- Department of Biochemistry and Cell Biology, State University of New York, Stony Brook, NY, USA
The SCF (SKP1-CUL1-F-box protein) ubiquitin ligase complex mediates polyubiquitination of proteins targeted for degradation, thereby controlling a plethora of biological processes in eukaryotic cells. Although this ubiquitination machinery is found and functional only in eukaryotes, many non-eukaryotic pathogens also encode F-box proteins, the critical subunits of the SCF complex. Increasing evidence indicates that such non-eukaryotic F-box proteins play an essential role in subverting or exploiting the host ubiquitin/proteasome system for efficient pathogen infection. A recent bioinformatic analysis has identified more than 70 F-box proteins in 22 different bacterial species, suggesting that use of pathogen-encoded F-box effectors in the host cell may be a widespread infection strategy. In this review, we focus on plant pathogen-encoded F-box effectors, such as VirF of Agrobacterium tumefaciens, GALAs of Ralstonia solanacearum, and P0 of Poleroviruses, and discuss the molecular mechanism by which plant pathogens use these factors to manipulate the host cell for their own benefit.
Introduction
Diverse pathogens have evolved virulence factors that mimic host cell functions (Elde and Malik, 2009). This molecular mimicry, presumably acquired through either divergent or convergent evolution, is indispensible for pathogens to exploit or subvert the host cellular processes for efficient infection. A fascinating example of the molecular mimicry is pathogen-encoded F-box proteins. As a component of the SCF (SKP1-CUL1-F-box protein) ubiquitin ligase complex, F-box proteins mediate polyubiquitination of target proteins and the subsequent proteasome-dependent protein degradation in eukaryotic cells (Petroski and Deshaies, 2005; Lechner et al., 2006; Vierstra, 2009; Hua and Vierstra, 2011). Surprisingly, an F-box-coding gene was also found in Agrobacterium tumefaciens, a bacterial pathogen that causes neoplastic growths and crown gall disease in plants (Schrammeijer et al., 2001). Considering that prokaryotes possess neither the ubiquitin/26S proteasome system (UPS) nor the functional SCF ubiquitin ligase complex, the Agrobacterium-encoded F-box protein presumably does not function in the bacterial cell. Rather, Agrobacterium translocates this F-box effector into plant cell (Vergunst et al., 2000, 2005) and hijacks the host SCF complex to facilitate bacterial infection (Tzfira et al., 2004). Since this discovery, similar F-box-like effector proteins have been described in many other viral and bacterial pathogens, including a human pathogen Legionella pneumophila (Price et al., 2009; Lomma et al., 2010). Furthermore, a recent bioinformatic analysis identified at least 74 putative F-box proteins encoded by 22 different bacterial species, most of which are known pathogens (Price and Kwaik, 2010). This finding further suggests the importance and widespread utilization of pathogen-derived F-box proteins for the infection strategy.
Like animals, plants face a challenge to adapt their development and growth to a rapidly changing environment. In particular, preparing for a potential threat from a wide array of pathogens is crucial for survival of plants. Increasing evidence suggests that plants utilize the UPS to recognize and combat pathogen invasion (Zeng et al., 2006; Citovsky et al., 2009). As a part of this defense strategy, plants often exploit the SCF ubiquitin ligase complexes, thereby targeting negative regulators of their own defense response and/or pathogen-derived proteins for degradation (Zeng et al., 2006; Citovsky et al., 2009). Notably, plants encode an unusually large number of F-box proteins, the substrate specificity module of the SCF complex (Gagne et al., 2002; Hua and Vierstra, 2011). For example, the model plant Arabidopsis thaliana possesses almost 700 F-box genes, which represent almost 2.3% of the protein-coding genes (Gagne et al., 2002; Hua and Vierstra, 2011). By comparison, fruit flies and humans encode only 27 and 69 F-box proteins, respectively (Hua and Vierstra, 2011). Furthermore, a maximum-likelihood analysis of codon evolution predicted that most of plant F-box genes are likely subject to positive selection specifically in the C-terminal substrate-binding domains (Thomas, 2006). Such site-specific positive selection in plant F-box genes as well as their high degree of diversity are reminiscent of the major histocompatibility complex (MHC) molecules (Hughes and Nei, 1988, 1989a,b), the membrane-associated proteins that activate immune responses in vertebrates by binding fragments of foreign proteins (i.e., antigens). Based on these evolutionary features shared by plant F-box genes and MHC molecules, it is tempting to speculate that plants may have evolved a diverse array of F-box proteins (hence, a wide variety of different SCF complexes) to enable broad protection against innumerable invading pathogens. If this is the case, it is not surprising that plant pathogens, in turn, have evolved a counter-defense strategy using a molecular mimic of F-box proteins to disrupt or co-opt the defense-associated SCF machinery of the host plants. Although only a few phytopathogen-encoded F-box proteins have been intensively studied thus far, understanding the role of such an F-box effector during the corresponding pathogen infection process is highly informative to illustrate the molecular arms race between host plants and pathogens. Here, we summarize the basic concepts of the UPS as well as the SCF ubiquitin ligase complex, and provide several case studies on plant pathogen-derived F-box proteins, including VirF of A. tumefaciens, GALAs of Ralstonia solanacearum, and P0 of Poleroviruses.
The Ubiquitin/26S Proteasome System
Post-translational modifications regulate the molecular function of target proteins by modulating their activity, stability, localization, and affinity to other molecules. Among many known post-translational modifications, addition of ubiquitin moieties (i.e., ubiquitination) is known to be a highly complex process regulated by numerous cellular factors, and plays an essential role in diverse biological processes.
