- 1 Department of Energy-Plant Research Laboratory, Michigan State University, East Lansing, MI, USA
- 2 Department of Biochemistry and Molecular Biology, Michigan State University, East Lansing, MI, USA
The oxylipin hormone jasmonate controls myriad processes involved in plant growth, development, and immune function. The discovery of jasmonoyl-L-isoleucine (JA-Ile) as the major bioactive form of the hormone highlights the need to understand biochemical and cell biological processes underlying JA-Ile homeostasis. Among the major metabolic control points governing the accumulation of JA-Ile in plant tissues are the availability of jasmonic acid, the immediate precursor of JA-Ile, and oxidative enzymes involved in catabolism and deactivation of the hormone. Recent studies indicate that JA-Ile turnover is mediated by a ω-oxidation pathway involving members of the CYP94 family of cytochromes P450. This discovery opens new opportunities to genetically manipulate JA-Ile levels for enhanced resistance to environmental stress, and further highlights ω-oxidation as a conserved pathway for catabolism of lipid-derived signals in plants and animals. Functional characterization of the full complement of CYP94 P450s promises to reveal new pathways for jasmonate metabolism and provide insight into the evolution of oxylipin signaling in land plants.
Introduction
Plants use a wide variety of lipid-based signals to control fundamental aspects of growth, development, and responses to environmental stress. Among the most intensively studied of these signals are members of the jasmonate family of oxylipins, collectively referred to as JAs. JAs are biochemically defined as cyclopente(a)none compounds derived from lipoxygenase-dependent oxidation of polyunsaturated fatty acids. Jasmonic acid (JA) and its derivatives are well known for their role in orchestrating immune responses to a broad spectrum of arthropod herbivores and microbial pathogens (Glazebrook, 2005; Wasternack, 2007; Browse and Howe, 2008; Howe and Jander, 2008; Wu and Baldwin, 2010). JAs also serve important roles in plant growth and development, including sexual reproduction, growth control, and secondary metabolism (McConn and Browse, 1996; Li et al., 2004; Yan et al., 2007; Pauwels et al., 2008; Zhang and Turner, 2008). Increasing evidence indicates that the JA pathway participates in extensive crosstalk with other hormones that mediate developmental plasticity (Dombrecht et al., 2007; Moreno et al., 2009; Pieterse et al., 2009; Hou et al., 2010; Robson et al., 2010; Ballare, 2011; Kazan and Manners, 2011; Zhu et al., 2011). Collectively, these studies support the view that JAs control resource allocation between growth- and defense-related processes, thus allowing plants to rapidly adapt to changing environmental conditions. A greater understanding of JA homeostasis is therefore relevant to many areas of plant physiology (Howe, 2010; Kazan and Manners, 2012).
Significant recent progress has been made in understanding the molecular mechanism by which JAs control large-scale changes in gene expression in response to stress. In unstressed cells containing low levels of the hormone, transcription factors (e.g., MYC2) that promote expression of JA-responsive genes are repressed by members of the JASMONATE ZIM-domain (JAZ) protein family (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007). In response to stress-related cues that trigger JA accumulation, bioactive forms of the hormone stimulate JAZ binding to CORONATINE INSENSITIVE1 (COI1), which is the F-box protein component of the E3 ubiquitin ligase SCFCOI1 (Xie et al., 1998; Thines et al., 2007; Katsir et al., 2008b; Melotto et al., 2008; Fonseca et al., 2009; Yan et al., 2009). Proteolytic degradation of JAZ via the ubiquitin/26S proteasome pathway releases JAZ-bound transcription factors from repression, thereby allowing expression of JA-responsive genes.
A major challenge in the field of plant lipid signaling is to identify biologically active oxylipins and their cognate receptors. Significant insight into this question came from the discovery that the interaction of COI1 with its JAZ substrates is promoted in a highly specific manner by the isoleucine-conjugated form of JA (jasmonoyl-L-isoleucine, JA-Ile), but not by free JA or other non-conjugated JAs (Thines et al., 2007; Katsir et al., 2008b; Melotto et al., 2008; Fonseca et al., 2009; Yan et al., 2009). X-ray crystallography studies demonstrated that the JA and Ile moieties of JA-Ile serve critical roles in the assembly of COI1–JAZ receptor complexes, as does an inositol pentakisphosphate cofactor that interacts with COI1 and JAZ near the hormone-binding pocket (Sheard et al., 2010). The key role of JA-Ile in promoting JAZ degradation is consistent with genetic studies that established a role for the JA-conjugating enzyme JAR1 in JA-mediated physiological processes (Staswick et al., 1998; Staswick and Tiryaki, 2004; Kang et al., 2006; Koo et al., 2009).
