- 1 Faculty of Life Sciences, University of Manchester, Manchester, UK
- 2 Children’s Nutrition Research Center, Baylor College of Medicine, Houston, TX, USA
Cation transport is a critical process in all organisms and is essential for mineral nutrition, ion stress tolerance, and signal transduction. Transporters that are members of the Ca2+/cation antiporter (CaCA) superfamily are involved in the transport of Ca2+ and/or other cations using the counter exchange of another ion such as H+ or Na+. The CaCA superfamily has been previously divided into five transporter families: the YRBG, Na+/Ca2+ exchanger (NCX), Na+/Ca2+, K+ exchanger (NCKX), H+/cation exchanger (CAX), and cation/Ca2+ exchanger (CCX) families, which include the well-characterized NCX and CAX transporters. To examine the evolution of CaCA transporters within higher plants and the green plant lineage, CaCA genes were identified from the genomes of sequenced flowering plants, a bryophyte, lycophyte, and freshwater and marine algae, and compared with those from non-plant species. We found evidence of the expansion and increased diversity of flowering plant genes within the CAX and CCX families. Genes related to the NCX family are present in land plant though they encode distinct MHX homologs which probably have an altered transport function. In contrast, the NCX and NCKX genes which are absent in land plants have been retained in many species of algae, especially the marine algae, indicating that these organisms may share “animal-like” characteristics of Ca2+ homeostasis and signaling. A group of genes encoding novel CAX-like proteins containing an EF-hand domain were identified from plants and selected algae but appeared to be lacking in any other species. Lack of functional data for most of the CaCA proteins make it impossible to reliably predict substrate specificity and function for many of the groups or individual proteins. The abundance and diversity of CaCA genes throughout all branches of life indicates the importance of this class of cation transporter, and that many transporters with novel functions are waiting to be discovered.
Introduction
The importance of transporters as “gatekeepers” of the cell is exemplified in recent genomic studies. For example, the central role of transporters in controlling cell growth was indicated in a study showing that increased yeast growth correlates with increased cell surface area, and thus increased nutrient uptake (Groeneveld et al., 2009). Indeed, of the 1312 distinct reaction steps in a reconstruction of yeast metabolism, 401 involve transport (Herrgard et al., 2008). Likewise, the analysis of fully sequenced photosynthetic eukaryotes finds that transporters consistently make up a significant proportion (∼5%) of the genome (Mäser et al., 2001; Merchant et al., 2007; Rensing et al., 2008). Calcium (Ca2+) is a critical element in all organisms; it is an essential nutrient which also has a conserved signaling role (Hirschi, 2004; Case et al., 2007). Transporters that mediate the movement of this ion are therefore particularly important. These Ca2+ transporters impact upon cell division, growth, development, and adaptation to environmental conditions. Three classes of membrane transporters help mediate Ca2+ flux across a membrane and regulate cytosolic Ca2+ levels: Ca2+-permeable channels, Ca2+-ATPases, and Ca2+/cation antiporters (CaCAs). Biochemical identification and functional analysis of CaCAs in an array of organisms have helped conceptualize the functional properties and important physiological roles of these exchangers in cellular ion homeostasis (Lytton, 2007; Manohar et al., 2011).
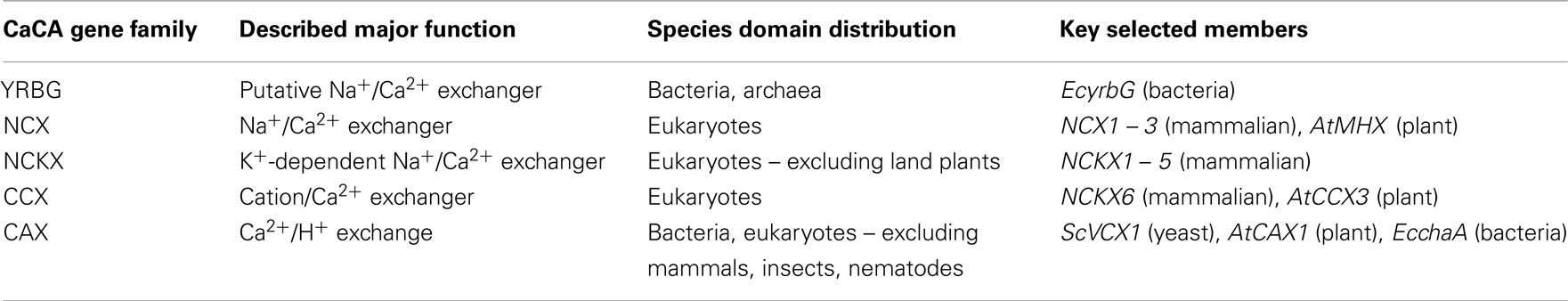
The majority of the ion-coupled Ca2+ exchange activity within a cell is encoded by members of the Ca2+/cation antiporter (CaCA) gene superfamily which are present widely in archaea, bacteria, fungi, plants, and animals (Saier et al., 1999; Cai and Lytton, 2004a; Table 1). These proteins serve as essential components in Ca2+ cycling systems. CaCA proteins promote Ca2+ efflux across membranes, normally against its concentration gradient, by using a counter-electrochemical gradient of other ions such as H+, Na+, or K+ to energize the process. Animal proteins principally use Na+ gradients as the driving force while plant and bacterial exchangers exclusively utilize H+. Based on functional and phylogenetic analysis, the CaCA superfamily is composed of at least five families (Cai and Lytton, 2004a): the Na+/Ca2+ exchanger (NCX) family, the Na+/Ca2+, K+ exchanger (NCKX) family and the CCX family, the H+/cation exchanger (CAX) family, and the YRBG family named after the Escherichia coli gene yrbG (Table 1). Detailed characterization of selected isoforms from each of these families has begun to shed light on their functions (summarized in Table 2).
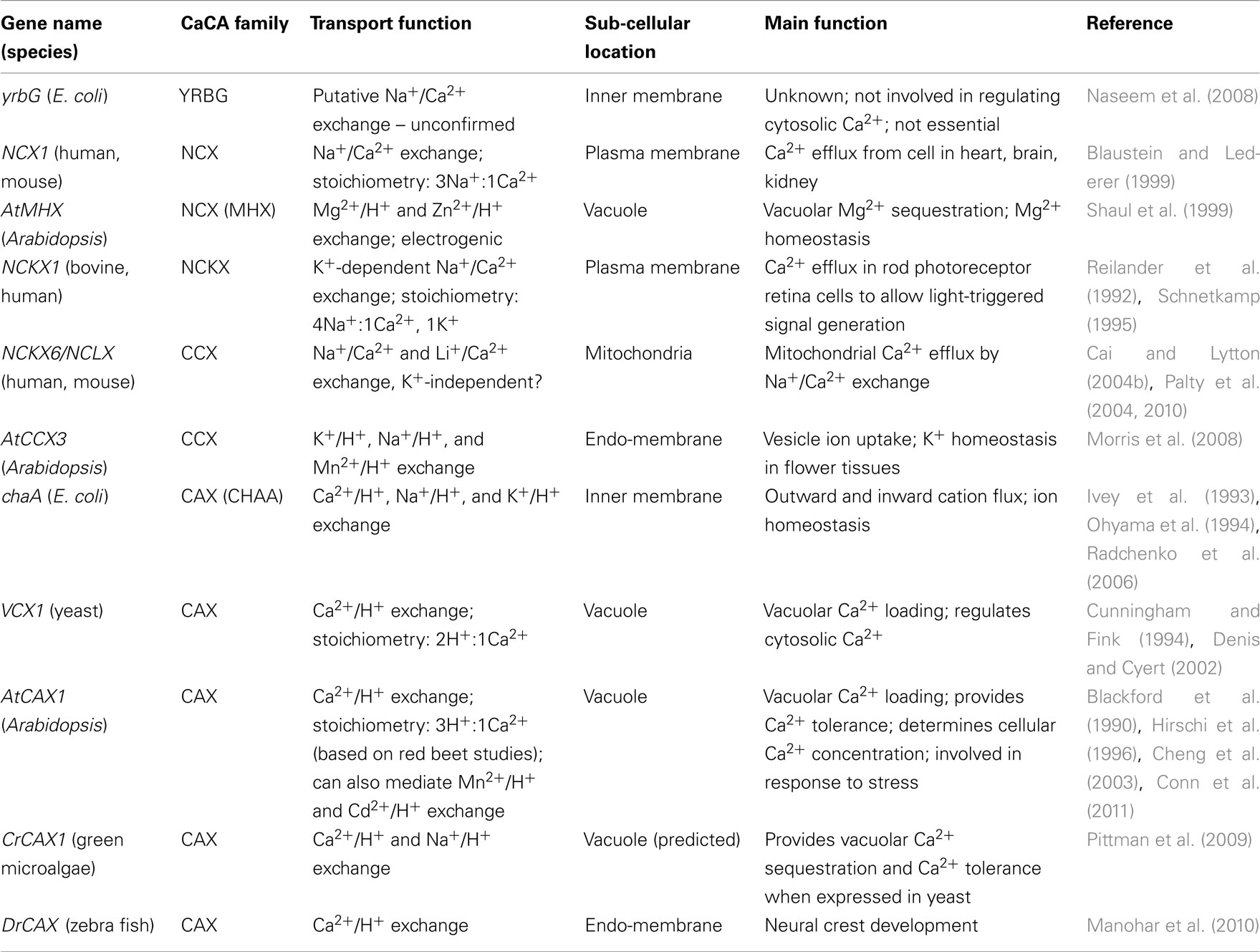
The YRBG family has been the least characterized. Sequence similarity suggests E. coli YrbG may function as a NCX but the transporter is not essential for bacterial growth and it is unknown whether YrbG is a Ca2+ transporter (Cai and Lytton, 2004a; Naseem et al., 2008). In E. coli, pH and monovalent cations can regulate cytosolic free Ca2+ through Ca2+ influx and efflux, but direct measurement of cytosolic Ca2+ levels using the Ca2+ reporter aequorin suggests that YrbG does not play a role in regulating cytosolic Ca2+ (Naseem et al., 2008). Orthologs of yrbG are present in other prokaryotes but do not appear to be present in eukaryotes (Cai and Lytton, 2004a).
Na+/Ca2+ exchangers, present at the plasma membrane of most animal cells, are a fast and high-capacity Ca2+ transport system that are important regulators of cellular Ca2+ homeostasis by causing efflux of Ca2+ from the cell. Detailed functional and molecular studies have revealed two distinct families of Na+/Ca2+ exchange proteins (Lytton, 2007). The first of these, the NCX transporters are composed of three distinct types in mammals: NCX1, NCX2, and NCX3. All three exchangers share about 70% overall amino acid identity and have identical transport function. Invertebrates have a single NCX gene, whereas vertebrate species have multiple NCX genes as a result of at least two duplication events (On et al., 2008). The presence of NCX genes in all animal species’ genomes allowed the construction of a phylogenetic tree that correlated to animal evolution and revealed that the origin of NCX duplication was initiated at the emergence of vertebrate organisms (On et al., 2008). A plant Mg2+/H+ exchanger termed Arabidopsis thaliana Mg2+/H+ exchanger (AtMHX), is also a member of the NCX family. This transporter concentrates Mg2+ into vacuoles and has no apparent Na+/Ca2+ exchange activity (Shaul et al., 1999). Another putative Mg2+/H+ exchanger (XNTA) has been discovered in Paramecium, but although it appears to be a member of the CaCA superfamily, it is distinct from AtMHX and does not appear to fall within the NCX clade (Haynes et al., 2002).
The second Na+/Ca2+-transporting exchanger family, the NCKX, operate in cellular Ca2+ efflux by extruding one Ca2+ ion and one K+ ion in exchange for four Na+ ions (Lytton, 2007). Like the NCX transporters, the NCKX play an important role in Ca2+ homeostasis, notably in retinal and neuronal cells, and are important in various roles including photoreceptor function and skin pigmentation (Table 2). Also like NCX transporters, NCKX are reversible and can import Ca2+ into the cell upon reversal of the transmembrane Na+ gradient. NCKX proteins differ from NCX proteins in their requirement for K+, their Ca2+ kinetics, and sequence divergence. To date there has been no evidence of NCKX activity or NCKX-like genes present in any plants.
