- Department of Plant and Soil Sciences, University of Kentucky, Lexington, KY, USA
Alternative poly(A) site choice (also known as alternative polyadenylation, or APA) has the potential to affect gene expression in qualitative and quantitative ways. APA may affect as many as 82% of all expressed genes in a plant. The consequences of APA include the generation of transcripts with differing 3′-UTRs (and thus differing regulatory potential) and of transcripts with differing protein-coding potential. Genome-wide studies of possible APA suggest a linkage with pre-mRNA splicing, and indicate a coincidence of and perhaps cooperation between RNA regulatory elements that affect splicing efficiency and the recognition of novel intronic poly(A) sites. These studies also raise the possibility of the existence of a novel class of polyadenylation-related cis elements that are distinct from the well-characterized plant polyadenylation signal. Many potential APA events, however, have not been associated with identifiable cis elements. The present state of the field reveals a broad scope of APA, and also numerous opportunities for research into mechanisms that govern both choice and regulation of poly(A) sites in plants.
Overview
Gene expression can be regulated at numerous steps that together represent virtually every stage in the biogenesis of a polypeptide. Among these steps is that which entails the processing and polyadenylation of mRNAs in the nucleus of eukaryotic cells. The chief means by which regulation is effected via polyadenylation is through the choice of one of several potential poly(A) sites carried on a precursor RNA. This process – alternative polyadenylation (APA) – affords a direct linkage between RNA processing and control of mRNA function. The latter can be altered by APA via loss of or changes in the exonic contents of mRNAs, resulting in changes in the protein-coding potential of the mRNA. APA can also affect the responsiveness of an mRNA to control by RNA regulatory elements. This latter phenomenon is illustrated in the numerous recent reports that show both correlation and causation between the regulatory element content of an mRNA [a consequence of the choice of poly(A) site during the biogenesis of the mRNA] and the functionality of the mRNA in promoting cellular growth and/or differentiation in animals (Ji and Tian, 2009; Ji et al., 2009; Mayr and Bartel, 2009), and in other reports that document global changes in poly(A) site choice associated with various aspects of development or differentiation in animals (Zhang et al., 2005; Macdonald and McMahon, 2010; Mangone et al., 2010; Shepard et al., 2011).
Historically, plant 3′-UTRs have been known to possess significant heterogeneity (Dean et al., 1986), and in one case 14 different poly(A) sites were found in one gene (Klahre et al., 1995); thus, the possibility of APA in plants has been acknowledged for some time. In this review, the scope and consequences of APA will be discussed, as will be the interplay between APA and RNA regulatory elements.
The Scope of Alternative Polyadenylation in Plants
By any standard, the extent of APA in plants seems to be extensive. Detailed analyses of large EST and cDNA collections in Arabidopsis suggests that the scope of APA is substantial, with 10% or more of all genes possessing alternative 3′ ends that in turn change the exonic content of the mRNA (Haas et al., 2003; Iida et al., 2004; Nagasaki et al., 2005). Meyers et al. (2004) found massively parallel signature sequence (MPSS) tags within upstream introns and non-terminal exons in approximately 25% of all genes in Arabidopsis. In an analysis of a dataset composed of 55,000 in silico authenticated poly(A) sites from rice ESTs, it was found that over 50% of expressed genes possess at least two poly(A) sites (Shen et al., 2008a). A similar extent of APA in Chlamydomonas reinhardtii was inferred from analyses of EST sequences (Shen et al., 2008b). Shen et al. (2011) analyzed combined datasets derived from MPSS and Illumina-based (SBS) sequences, both obtained from cDNA tags that query sequences adjacent to the DpnII site nearest the poly(A) site, and found that 60% of Arabidopsis genes and between 47 and 82% of rice genes possess more than one poly(A) site. A recent large-scale sequencing effort focused precisely on the mRNA–poly(A) junction confirms these meta-analyses (Wu et al., 2011). In this study, more than 74% of Arabidopsis genes whose expression could be ascertained in leaves and seeds were found to possess two or more “poly(A) site clusters,” or groups of closely spaced poly(A) sites.