For the purposes of this review, we define bioactive JAs as derivatives that promote COI1–JAZ interactions to affect transcriptional responses. Non-bioactive JAs are either precursors or deactivated (or less active) forms of bioactive JAs. Below, we summarize the current understanding of metabolic pathways involved in catabolism and deactivation of JA-Ile, with special attention given to the recently discovered cytochrome P450-mediated ω-oxidation pathway.
Overview of JA-Ile Metabolism
The initial stage of JA biosynthesis occurs in the plastid and involves the conversion of α-linolenic acid (18:3) to 12-oxo-phytodienoic acid (OPDA; Figure 1). Following transport to the peroxisome, OPDA is reduced to its cyclopentanone derivative (OPC-8:0) and subjected to three cycles of β-oxidation to yield the 3R,7S stereoisomer of JA [also known as (+)-7-iso-JA]. JA is transported to the cytosol where it is conjugated specifically to Ile by the enzyme JAR1 (Staswick and Tiryaki, 2004; Suza and Staswick, 2008). JA-Ile synthesized in the cytosol presumably diffuses into the nucleus where it binds COI1–JAZ receptor complexes to activate gene expression (Figure 1).
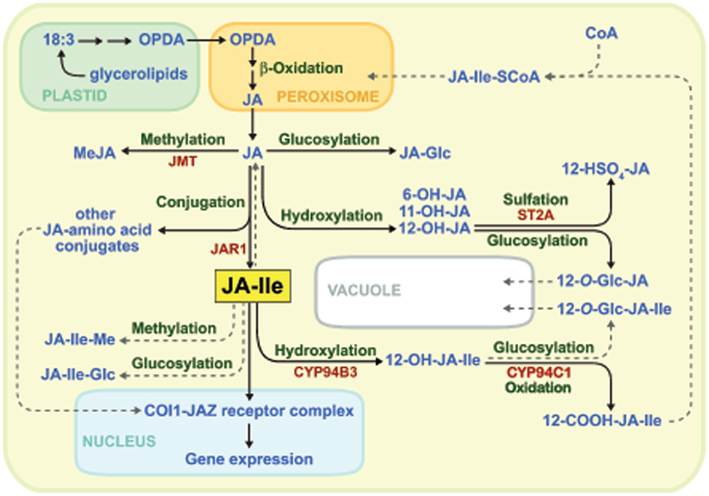
Figure 1. Major pathways for the biosynthesis and catabolism of JA-Ile. Confirmed biochemical pathways and products are indicated by solid lines, whereas dashed lines denote hypothetical pathways for which there is currently little or no evidence. See text for details.
The biosynthesis of JA-Ile is tightly controlled by developmental and environmental cues (Creelman and Mullet, 1997; Wasternack, 2007; Koo and Howe, 2009). Consistent with their role in regulating induced defenses in vegetative tissues, JA and JA-Ile typically accumulate in response to various biotic and abiotic stresses. Mechanical wounding of Arabidopsis leaves, for example, effectively triggers de novo accumulation of JA/JA-Ile within minutes of tissue damage (Chung et al., 2008; Glauser et al., 2008; Suza and Staswick, 2008; Koo and Howe, 2009; Koo et al., 2009; Suza et al., 2010). The rapidity of this response indicates that all JA-Ile biosynthetic enzymes, including JAR1, are present in unstressed cells prior to stimulation. This view is consistent with studies showing that the major rate-limiting step in JA/JA-Ile synthesis is lipase-mediated release of fatty acyl substrates from plastid glycerolipids (Ishiguro et al., 2001; Stenzel et al., 2003; Wasternack, 2007; Kallenbach et al., 2010; Bonaventure et al., 2011). The mechanism by which extracellular signals activate plastidic lipases to trigger JA-Ile formation is a major unanswered question. Genes encoding many of the enzymes involved in JA-Ile biosynthesis are coordinately up-regulated in response to environmental signals that activate production of the hormone (Reymond et al., 2000; Sasaki et al., 2001; Sasaki-Sekimoto et al., 2005; Koo et al., 2006; Ralph et al., 2006; Pauwels et al., 2008). This transcriptional response presumably constitutes a positive feedback mechanism to amplify the cell’s capacity for JA metabolism. At a practical level, this co-expression phenomenon has proven useful for the identification of novel components in the JA metabolic and signaling pathways (Koo et al., 2006, 2011; Thines et al., 2007; Heitz et al., 2012).