Initial analyses of the CaCA superfamily did not identify the CCX family and this family has only recently been created and considered a stand-alone group. A novel gene from mammals, originally thought to be a NCKX gene, named NCKX6, has significant sequence variation from other known NCX and NCKX genes (Cai and Lytton, 2004b). Subsequent phylogenetic analysis has grouped NCKX6 separately from the NCX and NCKX family in the CCX family, which includes members from vertebrates, invertebrates, fungi, protozoa, and plants (Cai and Lytton, 2004a). NCKX6 (also named NCLX) appears to be a bona fide NCX but it is slightly contentious as to whether it is K+-dependent or not; other studies have suggested that this transporter is K+-independent and can also transport Ca2+ by Li+ exchange (Palty et al., 2004; Table 2). Several transporters in plants, some formerly referred to as CAX, have been reassigned to the CCX family (Shigaki et al., 2006). For example, Arabidopsis CCX3 (formerly CAX9) is a H+-dependent cation transporter but with no apparent Ca2+ transport properties (Morris et al., 2008).
Members of the CAX family have been identified from many species including plants, algae, some invertebrate animals, fungi, protozoa, and bacteria, but are absent from the genomes of mammals, insects, and nematodes (Shigaki et al., 2006; Manohar et al., 2011). CAXs have been classified into three major categories based on phylogenetic analysis: Type 1, 2, and 3 (Shigaki et al., 2006). Several Type 1 and 3 CAXs have been characterized across species, particularly in plants and yeast, and have been shown to be important for Ca2+/H+ exchange, predominantly at the vacuolar membrane (Manohar et al., 2011; Pittman, 2011). In particular, many plant and fungal CAXs are important for providing tolerance to excess Ca2+, for regulating Ca2+ homeostasis and nutrition, and the modulation of environmental or developmental events via Ca2+ signal generation (McAinsh and Pittman, 2009; Cunningham, 2011). However, to date only two Type 2 CAXs, the S. cerevisiae Vnx1 and zebrafish DrCax1 have been characterized (Cagnac et al., 2007, 2010; Manohar et al., 2010).
The Type 1 plant CAXs have been further divided into two phylogenetically distinct groups (Type 1A and Type 1B; Shigaki et al., 2006). This distinction suggests functional differentiation between the two groups; however, no clear-cut functional differences have so far been identified. Arabidopsis and rice (Oryza sativa) each have six CAX ORFs equally divided between the two phylogenetic groups. One suggestion has been that there are differences in ion substrate selectivity between members of the two groups; for example, Type 1B proteins such as AtCAX2 and AtCAX5 have been shown to transport multiple ions including Mn2+ and Cd2+ in addition to Ca2+ (Hirschi et al., 2000; Shigaki et al., 2003; Edmond et al., 2009) while Type 1A members such as AtCAX1 and AtCAX3 are important for Ca2+ homeostasis and were originally thought to be specific for Ca2+ (Shigaki et al., 2003; Conn et al., 2011). However, Type 1A CAXs can mediate the transport of multiple ions in planta and when heterologously expressed in yeast (Kamiya et al., 2005; Shigaki et al., 2005, 2010; Koren’kov et al., 2007; Mei et al., 2009). Thus plant CAXs have a broad cation specificity and hence the CAX family has more recently been referred to as H+/cation exchangers rather than just H+/Ca2+ exchangers. The Type 1B sub-group may contain more diverse CAX proteins. Two non-canonical Type 1B CAXs with an additional transmembrane (TM) insertion between TM4 and TM5 were recently reported from plants in the Asteraceae family (Jain et al., 2009), although the relevance of this alteration is unknown. Recently, a CAX gene from the chlorophyte microalgae Chlamydomonas reinhardtii was identified and suggested to group within the Type 1C sub-group alongside protozoan CAX genes (Pittman et al., 2009). Like the higher plant CAXs, CrCAX1 appears to have broad substrate specificity, and in addition to Ca2+ can also transport the monovalent ion Na+.
All CaCA proteins share a similar topological model: on average 10 TM helices are predicted to form a Ca2+ transduction pathway which includes two conserved α-repeats regions within TM helices 2–3 and 7–8. These two α-repeats are important for ion selectivity, binding, and transport in diverse CaCA members (Kamiya and Maeshima, 2004; Ottolia et al., 2005; Shigaki et al., 2005) and are predicted from topology models to be orientated on opposite sides of the membrane (Figure 1), and in the folded structure of the protein face toward each other to form an ion conductance channel (Nicoll et al., 2007). This topology is exemplified by NCX (Figure 1). The sizes of individual members of the CaCA superfamily vary from ∼300 to 1000 amino acids, and although the core topological structure is conserved there are some exceptions; for example, the CCX proteins have additional TM domains within the C-terminal half of the protein (Cai and Lytton, 2004b). The clusters of 5 + 5 TM helices within both halves of a typical CaCA are separated by a large cytosolic loop (Figure 1) that regulates exchange activity in some members. It is likely that both halves of the protein formed following an ancient duplication event. While split halves of NCX or CAX proteins are not able to function alone (Ottolia et al., 2001; Zhao et al., 2009), it is not inconceivable that an ancestral form was useful as part of a related protein domain if this event occurred very early in the evolution of the gene family.
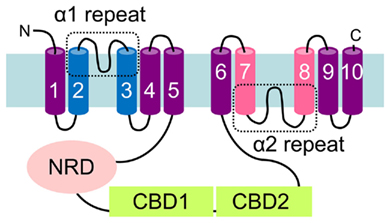
Figure 1. Topological model of the Na+/Ca2+ exchanger (NCX) protein. Barrel structures denote the transmembrane spanning domains identified by hydropathy analysis using TMHMM version 2. The conserved α1- and α2-repeat regions which span TM 2 and 3, and TM 7 and 8 are highlighted. The Ca2+-binding domains (CBD) and the predicted Na+ regulatory domain (NRD) present on the large central cytosolic loop are highlighted.
In most genomes putative membrane transporters abound with few transporters representing unique sequences in the genome (Bock et al., 2006). Despite fundamental differences in organismal complexity, an emerging theme is that diverse species have similar protein-coding potentials and alterations in CaCA transporters among taxa may be associated with modifications in function. The recent sequencing of genomes from diverse photosynthetic eukaryotes, including various higher plants, bryophytes, lycophytes, unicellular and multi-cellular green, red, and brown alga, and diatoms, act as powerful genomic resources for continued comparative analyses of CaCA proteins. Evolutional relationships among the characterized and uncharacterized CaCAs will lead to a better understanding of their potential roles. In this study we have performed a multi-genome identification of CaCA genes and a phylogenetic analysis that have provided insights into the evolution of CaCA transporters within higher plants and the green plant lineage.
Materials and Methods
Data Sources
Predicted protein-coding genes from complete and near complete genome sequences were downloaded for 74 species in May 2011 from a variety of databases (Table S1 in Supplementary Material). Species were selected for their broad taxonomic range in order to capture the full diversity of CaCA sequences, and where possible, photosynthetic organisms were included. The species examined included 12 land plants (including one lycophyte and one bryophyte), and 12 algae of various phyla and class (Table S1 in Supplementary Material), four fungi, eight other unicellular eukaryotes (including a variety of protists and oomycetes), seven animals (including two mammals), 21 bacteria (of which 10 were members of the cyanobacteria), and 10 archaea.
CaCA Identification
Putative CaCA proteins were identified using the basic local alignment search tool (BLAST; Altschul et al., 1997). A local database was constructed from the predicted protein sequences for the 74 species examined. The database was queried with 147 previously identified CaCA protein-coding sequences (Cai and Lytton, 2004a) including all known CaCA proteins from A. thaliana. The initial results contained a variety of putative ion transporter sequences from the CaCA superfamily and from other families, in particular those with known functions in Na+/H+, arginine, and citrate transport, which were discarded. To obtain sequences specific to the CaCA transporter family, only those matches with an e-value of <1, that spanned a region of >100 bp in length, with no less than 15% sequence identity, and where the predicted coding sequence corresponding to the match was >280 bp in length were considered for further analysis. The remaining 1328 candidate CaCA homologs were then screened by hydropathy analysis using TMHMM v2.01 (Krogh et al., 2001) to remove all non-TM proteins and to remove those whose hydropathy profile was clearly dissimilar to a selection of representative CaCA genes from the NCX, NKCX, CCX, CAX, and YRBG families from human, A. thaliana, O. sativa, S. cerevisiae, E. coli, and Synechocystis sp. Further screening of candidate sequences was performed by aligning them with a small selection of well-characterized representative CaCA proteins, and visual inspection was used to remove those that were clearly not members of the CaCA family. Finally 308 putative CaCA homologs were retained for phylogenetic analysis, plus 34 novel CaCA-like proteins (named EF–CAX) that were shown by Pfam analysis2 (Finn et al., 2010) to contain putative Ca2+-binding EF-hand domains (Pfam ID: PF00036) and so-called “Na_Ca_ex” (consensus CaCA) domains (Pfam ID: PF01699; Table S2 in Supplementary Material).
Phylogenetic Analyses
Phylogenetic analysis of full-length CaCA proteins was not suitable because their diversity makes sequence alignment difficult. Several steps were taken in order to obtain reasonable alignments. Firstly, poorly conserved and extended N- and C-terminal regions of the predicted proteins were removed. TMHMM was then used to identify and extract putative TM domains for each protein. Furthermore, the greatest sequence distance between two TM domains was recorded and inferred to be the length of the central loop region. Visual inspection of preliminary alignments suggested that the number of TM domains can be variable between species, which may affect the accuracy of sequence alignment. To account for these differences we used MAFFT v6.0 (Katoh et al., 2002) under the E-INS-i option, which is specifically designed to cope with homologous regions separated by unalignable regions. Other approaches were also investigated for aligning these regions, and all yielded comparable results. The final phylogeny was estimated using maximum likelihood under the WAG + F model of amino acid substitution with γ-distributed rates across sites, as implemented in RAxML v7.1 (Stamatakis, 2006). Other substitution models were examined and found to yield very similar or substantially worse log-likelihood values. Confidence in the tree was assessed using the fast bootstrap approach (Stamatakis et al., 2008). The tree was viewed using the FigTree program3. The resulting phylogeny appeared to be broadly consistent with previous findings, with five major clades corresponding to the CAX, CCX, NCX, NCKX, and YRBG genes. Thus gene products were classified according to their position in the phylogeny. For the phylogenetic analysis of the EF–CAX, due to poor sequence similarity between the N-terminal half of these proteins with N-terminal sequence of the other CaCA proteins, the conserved α2-repeat region sequence within the C-terminal half of CaCA and EF–CAX proteins was used instead. This sequence was identified using the consensus α2-repeat region sequence as determined previously for CaCA proteins (Cai and Lytton, 2004a), and extracted from each CaCA protein. The α2-repeat sequences were realigned to confirm that the correct consensus sequence was obtained and were then used to generate trees using maximum likelihood, as described above.
Amino Acid Sequence Comparisons
For the visual analysis and comparison of CaCA protein sequences, multiple sequence alignments were performed using ClustalW24 (Larkin et al., 2007) and shaded using the BOXSHADE v.3.2 viewer and manually annotated.