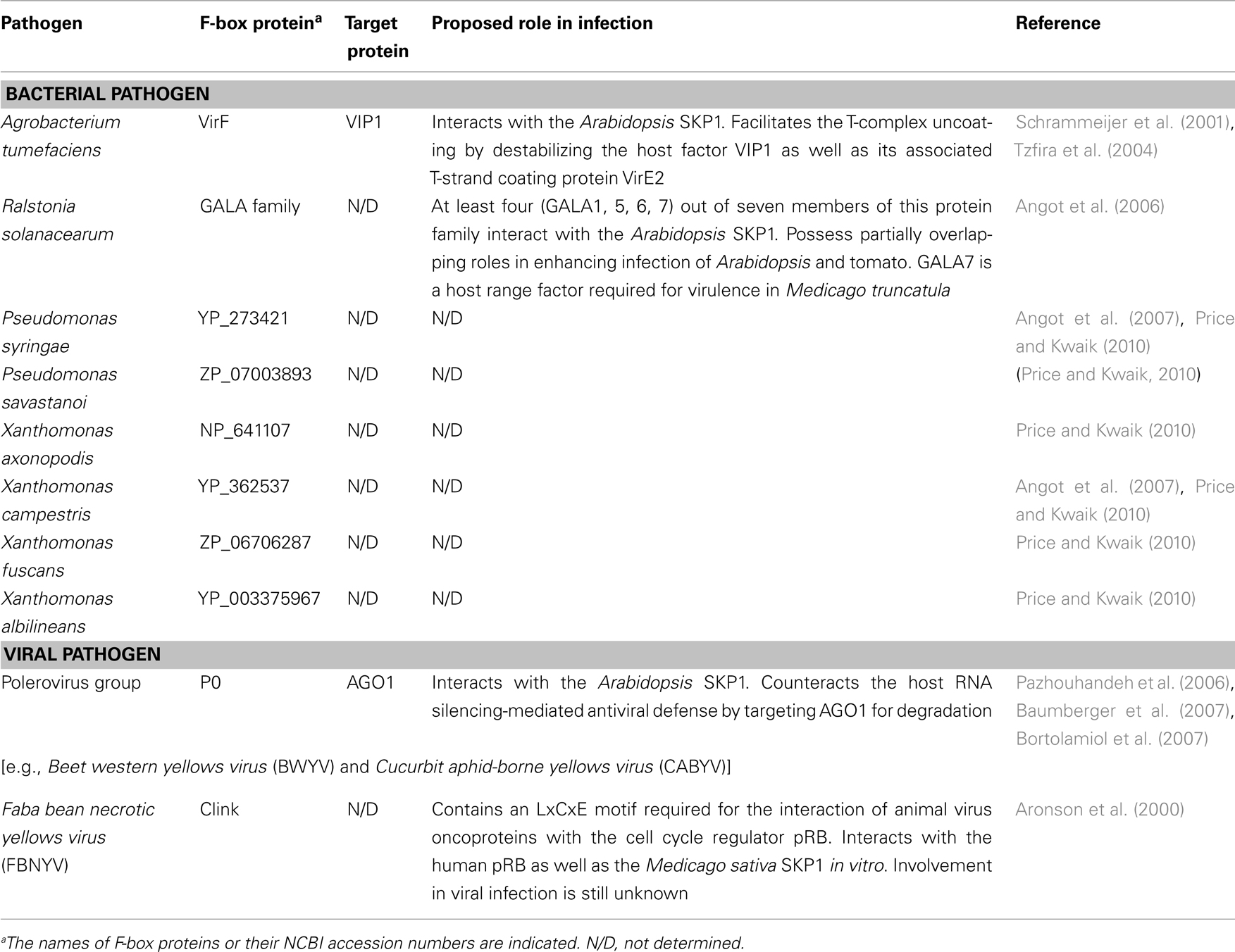
Ubiquitin is a 76-amino acid polypeptide that is highly conserved among eukaryotes. As its name suggests, ubiquitin is ubiquitously expressed in virtually all types of eukaryotic cells. Covalent attachment of ubiquitin to its target protein is mediated by a reaction cascade involving three classes of enzymes: ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin ligases (E3) (Petroski and Deshaies, 2005; Vierstra, 2009; Hua and Vierstra, 2011; Figure 1). The sequential reactions start with ATP-dependent activation of ubiquitin by an E1 enzyme, resulting in formation of a high energy thioester linkage between a cysteine residue present in E1 and the C-terminal glycine residue of ubiquitin. This activated ubiquitin is then transferred to another cysteine residue in an E2 enzyme, and subsequently conjugated to a lysine residue in the target protein with the help of a substrate-specific E3 ligase. E3 ligases are the largest and most diverse class of ubiquitinating enzymes, which is prominent especially in plants. For example, A. thaliana possesses more than 1,500 different E3 enzymes (Hua and Vierstra, 2011), suggesting that E3 ligases have evolved to regulate a wide spectrum of endogenous and foreign proteins through ubiquitination. E3 ligases are classified into two major types: the HECT-type and the RING finger-type ubiquitin ligases (Figure 1). The HECT-type ligases accomplish ubiquitin ligation by two steps, where the activated ubiquitin first becomes covalently attached to a cysteine residue in the HECT domain of the E3 ligase and then transferred to the final substrate. On the other hand, the RING finger-type ligases, which include the SCF complex, transfer the activated ubiquitin directly from the E2 enzyme to the target protein.
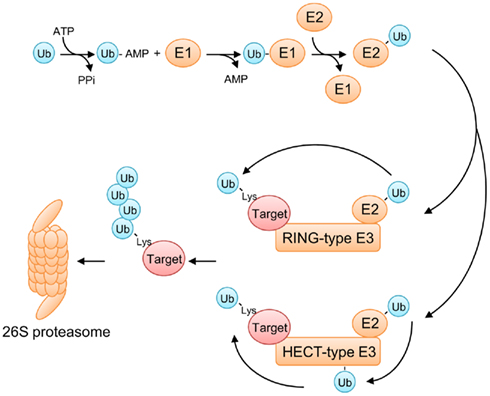
Figure 1. The ubiquitin/26S proteasome system (UPS). Ubiquitination is accomplished by sequential reactions involving three classes of enzymes (E1, E2, and E3). First, a ubiquitin-activating enzyme (E1) activates a ubiquitin molecule in an ATP-dependent manner, resulting in formation of a high energy thioester linkage between E1 and the C-terminal glycine residue of ubiquitin. The activated ubiquitin is then transferred to a ubiquitin-conjugating enzyme (E2), and subsequently conjugated to a lysine residue in the target protein with the help of a substrate-specific ubiquitin ligase (E3). E3 ligases are classified into two groups, depending on their mode of action. RING finger-type E3 ligases transfer a ubiquitin moiety directly from E2 to a lysine residue of the target protein. HECT-type E3 ligases conjugate ubiquitin in two steps: first, a ubiquitin moiety is transferred from E2 to the HECT domain of a HECT-type E3 ligase and, then, to a lysine residue of the target protein. Lys11- or Lys48-linked polyubiquitination targets the substrate for 26S proteasome-dependent protein degradation.