In addition to the availability of plastid-derived fatty acyl substrates, there is evidence that the rate of JA-Ile biosynthesis is influenced by metabolic pathways that compete with JAR1 for cytosolic pools of JA. This idea is consistent with the fact that stress-induced levels of JA-Ile are typically well below (∼10%) that of JA (Kang et al., 2006; Suza and Staswick, 2008; Koo et al., 2009). Among the metabolic pathways that potentially divert JA from JA-Ile biosynthesis are those involved in formation of JA-glucose esters (Swiatek et al., 2004), 12-hydroxy-JA (12-OH-JA) and its sulfated and glycosylated derivatives (Gidda et al., 2003; Miersch et al., 2008), volatile methyl-JA (MeJA), and JA-amino acid conjugates other than JA-Ile (Wang et al., 2007; Figure 1). Studies involving ectopic expression of an Arabidopsis JA carboxyl methyltransferase (JMT) in Nicotiana attenuata provided genetic evidence that increased flux of JA into MeJA has predicted negative effects on JA-Ile formation and JA-Ile-mediated physiological process (Stitz et al., 2011). These findings, together with the inability of JA and MeJA to promote COI1–JAZ binding (Thines et al., 2007), provide convincing evidence that JA and MeJA are non-bioactive precursors of JA-Ile. The ability of exogenous JA and MeJA to potently activate hormonal responses can be attributed to their in vivo conversion to JA-Ile and subsequent action through the COI1–JAZ receptor system (Tamogami et al., 2008; Wu et al., 2008).
The transient accumulation of JA-Ile in injured leaves is tightly correlated with the expression of primary JA-response genes (Chung et al., 2008; Koo et al., 2009). This observation implies the existence of pathways for catabolism and deactivation of the hormone. As is the case for most stress hormones in plants and animals, such pathways serve important roles in attenuating physiological outputs during the stress response or for switching off the response when stress levels have subsided. The ω-oxidation pathway, in which JA-Ile is converted to 12-hydroxy-JA-Ile (12-OH-JA-Ile) and then further oxidized to dicarboxy-JA-Ile (12-COOH-JA-Ile), is now recognized as a major route for catabolism of the hormone (Figures 1 and 2; Guranowski et al., 2007; Glauser et al., 2008; Miersch et al., 2008; Kitaoka et al., 2011; Koo et al., 2011; VanDoorn et al., 2011; Heitz et al., 2012). Genetic evidence (see below), together with the finding that 12-OH-JA-Ile is less effective than JA-Ile in promoting COI1–JAZ interaction in vitro (Koo et al., 2011), firmly establishes a functional role for ω-oxidation in attenuating JA-Ile-mediated signaling. Pathways involved in further metabolism of oxidized JA-Ile derivatives are likely to exist but have not yet been reported. One possibility is formation of O-linked glucosyl derivatives of 12-OH-JA-Ile, which may be rendered ineffective as a ligand for the COI1–JAZ receptor, or perhaps sequestered in the vacuole. It is also possible that the newly formed carboxyl group of 12-COOH-JA-Ile is esterified to CoA and subsequently subject to β-oxidization in the peroxisome (Figures 1 and 2). This hypothesis is supported by the existence of β-oxidization enzymes that metabolize unusual fatty acid substrates, including those containing unsaturated bonds such as that found in the pentenyl side chain of JA-Ile (Goepfert and Poirier, 2007).