Results and Discussion
Identification of Genes Encoding CaCA Homologs in Photosynthetic Eukaryotes
To examine the evolution and potential diversification of the CaCA superfamily in photosynthetic eukaryotes and throughout the land plant lineage, we screened the sequenced genomes of 10 flowering plant species. These included six dicots: A. thaliana, a tree species – poplar (Populus trichocarpa), grape (Vitis vinifera); and three legumes: soybean (Glycine max), Medicago truncatula, and Lotus japonica, and four monocots: rice (O. sativa), corn (Zea mays), sorghum (Sorghum bicolor), and Brachypodium distachyon. The genomes of two non-flowering land plants were available for analysis; the bryophyte moss Physcomitrella patens, and the lycophyte spikemoss Selaginella moellendorffii. A range of diverse algal species were also chosen. These comprised four freshwater algae: the model unicellular chlorophyte alga C. reinhardtii, the unicellular chlorophyte Coccomyxa sp. C-169 (formerly named Chlorella sp. C-169), and the multi-cellular chlorophyte Volvox carteri; and a selection of marine algae: the prasinophytes Ostreococcus tauri and Micromonas pusilla, the rhodophyte Cyanidioschyzon merolae, the haptophytes Emiliania huxleyi, the brown alga Ectocarpus siliculosus, the pelagophyte Aureococcus anophagefferens, and photosynthetic members of the stramenopiles, including the diatoms Thalassiosira pseudonana, Phaeodactylum tricornutum, and Fragilariopsis cylindrus. In addition, two non-photosynthetic stramenopiles, the oomycetes Phytophthora sojae, and Pythium ultimum were examined. A selection of mammalian, invertebrate, fungal, protozoan, bacterial, cyanobacterial, and archaeal species with completed genome sequences were chosen that provide a cross section of life for comparison with the plant and algal species (Table S1 in Supplementary Material). Using a dataset of previously identified CaCA query sequences, the genomes of all these species were screened by BLAST analysis and the results were further screened by hydrophobicity analysis, sequence alignment, and individual protein BLAST to validate each of the outputs and generate a final list of putative CaCA proteins (see Materials and Methods). In 13 of the species screened (including five bacteria and seven archaea) no obvious CaCA genes were identified (Table S1 in Supplementary Material). We were also unable to identify a clear-cut CaCA gene from the eukaryotic protozoan parasite Giardia lamblia. In contrast, the remaining 18 prokaryote species and all other eukaryotes examined including all of the plant and algal species possessed CaCA gene members (Figure 2). In all 342 CaCA genes were identified from 74 species (including the 13 species with no CaCA genes), giving an average of 4.6 CaCA genes per species. From the 12 land plants, 159 genes were determined (13.25 genes per species) and from the 12 algal species, 70 genes were identified (5.8 genes per species). The search method used multiple queries for each genome examined with the robust search method employed suggesting confidence that the search returned all expected genes and that CaCA genes are truly lacking in 13 of the species.
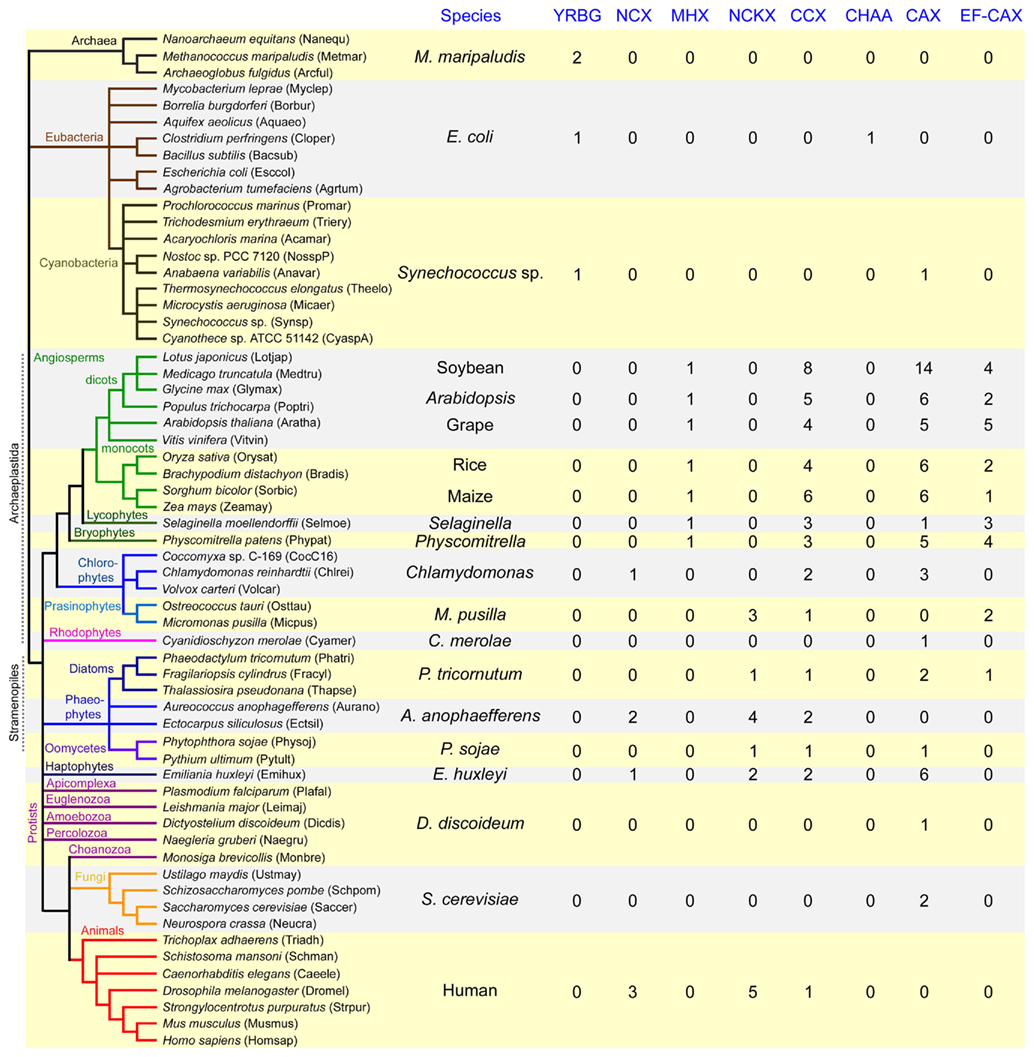
Figure 2. Variation in the distribution of genes within each of the major CaCA gene families (YRBG, NCX, NCKX, CCX, CHAA, CAX) and sub-families (MHX, EF–CAX) across eukaryotic and prokaryotic organisms. Gene numbers determined by phylogenetic analysis from representative species for each of the classes of organisms examined are shown. The species from which CaCA genes were identified are listed (left hand side) with the species name abbreviation in parentheses. The evolutionary relationships of the species is shown, generated using iTOL (http://itol.embl.de) and based on the NCBI taxonomy tree. For the full gene distribution for each of the species analyzed see Table S1 in Supplementary Material.
The classification of the genes into one of the five CaCA families was determined by phylogenetic analysis of the full superfamily. Initial amino acid sequence alignments using full-length sequence generated poor alignments and resulted in a low confidence tree. To improve the sequence alignments and generate the tree, the highly variable N- and C-terminal tail sequences and the hydrophilic loop sequences were therefore removed. The predicted size of the central loop region, the longest hydrophilic loop region of the CaCA protein, situated between TM5 and 6 of the consensus NCX (Figure 1), NCKX, CCX, and YRBG proteins, and between TM6 and 7 of the consensus CAX proteins, varied largely between the CaCA sequences, ranging from 19 amino acids in one of the CAX proteins to 602 amino acids in one of the CCX proteins. The NCX, NCKX, and CCX proteins all had on average a longer central loop region than the CAX and YRBG proteins; however, within all of these families there was significant variation in central loop length and the predicted number of TM domains (Table S3 in Supplementary Material). In particular, there was large variation within the NCX, NCKX, CCX, and CAX families. The significant variation within the CAX family was caused in part by the topological variation and variation in protein size between the Type 1 and Type 2 CAX protein sub-types (Shigaki et al., 2006). There was also large variation in the size of the N-terminal regions before the first TM span. Many of the animal NCX, NCKX, and CCX proteins are predicted to possess an N-terminal signal peptide (Lytton, 2007), while many of the plant CAX proteins possess a long N-terminal tail that has been shown to play a post-translational regulatory role (Pittman et al., 2002; Pittman, 2011), but which is absent in the fungal and bacterial CAX proteins.
The resulting tree indicates the presence of the five main CaCA families identified in previous analysis (Cai and Lytton, 2004a) although the bootstrap values at the base of the NCKX, NCX, and CCX branches were low (Figure 3). Likewise, some of the exact details for this tree and the individual family trees should be viewed with caution because long branches could lead to phylogenetic artifacts such as long branch attraction. The YRBG family contains only prokaryotic genes, including E. coli yrbG and genes from other bacteria (including cyanobacteria), but also archaea (Figure 3). None of the prokaryotic species analyzed here contained genes that fell within the NCX, NCKX, and CCX families. These families possess higher plant (in the case of NCX and CCX), algal, and animal genes, with some fungal-derived genes in the CCX family. The algal genes are widely dispersed amongst these three families and it is mostly the algal gene product sequences that yield the longest branch lengths within the tree indicating significant sequence variation. Furthermore, algal genes were also identified which were basal to the NCX and NCKX groups and could not be distinguished between the two families based on phylogeny (Figure 3). The largest proportion of CaCA genes identified was from the CAX family (120 of the 342 genes, which is 1.6 CAX genes per species; Table S1 in Supplementary Material). These genes were from all the classes of organisms examined including some invertebrate animals, as shown previously (Shigaki et al., 2006), but there were no mammalian or insect CAX genes. The CAX family included some bacterial and cyanobacterial genes but none from any of the archaeal species examined. As with the CCX family, the size and branching of the CAX family indicates significant sequence variation and potential diversification. The E. coli protein ChaA, originally identified as a H+/Ca2+ exchanger (Ivey et al., 1993) and subsequently suggested to function as a H+/Na+ exchanger (Ohyama et al., 1994), was not identified as a CaCA homolog by the BLAST search performed by Cai and Lytton (2004a). These authors commented that the ChaA protein had limited sequence similarity with other CaCA proteins. More recent phylogenetic analysis of the CAX family suggested that chaA falls within the Type 3C branch of the CAX tree (Shigaki et al., 2006). The BLAST search in this study did identify E. coli chaA and our global CaCA phylogenetic analysis indicates that chaA and other related genes from Agrobacterium tumefaciens and Mycobacterium leprae are distinct from the other CAX genes and form a separate group which we have named here CHAA (Figure 3). Like the YRBG group, the CHAA group is specifically prokaryotic.
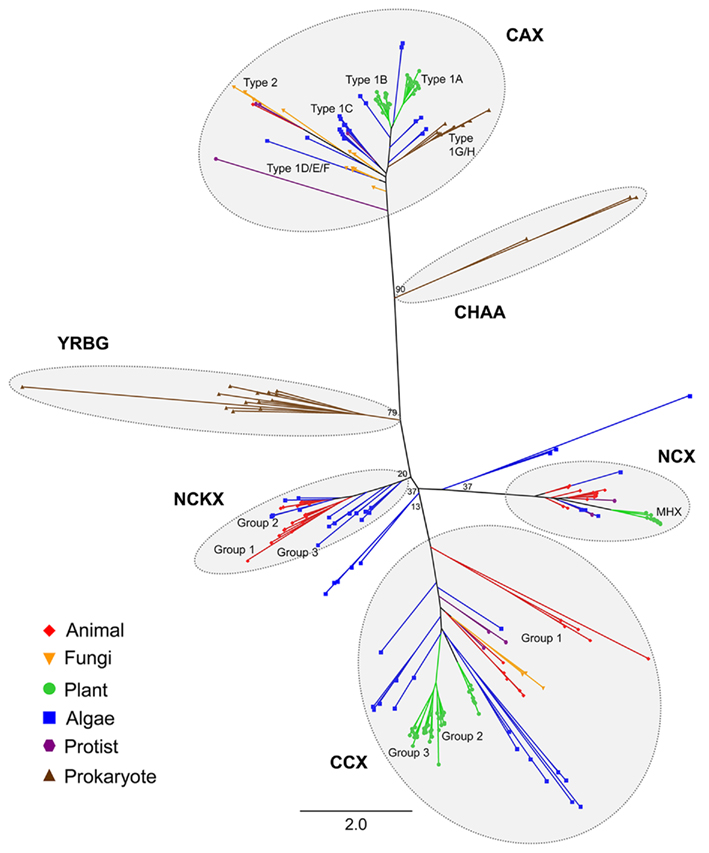
Figure 3. A phylogenetic tree of the CaCA superfamily. The tree was constructed using the maximum likelihood method and derived from alignments of conserved hydrophobic region sequences identified from the genomes of selected animal, fungal, land plant, algae, protist, bacterial, and archaebacterial species, listed in Figure 2. Six major groups are highlighted. The sub-groups determined for the CAX, CCX, NCX, and NCKX families as shown in Figures 4, 6, 7, and 9, are also given. Line colors and symbols denote the species class. Bootstrap values are indicated at the nodes of major branches. The branch length scale bar indicates the evolutionary distance of two amino acid substitutions per site.