There is considerable variation in the estimates that have been published over the past 8 years or so. The early studies of EST and cDNA sequences was focused on events that would alter the coding capacity of the respective RNAs; as such, heterogeneity within 3′-UTRs and other redundant APA events may have been overlooked. Later studies of MPSS and sequence tag signatures includes these latter sorts of poly(A) site variability. The range of estimates in the latter instances is also somewhat broad, with 50–74% of all genes possessing more than one poly(A) site. A likely source of differences in these instances is the “depth” of coverage afforded by the different strategies; in general, as more data is considered, more instances of APA are uncovered, as might be expected. Other considerations also factor into the estimates and their differences. These include the actual definition of a poly(A) site (is every 3′ end a distinct site, or are closely spaced clusters considered to be single sites?) and the extent to which overlapping transcription units may lead to mis-identification of multiple sites. The technical aspects of the methods used to identify poly(A) sites may also contribute to differences in estimates of genome-wide APA. For example, internal priming by reverse transcriptase is unavoidable and likely to be variable from study to study. Regardless of the modest uncertainties, it is clear that a majority of genes in plants possess multiple poly(A) sites.
The Consequences of Alternative Polyadenylation in Plants
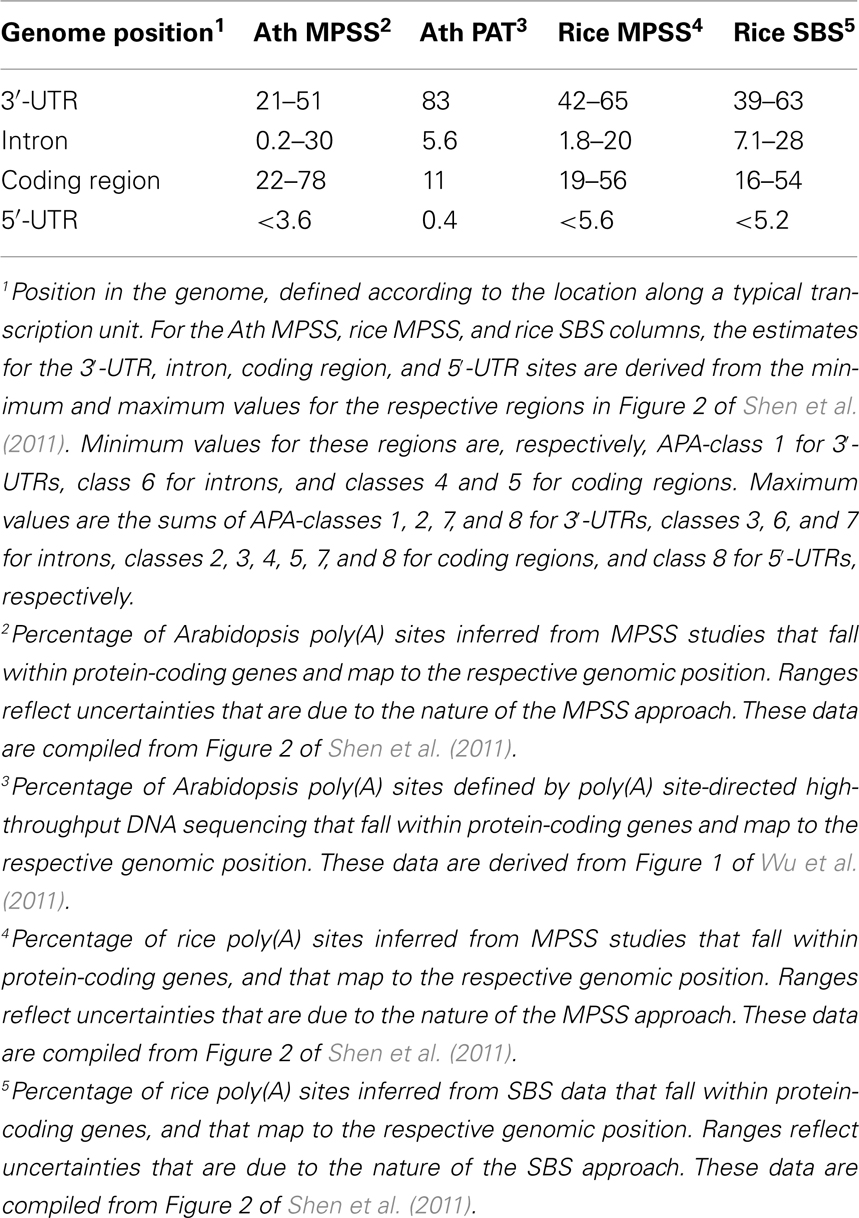
Beyond the prevalence of APA in plants, there is the matter of the consequences of alternative poly(A) site choice, and specifically how APA may re-model the content and functionality of an mRNA. APA may involve utilization of sites that lie within 3′-UTRs, introns, 5′-UTRs, and even protein-coding regions; each of these possibilities carries the potential to affect mRNA functionality. Two recent studies provide an estimate of the scopes of events that affect these different regions. In the study by Shen et al. (2011), poly(A) sites in genes with more than one distinct MPSS or SBS signature were classified according to the genomic region in which corresponding signature fell, as well as to the location of the nearest DpnII site [that determined proximity to an authentic poly(A) site]; the focus on genes with more than one unique signature allowed an examination of the nature of alternative poly(A) sites. The ranges for locations of these possible alternative sites are summarized in Table 1; the spread in values reflects some uncertainty in exact locations of sites that are defined by MPSS and SBS tags. As many as half of all of the alternative sites in Arabidopsis fell within 3′-UTRs, and between 39 and 65% of such sites fell within 3′-UTRs in rice. Remarkably, between 22 and78% of all alternative sites in Arabidopsis and between 16 and 56% of such sites in rice mapped to protein-coding regions. Fewer than 30% of such sites fell within introns or 5′-UTRs in Arabidopsis, and fewer than 28% of all rice alternative sites mapped to these genomic regions.