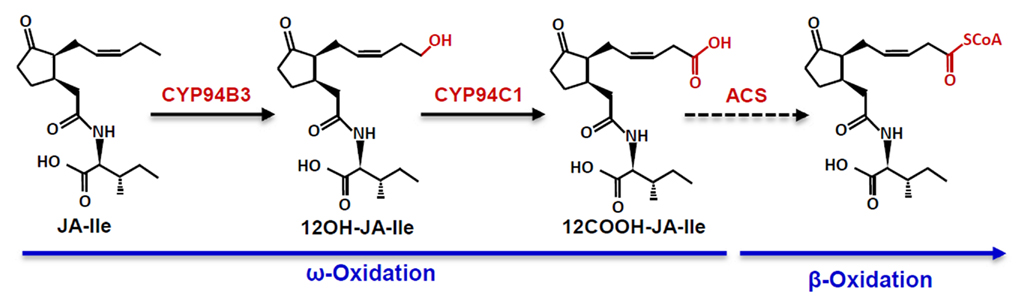
Figure 2. The CYP94-mediated ω-oxidation pathway for JA-Ile catabolism. The dashed line denoting formation of the CoA ester of 12COOH-JA-Ile by an acyl-CoA synthetase (ACS) is hypothetical.
The turnover of endogenous JA-Ile in mutants that are impaired in JA-Ile 12-hydroxylation suggests that additional pathways may participate in JA-Ile metabolism (Figure 1; Koo et al., 2011). Among the possible alternative metabolic routes of hormone deactivation are formation of JA-Ile glucose esters and, by analogy to the role of amidohydrolases in auxin homeostasis (LeClere et al., 2002; Woodward and Bartel, 2005), hydrolysis of JA-Ile to free JA and Ile. It has also been suggested that esterification of JA-Ile to JA-Ile-Me, as well as epimerization of (3R,7S)-JA-Ile to the less active (3R,7R) isomer, may be endogenous mechanisms to reduce the activity of the hormone; both of these products are largely inactive in promoting COI1–JAZ interaction in vitro (Fonseca et al., 2009). Recent in vivo studies, however, indicate that epimerization of (3R,7S)-JA-Ile is unlikely to play a significant role in deactivation of JA-Ile (Suza et al., 2010).
Role of CYP94 P450s in Catabolism of Bioactive JAs
In contrast to detailed knowledge of nearly all genes encoding the core set of JA-Ile biosynthetic enzymes (Wasternack, 2007; Schaller and Stintzi, 2009), understanding the genetic basis of JA-Ile catabolism is still a largely unexplored area of plant hormone research. The pervasive role of cytochrome P450 hydroxylases in the deactivation of small-molecule hormones suggest that P450s also participate in catabolism of JA-Ile. Based on this assumption, Koo et al. (2011) focused attention on members of the CYP86 clan of P450s in Arabidopsis that play a prominent role in fatty acid hydroxylation, including ω-hydroxylation. Gene co-expression analysis was used to narrow the list of candidates to three CYP94 genes (CYP94B1, CYP94B3, and CYP94C1) whose expression is induced in response to wounding and JA treatment. Various members of this group were previously shown to be regulated by the JA pathway, and to encode enzymes that hydroxylate straight-chain fatty acids in vitro (Duan and Schuler, 2005; Benveniste et al., 2006; Kandel et al., 2007; Ehlting et al., 2008). Analysis of T-DNA-tagged mutants showed that disruption of CYP94B3, but not CYP94B1 or CYP94C1, results in a metabolic phenotype indicative of a defect in JA-Ile 12-hydroxylase activity; in response to wounding, cyp94b3 mutants hyperaccumulate JA-Ile and are also deficient in 12-OH-JA-Ile. The biochemical function of CYP94B3 was confirmed by experiments showing that the recombinant enzyme has JA-Ile 12-hydroxylase activity (Koo et al., 2011). Independent studies confirmed the identity of CYP94B3 as a JA-Ile 12-hydroxylase and further demonstrated that CYP94C1 plays a major role in conversion of 12-OH-JA-Ile to 12-COOH-JA-Ile (Kitaoka et al., 2011; Heitz et al., 2012). These collective studies establish a central role for CYP94s in the ω-oxidation pathway for turnover and deactivation of JA-Ile (Figure 2). Interestingly, CYP P450-mediated ω-oxidation (and subsequent β-oxidation) pathways also mediate catabolism and deactivation of eicosanoid signals in animal cells (Hardwick, 2008; Buczynski et al., 2009). The discovery of the ω-oxidation pathway for JA-Ile catabolism therefore highlights a conserved feature of lipid-based signaling in plants and animals.