NCKX and “Animal-Type” NCX Genes are Absent in Land Plants but Present in Algae
Our analysis of the selected genomes indicated that NCX and NCKX genes appear to be ubiquitous in vertebrate and invertebrate species, as previously determined (Cai and Lytton, 2004a; On et al., 2008). In contrast, no genes from any of the higher plants, bryophyte, or lycophyte species were identified that group within the NCKX family (Figure 3). Likewise, no genes from any of the land plants were found to cluster closely with the NCX genes from animals (Figure 4), although more distantly related NCX-like genes were present (see below). Higher plants probably lack sufficient transmembrane Na+ gradients to energize the counter transport of Ca2+ or other cations and hence have retained Ca2+/H+ exchangers instead of Na+/Ca2+ exchange activity (Hirschi, 2001). NCX genes were however, detected in algae including the chlorophyte Chlamydomonas, the pelagophyte A. anophagefferens, and the haptophyte E. huxleyi (Figure 4). These algal NCX homologs have high sequence similarity with mammalian NCX proteins including conservation of the key residues in the α1- and α2-repeat regions (Figure 5) that are critical for Na+/Ca2+ exchange activity (Lytton, 2007). Furthermore, the genes from both of the marine algae, which are more distantly related from the land plants and chlorophyte algae, cluster more closely with animal NCX genes (Figure 4), and we would predict them to have a similar function.
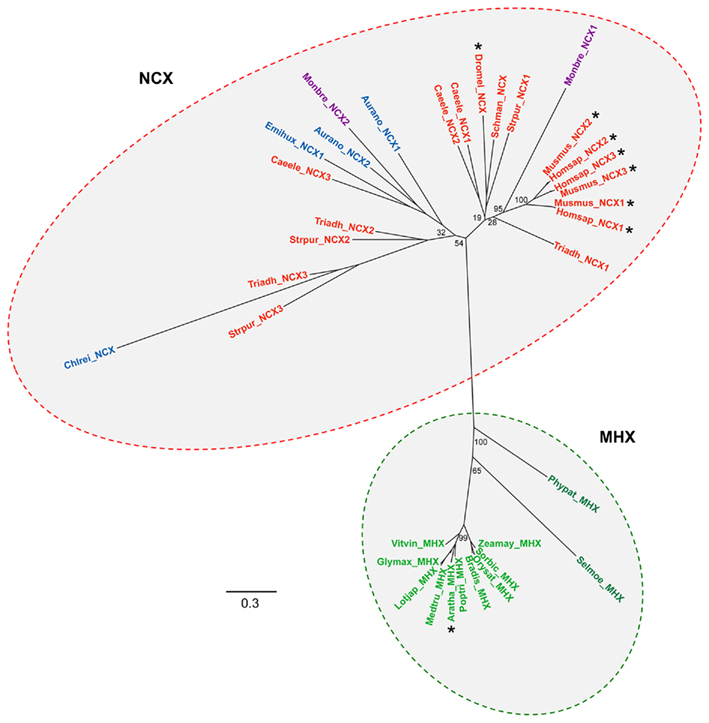
Figure 4. Na+/Ca2+ exchanger family phylogenetic tree. The tree was constructed using genes identified in the NCX group of the full CaCA tree (Figure 3). A separate NCX sub-group named MHX is highlighted which includes all the plant genes including Arabidopsis MHX. Branch labels include the species name abbreviation defined in Figure 2 and are colored according to the species class using the same color code as in Figure 3. Bootstrap values are indicated at the nodes of major branches. The branch length scale bar indicates the evolutionary distance of 0.3 amino acid substitutions per site. Genes that are highlighted with an asterisk encode transporters for which the ion specificity has been determined.
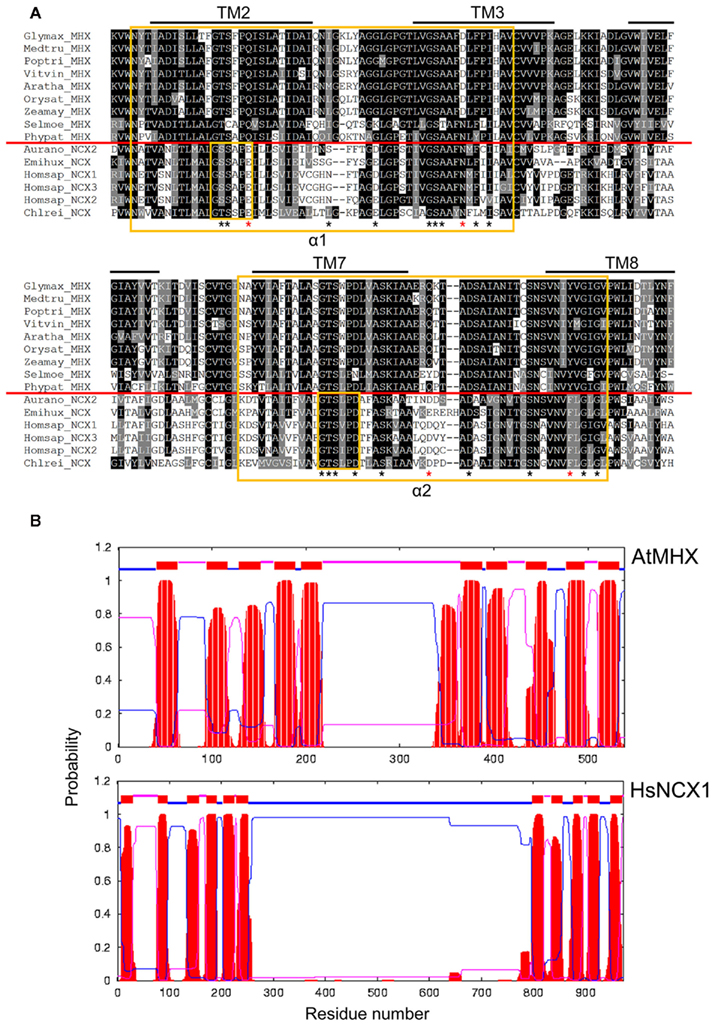
Figure 5. Sequence and structural variation of NCX and MHX homologs. (A) Multiple amino acid sequence alignments of the α1- and α2-repeat region sequence from selected plant MHX, human NCX, and algal NCX proteins. Alignments were performed using ClustalW. Amino acids that are identical or similar are shaded black or gray, respectively. Predicted hydrophobic regions and putative transmembrane spans are over-lined. The α-repeat regions and signature residues are boxed in yellow. The red line separates the MHX and NCX sequences. Asterisks indicate residues shown to be important for Na+/Ca2+ exchange activity in NCX1, with those in red being not conserved in the MHX sequences. (B) Topology models of AtMHX and HsNCX1 generated by TMHMM. Red areas indicate predicted TM spans.
The NCKX tree also contains genes from various algal species (Figure 6). The NCKX tree has three main groups with Group 1 being animal-specific, containing invertebrate, insect, and mammalian genes, including human NCKX3, NCKX4, and NCKX5. Group 2 has animal genes including human NCKX1 and NCKX2. Algal NCKX genes from the two Stramenopile species A. anophagefferens and P. tricornutum, and the prasinophyte Micromonas cluster close to Group 2 but the lower bootstrap values and longer branch lengths of these genes do not allow us to confidently assign them to Group 2 rather than Group 3. Group 3 is an algal NCKX group, including another Micromonas gene and two genes from the related O. tauri picoalgae, two genes from E. huxleyi, and two genes from the brown alga E. siliculosus, plus genes from the two oomycete species, that are related to the Stramenopile algae (Figure 6). It is notable that all of the algal NCKX genes are from marine species and from non of the freshwater species, suggesting a potential link between the high Na+ and K+ concentration in seawater and the presence of a putative K+-regulated Na+-coupled Ca2+ transporter. Indeed all of the marine algae examined here, with the exception of the red algae C. merolae, either possessed an NCKX gene or a gene that was basal to the NCKX group (Figure 2), suggesting that these transporters may be critical for life in a marine environment. The apparent absence of NCX and NCKX genes from all land plants but their presence in various classes of algae, including the more closely related chlorophytes and prasinophytes, may suggest that they have been lost in all plants. However, it is also possible that these genes were also absent from the common ancestor species in the green eukaryote lineage and that the algae lineage subsequently acquired these genes through horizontal transfer.
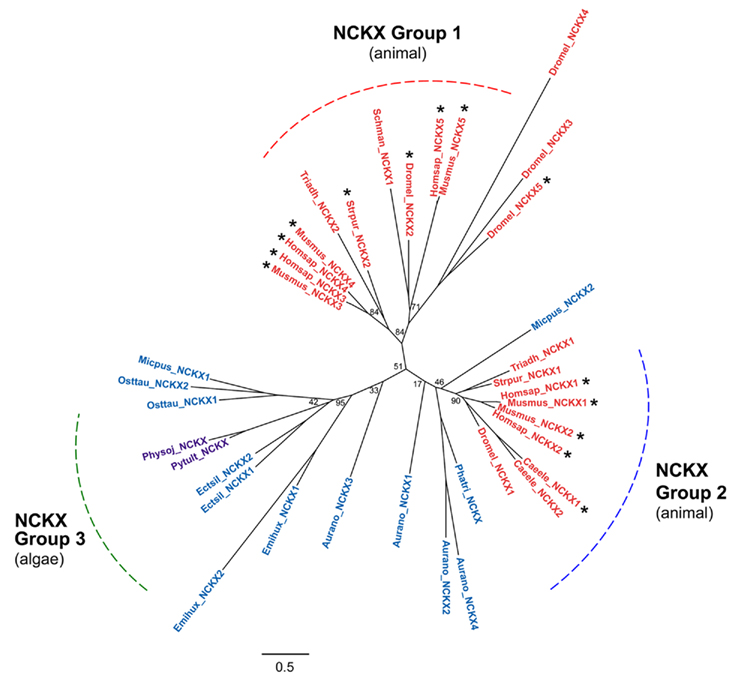
Figure 6. Na+/Ca2+/K+ exchanger family phylogenetic tree. The tree was constructed using genes identified in the NCKX group of the full CaCA tree (Figure 3). Three NCKX sub-groups are highlighted. Branch labels include the species name abbreviation defined in Figure 2 and are colored according to the species class using the same color code as in Figure 3. Bootstrap values are indicated at the nodes of major branches. The branch length scale bar indicates the evolutionary distance of 0.5 amino acid substitutions per site. Genes that are highlighted with an asterisk encode transporters for which the ion specificity has been determined.
Algal NCX and NCKX proteins have not yet been cloned and characterized, further work is needed to determine if these transporters encode Na+/Ca2+ and K+-dependent Na+/Ca2+ exchange activities. However, plasma membrane Na+/Ca2+ exchange activity has been detected in some algae, such as the marine chlorophyte Dunaliella, and in the freshwater chlorophyte Chlamydomonas when assayed in the presence of external Na+, with a predicted ratio of 3 Na+:1 Ca2+ (Karimova et al., 2000), equivalent to the stoichiometry of animal NCX. While the presence of a high external Na+ concentration in marine algae and therefore a steep Na+ gradient across the plasma membrane might be expected to make the presence of a Na+-coupled ion transporter energetically favorable, it is perhaps somewhat surprising that a freshwater alga like Chlamydomonas possesses a putative NCX in addition to CAX genes. However, Chlamydomonas is also a soil-living microorganism and may encounter saline conditions in some environments. NCX and NCKX genes did not appear to be ubiquitous in all classes of algae, particularly other freshwater algae. They could not be identified in the freshwater chlorophytes V. carteri and Coccomyxa sp., or in the genome of the red alga C. merolae, although future analysis of these genomes will be needed to confirm that these gene losses and not due to poor genome sequence quality.
