%. GO analysis was carried out using AgriGO (Du et al., 2010).
Quantitative Real-Time PCR
Total RNA was extracted from apices of plants grown in an independent experiment using RNeasy Mini columns with on-column DNAse digestion (Qiagen). Reverse transcription was performed with 1 μg of total RNA, using a Reverse Transcription Kit (Fermentas). PCR amplification was carried out in the presence of the double-strand DNA-specific dye SYBR Green (Molecular Probes) using intron spanning primers (Andersen et al., 2008). Amplification was monitored in real-time with the Opticon Continuous Fluorescence Detection System (MJR). BETA-TUBULIN-2 transcript levels served to normalize mRNA measurements.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Patrice Salome for seeds of the gi-201 mutant, Martin Vötsch for support in preparing figures, Sascha Laubinger for discussion and Gerd Jürgens and Detlef Weigel for continuous support and discussion. This work was supported by fellowships from the Konrad Adenauer Stiftung (Annette T. Maier) DFG-AFGN grant LO1450/2-1 (Jan U. Lohmann), as well as funds from the EMBO Young Investigator Program and the HFSP Career Development Award to Jan U. Lohmann, and the Max Planck Society.
References
Andersen, S. U., Buechel, S., Zhao, Z., Ljung, K., Novak, O., Busch, W., Schuster, C., and Lohmann, J. U. (2008). Requirement of B2-type cyclin-dependent kinases for meristem integrity in Arabidopsis thaliana. Plant Cell 20, 88–100.
Barton, M., and Poethig, R. (1993). Formation of the shoot apical meristem in Arabidopsis thaliana – an analysis of development in the wild-type and in the shoot meristemless mutant. Development 119, 823–831.
Barton, M. K. (2010). Twenty years on: the inner workings of the shoot apical meristem, a developmental dynamo. Dev. Biol. 341, 95–113.
Bartrina, I., Otto, E., Strnad, M., Werner, T., and Schmülling, T. (2011). Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 23, 69–80.
Brand, U., Fletcher, J. C., Hobe, M., Meyerowitz, E. M., and Simon, R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619.
Breitling, R., Armengaud, P., Amtmann, A., and Herzyk, P. (2004). Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573, 83–92.
Buechel, S., Leibfried, A., To, J. P. C., Zhao, Z., Andersen, S. U., Kieber, J. J., and Lohmann, J. U. (2010). Role of A-type Arabidopsis response regulators in meristem maintenance and regeneration. Eur. J. Cell Biol. 89, 279–284.
Busch, M. A., Bomblies, K., and Weigel, D. (1999). Activation of a floral homeotic gene in Arabidopsis. Science 285, 585–587.
Busch, W., Miotk, A., Ariel, F. D., Zhao, Z., Forner, J., Daum, G., Suzaki, T., Schuster, C., Schultheiss, S. J., Leibfried, A., Haubeiss, S., Ha, N., Chan, R. L., and Lohmann, J. U. (2010). Transcriptional control of a plant stem cell niche. Dev. Cell 18, 849–861.
Chuang, C. F., Running, M. P., Williams, R. W., and Meyerowitz, E. M. (1999). The PERIANTHIA gene encodes a bZIP protein involved in the determination of floral organ number in Arabidopsis thaliana. Genes Dev. 13, 334–344.
Clark, S. E., Running, M. P., and Meyerowitz, E. M. (1993). CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119, 397–418.
Clark, S. E., Running, M. P., and Meyerowitz, E. M. (1995). CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121, 2057–2067.
Das, P., Ito, T., Wellmer, F., Vernoux, T., Dedieu, A., Traas, J., and Meyerowitz, E. M. (2009). Floral stem cell termination involves the direct regulation of AGAMOUS by PERIANTHIA. Development 136, 1605–1611.
Du, Z., Zhou, X., Ling, Y., Zhang, Z., and Su, Z. (2010). agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38, W64–W70.
Fowler, S., Lee, K., Onouchi, H., Samach, A., Richardson, K., Morris, B., Coupland, G., and Putterill, J. (1999). GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18, 4679–4688.
Furutani, M., Vernoux, T., Traas, J., Kato, T., Tasaka, M., and Aida, M. (2004). PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development 131, 5021–5030.
Gälweiler, L., Guan, C., Müller, A., Wisman, E., Mendgen, K., Yephremov, A., and Palme, K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282, 2226–2230.
Geier, F., Lohmann, J. U., Gerstung, M., Maier, A. T., Timmer, J., and Fleck, C. (2008). A quantitative and dynamic model for plant stem cell regulation. PLoS ONE 3, e3553. doi: 10.1371/journal.pone.0003553
Hepworth, S. R., Zhang, Y., McKim, S., Li, X., and Haughn, G. W. (2005). BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell 17, 1434–1448.
Jakoby, M., Weisshaar, B., Dröge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., Parcy, F., and bZIP Research Group. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111.
Jasinski, S., Piazza, P., Craft, J., Hay, A., Woolley, L., Rieu, I., Phillips, A., Hedden, P., and Tsiantis, M. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15, 1560–1565.