The phenotype of mutants affected in CYP94B3 expression demonstrates the importance of JA-Ile catabolism in plant growth, development, and defense. CYP94B3-overexpressing plants, which are severely deficient in wound-induced JA-Ile accumulation, display hallmark signs of JA-Ile deficiency, including defects in anther and pollen development, resistance to JA-mediated root growth inhibition, and increased susceptibility to insect herbivores (Koo et al., 2011). In contrast, hyperaccumulation of JA-Ile in cyp94b3 null mutants results in increased sensitivity to the hormone. Among the phenotypes reported for cyp94b3 single and cyp94b3cyp94c1 double mutants are enhanced expression of JA-response genes and increased sensitivity to JA-mediated root growth inhibition (Kitaoka et al., 2011; Koo et al., 2011; Heitz et al., 2012). It was also reported that cyp94b3 mutants are more susceptible to a virulent strain of Pseudomonas syringae (Hwang and Hwang, 2010), which may reflect the fact that increased signaling through the JA-Ile receptor compromises host resistance to this pathogen by suppressing the salicylate-mediated branch of immunity (Glazebrook, 2005; Katsir et al., 2008b). These collective results demonstrate that CYP94B3 and CYP94C1 function to reduce the active pool of JA-Ile, and that this route of hormone catabolism affects physiological processes in roots, leaves, and reproductive tissues. This conclusion is consistent with the emerging view that JA-Ile is the active signal for many, if not most, jasmonate responses.
Mining the CYP94ome
Cytochrome P450s constitute the most metabolically diverse group of enzymes in the plant kingdom (Nelson and Werck-Reichhart, 2011). The recent characterization of CYP94B3 and CYP94C1 (Kitaoka et al., 2011; Koo et al., 2011; Heitz et al., 2012) highlight the widespread role of P450s in plant hormone metabolism (Mizutani and Ohta, 2010) and shed light on the physiological function of the broader CYP94 family. Arabidopsis contains six CYP94 members belonging to three subfamilies: CYP94B1, CYP94B2, CYP94B3, CYP94C1, CYP94D1, and CYP94D2 (Figure 3). That various CYP94 subfamilies have undergone significant expansion and contraction in particular angiosperm lineages suggests the existence of rapidly evolving metabolic pathways involving fatty acid-derived compounds (Guttikonda et al., 2010; Nelson and Werck-Reichhart, 2011). P450 phylogenies also indicate that the CYP94 family is conserved in non-vascular plants (e.g., Physcomitrella patens) but is not present in photosynthetic aquatic organisms. This distribution of CYP94s coincides with the occurrence in these lineages of genes encoding various components of the JA biosynthesis and signaling pathways (Chico et al., 2008; Katsir et al., 2008a; Browse, 2009). Interestingly, P. patens contains plastidic enzymes for the synthesis of OPDA but appears to lack the ability to produce JA and JA-Ile (Stumpe et al., 2010). Functional characterization of CYP94 P450s in P. patens may thus provide insight into the evolutionary origins of oxylipin signaling and metabolism.
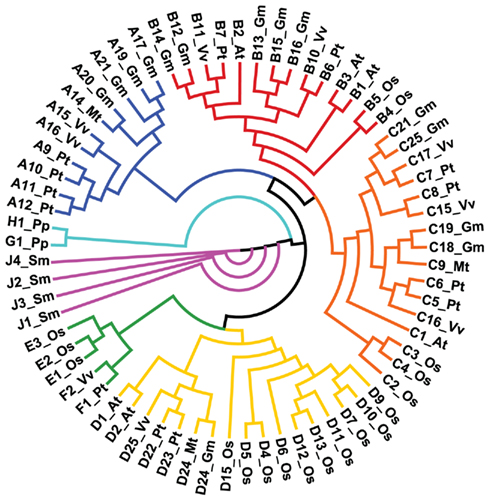
Figure 3. Phylogeny of the CYP94 family of cytochrome P450s in the land plants. A total of 70 deduced full-length amino acid sequences were aligned by Clustal X and displayed as a circular cladogram using Phylip 3.68 and DENDROSCOPE v3.0.13beta (Huson et al., 2007). Abbreviations and number of sequences: Arabidopsis thaliana (At: 6), Oryza sativa (Os: 18), Glycine max (Gm: 14), Medicago truncatula (Mt: 4), Vitis vinifera (Vv: 9), Populus trichocarpa (Pt: 13), Physcomitrella patens (Pp: 2), Selaginella moellendorffii (Sm: 4).