In many cases, ubiquitin polymers are assembled by reiterative rounds of ubiquitination, where the first ubiquitin monomer is conjugated to a lysine residue of the target protein, followed by attachment of additional ubiquitin monomers to any of 7 lysine residues available in the previously attached ubiquitin molecule (Figure 1). The type of linkage in a polyubiquitin chain determines the fate of the substrate protein. The best characterized polyubiquitin chains thus far are Lys11-linked and Lys48-linked chains. Substrates with these polyubiquitin chains are directly recognized and degraded by the 26S proteasome, a very large protein complex of 2.5 MDa that catalyzes proteolysis (Figure 1). This UPS not only facilitates removal of misfolded proteins, but also regulates a plethora of cellular processes by targeting regulatory proteins for selective degradation. Furthermore, the UPS is also known to play a key role in host immune responses against invading pathogens (Zeng et al., 2006; Citovsky et al., 2009). For instance, some plant virus-encoded movement proteins (MPs), the essential factors for the cell-to-cell spread of viral infection, are targeted for degradation by the UPS (Reichel and Beachy, 2000; Drugeon and Jupin, 2002). Similarly, the viral RNA-dependent RNA polymerase (RdRp), which is required for genome replication of positive-strand RNA viruses, is also degraded by the UPS in plant cells (Camborde et al., 2010). These examples illustrate the importance of the defensive role of the UPS in host–pathogen interactions and imply that the host UPS represents a challenge that pathogens need to overcome for successful infection.
The SCF Complex
The role of E3 ubiquitin ligases in the UPS is substantial as these enzymes determine substrate specificity by selectively recruiting target proteins to the ubiquitination machinery. Among the E3 families, the SCF complex is by far the largest and best characterized class of E3 ligases. The SCF complex is comprised of cullin 1 (CUL1), S-phase kinase-associated protein 1 (SKP1), RING-box 1 (RBX1), and an F-box protein (Petroski and Deshaies, 2005; Lechner et al., 2006; Vierstra, 2009; Hua and Vierstra, 2011; Figure 2). In this multi-protein complex, CUL1 serves as a scaffold that tethers SKP1 and RBX1 to its N-terminal and C-terminal domains, respectively. An F-box protein, which confers substrate specificity to the SCF complex, directly interacts with SKP1 via its F-box domain whereas RBX1 functions as the docking site for E2 ubiquitin-conjugating enzymes. Most, but not all, F-box proteins harbor an additional interaction domain that brings specific target proteins in close proximity to the catalytic core of the SCF complex (Gagne et al., 2002).
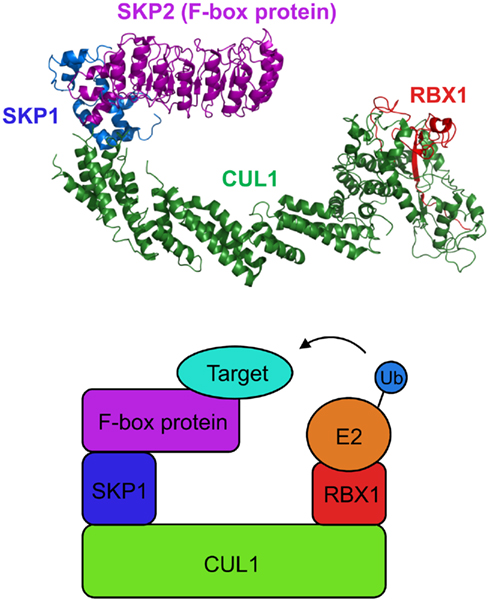
Figure 2. Structure of the SCF complex. The crystal structure of the yeast SCFSKP2 complex (top; Zheng et al., 2002; Hao et al., 2005; assembled from Protein Data Bank files 1LDK and 2ASS) and a simplified cartoon of the SCF complex (bottom). The N-terminal half and the C-terminal half of CUL1 interact with SKP1 and RBX1, respectively. RBX1 serves as a docking site for an E2 enzyme. An F-box protein interacts with SKP1 via its F-box domain and recruits the target protein to the ubiquitin ligase complex.
In plants, the SCF complex has been shown to regulate a multitude of developmental processes (Lechner et al., 2006; Vierstra, 2009; Hua and Vierstra, 2011). Indeed, different SCF complexes are involved in signaling pathways of almost all plant hormones, including auxin, gibberellins, jasmonic acid, etc. (Santner and Estelle, 2010). In addition, the SCF complex plays a critical role in plant pathogen interactions (Zeng et al., 2006; Citovsky et al., 2009). One such example is the involvement of the ACIF1 (Avr9/Cf-9-Induced F-box1)-containing SCF complex (SCFACIF1) in disease resistance in tomato and tobacco (van den Burg et al., 2008). Silencing of the ACIF1 gene in tobacco compromises the host defense responses to Tobacco mosaic virus (TMV) as well as to the bacterial pathogen Pseudomonas syringae (van den Burg et al., 2008). In tomato, knockdown of ACIF1 attenuates resistance against the pathogenic fungus Cladosporium fulvum (van den Burg et al., 2008). The Arabidopsis homologs of ACIF1 are also implicated for defense responses as simultaneous knockdown of three ACIF1 homologs alters expression of defense-responsive genes, including those coding for pathogenesis-related (PR) proteins (van den Burg et al., 2008). Thus, the F-box protein ACIF1 and the SCFACIF1 complex may serve as a conserved, integral part of plant defense against various pathogens, from viruses to bacteria to fungi.
The role of the SCF complex in plant pathogen interactions is also exemplified by the rice defense-related F-box (OsDRF1) protein (Cao et al., 2008). Expression of the OsDRF1 transcripts is induced by infection of the rice blast fungus Magnaporthe grisea, as well as by treatment of benzothiadiazole (BTH), a chemical inducer of plant defense responses (Cao et al., 2008). Interestingly, overexpression of OsDRF1 in tobacco transgenic plants leads to enhanced disease resistance to Tomato mosaic virus (ToMV) and P. syringae (Cao et al., 2008). These results suggest that the potential OsDRF1-containing SCF complex (SCFOsDRF1) may positively regulate plant defense responses against diverse pathogens.
The SCF complex functions not only as a positive regulator but also as a negative regulator of plant defense responses. For instance, the Arabidopsis F-box protein CPR1 (Constitutive Expresser of PR Genes 1, also known as CPR30) is involved in negative regulation of plant immunity via the UPS (Gou et al., 2009, 2011; Cheng et al., 2011). The CPR1-containing SCF complex (SCFCPR1) mediates degradation of at least two resistance (R) proteins, SNC1 (Suppressor of npr1-1 Constitutive 1) and RPS2 (Resistance to P. syringae 2), both of which are critical immune receptors that recognize pathogen invasion (Cheng et al., 2011; Gou et al., 2011). Consistent with this negative role of SCFCPR1, loss-of-function mutations in CPR1 lead to enhanced resistance to P. syringae (Gou et al., 2009, 2011) as well as the oomycete pathogen Hyaloperonospora arabidopsidis (Cheng et al., 2011).