Adaptive Evolution of MHX Genes in Land Plants
While no obvious NCX genes were detected across the plant genes analyzed, distantly related NCX-like genes were detected in all plant species, and formed a tight monophyletic group which includes the A. thaliana AtMHX gene, hence we named this group MHX (Figure 4). AtMHX was previously found to be closely related to mammalian NCXs but was shown to function as a H+-coupled Mg2+ and Zn2+ exchanger and not a NCX (Shaul et al., 1999). A single AtMHX homolog was identified from the closely related zinc-hyperaccumulator species Arabidopsis halleri (Elbaz et al., 2006) but the extent of MHX genes within plants and other species has not been previously examined. The putative Paramecium Mg2+ exchanger XNTA appears to be a CaCA homolog but has significant divergence from the NCX gene family and therefore is not regarded as a MHX homolog (Haynes et al., 2002). An NCX-like gene was identified in each of the flowering plant genomes examined. Each of these gene sequences had high identity to AtMHX and formed a clear MHX sub-group within the NCX family (Figure 4). None of the plants examined had more than one MHX gene in contrast to the variation in numbers of CAX and CCX genes between plant species. Single MHX genes were also present in the lower land plants Physcomitrella and Selaginella but MHX genes do not appear to be present in algae as none of the NCX or NCX-related genes from any of the algal species grouped with the plant MHX genes (Figures 3 and 4). Furthermore, MHX-like genes were not present in any non-plant species. The birth of MHX genes appears to be an example of adaptive evolution which has taken place most probably in the common ancestor of land plants.
There is 20–22% amino acid sequence identity (31–33% similarity) between the plant MHX sequences and human NCX1, and both sets of proteins share a number of conserved residues particularly within the hydrophobic regions and in the α1- and α2-repeat regions, yet there are clear distinctions (Figure 5A). Many of the residues that have been shown to critical for Na+/Ca2+ exchange activity in NCX1 (Nicoll et al., 1996; Iwamoto et al., 2000; Philipson and Nicoll, 2000; Ottolia et al., 2005) are conserved in the MHX sequences but there are some specific differences. Key residues Glu-113 and Asp-130 in the α1-repeat region of NCX1, and Asp-825 and Phe-844 in the NCX1 α2-repeat region, are substituted in the equivalent positions in each of the plant MHX homologs (Figure 5A). For example, Glu-113 is replaced by a Gln residue in each MHX protein. Such a G113Q substitution was shown to abolish Na+-dependent Ca2+ transport activity of NCX1 (Nicoll et al., 1996). Mutations of Asp-130 and Asp-825 of NCX1 were found to substantially reduce the affinity of the exchanger for Ca2+ (Iwamoto et al., 2000). Asn-143 of NCX1 is also a critical residue and is substituted by Asp in the higher plant MHX proteins but not the lower plant MHXs (Figure 5A). Interestingly, the N143D mutation of NCX1 showed wild type activity (Ottolia et al., 2005). These amino acid changes in the α1-repeat region of AtMHX compared to NCX1 therefore appear to explain the lack of Na+/Ca2+ exchange activity by AtMHX and probably by the other plant MHX transporters. The MHX genes also encode proteins with a much shorter central loop than the NCX genes (Figure 5B; Table S3 in Supplementary Material) and lack the Ca2+ binding domain sequence motifs present on the NCX proteins in this region (Lytton, 2007), further explaining the loss of Ca2+ activity and regulation by the AtMHX transporter. The residues in the α1-repeat region are thought to be involved in ion selectivity (Shigekawa et al., 2002; Ottolia et al., 2005) and therefore we might presume that the residues within the MHX α1-repeat region encode the Mg2+ and Zn2+/H+ exchange activity observed for AtMHX. Whether each of the plant MHX transporters is involved in Mg2+ transport or in the transport of other cations remains to be seen.
The Diversification of CCX Genes in Land Plants
To date only two CCX members have been functionally characterized in depth, mammalian NCKX6/NCLX, which has been shown to have Na+(Li+)/Ca2+ exchange activity (Cai and Lytton, 2004b; Palty et al., 2004) and CCX3 from Arabidopsis which appears to function as a H+/K+ exchanger and which can also transport Na+ and Mn2+ but not Ca2+ (Morris et al., 2008). Thus, there is clearly variation in function within the CCX transporter family as indicated by the CCX tree (Figure 7). Apart from the duplication of insect and nematode CCX genes (four to five genes), single genes are present in mammal and fungi species, and one or two CCX genes were present in each algal species examined, except in Coccomyxa sp. and C. merolae where CCX genes could not be identified. In contrast, there has been a significant diversification in CCX genes within land plants, with the bryophyte and lycophyte species each possessing three genes, and individual flowering plant species possessing three to eight CCX genes (Figure 2), suggesting that these are important genes within plants. There is significant variation between these plant CCX genes which are found within each of the three main groups of this tree (Figure 7). There are two plant-specific groups; Group 3 contains solely angiosperm genes and includes Arabidopsis CCX1 and CCX2, while Group 2, which includes Arabidopsis CCX3 and CCX4, also contains CCX genes from Physcomitrella and Selaginella. Each of the 10 flowering plant species analyzed has at least one member in each of these two groups. There is evidence of recent gene duplication within both groups, particularly within Group 3 where there are mostly two genes for each dicot species and three genes for each monocot species, while within Group 2 three of the dicot species (Arabidopsis, soybean, and poplar), plus Physcomitrella and Selaginella, have two CCX genes. Group 1 contains all the algal and non-plant genes, including NCKX6/NCLX, single CCX genes from Physcomitrella and Selaginella, and single CCX genes each from seven of the 10 higher plant species, including CCX5 from Arabidopsis, but CCX5-like genes from the remaining three monocot species (Brachypodium, rice, and sorghum) appear to have been lost (Figure 7). There is considerable variation among the algal CCX genes which are widely dispersed throughout Group 1, as indicated by their long tree branch lengths, but as yet nothing is known about these genes or their function to explain this variation.
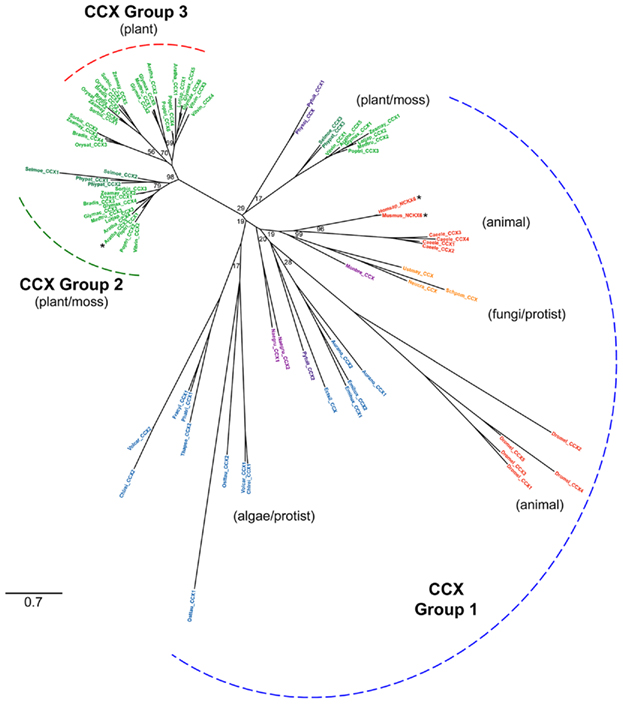
Figure 7. Cation/Ca2+ exchanger family phylogenetic tree. The tree was constructed using genes identified in the CCX group of the full CaCA tree (Figure 3). Three CCX sub-groups are highlighted. Branch labels include the species name abbreviation defined in Figure 2 and are colored according to the species class using the same color code as in Figure 3. Bootstrap values are indicated at the nodes of major branches. The branch length scale bar indicates the evolutionary distance of 0.7 amino acid substitutions per site. Genes that are highlighted with an asterisk encode transporters for which the ion specificity has been determined.
Further biochemical analysis of the plant CCX proteins is required before we can discern whether the CCX members from each group differ in substrate specificity. However, it is tempting to speculate that members of Group 2 may share AtCCX3 H+/K+ exchange activity and that the plant members of Group 1 like AtCCX5 may be more likely to have Ca2+ transport activity as they are more closely grouped to NCKX6/NCLX. A closer look at the amino acid sequence of selected plant, algal, and human CCX sequences from each of the three groups shows that they all have strong sequence conservation within the α1- and α2-repeat regions, including 100% identity in the α1 signature motif GNGAPD and the α2 signature motif G(N/D)SxGD (Figure 8). Yet there are still some obvious sequence differences in both α-repeat regions between the Group 1 and the Group 2/3 CCX proteins, in particular between the Ca2+ transporting NCKX6/NCLX and the non-Ca2+ transporting AtCCX3, that might determine cation specificity.
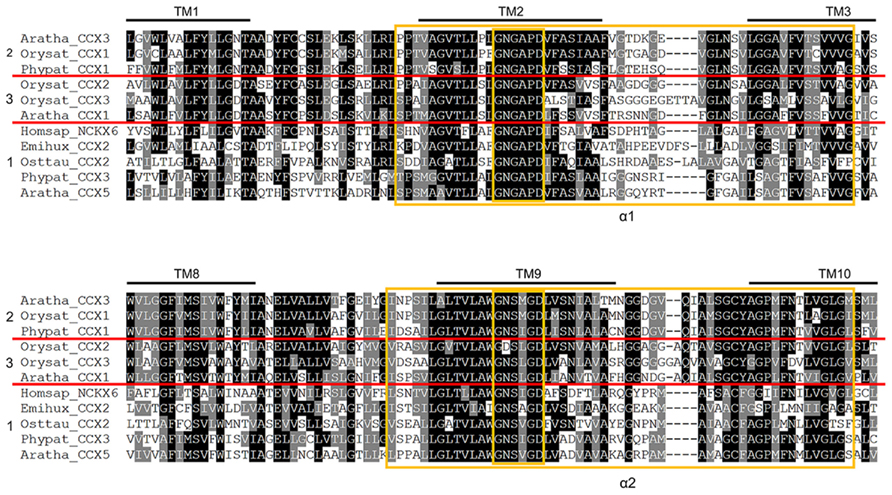
Figure 8. Sequence variation of CCX homologs. Multiple amino acid sequence alignments of the α1- and α2-repeat region sequence from selected plant, moss, algal, and human CCX proteins. Alignments were performed using ClustalW. Amino acids that are identical or similar are shaded black or gray, respectively. Predicted transmembrane spans are over-lined. The α-repeat regions and signature residues are boxed in yellow. The red lines separate the Group 1, 2, and 3 CCX sequences.
Expansion of the CAX Gene Family Throughout the Plant Kingdom
Previous analysis has examined the diversification and evolution of CAX genes and identified distinct phylogenetic groups that encompass genes from bacterial, fungal, plant, and invertebrate species (Shigaki et al., 2006). Here we have screened through a wider range of flowering plant genomes and a wide selection of algal species, from which CAX genes have not been examined in detail. Shigaki et al. (2006) identified a number of distinct Type 1 phylogenetic groups, many of which were comprised of fungal (Type 1D, 1E, and 1F) and bacterial/cyanobacterial (Type 1G and 1H) CAX genes. As this study has focused predominantly on plant and algae species, only a small number of selected fungal and bacterial species were examined and therefore the CAX tree generated here lacks distinction between the fungal and bacterial Type 1 groups (Figure 9A). The Leishmania parasitic protist gene stands out with an unusually long branch that may be reflective of the effect of the parasitic lifestyle on molecular evolution. The Type 2 group which contains distinct CAX genes including yeast VNX1 and invertebrate CAX genes was also clearly observed in this tree, but this group contains no plant or algal genes. A number of CAX genes were identified from various algal species, although three of the species, O. tauri, M. pusilla, and A. anophagefferens appeared to lack CAX homologs (Figure 2). A previously cloned CAX gene from Chlamydomonas was shown to cluster within the Type 1C group which includes CAX genes from parasitic protists such as Plasmodium falciparum (Pittman et al., 2009). Genes from other freshwater chlorophyte algae including a second gene from Chlamydomonas, likewise fall within this group, plus genes from some of the marine algae and the oomycetes. However, five CAX genes from the haptophyte alga E. huxleyi, two stramenopile algae genes, and two chlorophyte algae genes from Chlamydomonas and Volvox, cluster more closely to the land plant CAX genes and might be considered as Type 1B-like genes. It might therefore be expected that these algal CAX homologs share some functional characteristics of the plant Type 1B CAX transporters. Despite the sequence divergence between Chlamydomonas CAX1 in the Type 1C group and Arabidopsis CAX1 in the Type 1A group, both of these transporters have very similar function; when expressed in a yeast heterologous expression system both function as vacuolar-localized H+/Ca2+ exchangers with equivalent regulatory characteristics (Pittman et al., 2009). The main functional difference appears to be that CrCAX1 can also transport Na+. To date all of the biochemically characterized CAX transporters show Ca2+ transport activity, including those in the Type 2 group (Manohar et al., 2010), therefore it is not clear why there is so much sequence diversity within the CAX family compared to the NCX or NCKX families which yield much tighter families (Figure 3).