The role of CYP94B3 and CYP94C1 in JA-Ile ω-oxidation suggests that other CYP94s serve similar roles in oxylipin metabolism. For example, the inability of CYP94B3 and C1 to hydroxylate JA in vitro (Kitaoka et al., 2011; Heitz et al., 2012) raises the possibility that formation of 12-OH-JA is catalyzed by another CYP94 member. The presence of residual amounts of 12-OH-JA-Ile in cyp94b3 and cyp94b3cyp94c1 mutants (Kitaoka et al., 2011; Koo et al., 2011; Heitz et al., 2012) further suggests that additional enzymes participate in 12-hydroxylation of JA-Ile. Given the broad specificity of many CYP94s for fatty acyl substrates in vitro (Benveniste et al., 2006; Kandel et al., 2007; Pinot and Beisson, 2011; Heitz et al., 2012), functional characterization of additional family members will clearly benefit from analysis of metabolic phenotypes of cyp94 mutants.
Summary and Future Perspectives
It is becoming increasing clear that the JA signaling pathway is embedded in a complex phytohormone network that controls myriad aspects of plant growth and development, as well as the way in which plants adapt to their environment. Current models of JA signal transduction support the view that the intracellular level of JA-Ile plays a major role in controlling the strength of JA responses. Recent studies with mutants that are altered in CYP94 expression demonstrate the importance of JA-Ile catabolism in attenuating this hormone response pathway (Kitaoka et al., 2011; Koo et al., 2011; Heitz et al., 2012). Negative feedback control of JA responses also depends on other mechanisms, including JA-induced expression of stabilized JAZ isoforms that fail to efficiently bind COI1 in the presence of JA-Ile (Chung and Howe, 2009; Chung et al., 2010; Shyu et al., 2012). Further elucidation of these negative feedback pathways may facilitate biotechnological efforts aimed at exploiting the JA pathway for enhanced resistance of crop plants to insects and pathogens, and for producing economically important plant compounds (e.g., paclitaxel) whose expression is controlled by the JA pathway. Several additional gaps in our understanding of how JA-Ile levels are regulated also remain to be addressed. For example, there is a pressing need to understand the early signaling events that trigger JA-Ile synthesis, and to fully elucidate metabolic pathways that govern deactivation of the hormone. Progress toward the latter goal will undoubtedly be facilitated by further characterization of the “CYP94ome.” Given the high degree of sequence similarity between CYP94s and CYP86 P450s that catalyze the formation of cutin and wax polymers of the epidermal cuticle (Pinot and Beisson, 2011), this line of research may also provide insight into the evolutionary relationship between oxylipins that serve signaling versus structural roles in plant defense.
Another promising area of future research will be to discern the spatial and temporal distribution of JAs in different tissues and cell types. Existing approaches to study the tissue-specific accumulation (Hause et al., 2000; Glauser et al., 2008; Stitz et al., 2011) and non-cell autonomous action (Li et al., 2002; Koo et al., 2009) of JAs would benefit from new technologies to quantify hormone levels with cellular and sub-cellular resolution (Bermejo et al., 2011; Mielke et al., 2011). Such efforts are expected to lead to a better understanding of how bioactive and non-bioactive JAs are compartmentalized within cells and transported between cells. Given the plethora of JA derivatives produced in plants, it will also be important to identify additional bioactive JAs that exert physiological effects, either through the COI1–JAZ receptor system or via other mechanisms. For example, the identity of 12-OH-JAs as factors for leaf closing (Nakamura et al., 2011) and tuber induction (Yoshihara et al., 1989) provides tantalizing evidence that JAs evolved as lipid-derived signals for regulating specialized aspects of plant physiology.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Marlene Cameron for assistance with the production of Figures, and Dr. David Nelson (University of Tennessee) for providing CYP94 sequences used for phylogenetic analyses. This work was supported by the National Institutes of Health (grant R01GM57795) and the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (grant DE–FG02–91ER20021).
References
Ballare, C. L. (2011). Jasmonate-induced defenses: a tale of intelligence, collaborators and rascals. Trends Plant Sci. 16, 249–257.
Benveniste, I., Saito, T., Wang, Y., Kandel, S., Huang, H. W., Pinot, F., Kahn, R. A., Salaun, J. P., and Shimoji, M. (2006). Evolutionary relationship and substrate specificity of Arabidopsis thaliana fatty acid ω-hydroxylase. Plant Sci. 170, 326–338.