Utilization of the SCF complex in plant defense responses should exert selective pressure on pathogens toward evolution of counter-defense strategies by which they can subvert or co-opt the host SCF machinery. Indeed, increasing evidence suggests that several plant pathogens, such as A. tumefaciens, R. solanacearum, and Poleroviruses, hijack the host SCF complex for their own benefits by translocating or expressing their own F-box effectors in the host cell (Tzfira et al., 2004; Angot et al., 2006; Baumberger et al., 2007; Bortolamiol et al., 2007; Table 1). Moreover, recent bioinformatic analyses revealed that many other plant pathogens also encode F-box-like proteins (Angot et al., 2007; Price and Kwaik, 2010; Table 1), suggesting that hijacking of the host SCF machinery through this molecular mimicry may be a widespread infection strategy. To better understand the plant pathogen molecular arms race revolving around the SCF complex, we summarize recent advances in the studies of phytopathogen-encoded F-box effectors.
VirF of Agrobacterium
Agrobacterium tumefaciens-encoded VirF is the first F-box protein that was identified in prokaryotes and proved functional in eukaryotic host cells (Schrammeijer et al., 2001; Tzfira et al., 2004). Agrobacterium is a phytopathogenic soil bacterium that causes neoplastic growths (crown gall tumors) on various plant species by transferring and integrating its own genetic materials into the host genomes. This Agrobacterium-mediated genetic transformation has been considered to represent the only known natural example of trans-kingdom gene transfer until a recent study using cultured human cells showed that the zoonotic pathogen Bartonella henselae is also capable of mobilizing its DNA into the eukaryotic host (Schröder et al., 2011). Furthermore, under laboratory conditions, Agrobacterium can genetically transform virtually any eukaryotic species, from fungi to human cells (Bundock et al., 1995; Piers et al., 1996; Kunik et al., 2001; Lacroix et al., 2006).
The infection process of Agrobacterium is regulated by many bacterial factors as well as the host cellular components (Tzfira and Citovsky, 2006; Gelvin, 2010; Pitzschke and Hirt, 2010; Figure 3). First, Agrobacterium activates expression of virulence (Vir) proteins in response to phenolic compounds exuded from wounded plant tissues. Among the induced Vir proteins, VirD1 and VirD2 mediate generation of a single-stranded copy of transferred DNA (T-DNA), a specific DNA segment of the bacterial tumor-inducing (Ti) plasmid. The resulting mobilized single-stranded (ss) DNA molecule, termed T-strand, is then delivered into the host cell via the type IV secretion system composed of the VirB and VirD4 proteins. Within the plant cell, the T-strand is thought to exist as a nucleoprotein complex called T-complex, in which its 5′ end is covalently attached to one VirD2 molecule and the entire length of the T-strand is coated with numerous molecules of the ssDNA binding protein VirE2, another bacterial effector exported into the host cell (Sheng and Citovsky, 1996; Gelvin, 2000; Tzfira and Citovsky, 2000, 2002; Zupan et al., 2000). Furthermore, the host VirE2-Interacting Protein 1 (VIP1) directly binds to VirE2 and facilitates the nuclear import of T-DNA as well as its subsequent targeting to the host genome (Tzfira et al., 2001; Li et al., 2005; Lacroix et al., 2008).
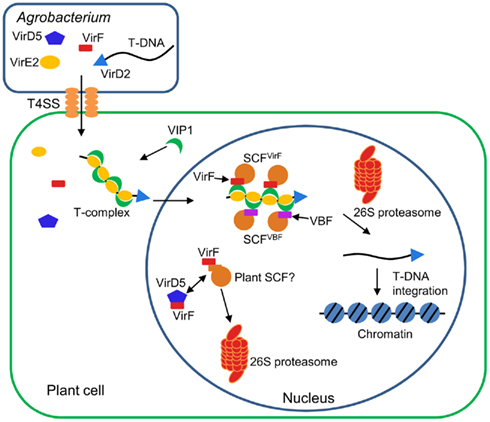
Figure 3. Involvement of F-box proteins in Agrobacterium infection. Agrobacterium exports a single-stranded copy of T-DNA (T-strand) as well as virulence (Vir) effector proteins into plant cell. Within plant cell, T-strand is assembled into a nucleoprotein complex (T-complex) in which one VirD2 molecule is attached to the 5′ end of the T-strand and multiple VirE2 molecules coat the entire length of the T-strand. In addition, the plant factor VIP1 directly interacts with VirE2 and guides the T-complex into the host cell nucleus. Once the T-complex enters the nucleus, it is presumably disassembled by an SCF complex containing VirF as an F-box component. The SCFVirF complex mediates polyubiquitination of VIP1, thereby targeting VIP1 as well as its associated VirE2 for 26S proteasome-dependent degradation. In plant species that do not require VirF for full virulence, Agrobacterium most likely also utilizes the host F-box protein VBF for the T-complex uncoating. As a defense strategy, the host plants destabilize VirF via the ubiquitin/26S proteasome system (UPS), presumably using an as yet unidentified plant SCF complex. Another exported effector, VirD5, counteracts this host-induced degradation of VirF by directly binding to and stabilizing VirF.
Once the T-complex reaches the host chromatin, the coating VirE2 and VIP1 proteins are most likely removed from the T-strand, resulting in release of the “naked” DNA molecule as a substrate for integration. Presumably, this uncoating process is mediated by VirF (Tzfira et al., 2004), another bacterial effector that is translocated into the host cell independently of the T-strand (Vergunst et al., 2000, 2005) and localizes to the host cell nucleus by as yet unknown mechanism (Tzfira et al., 2004; Figure 3). The Agrobacterium F-box effector VirF has been shown to interact with the plant SKP1 through the F-box domain (Schrammeijer et al., 2001), suggesting that VirF functions as a component of the SCF complex in plant cell (Schrammeijer et al., 2001; Tzfira et al., 2004). Although VirF does not possess any additional protein interaction domains, which are usually found in many F-box proteins, it directly binds VIP1 and targets it for 26S proteasome-dependent degradation in plant and yeast cells (Tzfira et al., 2004). In addition, VirE2, which is associated with VIP1, is indirectly destabilized by VirF at least in a heterologous yeast system (Tzfira et al., 2004). The degradation of VIP1 and VirE2 is indeed mediated by the VirF-containing SCF complex (SCFVirF) because this degradation does not occur in yeast mutant cells that do not express functional SKP1 (Tzfira et al., 2004). Together, these observations suggest that Agrobacterium hijacks the host SCF machinery for efficient infection with the help of the bacterial F-box effector. It remains unknown how Agrobacterium controls the timing of the SCFVirF-mediated T-complex uncoating, but the proposed role of VirF during Agrobacterium infection illustrates the strategic importance of molecular mimicry by pathogen-encoded F-box effectors.