The previously analyzed flowering plant CAX genes were found to cluster into either the Type 1A or Type 1B groups, and this was consistently seen here with all the flowering plant genomes examined. Within the Type 1A group, clear distinction between the genes from monocot and dicot plants was observed (Figure 9B) but this was not as obvious within the Type 1B group which showed greater overall divergence and also included the CAX genes from Physcomitrella and Selaginella (Figure 9C). This analysis indicates that there has been a diversification of CAX genes within land plants. Within the Chlorophyta, the species possess one to three CAX genes; in contrast, in the land plants each species has on average five to six CAX genes (Figure 2). This expansion may have occurred prior to the evolution of the flowering plants as the bryophyte Physcomitrella has five CAX genes. The most parsimonious explanation is that there was likely to have been at least one duplication event in the flowering plant ancestor. However, the evolution of CAX genes in plants seems to have been a dynamic process with many apparent instances of gains and losses, for example, the lycophyte Selaginella appears to have just a single CAX gene. With two exceptions, all the monocots and dicots analyzed here have between four and six CAX genes, generally equally divided between the Type 1A and Type 1B groups. 14 CAX genes were identified from soybean, also spread between both groups, but we could only identify two CAX genes from L. japonica, both within the Type 1A group. The genes in most of these species await future studies to see whether any novel functional traits will be uncovered or whether the range of characteristics determined for the Arabidopsis CAX proteins is conserved for the other flowering plants.
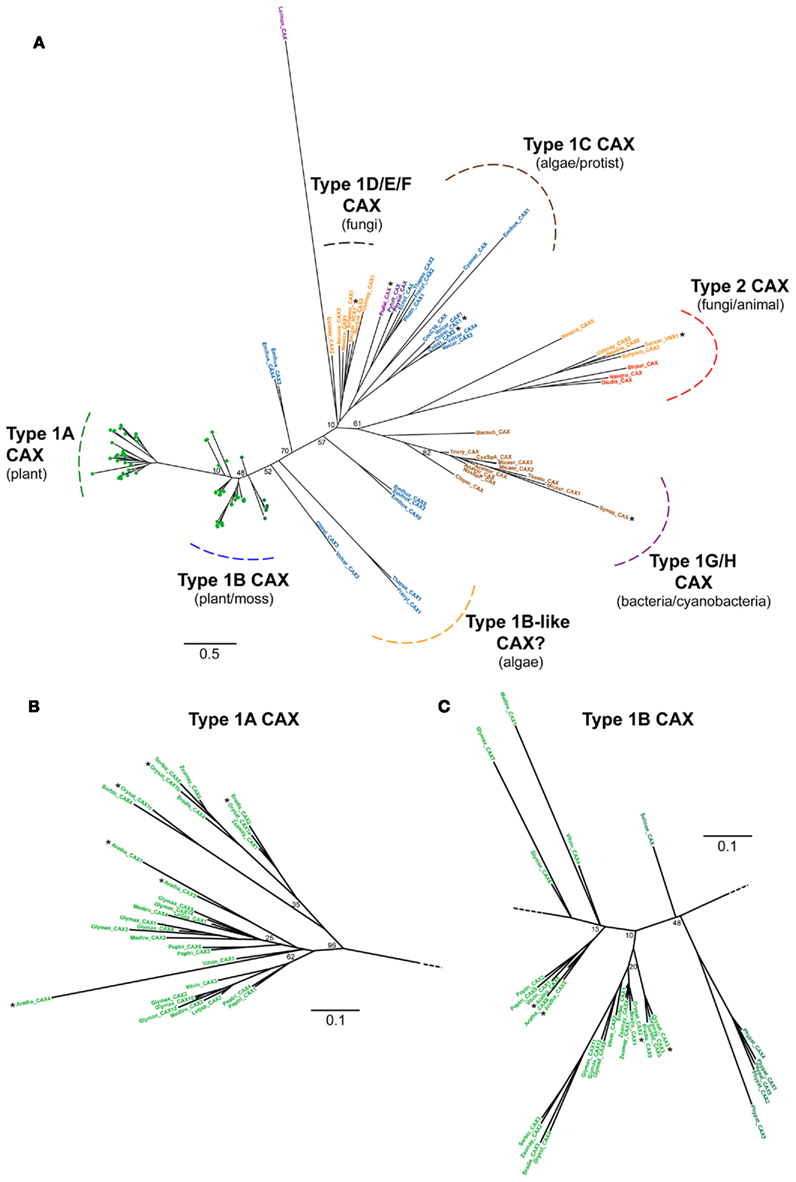
Figure 9. CAX family phylogenetic tree. (A) The tree was constructed using genes identified in the CAX group of the full CaCA tree (Figure 3). The Type 1A-H and Type 2 CAX sub-groups are highlighted and determined using the nomenclature described previously (Shigaki et al., 2006). Branch labels include the species name abbreviation defined in Figure 2 and are colored according to the species class using the same color code as in Figure 3. The individual gene labels for the Type 1A and 1B groups are replaced by circles and the gene name labels are given in the enlarged tree sections in (B,C). Bootstrap values are indicated at the nodes of major branches. The branch length scale bar indicates the evolutionary distance of 0.5 amino acid substitutions per site [0.1 amino acid substitutions per site in (B,C)]. Genes that are highlighted with an asterisk encode transporters for which the ion specificity has been determined.
Identification of Genes Encoding CAX-Like Proteins Containing Ef-Hand Domains in Some Plants and Algae
In addition to the CaCA genes described, the BLAST search identified a number of genes which were clearly distinct from the others. Further analysis indicated that they were most closely related to the plant CAX genes. Amino acid sequence alignment showed that these gene products have higher similarity with selected plant CAX sequences in the C-terminal half of the proteins but the sequences aligned poorly against the N-terminal half of the CAX proteins (Figure 10) despite having an overall CAX-like topology including 6 + 5 TM domains separated by a central loop, although this loop is longer than in most CAX proteins (Figure 11A). Pfam analysis (Finn et al., 2010) was performed on these proteins and confirmed that they contain a “Na_Ca_ex” domain (Pfam ID: PF01699) in the C-terminal half of the protein. This domain denotes the core consensus 5 TM hydrophobic region present in NCXs and in all other CaCA proteins5 including CAX proteins (Figure 11A). A closer look at the sequence of these proteins found that there was a high number of similar residues shared with the α2-repeat region of the CAX sequences including the presence of residues conserved throughout CaCA proteins (Cai and Lytton, 2004a) but poor sequence similarity with the α2-repeat region (Figure 10). In addition the CAX-like proteins possess one or more EF-hand-type Ca2+-binding domains (Pfam ID: PF00036) within the long central loop (Figure 11A). One of these genes from Arabidopsis (At1g53210) has been identified from a genome search (Shigaki et al., 2006), but as yet they have not been cloned or characterized. From all of the genomes analyzed here, 34 of these CAX/EF-hand (EF–CAX) genes were identified, predominantly from land plants (Arabidopsis, poplar, grape, soybean, Brachypodium, maize, rice, and sorghum) but also from Physcomitrella, Selaginella, and from some diverse algae species: the chlorophyte Coccomyxa sp., the prasinophyte M. pusilla, and three of the stramenopile algae (E. siliculosus, F. cylindrus, and P. tricornutum). Interestingly, Selaginella has more EF–CAX genes (three genes) than CAX genes (one gene), while M. pusilla, which has two EF–CAX genes, has no identifiable CAX genes. These genes were not identified from any of the bacterial, fungal, or animal genomes, but the presence of these genes in these select algae species suggests that they either evolved once in an ancestral organism from an ancestral CAX gene, or on at least two occasions, such as within the Stramenopiles and within the Archaeplastida. Using the α2-repeat region sequence alone to generate a phylogenetic tree, we confirmed that the EF–CAXs are more closely related to the CAX family members (Figure 11B) than to any of the other CaCA families (data not shown). There are some long branch lengths within the EF–CAX clade indicating a degree of divergence between these genes, although the divergence between plant and algae EF–CAX genes appears to be less than between plant and algae CAX genes. Furthermore, the analysis demonstrated that the EF–CAX genes are more distant from the plant Type 1A and Type 1B CAX genes than the plant Type 1A/1B CAX genes are from the non-plant CAX genes (Figure 11B).

Figure 10. Multiple sequence alignment of CAX and EF–CAX sequences. Full-length amino acid sequences of selected Arabidopsis, Physcomitrella, and Selaginella CAX and EF–CAX proteins were aligned. The alignment was performed using ClustalW. Amino acids that are identical or similar are shaded black/red or gray/pink, respectively. The α-repeat regions and signature residues are highlighted. Amino acids that are conserved in >90% of CaCA proteins (Cai and Lytton, 2004a) are highlighted (asterisk) within the α1- and α2-repeat regions.
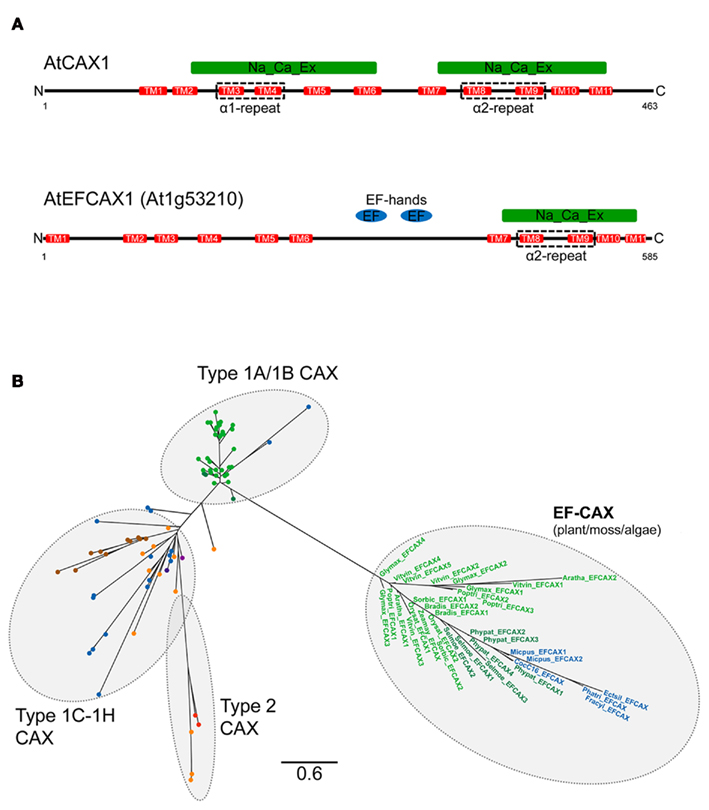
Figure 11. Structural and phylogenetic comparisons of CAX and EF–CAX proteins. (A) Schematic representation of the topology structure of a typical CAX protein (Arabidopsis CAX1) and a representative EF–CAX protein (Arabidopsis EFCAX1). The black line denotes the protein from N-terminus to C-terminus, red bars denote the TM domains as predicted by TMHMM, and the numbers indicate the first and last amino acid residues. The locations of the α1- and α2-repeat regions are highlighted. The identified Pfam domains are indicated above the protein schematic. (B) CAX gene family phylogenetic tree including the EF–CAX genes. The tree was constructed using the maximum likelihood method and derived from alignments of α2-repeat region sequences extracted from genes identified in the CAX group of the full CaCA tree (Figure 3) and the EF–CAX genes as determined by Pfam analysis. The CAX sub-groups are highlighted in addition to the EF–CAX sub-group. Branch labels include the species name abbreviation defined in Figure 2 or are replaced by a circle and are colored according to the species class using the same color code as in Figure 3. The branch length scale bar indicates the evolutionary distance of 0.6 amino acid substitutions per site.