The study by Wu et al. (2011) did not analyze just alternative sites per se. However, the genome-wide distribution of all sites described in this study provides additional insight into the scope and consequences of APA. In this study, about 83% of sites that map to annotated genes fell within 3′-UTRs. The second most “abundant” class of sites was that mapping to protein-coding regions (11%). About 6% fell within introns or 5′-UTRs. There is some degree of discrepancy in the absolute values for some of these classes – for example, the upper limit for the percentage of poly(A) sites that fall within protein-coding regions in Wu et al., 2011; 11%) is much lower than the lower limit (22%) seen for this class of sites in Shen et al. (2011). The sources of these differences are not entirely clear. The MPSS and SBS data analyzed in Shen et al. (2011) were not masked to eliminate signatures that map to genomic poly(A) tracts and thus may be affected by internal priming by reverse transcriptase. In contrast, the mapping reported by Wu et al. (2011) did include this masking. It is possible that this may explain some of the difference in the two studies. Another source of difference lies in the possibility that CDS-localized poly(A) sites may occur preferentially in genes with multiple sites; in this case, the analysis in Shen et al. (2011) would likely have been biased for an increased incidence of CDS-localized poly(A) sites. Regardless of these caveats, the observation that CDS-localized APA is predominant indicates it must be considered when weighing the possible impacts of APA.
RNA Sequence Signals that Control Alternative Polyadenylation in Plants
Several of these studies suggest that there may be some bias in the patterns of APA. Thus, the alternative 3′ ends seen in libraries prepared from cold-treated Arabidopsis plants had a statistically significant tendency to truncate the mRNA, compared with the longer alternative 3′ end (Iida et al., 2004). The usage of poly(A) sites within introns seems to increase dramatically in cold-treated rice plants (Shen et al., 2011), an observation that is consistent with the analysis of Arabidopsis ESTs. Other modes of APA seemed to be more prevalent in germinating Arabidopsis seedlings (Shen et al., 2011). More than 100 examples of poly(A) site switching [use of one poly(A) site in one sample, and of a different one in another] could be identified in a comparison of poly(A) site choice in genes expressed in Arabidopsis leaves and dry seed (Wu et al., 2011). These studies are far from comprehensive, but they indicate that patterns of poly(A) site choice can vary in plants, raising the possibility of regulatory changes in the functioning of different poly(A) sites in a transcription unit.
These regulatory changes are likely to be mediated through specific RNA sequence elements. For example, targets for sequence-specific RNA-binding proteins may promote polyadenylation at particular sites, thereby remodeling the transcriptional outputs of genes possessing such targets. Examples of this mode of regulation include the promotion of APA-mediated by FCA and FPA, two RNA-binding proteins that contribute to the regulation of flowering time in Arabidopsis (Simpson et al., 2003; Hornyik et al., 2010). Among other events, these proteins promote usage of a poly(A) site within the first introns of pre-mRNAs transcribed from the FCA and FPA genes, respectively, establishing a feedback regulatory mechanism for controlling FCA and FPA expression levels. The dynamic between the thiamine riboswitch, splicing, and polyadenylation is another such example (Wachter et al., 2007). The thiamine riboswitch inhibits splicing of an intron within the 3′-UTR of the THIC gene, thereby promoting usage of an intronic poly(A) site and production of a short, more highly expressed mRNA isoform. Thiamine binding by the riboswitch promotes the splicing of the intron, and thus the usage of a downstream poly(A) site.