The paradox of the infection strategy using VirF is, however, that F-box proteins are inherently unstable due to their own degradation mediated by the autocatalytic mechanism (Zhou and Howley, 1998; Galan and Peter, 1999) or other E3 ligases (Ayad et al., 2003; Guardavaccaro et al., 2003; Margottin-Goguet et al., 2003). Thus, Agrobacterium, which delivers only the VirF protein molecules but not the virF gene into the host cell, faces a challenge to protect VirF against detrimental degradation. Most likely, this is achieved by another translocated effector of Agrobacterium, VirD5, that directly binds to and stabilizes VirF, which otherwise undergoes rapid degradation via the host UPS (Magori and Citovsky, 2011; Figure 3). This degradation of VirF is likely mediated by an unknown plant E3 ligase, but not the autocatalytic mechanism because mutations in the F-box, a domain required for autoubiquitination, do not stabilize the VirF protein (Magori and Citovsky, 2011). Although it remains elusive whether the host-induced degradation of VirF is actually associated with bona fide plant defense responses, these observations illustrate a novel type of host–pathogen molecular arms race, in which both sides compete for the control of the stability of pathogen-encoded F-box effectors.
It should be noted that VirF was originally identified as a bacterial host range factor, the function of which is required for Agrobacterium infection in some, but not all, plant species (Hooykaas et al., 1984; Melchers et al., 1990; Jarchow et al., 1991; Regensburg-Tuink and Hooykaas, 1993). For example, Agrobacterium strains lacking the virF gene exhibit substantially attenuated infectivity on tomato and tree tobacco (Nicotiana glauca; Hooykaas et al., 1984; Melchers et al., 1990), while such strains still exhibit wild-type virulence on Arabidopsis (Zaltsman et al., 2010). Thus, in plant species that are susceptible to the VirF-lacking strains, Agrobacterium may utilize a host F-box protein as an alternative to VirF. Consistent with this notion, an Arabidopsis F-box protein, VIP1-Binding F-box protein (VBF), was shown to substitute for the VirF function during Agrobacterium infection (Zaltsman et al., 2010; Figure 3). Similarly to VirF, VBF interacts with the plant SKP1, and promotes proteasomal degradation of VIP1 as well as VirE2 (Zaltsman et al., 2010). Interestingly, the VBF transcripts are upregulated in Arabidopsis upon inoculation with Agrobacterium (Ditt et al., 2006) and fungal pathogens (Li et al., 2006). This, in turn, suggests that Agrobacterium may co-opt the plant defense responses to facilitate the T-complex uncoating and, hence, successful infection.
GALAs of Ralstonia
Another phytopathogenic bacterium R. solanacearum causes a lethal wilting disease on more than 200 plant species, which belong to as many as 50 families (Schell, 2000). This unusually wide host range may be related to a diverse array of effector proteins that Ralstonia potentially uses during the infection process. Indeed, genomic and gene expression studies have predicted that Ralstonia injects over 70 different effectors into the eukaryotic host cell via the type III secretion system (Salanoubat et al., 2002; Cunnac et al., 2004; Poueymiro and Genin, 2009). Among these translocated effectors, the GALA family plays an essential role in Ralstonia pathogenicity (Angot et al., 2006). This protein family is a group of seven members (GALA1 to 7), each of which contains an F-box domain as well as a C-terminal leucine-rich repeat (LRR) domain (Cunnac et al., 2004). The name “GALA” derives from a conserved GAxALA amino acid sequence in the LRR domain of these effectors (Cunnac et al., 2004). The evolutionary origin of the GALA family is unknown, but a phylogenetic analysis of various F-box domains suggested that the Ralstonia GALA genes may have been acquired from plants via horizontal gene transfer (Kajava et al., 2008).
Like VirF of Agrobacterium, at least four GALA proteins (GALA1, 5, 6, and 7) have been shown to interact with the Arabidopsis SKP1 (Angot et al., 2006), suggesting that translocated GALAs may be incorporated into the host SCF complex. Although mutating any single GALA gene does not alter Ralstonia infection of Arabidopsis or tomato (Cunnac et al., 2004), a strain harboring simultaneous mutations of all of the seven GALA genes shows almost no virulence in Arabidopsis and exhibits significantly attenuated infectivity in tomato (Angot et al., 2006). These observations suggest that the GALA family members have partially overlapping functions during Ralstonia infection of its plant hosts. However, among seven GALA proteins, at least GALA7 appears to possess a distinct host-specific function as its mutation alone does not affect the bacterial virulence in Arabidopsis or tomato, but leads to reduced disease symptoms in Medicago truncatula (Angot et al., 2006). It is tempting to speculate that the specific role of GALA7 in Medicago may be conferred by its C-terminal LRR domain, which potentially recruits Medicago-specific factors to the SCFGALA7 ubiquitin ligase complex. Similarly, the other GALA members may also target different and specific host factors in different plant species. The molecular mechanism by which the SCFGALA complex enhances Ralstonia infection remains elusive. Nevertheless, this bacterial F-box protein family can be a good model to understand not only how Ralstonia exploits the host SCF machinery for infection of specific hosts, but also how this plant pathogen has used this machinery to expand its host range.
P0 of Polerovirus
RNA silencing, also known as post-transcriptional gene silencing (PTGS), plays a central role in plant defense responses against viral pathogens (Vance and Vaucheret, 2001; Baulcombe, 2004; Voinnet, 2005; Diaz-Pendon and Ding, 2008). In this defense system, double-stranded RNA molecules derived from viral genomes or transcripts are processed into 21–25 nt small interfering RNA (siRNA) duplexes by the RNaseIII enzyme Dicer. Subsequently, a single-stranded siRNA is incorporated into the RNA-induced silencing complex (RISC), which then degrades viral RNAs that are complementary to the bound siRNA. Interestingly, many plant and animal viruses are known to encode suppressor proteins that suppress this host RNA silencing machinery (Vance and Vaucheret, 2001; Baulcombe, 2004; Voinnet, 2005; Diaz-Pendon and Ding, 2008). Among such viral suppressors, the Polerovirus P0 protein harbors an F-box motif that is required for the interaction of P0 with the plant SKP1 (Pazhouhandeh et al., 2006). Mutations in the F-box domain of P0 not only abolish the interaction between P0 and SKP1, but also significantly reduce accumulation of the viral RNAs in the host plants (Pazhouhandeh et al., 2006), indicating that P0 is a bona fide F-box protein essential for viral infectivity. The potential targets of the SCFP0 complex include ARGONAUTE1 (AGO1), the catalytic component of the RISC complex (Baumberger et al., 2007; Bortolamiol et al., 2007). P0 directly interacts with AGO1 and targets it for degradation in the host cell (Baumberger et al., 2007; Bortolamiol et al., 2007). Indeed, transient expression of P0 in Nicotiana benthamiana or Arabidopsis leads to substantial reduction in the AGO1 protein levels (Baumberger et al., 2007; Bortolamiol et al., 2007). Consistent with this result, ectopic expression of P0 in transgenic Arabidopsis plants elicits pleiotropic developmental abnormalities that are reminiscent of ago1 mutants (Bortolamiol et al., 2007). Together, these observations reveal a novel counter-defense strategy, in which viral pathogens disrupt the host RNA silencing through targeted degradation of AGO1. The P0 protein of Poleroviruses is the first evidence that not only bacterial pathogens but also viral pathogens have evolved to encode their own F-box proteins and hijack the host SCF machinery to enhance infection efficiency.