Kayes, J. M., and Clark, S. E. (1998). CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 125, 3843–3851.
Laux, T., Mayer, K. F., Berger, J., and Jürgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122, 87–96.
Leibfried, A., To, J. P. C., Busch, W., Stehling, S., Kehle, A., Demar, M., Kieber, J. J., and Lohmann, J. U. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438, 1172–1175.
Lohmann, J. U., Hong, R. L., Hobe, M., Busch, M. A., Parcy, F., Simon, R., and Weigel, D. (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105, 793–803.
Long, J. A., Moan, E. I., Medford, J. I., and Barton, M. K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379, 66–69.
Maier, A. T., Stehling-Sun, S., Wollmann, H., Demar, M., Hong, R. L., Haubeiss, S., Weigel, D., and Lohmann, J. U. (2009). Dual roles of the bZIP transcription factor PERIANTHIA in the control of floral architecture and homeotic gene expression. Development 136, 1613–1620.
Mayer, K. F., Schoof, H., Haecker, A., Lenhard, M., Jürgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815.
Mok, D. W., and Mok, M. C. (2001). Cytokinin metabolism and action. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 89–118.
Parcy, F., Nilsson, O., Busch, M. A., Lee, I., and Weigel, D. (1998). A genetic framework for floral patterning. Nature 395, 561–566.
Park, D. H., Somers, D. E., Kim, Y.-S., Choy, Y. H., Lim, H. K., Soh, M. S., Kim, H. J., Kay, S. A., and Nam, H. G. (1999). Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285, 1579–1582.
Reinhardt, D., Mandel, T., and Kuhlemeier, C. (2000). Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12, 507–518.
Running, M. P., and Meyerowitz, E. M. (1996). Mutations in the PERIANTHIA gene of Arabidopsis specifically alter floral organ number and initiation pattern. Development 122, 1261–1269.
Schmid, M., Davison, T. S., Henz, S. R., Pape, U. J., Demar, M., Vingron, M., Schölkopf, B., Weigel, D., and Lohmann, J. U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37, 501–506.
Schoof, H., Lenhard, M., Haecker, A., Mayer, K. F., Jürgens, G., and Laux, T. (2000). The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644.
To, J. P. C., Haberer, G., Ferreira, F. J., Deruère, J., Mason, M. G., Schaller, G. E., Alonso, J. M., Ecker, J. R., and Kieber, J. J. (2004). Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16, 658–671.
Weigel, D., Alvarez, J., Smyth, D. R., Yanofsky, M. F., and Meyerowitz, E. M. (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69, 843–859.
Weigel, D., and Glazebrook, J. (2002). Arabidopsis: A Laboratory Manual, 1st Edn. Cold Spring Harbor, NY: CSHL Press.
Werner, T., Motyka, V., Laucou, V., Smets, R., Van Onckelen, H., and Schmülling, T. (2003). Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15, 2532–2550.
Werner, T., and Schmülling, T. (2009). Cytokinin action in plant development. Curr. Opin. Plant Biol. 12, 527–538.
Wu, Z., Irizarry, R., Gentleman, R., Martinez-Murillo, F., and Spencer, F. (2004). A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 99, 909–917.
Yanai, O., Shani, E., Dolezal, K., Tarkowski, P., Sablowski, R., Sandberg, G., Samach, A., and Ori, N. (2005). Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 15, 1566–1571.
Zander, M., La Camera, S., Lamotte, O., Métraux, J.-P., and Gatz, C. (2010). Arabidopsis thaliana class-II TGA transcription factors are essential activators of jasmonic acid/ethylene-induced defense responses. Plant J. 61, 200–210.
Zhao, Y., Christensen, S. K., Fankhauser, C., Cashman, J. R., Cohen, J. D., Weigel, D., and Chory, J. (2001). A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291, 306–309.
Keywords: Arabidopsis, meristem regulation, stem cells, auxin, cytokinin, PERIANTHIA, type-A ARR, SHOOTMERISTEMLESS
Citation: Maier AT, Stehling-Sun S, Offenburger S-L and Lohmann JU (2011) The bZIP transcription factor PERIANTHIA: a multifunctional hub for meristem control. Front. Plant Sci. 2:79. doi: 10.3389/fpls.2011.00079
Received: 30 September 2011; Paper pending published: 17 October 2011;
Accepted: 26 October 2011; Published online: 21 November 2011.
Edited by:
Andreas P. M. Weber, University of Duesseldorf, GermanyReviewed by:
Yvonne Stahl, Heinrich-Heine University, GermanyKay Schneitz, Technische Universität München, Germany
Copyright: © 2011 Maier, Stehling-Sun, Offenburger and Lohmann. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Jan U. Lohmann, Department of Stem Cell Biology, Centre for Organismal Studies, University of Heidelberg, Im Neuenheimer Feld 230, D-69120 Heidelberg, Germany. e-mail: jlohmann@meristemania.org
†Present address: Sandra Stehling-Sun, University of Washington, Seattle, WA, USA.
 Annette T. Maier1,2
Annette T. Maier1,2