Interestingly, these examples involve polyadenylation at intronic locations. More than 4000 Arabidopsis genes may be subject to APA within introns. The affected introns in these genes are somewhat more inclined to possess sub-optimal 5′ splice sites but typical 3′-splice sites (Wu et al., 2011). Thus, mechanisms involving RNA–protein interactions that may affect splicing [such as changes in the levels or activities of proteins such as SR proteins (Lopato et al., 1999; Kalyna et al., 2003), UBP1 (Lambermon et al., 2000), or the cap-binding complex (Raczynska et al., 2010)] may in some instances alter poly(A) site choice. These considerations serve to emphasize a current theme, that splicing and polyadenylation may be temporally and physically linked. The consequence of this realization is that events that occur at the intron boundaries (such as recognition of splice sites by splicing factors) and within introns (such as recognition of specific sequences by RNA-binding proteins such as FCA or FPA) combine to yield patterns of poly(A) site choice.
The classical plant polyadenylation signal consists of three elements: a U-rich “far-upstream element” situated between 60 and 130 nts 5′ of the poly(A) site, an A-rich “near-upstream element” that is located between 10 and 30 nts upstream from the poly(A) site, and the U-rich cleavage element that includes the poly(A) site (Loke et al., 2005). In Arabidopsis, poly(A) sites that fall within introns, 5′-UTRs, and 3′-UTRs all possess sequence compositions that are indistinguishable from the classical polyadenylation signal (Wu et al., 2011). This is also true for 3′-UTR-localized sites that are utilized specifically in leaves or seeds (Wu et al., 2011). To date, no identifiable auxiliary element has been identified that can explain the differential utilization of these sites. However, it is reasonable to postulate that such motifs must exist, since it is otherwise difficult to explain the differential usage of these sites.
As mentioned above, a number of alternative poly(A) sites in plants fall within the protein-coding regions of mRNAs. These sites possess a distinctive poly(A) signal that consists largely of an extended A + G-rich region that flanks the actual cleavage site on both sides (Wu et al., 2011). This sequence signature is likely to be part of an RNA sequence element that controls CDS-localized polyadenylation. However, these signatures cannot be the entire element; this follows from the observation that a majority of such sequence signatures within protein-coding regions are not used as poly(A) sites (Wu et al., 2011). The means by which the A + G-rich region functions in polyadenylation are not known, nor are any hypothetical accompanying sequences that may function in concert with the A + G-rich region.
Alternative Polyadenylation and RNA-Mediated Regulation
It is evident that, in animals, APA has the potential to impact the functioning of other RNA regulatory elements and mechanisms, largely through the inclusion or exclusion of such elements owing to APA-mediated mRNA remodeling (Licatalosi and Darnell, 2010; Di Giammartino et al., 2011; Lutz and Moreira, 2011). However, the extent of the interplay between APA and RNA-mediated regulation in plants is less well-known. Generally speaking, APA provides two choices, the production of a short or a long transcript. The shorter transcript would be the one devoid of RNA regulatory elements and thus freed from the respective mode of regulation, while the longer transcript would be subject to control (be it degradation, inhibition, or enhancement of translation, or other mechanisms associated with the myriad of RNA regulatory elements that may be found in an mRNA). For as many as 65% of APA events in plants, those that involve different sites downstream from the translation termination codon, these possibilities are plausible. For cases involving APA elsewhere within the transcription unit, the outcomes are harder to distinguish. This is because APA that truncates a mRNA upstream from the normal translation termination codon will not only remove possible RNA regulatory elements but will also truncate the mRNA itself, in ways that will radically alter or eliminate the functionality of the mRNA. This is especially true for mRNAs that are polyadenylated within protein-coding regions; in most instances, such mRNAs will lack translation termination codons and should be subjected to quality-control mechanisms that limit the potential for such mRNAs to be translated into truncated and possibly toxic polypeptides (Vasudevan et al., 2002). Thus, even though APA involving upstream sites has the potential to “bracket” a large number of RNA regulatory elements, both hypothetical outcomes would yield mRNAs subject to forms of negative control.