Conclusion
Host–pathogen interactions represent a never-ending arms race between host organisms defending against unwanted invaders, and pathogens counteracting the host defense system. Due to their short generation times and large population sizes, bacterial, and viral pathogens are thought to evolve much faster than multicellular, eukaryotic hosts (Arber, 2000). In addition, horizontal gene transfer, a process in which different prokaryotic species exchange their genetic materials, further speeds up evolution of pathogens (Arber, 2000). Thus, it is not surprising that pathogens, rather than hosts, seem to dominate the battle by constantly updating their infection strategies. In particular, use of F-box effectors within host cells illustrates how elaborate pathogens are when infecting their hosts. Hijacking the host SCF machinery by pathogens makes biological sense because the SCF complex plays a key role in host defense responses in many cases. Furthermore, due to the essential function of the SCF complexes in diverse cellular processes, hosts cannot modify this ubiquitin ligase system just to evade pathogen attack, further allowing for evolution of pathogen-encoded F-box proteins. Ever since the discovery of the Agrobacterium VirF protein, many additional pathogen-encoded F-box effectors have been identified, and at least several of them have been demonstrated to play an indispensible role in pathogenicity. How widespread is this infection strategy? How have pathogens evolved such eukaryotic F-box effectors? How have the F-box effectors affected evolution of host defense systems? With the advent of the discovery and studies of pathogen-encoded F-box proteins, we are beginning to understand the true complexity of host–pathogen interactions.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Miki Itaya for her help with preparation of Figure 2. The work in our laboratory is supported by grants from USDA/NIFA, NIH, NSF, BARD, DOE, and BSF (to Vitaly Citovsky).
References
Angot, A., Peeters, N., Lechner, E., Vailleau, F., Baud, C., Gentzbittel, L., Sartorel, E., Genschik, P., Boucher, C., and Genin, S. (2006). Ralstonia solanacearum requires F-box-like domain-containing type III effectors to promote disease on several host plants. Proc. Natl. Acad. Sci. U.S.A. 103, 14620–14625.
Angot, A., Vergunst, A., Genin, S., and Peeters, N. (2007). Exploitation of eukaryotic ubiquitin signaling pathways by effectors translocated by bacterial type III and type IV secretion systems. PLoS Pathog. 3, e3. doi:10.1371/journal.ppat.0030003
Arber, W. (2000). Genetic variation: molecular mechanisms and impact on microbial evolution. FEMS Microbiol. Rev. 24, 1–7.
Aronson, M. N., Meyer, A. D., Gyorgyey, J., Katul, L., Vetten, H. J., Gronenborn, B., and Timchenko, T. (2000). Clink, a nanovirus-encoded protein, binds both pRB and SKP1. J. Virol. 74, 2967–2972.
Ayad, N. G., Rankin, S., Murakami, M., Jebanathirajah, J., Gygi, S., and Kirschner, M. W. (2003). Tome-1, a trigger of mitotic entry, is degraded during G1 via the APC. Cell 113, 101–113.
Baumberger, N., Tsai, C. H., Lie, M., Havecker, E., and Baulcombe, D. C. (2007). The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr. Biol. 17, 1609–1614.
Bortolamiol, D., Pazhouhandeh, M., Marrocco, K., Genschik, P., and Ziegler-Graff, V. (2007). The Polerovirus F box protein P0 targets ARGONAUTE1 to suppress RNA silencing. Curr. Biol. 17, 1615–1621.
Bundock, P., Den Dulk-Ras, A., Beijersbergen, A., and Hooykaas, P. J. (1995). Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 14, 3206–3214.
Camborde, L., Planchais, S., Tournier, V., Jakubiec, A., Drugeon, G., Lacassagne, E., Pflieger, S., Chenon, M., and Jupin, I. (2010). The ubiquitin-proteasome system regulates the accumulation of turnip yellow mosaic virus RNA-dependent RNA polymerase during viral infection. Plant Cell 22, 3142–3152.
Cao, Y., Yang, Y., Zhang, H., Li, D., Zheng, Z., and Song, F. (2008). Overexpression of a rice defense-related F-box protein gene OsDRF1 in tobacco improves disease resistance through potentiation of defense gene expression. Physiol. Plant 134, 440–452.
Cheng, Y. T., Li, Y., Huang, S., Huang, Y., Dong, X., Zhang, Y., and Li, X. (2011). Stability of plant immune-receptor resistance proteins is controlled by SKP1-CULLIN1-F-box (SCF)-mediated protein degradation. Proc. Natl. Acad. Sci. U.S.A. 108, 14694–14699.
Citovsky, V., Zaltsman, A., Kozlovsky, S. V., Gafni, Y., and Krichevsky, A. (2009). Proteasomal degradation in plant-pathogen interactions. Semin. Cell Dev. Biol. 20, 1048–1054.
Cunnac, S., Occhialini, A., Barberis, P., Boucher, C., and Genin, S. (2004). Inventory and functional analysis of the large Hrp regulon in Ralstonia solanacearum: identification of novel effector proteins translocated to plant host cells through the type III secretion system. Mol. Microbiol. 53, 115–128.
Diaz-Pendon, J. A., and Ding, S. W. (2008). Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis. Annu. Rev. Phytopathol. 46, 303–326.
Ditt, R. F., Kerr, K. F., De Figueiredo, P., Delrow, J., Comai, L., and Nester, E. W. (2006). The Arabidopsis thaliana transcriptome in response to Agrobacterium tumefaciens. Mol. Plant Microbe Interact. 19, 665–681.
Drugeon, G., and Jupin, I. (2002). Stability in vitro of the 69K movement protein of turnip yellow mosaic virus is regulated by the ubiquitin-mediated proteasome pathway. J. Gen. Virol. 83, 3187–3197.
Elde, N. C., and Malik, H. S. (2009). The evolutionary conundrum of pathogen mimicry. Nat. Rev. Microbiol. 7, 787–797.