Bermejo, C., Ewald, J. C., Lanquar, V., Jones, A. M., and Frommer, W. B. (2011). In vivo biochemistry: quantifying ion and metabolite levels in individual cells or cultures of yeast. Biochem. J. 438, 1–10.
Bonaventure, G., Schuck, S., and Baldwin, I. T. (2011). Revealing complexity and specificity in the activation of lipase-mediated oxylipin biosynthesis: a specific role of the Nicotiana attenuata GLA1 lipase in the activation of jasmonic acid biosynthesis in leaves and roots. Plant Cell Environ. 34, 1507–1520.
Browse, J. (2009). Jasmonate passes muster: a receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60, 183–205.
Browse, J., and Howe, G. A. (2008). Update on jasmonate signaling: new weapons and a rapid response against insect attack. Plant Physiol. 146, 832–838.
Buczynski, M. W., Dumlao, D. S., and Dennis, E. A. (2009). An integrated omics analysis of eicosanoid biology. J. Lipid Res. 50, 1015–1038.
Chico, J. M., Chini, A., Fonseca, S., and Solano, R. (2008). JAZ repressors set the rhythm in jasmonate signaling. Curr. Opin. Plant Biol. 11, 486–494.
Chini, A., Fonseca, S., Fernandez, G., Adie, B., Chico, J. M., Lorenzo, O., Garcia-Casado, G., Lopez-Vidriero, I., Lozano, F. M., Ponce, M. R., Micol, J. L., and Solano, R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671.
Chung, H. S., Cooke, T. F., Depew, C. L., Patel, L. C., Ogawa, N., Kobayashi, Y., and Howe, G. A. (2010). Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. Plant J. 63, 613–622.
Chung, H. S., and Howe, G. A. (2009). A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21, 131–145.
Chung, H. S., Koo, A. J. K., Gao, X., Jayanty, S., Thines, B., Jones, A. D., and Howe, G. A. (2008). Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 146, 952–964.
Creelman, R. A., and Mullet, J. E. (1997). Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 355–381.
Dombrecht, B., Xue, G. P., Sprague, S. J., Kirkegaard, J. A., Ross, J. J., Reid, J. B., Fitt, G. P., Sewelam, N., Schenk, P. M., Manners, J. M., and Kazan, K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19, 2225–2245.
Duan, H., and Schuler, M. A. (2005). Differential expression and evolution of the Arabidopsis CYP86A subfamily. Plant Physiol. 137, 1067–1081.
Ehlting, J., Sauveplane, V., Olry, A., Ginglinger, J. F., Provart, N. J., and Werck-Reichhart, D. (2008). An extensive (co-)expression analysis tool for the cytochrome P450 superfamily in Arabidopsis thaliana. BMC Plant Biol. 8, 47. doi:10.1186/1471-2229-8-47
Fonseca, S., Chini, A., Hamberg, M., Adie, B., Porzel, A., Kramell, R., Miersch, O., Wasternack, C., and Solano, R. (2009). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5, 344–350.
Gidda, S. K., Miersch, O., Levitin, A., Schmidt, J., Wasternack, C., and Varin, L. (2003). Biochemical and molecular characterization of a hydroxyjasmonate sulfotransferase from Arabidopsis thaliana. J. Biol. Chem. 278, 17895–17900.
Glauser, G., Grata, E., Dubugnon, L., Rudaz, S., Farmer, E. E., and Wolfender, J. L. (2008). Spatial and temporal dynamics of jasmonate synthesis and accumulation in Arabidopsis in response to wounding. J. Biol. Chem. 283, 16400–16407.
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227.
Goepfert, S., and Poirier, Y. (2007). (β-Oxidation in fatty acid degradation and beyond. Curr. Opin. Plant Biol. 10, 245–251.
Guranowski, A., Miersch, O., Staswick, P. E., Suza, W., and Wasternack, C. (2007). Substrate specificity and products of side-reactions catalyzed by jasmonate:amino acid synthetase (JAR1). FEBS Lett. 581, 815–820.