These considerations relate to RNA-based regulation of protein-coding genes. APA also has the possibility to affect the expression on non-coding RNAs, including the primary transcripts that are processed to yield microRNAs or trans-acting siRNAs. An exhaustive study of such phenomena has not been reported, but perusal of the results of high-throughput sequencing of poly(A) site-directed cDNA tags reveals that such a possibility is a viable one (Figure 1). Such possibilities allow for even more layers of regulation linked to RNA; in the example shown in Figure 1, APA would generate two transcripts from the Arabidopsis TAS3a locus (At3g17185), only one of which would be subject to miR390-mediated processing.
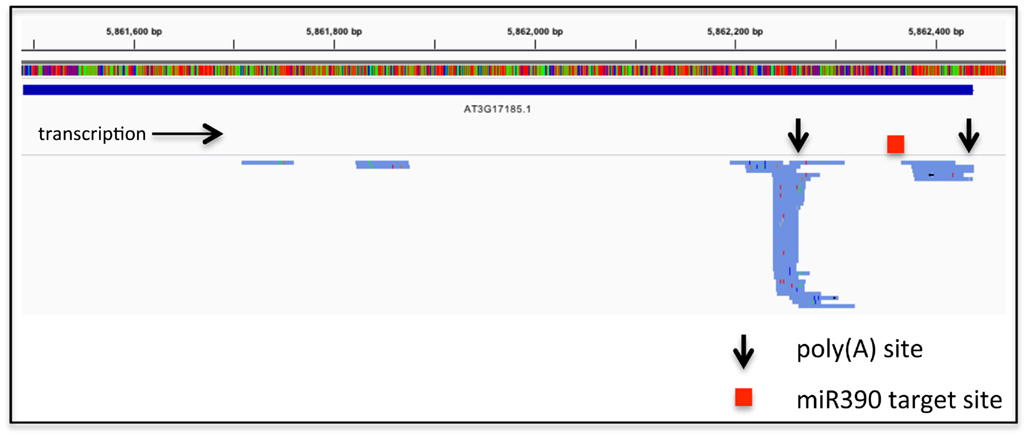
Figure 1. Alternative poly(A) site choice in the gene encoding TAS3a (At3g17185). The bar with the colored vertical lines represents the DNA sequence of the locus, color-coded using the default settings from the Integrated Genomics Viewer (IGV2.0). The extent of the RNA coding region is represented as the solid blue line beneath the sequence. Beneath this are shown the directions of transcription, locations of the two principal poly(A) sites, and the miR390 target site (Allen et al., 2005). Finally, the numerous blue lines below the gray separator represent individual poly(A) tags (taken from the Arabidopsis leaf set from Wu et al., 2011) that map to the At3g17185 locus. Colored tics within the tag representations signify differences in sequence from the known TAS3a sequence; these differences provide an illustration of the error rate inherent in the high-throughput PAT sequencing. The right-most extremity of each tag represents the 3′-end of the tag, and hence the mRNA–poly(A) junction. These tags were mapped to the Arabidopsis genome using CLC Genomics Workbench and displayed using the Integrated Genomics Viewer 2.0 (Robinson et al., 2011).
Alternative poly(A) site choice has the potential to affect the production of antisense RNAs via the alterations of 3′-UTRs of mRNAs encoded by paired of nearby, convergently transcribed genes. Analogous cis-antisense RNA and siRNA production can be induced via transcriptional induction of one or both members of such pairs of genes (e.g., Borsani et al., 2005; Jin et al., 2008). However, recent studies suggest that, for most gene pairs with overlapping 3′-UTRs that have the potential to generate cis-antisense RNAs, there is no clear negative correlation between expression levels and the possibility of formation of antisense RNA (Henz et al., 2007; Wu et al., 2011). Future research that is focused on the identification of possible inducible APA and mRNA remodeling, as it relates to gene pairs that may encode cis-antisense RNAs due to overlapping 3′-UTRs, is needed to better understand the possible contributions that APA makes to siRNA production.
Summary
With the advent of high-throughput approaches for sequencing and meta-analysis and the application of these methods to the study of polyadenylation, it has become apparent that APA is a widespread phenomenon that has the potential to affect a majority of expressed genes in plants. This field is relatively young, and most of the reports that pertain to the subject raise many more questions than they answer. For the foreseeable future, the study of APA in plants promises to yield many surprises and insights into the interplay between posttranscriptional controls and other molecular and physiological processes.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Research performed in the author’s laboratory and described in this review was supported by US National Science Foundation (IOS-0817818), Kentucky Science and Engineering Foundation (KSEF-2061-RDE-013), and the University of Kentucky Executive Vice President for Research.