Gagne, J. M., Downes, B. P., Shiu, S. H., Durski, A. M., and Vierstra, R. D. (2002). The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 99, 11519–11524.
Galan, J. M., and Peter, M. (1999). Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc. Natl. Acad. Sci. U.S.A. 96, 9124–9129.
Gelvin, S. B. (2000). Agrobacterium and plant genes involved in T-DNA transfer and integration. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 223–256.
Gelvin, S. B. (2010). Plant proteins involved in Agrobacterium-mediated genetic transformation. Annu. Rev. Phytopathol. 48, 45–68.
Gou, M., Shi, Z., Zhu, Y., Bao, Z., Wang, G., and Hua, J. (2011). The F-box protein CPR1/CPR30 negatively regulates R protein SNC1 accumulation. Plant J. doi: 10.1111/j.1365-313X.2011.04799.x
Gou, M., Su, N., Zheng, J., Huai, J., Wu, G., Zhao, J., He, J., Tang, D., Yang, S., and Wang, G. (2009). An F-box gene, CPR30, functions as a negative regulator of the defense response in Arabidopsis. Plant J. 60, 757–770.
Guardavaccaro, D., Kudo, Y., Boulaire, J., Barchi, M., Busino, L., Donzelli, M., Margottin-Goguet, F., Jackson, P. K., Yamasaki, L., and Pagano, M. (2003). Control of meiotic and mitotic progression by the F box protein β-Trcp1 in vivo. Dev. Cell 4, 799–812.
Hao, B., Zheng, N., Schulman, B. A., Wu, G., Miller, J. J., Pagano, M., and Pavletich, N. P. (2005). Structural basis of the Cks1-dependent recognition of p27(Kip1) by the SCF(Skp2) ubiquitin ligase. Mol. Cell 20, 9–19.
Hooykaas, P. J., Hofker, M., Den Dulk-Ras, H., and Schilperoort, R. A. (1984). A comparison of virulence determinants in an octopine Ti plasmid, a nopaline Ti plasmid, and an Ri plasmid by complementation analysis of Agrobacterium tumefaciens mutants. Plasmid 11, 195–205.
Hua, Z., and Vierstra, R. D. (2011). The cullin-RING ubiquitin-protein ligases. Annu. Rev. Plant Biol. 62, 299–334.
Hughes, A. L., and Nei, M. (1988). Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335, 167–170.
Hughes, A. L., and Nei, M. (1989a). Evolution of the major histocompatibility complex: independent origin of nonclassical class I genes in different groups of mammals. Mol. Biol. Evol. 6, 559–579.
Hughes, A. L., and Nei, M. (1989b). Nucleotide substitution at major histocompatibility complex class II loci: evidence for overdominant selection. Proc. Natl. Acad. Sci. U.S.A. 86, 958–962.
Jarchow, E., Grimsley, N. H., and Hohn, B. (1991). virF, the host-range-determining virulence gene of Agrobacterium tumefaciens, affects T-DNA transfer to Zea mays. Proc. Natl. Acad. Sci. U.S.A. 88, 10426–10430.
Kajava, A. V., Anisimova, M., and Peeters, N. (2008). Origin and evolution of GALA-LRR, a new member of the CC-LRR subfamily: from plants to bacteria? PLoS ONE 3, e1694. doi:1610.1371/journal.pone.0001694
Kunik, T., Tzfira, T., Kapulnik, Y., Gafni, Y., Dingwall, C., and Citovsky, V. (2001). Genetic transformation of HeLa cells by Agrobacterium. Proc. Natl. Acad. Sci. U.S.A. 98, 1871–1876.
Lacroix, B., Loyter, A., and Citovsky, V. (2008). Association of the Agrobacterium T-DNA-protein complex with plant nucleosomes. Proc. Natl. Acad. Sci. U.S.A. 105, 15429–15434.
Lacroix, B., Tzfira, T., Vainstein, A., and Citovsky, V. (2006). A case of promiscuity: Agrobacterium’s endless hunt for new partners. Trends Genet. 22, 29–37.
Lechner, E., Achard, P., Vansiri, A., Potuschak, T., and Genschik, P. (2006). F-box proteins everywhere. Curr. Opin. Plant Biol. 9, 631–638.
Li, J., Brader, G., Kariola, T., and Palva, E. T. (2006). WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 46, 477–491.
Li, J., Krichevsky, A., Vaidya, M., Tzfira, T., and Citovsky, V. (2005). Uncoupling of the functions of the Arabidopsis VIP1 protein in transient and stable plant genetic transformation by Agrobacterium. Proc. Natl. Acad. Sci. U.S.A. 102, 5733–5738.
Lomma, M., Dervins-Ravault, D., Rolando, M., Nora, T., Newton, H. J., Samson, F. M., Sahr, T., Gomez-Valero, L., Jules, M., Hartland, E. L., and Buchrieser, C. (2010). The Legionella pneumophila F-box protein Lpp2082 (AnkB) modulates ubiquitination of the host protein parvin B and promotes intracellular replication. Cell. Microbiol. 12, 1272–1291.
Magori, S., and Citovsky, V. (2011). Agrobacterium counteracts host-induced degradation of its effector F-box protein. Sci. Signal. 4, pii: ra69.
Margottin-Goguet, F., Hsu, J. Y., Loktev, A., Hsieh, H. M., Reimann, J. D., and Jackson, P. K. (2003). Prophase destruction of Emi1 by the SCFβTrCP/Slimb ubiquitin ligase activates the anaphase promoting complex to allow progression beyond prometaphase. Dev. Cell 4, 813–826.
Melchers, L. S., Maroney, M. J., Den Dulk-Ras, A., Thompson, D. V., Van Vuuren, H. A., Schilperoort, R. A., and Hooykaas, P. J. (1990). Octopine and nopaline strains of Agrobacterium tumefaciens differ in virulence; molecular characterization of the virF locus. Plant Mol. Biol. 14, 249–259.
Pazhouhandeh, M., Dieterle, M., Marrocco, K., Lechner, E., Berry, B., Brault, V., Hemmer, O., Kretsch, T., Richards, K. E., Genschik, P., and Ziegler-Graff, V. (2006). F-box-like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc. Natl. Acad. Sci. U.S.A. 103, 1994–1999.
Petroski, M. D., and Deshaies, R. J. (2005). Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20.
Piers, K. L., Heath, J. D., Liang, X., Stephens, K. M., and Nester, E. W. (1996). Agrobacterium tumefaciens-mediated transformation of yeast. Proc. Natl. Acad. Sci. U.S.A. 93, 1613–1618.
Pitzschke, A., and Hirt, H. (2010). New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation. EMBO J. 29, 1021–1032.