The functions of the EF–CAX proteins are unknown and future work will be need to determine if these proteins function as a transporter or merely as a Ca2+ binding protein. It is unclear how effective a CaCA transporter would be with a single α-repeat region. Experiments with animal NCX proteins indicates that both α-repeat regions are required for transport function and it has been suggested that together these regions interact within the tertiary structure to form an ion conductance pore (Nicoll et al., 2007). For the Arabidopsis CAX proteins, it has been shown that separated N- or C-terminal halves of CAX1 or a mixture of N-terminal CAX1 with N-terminal CAX3 lack transport activity (Zhao et al., 2009). In contrast, a mutated NCKX6/NCLX protein containing either a single α1- or α2-repeat region appears to function; however, oligomerization between two copies of these α1- or α2-repeat proteins thus yielding a protein with two α-repeat regions is required (Palty et al., 2006). If the EF-CAX proteins were to function as a transporter, these proteins may need to form oligomers to allow transport activity to occur. Alternatively, one possibility might be that the N-terminal half of the EF-CAX protein contains a region which can function as an α-repeat region but that the sequence has changed significantly from the consensus α-repeat sequence and is no longer recognizable. Clearly future functional analysis is needed to understand these novel proteins.
Conclusion
Ca2+ homeostasis is critical for all organisms and therefore a suite of Ca2+ regulators including transporters must be present. The CaCA superfamily represents an ancient class of genes which have members present in most species. A major function of these transporters is in Ca2+ transport as is evident from the functional analysis of CaCA gene products from different classes of organisms and from each of the different CaCA families: for example, E. coli yrbG, Drosophila NCX, C. elegans NCKX1, human NCKX6, and rice CAX1a all appear to mediate Ca2+ transport activity (Schwarz and Benzer, 1997; Szerencsei et al., 2000; Cai and Lytton, 2004b; Kamiya and Maeshima, 2004; Naseem et al., 2008), yet the analysis here shows that they have minimal sequence conservation. It is therefore difficult to predict the function and ion specificity of these many putative transporters, yet some of them are certainly likely to function in the transport of ions other than Ca2+. This divergence in function is particularly apparent in higher plants, such that the Arabidopsis NCX and CCX proteins AtMHX and AtCCX3 possess Mg2+ transport and K+ transport activity, respectively, rather than Ca2+ transport (Shaul et al., 1999; Morris et al., 2008). Overall, the CaCA complement and function in plants is different than in most animals. The plants possess a wide diversity of CAX genes, some or all of which encode proteins that will have Ca2+ transport and probably other cation transport function, a diversity of CCX genes with mostly unknown function, and a single MHX genes. This diversification appears to mirror the evolution of plants on land, as shown by the analysis of Physcomitrella and Selaginella in this study, which also multiple CAX and CCX genes plus single MHX genes. In contrast, many of the algal species have few or no CAX and CCX genes, which are divergent from those of the land plants, and animal-like NCX and NCKX genes. Recent analysis of Ca2+ channels in algal species has likewise found that many of the animal Ca2+ channel genes that are lost in land plants are present in algae (Wheeler and Brownlee, 2008; Verret et al., 2010), indicating that many algal species possess animal-like systems for Ca2+ signaling and suggesting that there are significant differences in Ca2+ signaling and homeostasis mechanisms between algae and land plants. The motile lifestyle of most algae species compared to the sessile land plants plus the significant differences in environmental conditions between the sets of organisms may in part explain the presence of very different Ca2+ regulatory mechanisms. Algae may therefore represent an excellent model for studying the evolution of Ca2+ signaling and aspects of animal-like signaling.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Toshiro Shigaki for comments on the manuscript and Mousheng Wu for help preparing Figure 1.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Plant_Physiology/10.3389/fpls.2012.00001/abstract
Footnotes
References
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J. H., Zhang, Z., Miller, W., and Lipman, D. J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402.
Blackford, S., Rea, P. A., and Sanders, D. (1990). Voltage sensitivity of H+/Ca2+ antiport in higher-plant tonoplast suggests a role in vacuolar calcium accumulation. J. Biol. Chem. 265, 9617–9620.
Blaustein, M. P., and Lederer, W. J. (1999). Sodium/calcium exchange: its physiological implications. Physiol. Rev. 79, 763–854.
Bock, K. W., Honys, D., Ward, J. M., Padmanaban, S., Nawrocki, E. P., Hirschi, K. D., Twell, D., and Sze, H. (2006). Integrating membrane transport with male gametophyte development and function through transcriptomics. Plant Physiol. 140, 1151–1168.
Cagnac, O., Aranda-Sicilia, M. N., Leterrier, M., Rodriguez-Rosales, M.-P., and Venema, K. (2010). Vacuolar cation/H+ antiporters of Saccharomyces cerevisiae. J. Biol. Chem. 285, 33914–33922.
Cagnac, O., Leterrier, M., Yeager, M., and Blumwald, E. (2007). Identification and characterization of Vnx1p, a novel type of vacuolar monovalent cation/H+ antiporter of Saccharomyces cerevisiae. J. Biol. Chem. 282, 24284–24293.
Cai, X. J., and Lytton, J. (2004a). The cation/Ca2+ exchanger superfamily: phylogenetic analysis and structural implications. Mol. Biol. Evol. 21, 1692–1703.
Cai, X. J., and Lytton, J. (2004b). Molecular cloning of a sixth member of the K+-dependent Na+/Ca2+ exchanger gene family, NCKX6. J. Biol. Chem. 279, 5867–55876.
Case, R. M., Eisner, D., Gurney, A., Jones, O., Muallem, S., and Verkhratsky, A. (2007). Evolution of calcium homeostasis: from birth of the first cell to an omnipresent signalling system. Cell Calcium 42, 345–350.
Cheng, N. H., Pittman, J. K., Barkla, B. J., Shigaki, T., and Hirschi, K. D. (2003). The Arabidopsis cax1 mutant exhibits impaired ion homeostasis, development, and hormonal responses and reveals interplay among vacuolar transporters. Plant Cell 15, 347–364.
Conn, S. J., Gilliham, M., Athman, A., Schreiber, A. W., Baumann, U., Moller, I., Cheng, N. H., Stancombe, M. A., Hirschi, K. D., Webb, A. A. R., Burton, R., Kaiser, B. N., Tyerman, S. D., and Leigh, R. A. (2011). Cell-specific vacuolar calcium storage mediated by CAX1 regulates apoplastic calcium concentration, gas exchange, and plant productivity in Arabidopsis. Plant Cell 23, 240–257.
Cunningham, K. W. (2011). Acidic calcium stores of Saccharomyces cerevisiae. Cell Calcium 50, 129–138.
Cunningham, K. W., and Fink, G. R. (1994). Calcineurin-dependent growth-control in Saccharomyces cerevisiae mutants lacking Pmc1, a homolog of plasma-membrane Ca2+ ATPases. J. Cell Biol. 124, 351–363.
Denis, V., and Cyert, M. S. (2002). Internal Ca2+ release in yeast is triggered by hypertonic shock and mediated by a TRP channel homologue. J. Cell Biol. 156, 29–34.
Edmond, C., Shigaki, T., Ewert, S., Nelson, M., Connorton, J., Chalova, V., Noordally, Z., and Pittman, J. K. (2009). Comparative analysis of CAX2-like cation transporters indicates functional and regulatory diversity. Biochem. J. 418, 145–154.
Elbaz, B., Shoshani-Knaani, N., David-Assael, O., Mizrachy-Dagri, T., Mizrahi, K., Saul, H., Brook, E., Berezin, I., and Shaul, O. (2006). High expression in leaves of the zinc hyperaccumulator Arabidopsis halleri of AhMHX, a homolog of an Arabidopsis thaliana vacuolar metal/proton exchanger. Plant Cell Environ. 29, 1179–1190.
Finn, R. D., Mistry, J., Tate, J., Coggill, P., Heger, A., Pollington, J. E., Gavin, O. L., Gunasekaran, P., Ceric, G., Forslund, K., Holm, L., Sonnhammer, E. L. L., Eddy, S. R., and Bateman, A. (2010). The Pfam protein families database. Nucleic Acids Res. 38, D211–D222.
Groeneveld, P., Stouthamer, A. H., and Westerhoff, H. V. (2009). Super life – how and why ‘cell selection’ leads to the fastest-growing eukaryote. FEBS J. 276, 254–270.
Haynes, W. J., Kung, C., Saimi, Y., and Preston, R. R. (2002). An exchanger-like protein underlies the large Mg2+ current in Paramecium. Proc. Natl. Acad. Sci. U.S.A. 99, 15717–15722.
Herrgard, M. J., Swainston, N., Dobson, P., Dunn, W. B., Arga, K. Y., Arvas, M., Bluthgen, N., Borger, S., Costenoble, R., Heinemann, M., Hucka, M., Le Novere, N., Li, P., Liebermeister, W., Mo, M. L., Oliveira, A. P., Petranovic, D., Pettifer, S., Simeonidis, E., Smallbone, K., Spasic, I., Weichart, D., Brent, R., Broomhead, D. S., Westerhoff, H. V., Kirdar, B., Penttila, M., Klipp, E., Palsson, B. O., Sauer, U., Oliver, S. G., Mendes, P., Nielsen, J., and Kell, D. B. (2008). A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nat. Biotechnol. 26, 1155–1160.
Hirschi, K. (2001). Vacuolar H+/Ca2+ transport: who’s directing the traffic? Trends Plant Sci. 6, 100–104.
Hirschi, K. D. (2004). The calcium conundrum. Both versatile nutrient and specific signal. Plant Physiol. 136, 2438–2442.
Hirschi, K. D., Korenkov, V. D., Wilganowski, N. L., and Wagner, G. J. (2000). Expression of Arabidopsis CAX2 in tobacco. Altered metal accumulation and increased manganese tolerance. Plant Physiol. 124, 125–133.
Hirschi, K. D., Zhen, R. G., Cunningham, K. W., Rea, P. A., and Fink, G. R. (1996). CAX1, an H+/Ca2+ antiporter from Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 93, 8782–8786.
Ivey, D. M., Guffanti, A. A., Zemsky, J., Pinner, E., Karpel, R., Padan, E., Schuldiner, S., and Krulwich, T. A. (1993). Cloning and characterization of a putative Ca2+/H+ antiporter gene from Escherichia coli upon functional complementation of Na+/H+ antiporter-deficient strains by the overexpressed gene. J. Biol. Chem. 268, 11296–11303.
Iwamoto, T., Uehara, A., Imanaga, I., and Shigekawa, M. (2000). The Na+/Ca2+ exchanger NCX1 has oppositely oriented reentrant loop domains that contain conserved aspartic acids whose mutation alters its apparent Ca2+ affinity. J. Biol. Chem. 275, 38571–38580.
Jain, N., Nadgauda, R., and Shigaki, T. (2009). Mining cation (CAX) transporter diversity for nutrition-enhanced crops and phytoremediation. Int. J. Integr. Biol. 7, 22–25.
Kamiya, T., Akahori, T., and Maeshima, M. (2005). Expression profile of the genes for rice cation/H+ exchanger family and functional analysis in yeast. Plant Cell Physiol. 46, 1735–1740.
Kamiya, T., and Maeshima, M. (2004). Residues in internal repeats of the rice cation/H+ exchanger are involved in the transport and selection of cations. J. Biol. Chem. 279, 812–819.
Karimova, F. G., Kortchouganova, E. E., Tarchevsky, I. A., and Iagoucheva, M. R. (2000). The oppositely directed Ca2+ and Na+ transmembrane transport in algal cells. Protoplasma 213, 93–98.