References
Allen, E., Xie, Z., Gustafson, A. M., and Carrington, J. C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221.
Borsani, O., Zhu, J., Verslues, P. E., Sunkar, R., and Zhu, J. K. (2005). Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123, 1279–1291.
Dean, C., Tamaki, S., Dunsmuir, P., Favreau, M., Katayama, C., Dooner, H., and Bedbrook, J. (1986). mRNA transcripts of several plant genes are polyadenylated at multiple sites in vivo. Nucleic Acids Res. 14, 2229–2240.
Di Giammartino, D. C., Nishida, K., and Manley, J. L. (2011). Mechanisms and consequences of alternative polyadenylation. Mol. Cell 43, 853–866.
Haas, B. J., Delcher, A. L., Mount, S. M., Wortman, J. R., Smith, R. K. Jr., Hannick, L. I., Maiti, R., Ronning, C. M., Rusch, D. B., Town, C. D., Salzberg, S. L., and White, O. (2003). Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31, 5654–5666.
Henz, S. R., Cumbie, J. S., Kasschau, K. D., Lohmann, J. U., Carrington, J. C., Weigel, D., and Schmid, M. (2007). Distinct expression patterns of natural antisense transcripts in Arabidopsis. Plant Physiol. 144, 1247–1255.
Hornyik, C., Terzi, L. C., and Simpson, G. G. (2010). The spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Dev. Cell 18, 203–213.
Iida, K., Seki, M., Sakurai, T., Satou, M., Akiyama, K., Toyoda, T., Konagaya, A., and Shinozaki, K. (2004). Genome-wide analysis of alternative pre-mRNA splicing in Arabidopsis thaliana based on full-length cDNA sequences. Nucleic Acids Res. 32, 5096–5103.
Ji, Z., Lee, J. Y., Pan, Z., Jiang, B., and Tian, B. (2009). Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc. Natl. Acad. Sci. U.S.A. 106, 7028–7033.
Ji, Z., and Tian, B. (2009). Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS ONE 4, e8419. doi:10.1371/journal.pone.0008419
Jin, H., Vacic, V., Girke, T., Lonardi, S., and Zhu, J. K. (2008). Small RNAs and the regulation of cis-natural antisense transcripts in Arabidopsis. BMC Mol. Biol. 9, 6. doi:10.1186/1471-2199-9-6
Kalyna, M., Lopato, S., and Barta, A. (2003). Ectopic expression of atRSZ33 reveals its function in splicing and causes pleiotropic changes in development. Mol. Biol. Cell 14, 3565–3577.
Klahre, U., Hemmings-Mieszczak, M., and Filipowicz, W. (1995). Extreme heterogeneity of polyadenylation sites in mRNAs encoding chloroplast RNA-binding proteins in Nicotiana plumbaginifolia. Plant Mol. Biol. 28, 569–574.
Lambermon, M. H., Simpson, G. G., Wieczorek Kirk, D. A., Hemmings-Mieszczak, M., Klahre, U., and Filipowicz, W. (2000). UBP1, a novel hnRNP-like protein that functions at multiple steps of higher plant nuclear pre-mRNA maturation. EMBO J. 19, 1638–1649.
Licatalosi, D. D., and Darnell, R. B. (2010). RNA processing and its regulation: global insights into biological networks. Nat. Rev. Genet. 11, 75–87.
Loke, J. C., Stahlberg, E. A., Strenski, D. G., Haas, B. J., Wood, P. C., and Li, Q. Q. (2005). Compilation of mRNA polyadenylation signals in Arabidopsis revealed a new signal element and potential secondary structures. Plant Physiol. 138, 1457–1468.
Lopato, S., Kalyna, M., Dorner, S., Kobayashi, R., Krainer, A. R., and Barta, A. (1999). atSRp30, one of two SF2/ASF-like proteins from Arabidopsis thaliana, regulates splicing of specific plant genes. Genes Dev. 13, 987–1001.
Lutz, C. S., and Moreira, A. (2011). Alternative mRNA polyadenylation in eukaryotes: an effective regulator of gene expression. Wiley Interdiscip. Rev. RNA 2, 22–31.