Poueymiro, M., and Genin, S. (2009). Secreted proteins from Ralstonia solanacearum: a hundred tricks to kill a plant. Curr. Opin. Microbiol. 12, 44–52.
Price, C. T., Al-Khodor, S., Al-Quadan, T., Santic, M., Habyarimana, F., Kalia, A., and Kwaik, Y. A. (2009). Molecular mimicry by an F-box effector of Legionella pneumophila hijacks a conserved polyubiquitination machinery within macrophages and protozoa. PLoS Pathog. 5, e1000704. doi:1000710.1001371/journal.ppat.1000704
Price, C. T., and Kwaik, Y. A. (2010). Exploitation of host polyubiquitination machinery through molecular mimicry by eukaryotic-like bacterial F-box effectors. Front. Microbiol. 1:122. doi:110.3389/fmicb.2010.00122
Regensburg-Tuink, A. J., and Hooykaas, P. J. (1993). Transgenic N. glauca plants expressing bacterial virulence gene virF are converted into hosts for nopaline strains of A. tumefaciens. Nature 363, 69–71.
Reichel, C., and Beachy, R. N. (2000). Degradation of tobacco mosaic virus movement protein by the 26S proteasome. J. Virol. 74, 3330–3337.
Salanoubat, M., Genin, S., Artiguenave, F., Gouzy, J., Mangenot, S., Arlat, M., Billault, A., Brottier, P., Camus, J. C., Cattolico, L., Chandler, M., Choisne, N., Claudel-Renard, C., Cunnac, S., Demange, N., Gaspin, C., Lavie, M., Moisan, A., Robert, C., Saurin, W., Schiex, T., Siguier, P., Thebault, P., Whalen, M., Wincker, P., Levy, M., Weissenbach, J., and Boucher, C. A. (2002). Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415, 497–502.
Santner, A., and Estelle, M. (2010). The ubiquitin-proteasome system regulates plant hormone signaling. Plant J. 61, 1029–1040.
Schell, M. A. (2000). Control of virulence and pathogenicity genes of Ralstonia solanacearum by an elaborate sensory network. Annu. Rev. Phytopathol. 38, 263–292.
Schrammeijer, B., Risseeuw, E., Pansegrau, W., Regensburg-Tuink, T. J., Crosby, W. L., and Hooykaas, P. J. (2001). Interaction of the virulence protein VirF of Agrobacterium tumefaciens with plant homologs of the yeast Skp1 protein. Curr. Biol. 11, 258–262.
Schröder, G., Schuelein, R., Quebatte, M., and Dehio, C. (2011). Conjugative DNA transfer into human cells by the VirB/VirD4 type IV secretion system of the bacterial pathogen Bartonella henselae. Proc. Natl. Acad. Sci. U.S.A. 108, 14643–14648.
Sheng, J., and Citovsky, V. (1996). Agrobacterium-plant cell DNA transport: have virulence proteins, will travel. Plant Cell 8, 1699–1710.
Thomas, J. H. (2006). Adaptive evolution in two large families of ubiquitin-ligase adapters in nematodes and plants. Genome Res. 16, 1017–1030.
Tzfira, T., and Citovsky, V. (2000). From host recognition to T-DNA integration: the function of bacterial and plant genes in the Agrobacterium-plant cell interaction. Mol. Plant Pathol. 1, 201–212.
Tzfira, T., and Citovsky, V. (2002). Partners-in-infection: host proteins involved in the transformation of plant cells by Agrobacterium. Trends Cell Biol. 12, 121–129.
Tzfira, T., and Citovsky, V. (2006). Agrobacterium-mediated genetic transformation of plants: biology and biotechnology. Curr. Opin. Biotechnol. 17, 147–154.
Tzfira, T., Vaidya, M., and Citovsky, V. (2001). VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J. 20, 3596–3607.
Tzfira, T., Vaidya, M., and Citovsky, V. (2004). Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature 431, 87–92.
van den Burg, H. A., Tsitsigiannis, D. I., Rowland, O., Lo, J., Rallapalli, G., Maclean, D., Takken, F. L., and Jones, J. D. (2008). The F-box protein ACRE189/ACIF1 regulates cell death and defense responses activated during pathogen recognition in tobacco and tomato. Plant Cell 20, 697–719.
Vance, V., and Vaucheret, H. (2001). RNA silencing in plants – defense and counterdefense. Science 292, 2277–2280.
Vergunst, A. C., Schrammeijer, B., Den Dulk-Ras, A., De Vlaam, C. M., Regensburg-Tuink, T. J., and Hooykaas, P. J. (2000). VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290, 979–982.
Vergunst, A. C., Van Lier, M. C., Den Dulk-Ras, A., Stuve, T. A., Ouwehand, A., and Hooykaas, P. J. (2005). Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc. Natl. Acad. Sci. U.S.A. 102, 832–837.
Vierstra, R. D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10, 385–397.
Voinnet, O. (2005). Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6, 206–220.
Zaltsman, A., Krichevsky, A., Loyter, A., and Citovsky, V. (2010). Agrobacterium induces expression of a host F-box protein required for tumorigenicity. Cell Host Microbe 7, 197–209.
Zeng, L. R., Vega-Sanchez, M. E., Zhu, T., and Wang, G. L. (2006). Ubiquitination-mediated protein degradation and modification: an emerging theme in plant-microbe interactions. Cell Res. 16, 413–426.
Zheng, N., Schulman, B. A., Song, L., Miller, J. J., Jeffrey, P. D., Wang, P., Chu, C., Koepp, D. M., Elledge, S. J., Pagano, M., Conaway, R. C., Conaway, J. W., Harper, J. W., and Pavletich, N. P. (2002). Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416, 703–709.
Zhou, P., and Howley, P. M. (1998). Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol. Cell 2, 571–580.
Keywords: F-box, SCF complex, ubiquitin, protein degradation, Agrobacterium, Ralstonia, Polerovirus
Citation: Magori S and Citovsky V (2011) Hijacking of the host SCF ubiquitin ligase machinery by plant pathogens. Front. Plant Sci. 2:87. doi: 10.3389/fpls.2011.00087
Received: 18 October 2011; Paper pending published: 01 November 2011;
Accepted: 06 November 2011; Published online: 22 November 2011.
Edited by:
Xin Li, University of British Columbia, CanadaReviewed by:
Keiko Yoshioka, University of Toronto, CanadaJacqueline Monaghan, The Sainsbury Laboratory, UK
Copyright: © 2011 Magori and Citovsky. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Shimpei Magori, Department of Biochemistry and Cell Biology, State University of New York, Stony Brook, 445 Life Sciences Building, Stony Brook, NY, 11794-5215, USA. e-mail: smagori@notes.cc.sunysb.edu