Katoh, K., Misawa, K., Kuma, K., and Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066.
Koren’kov, V., Park, S., Cheng, N. H., Sreevidya, C., Lachmansingh, J., Morris, J., Hirschi, K., and Wagner, G. J. (2007). Enhanced Cd2+-selective root-tonoplast-transport in tobaccos expressing Arabidopsis cation exchangers. Planta 225, 403–411.
Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580.
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J., and Higgins, D. G. (2007). Clustal W and Clustal X version 2.0 . Bioinformatics 23, 2947–2948.
Lytton, J. (2007). Na+/Ca2+ exchangers: three mammalian gene families control Ca2+ transport. Biochem. J. 406, 365–382.
Manohar, M., Mei, H., Franklin, A. J., Sweet, E. M., Shigaki, T., Riley, B. B., MacDiarmid, C. W., and Hirschi, K. (2010). Zebrafish (Danio rerio) endomembrane antiporter similar to a yeast cation/H+ transporter is required for neural crest development. Biochemistry 49, 6557–6566.
Manohar, M., Shigaki, T., and Hirschi, K. D. (2011). Plant cation/H+ exchangers (CAXs): biological functions and genetic manipulations. Plant Biol. 13, 561–569.
Mäser, P., Thomine, S., Schroeder, J. I., Ward, J. M., Hirschi, K., Sze, H., Talke, I. N., Amtmann, A., Maathuis, F. J. M., Sanders, D., Harper, J. F., Tchieu, J., Gribskov, M., Persans, M. W., Salt, D. E., Kim, S. A., and Guerinot, M. L. (2001). Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 126, 1646–1667.
Mei, H., Cheng, N. H., Zhao, J., Park, P., Escareno, R. A., Pittman, J. K., and Hirschi, K. D. (2009). Root development under metal stress in Arabidopsis thaliana requires the H+/cation antiporter CAX4. New Phytol. 183, 95–105.
Merchant, S. S., Prochnik, S. E., Vallon, O., Harris, E. H., Karpowicz, S. J., Witman, G. B., Terry, A., Salamov, A., Fritz-Laylin, L. K., Marechal-Drouard, L., Marshall, W. F., Qu, L. H., Nelson, D. R., Sanderfoot, A. A., Spalding, M. H., Kapitonov, V. V., Ren, Q. H., Ferris, P., Lindquist, E., Shapiro, H., Lucas, S. M., Grimwood, J., Schmutz, J., Grigoriev, I. V., Rokhsar, D. S., Grossman, A. R., Annotation, C., and Team, J. G. I. A. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–251.
Morris, J., Tian, H., Park, S., Sreevidya, C. S., Ward, J. M., and Hirschi, K. D. (2008). AtCCX3 is an Arabidopsis endomembrane H+-dependent K+ transporter. Plant Physiol. 148, 1474–1486.
Naseem, R., Holland, I. B., Jacq, A., Wann, K. T., and Campbell, A. K. (2008). pH and monovalent cations regulate cytosolic free Ca2+ in E. coli. Biochim. Biophys. Acta 1778, 1415–1422.
Nicoll, D. A., Hryshko, L. V., Matsuoka, S., Frank, J. S., and Philipson, K. D. (1996). Mutation of amino acid residues in the putative transmembrane segments of the cardiac sarcolemmal Na+-Ca2+ exchanger. J. Biol. Chem. 271, 13385–13391.
Nicoll, D. A., Ren, X. Y., Ottolia, M., Phillips, M., Paredes, A. R., Abramson, J., and Philipson, K. D. (2007). What we know about the structure of NCX1 and how it relates to its function. Ann. N. Y. Acad. Sci. 1099, 1–6.
Ohyama, T., Igarashi, K., and Kobayashi, H. (1994). Physiological role of the chaA gene in sodium and calcium circulations at a high pH in Escherichia coli. J. Bacteriol. 176, 4311–4315.
On, C., Marshall, C. R., Chen, N., Moyes, C. D., and Tibbits, G. F. (2008). Gene structure evolution of the Na+/Ca2+ exchanger (NCX) family. BMC Evol. Biol. 8, 127.
Ottolia, M., John, S., Qiu, Z. Y., and Philipson, K. D. (2001). Split Na+-Ca2+ exchangers – implications for function and expression. J. Biol. Chem. 276, 19603–19609.
Ottolia, M., Nicoll, D. A., and Philipson, K. D. (2005). Mutational analysis of the alpha-1 repeat of the cardiac Na+-Ca2+ exchanger. J. Biol. Chem. 280, 1061–1069.
Palty, R., Hershfinkel, M., Yagev, O., Saar, D., Barkalifa, R., Khananshvili, D., Peretz, A., Grossman, Y., and Sekler, I. (2006). Single R-domain constructs of the Na+/Ca2+ exchanger, NCLX, oligomerize to form a functional exchanger. Biochemistry 45, 11856–11866.
Palty, R., Ohana, E., Hershfinkel, M., Volokita, M., Elgazar, V., Beharier, O., Silverman, W. F., Argaman, M., and Sekler, I. (2004). Lithium-calcium exchange is mediated by a distinct potassium-independent sodium-calcium exchanger. J. Biol. Chem. 279, 25234–25240.
Palty, R., Silverman, W. F., Hershfinkel, M., Caporale, T., Sensi, S. L., Parnis, J., Nolte, C., Fishman, D., Shoshan-Barmatz, V., Herrmann, S., Khananshvili, D., and Sekler, I. (2010). NCLX is an essential component of mitochondrial Na+/Ca2+ exchange. Proc. Natl. Acad. Sci. U.S.A. 107, 436–441.
Philipson, K. D., and Nicoll, D. A. (2000). Sodium-calcium exchange: a molecular perspective. Annu. Rev. Physiol. 62, 111–133.
Pittman, J. K., Edmond, C., Sunderland, P. A., and Bray, C. M. (2009). A cation-regulated and proton gradient-dependent cation transporter from Chlamydomonas reinhardtii has a role in calcium and sodium homeostasis. J. Biol. Chem. 284, 525–533.
Pittman, J. K., Shigaki, T., Cheng, N. H., and Hirschi, K. D. (2002). Mechanism of N-terminal autoinhibition in the Arabidopsis Ca2+/H+ antiporter CAX1. J. Biol. Chem. 277, 26452–26459.
Radchenko, M. V., Tanaka, K., Waditee, R., Oshimi, S., Matsuzaki, Y., Fukuhara, M., Kobayashi, H., Takabe, T., and Nakamura, T. (2006). Potassium/proton antiport system of Escherichia coli. J. Biol. Chem. 281, 19822–19829.
Reilander, H., Achilles, A., Friedel, U., Maul, G., Lottspeich, F., and Cook, N. J. (1992). Primary structure and functional expression of the Na/Ca,K-exchanger from bovine rod photoreceptors. EMBO J. 11, 1689–1695.
Rensing, S. A., Lang, D., Zimmer, A. D., Terry, A., Salamov, A., Shapiro, H., Nishiyama, T., Perroud, P. F., Lindquist, E. A., Kamisugi, Y., Tanahashi, T., Sakakibara, K., Fujita, T., Oishi, K., Shin-I, T., Kuroki, Y., Toyoda, A., Suzuki, Y., Hashimoto, S., Yamaguchi, K., Sugano, S., Kohara, Y., Fujiyama, A., Anterola, A., Aoki, S., Ashton, N., Barbazuk, W. B., Barker, E., Bennetzen, J. L., Blankenship, R., Cho, S. H., Dutcher, S. K., Estelle, M., Fawcett, J. A., Gundlach, H., Hanada, K., Heyl, A., Hicks, K. A., Hughes, J., Lohr, M., Mayer, K., Melkozernov, A., Murata, T., Nelson, D. R., Pils, B., Prigge, M., Reiss, B., Renner, T., Rombauts, S., Rushton, P. J., Sanderfoot, A., Schween, G., Shiu, S. H., Stueber, K., Theodoulou, F. L., Tu, H., Van de Peer, Y., Verrier, P. J., Waters, E., Wood, A., Yang, L. X., Cove, D., Cuming, A. C., Hasebe, M., Lucas, S., Mishler, B. D., Reski, R., Grigoriev, I. V., Quatrano, R. S., and Boore, J. L. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69.
Saier, M. H., Eng, B. H., Fard, S., Garg, J., Haggerty, D. A., Hutchinson, W. J., Jack, D. L., Lai, E. C., Liu, H. J., Nusinew, D. P., Omar, A. M., Pao, S. S., Paulsen, I. T., Quan, J. A., Sliwinski, M., Tseng, T. T., Wachi, S., and Young, G. B. (1999). Phylogenetic characterization of novel transport protein families revealed by genome analyses. Biochim. Biophys. Acta 1422, 1–56.
Schnetkamp, P. P. M. (1995). Calcium homeostasis in vertebrate retinal rod outer segments. Cell Calcium 18, 322–330.
Schwarz, E. M., and Benzer, S. (1997). Calx, a Na-Ca exchanger gene of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 94, 10249–10254.
Shaul, O., Hilgemann, D. W., de-Almeida-Engler, J., Van Montagu, M., Inze, D., and Galili, G. (1999). Cloning and characterization of a novel Mg2+/H+ exchanger. EMBO J. 18, 3973–3980.
Shigaki, T., Barkla, B. J., Miranda-Vergara, M. C., Zhao, J., Pantoja, O., and Hirschi, K. D. (2005). Identification of a crucial histidine involved in metal transport activity in the Arabidopsis cation/H+ exchanger CAX1. J. Biol. Chem. 280, 30136–30142.
Shigaki, T., Mei, H., Marshall, J., Li, X., Manohar, M., and Hirschi, K. D. (2010). The expression of the open reading frame of Arabidopsis CAX1, but not its cDNA, confers metal tolerance in yeast. Plant Biol. 12, 935–939.
Shigaki, T., Pittman, J. K., and Hirschi, K. D. (2003). Manganese specificity determinants in the Arabidopsis metal/H+ antiporter CAX2. J. Biol. Chem. 278, 6610–6617.
Shigaki, T., Rees, I., Nakhleh, L., and Hirschi, K. D. (2006). Identification of three distinct phylogenetic groups of CAX cation/proton antiporters. J. Mol. Evol. 63, 815–825.
Shigekawa, M., Iwamoto, T., Uehara, A., and Kita, S. (2002). Probing ion binding sites in the Na+/Ca2+ exchanger. Ann. N. Y. Acad. Sci. 976, 19–30.
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690.
Stamatakis, A., Hoover, P., and Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771.
Szerencsei, R. T., Tucker, J. E., Cooper, C. B., Winkfein, R. J., Farrell, P. J., Iatrou, K., and Schnetkamp, P. P. M. (2000). Minimal domain requirement for cation transport by the potassium-dependent Na/Ca-K exchanger – comparison with an NCKX paralog from Caenorhabditis elegans. J. Biol. Chem. 275, 669–676.
Verret, F., Wheeler, G., Taylor, A. R., Farnham, G., and Brownlee, C. (2010). Calcium channels in photosynthetic eukaryotes: implications for evolution of calcium-based signalling. New Phytol. 187, 23–43.
Wheeler, G. L., and Brownlee, C. (2008). Ca2+ signalling in plants and green algae – changing channels. Trends Plant Sci. 13, 506–514.
Keywords: calcium transport, cation transport, evolution, H+/Ca2+ exchanger, Na+/Ca2+ exchanger, phylogeny, CaCA
Citation: Emery L, Whelan S, Hirschi KD and Pittman JK (2012) Protein phylogenetic analysis of Ca2+/cation antiporters and insights into their evolution in plants. Front. Plant Sci. 3:1. doi: 10.3389/fpls.2012.00001
Received: 30 September 2011; Accepted: 01 January 2012;
Published online: 13 January 2012.
Edited by:
Heven Sze, University of Maryland, USACopyright: © 2012 Emery, Whelan, Hirschi and Pittman. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Jon K. Pittman, Faculty of Life Sciences, University of Manchester, Michael Smith Building, Oxford Road, Manchester M13 9PT, UK. e-mail: jon.pittman@manchester.ac.uk
 Laura Emery1
Laura Emery1