Macdonald, C. C., and McMahon, K. W. (2010). Tissue-specific mechanisms of alternative polyadenylation: testis, brain, and beyond. Wiley Interdiscip. Rev. RNA 1, 494–501.
Mangone, M., Manoharan, A. P., Thierry-Mieg, D., Thierry-Mieg, J., Han, T., Mackowiak, S. D., Mis, E., Zegar, C., Gutwein, M. R., Khivansara, V., Attie, O., Chen, K., Salehi-Ashtiani, K., Vidal, M., Harkins, T. T., Bouffard, P., Suzuki, Y., Sugano, S., Kohara, Y., Rajewsky, N., Piano, F., Gunsalus, K. C., and Kim, J. K. (2010). The landscape of C. elegans 3′UTRs. Science 329, 432–435.
Mayr, C., and Bartel, D. P. (2009). Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138, 673–684.
Meyers, B. C., Vu, T. H., Tej, S. S., Ghazal, H., Matvienko, M., Agrawal, V., Ning, J., and Haudenschild, C. D. (2004). Analysis of the transcriptional complexity of Arabidopsis thaliana by massively parallel signature sequencing. Nat. Biotechnol. 22, 1006–1011.
Nagasaki, H., Arita, M., Nishizawa, T., Suwa, M., and Gotoh, O. (2005). Species-specific variation of alternative splicing and transcriptional initiation in six eukaryotes. Gene 364, 53–62.
Raczynska, K. D., Simpson, C. G., Ciesiolka, A., Szewc, L., Lewandowska, D., McNicol, J., Szweykowska-Kulinska, Z., Brown, J. W., and Jarmolowski, A. (2010). Involvement of the nuclear cap-binding protein complex in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 38, 265–278.
Robinson, J. T., Thorvaldsdottir, H., Winckler, W., Guttman, M., Lander, E. S., Getz, G., and Mesirov, J. P. (2011). Integrative genomics viewer. Nat. Biotechnol. 29, 24–26.
Shen, Y., Ji, G., Haas, B. J., Wu, X., Zheng, J., Reese, G. J., and Li, Q. Q. (2008a). Genome level analysis of rice mRNA 3′-end processing signals and alternative polyadenylation. Nucleic Acids Res. 36, 3150–3161.
Shen, Y., Liu, Y., Liu, L., Liang, C., and Li, Q. Q. (2008b). Unique features of nuclear mRNA poly(A) signals and alternative polyadenylation in Chlamydomonas reinhardtii. Genetics 179, 167–176.
Shen, Y., Venu, R. C., Nobuta, K., Wu, X., Notibala, V., Demirci, C., Meyers, B. C., Wang, G. L., Ji, G., and Li, Q. Q. (2011). Transcriptome dynamics through alternative polyadenylation in developmental and environmental responses in plants revealed by deep sequencing. Genome Res. 21, 1478–1486.
Shepard, P. J., Choi, E. A., Lu, J., Flanagan, L. A., Hertel, K. J., and Shi, Y. (2011). Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA 17, 761–772.
Simpson, G. G., Dijkwel, P. P., Quesada, V., Henderson, I., and Dean, C. (2003). FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113, 777–787.
Vasudevan, S., Peltz, S. W., and Wilusz, C. J. (2002). Non-stop decay – a new mRNA surveillance pathway. Bioessays 24, 785–788.
Wachter, A., Tunc-Ozdemir, M., Grove, B. C., Green, P. J., Shintani, D. K., and Breaker, R. R. (2007). Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant Cell 19, 3437–3450.
Wu, X., Liu, M., Downie, B., Liang, C., Ji, G., Li, Q. Q., and Hunt, A. G. (2011). Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc. Natl. Acad. Sci. U.S.A. 108, 12533–12538.
Keywords: alternative polyadenylation, splicing, introns, exons, UTRs
Citation: Hunt AG (2012) RNA regulatory elements and polyadenylation in plants. Front. Plant Sci. 2:109. doi: 10.3389/fpls.2011.00109
Received: 31 October 2011; Paper pending published: 13 November 2011;
Accepted: 17 December 2011; Published online: 04 January 2012.
Edited by:
Anireddy S. N. Reddy, Colorado State University, USACopyright: © 2012 Hunt. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Arthur G. Hunt, Department of Plant and Soil Sciences, University of Kentucky, Lexington, KY 40546-0312, USA. e-mail: aghunt00@uky.edu