Guttikonda, S. K., Trupti, J., Bisht, N. C., Chen, H., An, Y. Q. C., Pandey, S., Xu, D., and Yu, O. (2010). Whole genome co-expression analysis of soybean cytochrome P450 genes identifies nodulation-specific P450 monooxygenases. BMC Plant Biol. 10, 243. doi:10.1186/1471-2229-10-243
Hardwick, J. P. (2008). Cytochrome P450 omega hydroxylase (CYP4) function in fatty acid metabolism and metabolic diseases. Biochem. Pharmacol. 75, 2263–2275.
Hause, B., Stenzel, I., Miersch, O., Maucher, H., Kramell, R., Ziegler, J., and Wasternack, C. (2000). Tissue-specific oxylipin signature of tomato flowers: allene oxide cyclase is highly expressed in distinct flower organs and vascular bundles. Plant J. 24, 113–126.
Heitz, T., Widemann, E., Lugan, R., Miesch, L., Ullmann, P., Désaubry, L., Holder, E., Grausem, B., Kandel, S., Miesch, M., Werck-Reichhart, D., and Pinot, F. (2012). Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of the plant hormone jasmonoyl-isoleucine for catabolic turnover. J. Biol. Chem. PMID: 22215670. [Epub ahead of print].
Hou, X., Lee, L. Y., Xia, K., Yan, Y., and Yu, H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19, 884–894.
Howe, G., and Jander, G. (2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59, 41–66.
Howe, G. A. (2010). Ubiquitin ligase-coupled receptors extend their reach to jasmonate. Plant Physiol. 154, 471–474.
Huson, D. H., Richter, D. C., Rausch, C., Dezulian, T., Franz, M., and Rupp, R. (2007). Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics 8, 460. doi:10.1186/1471-2105-8-460
Hwang, I. S., and Hwang, B. K. (2010). Role of the pepper cytochrome P450 gene CaCYP450A in defense responses against microbial pathogens. Planta 232, 1409–1421.
Ishiguro, S., Kawai-Oda, A., Ueda, J., Nishida, I., and Okada, K. (2001). The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13, 2191–2209.
Kallenbach, M., Alagna, F., Baldwin, I. T., and Bonaventure, G. (2010). Nicotiana attenuata SIPK, WIPK, NPR1, and fatty acid-amino acid conjugates participate in the induction of jasmonic acid biosynthesis by affecting early enzymatic steps in the pathway. Plant Physiol. 152, 96–106.
Kandel, S., Sauveplane, V., Compagnon, V., Franke, R., Millet, Y., Schreiber, L., Werck-Reichhart, D., and Pinot, F. (2007). Characterization of a methyl jasmonate and wounding-responsive cytochrome P450 of Arabidopsis thaliana catalyzing dicarboxylic fatty acid formation in vitro. FEBS J. 274, 5116–5127.
Kang, J. H., Wang, L., Giri, A., and Baldwin, I. T. (2006). Silencing threonine deaminase and JAR4 in Nicotiana attenuata impairs jasmonic acid-isoleucine-mediated defenses against Manduca sexta. Plant Cell 18, 3303–3320.
Katsir, L., Chung, H. S., Koo, A. J. K., and Howe, G. A. (2008a). Jasmonate signaling: a conserved mechanism of hormone sensing. Curr. Opin. Plant Biol. 11, 428–435.
Katsir, L., Schilmiller, A. L., Staswick, P. E., He, S. Y., and Howe, G. A. (2008b). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. U.S.A. 105, 7100–7105.
Kazan, K., and Manners, J. M. (2011). The interplay between light and jasmonate signalling during defence and development. J. Exp. Bot. 62, 4087–4100.
Kazan, K., and Manners, J. M. (2012). JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci. 17, 22–31.
Kitaoka, N., Matsubara, T., Sato, M., Takahashi, K., Wakuta, S., Kawaide, H., Matsui, H., Nabeta, K., and Matsuura, H. (2011). Arabidopsis CYP94B3 encodes jasmonyl-l-isoleucine 12-hydroxylase, a key enzyme in the oxidative catabolism of jasmonate. Plant Cell Physiol. 52, 1757–1765.
Koo, A. J., Cooke, T. F., and Howe, G. A. (2011). Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-L-isoleucine. Proc. Natl. Acad. Sci. U.S.A. 108, 9298–9303.
Koo, A. J. K., Chung, H. S., Kobayashi, Y., and Howe, G. A. (2006). Identification of a peroxisomal acyl-activating enzyme involved in the biosynthesis of jasmonic acid in Arabidopsis. J. Biol. Chem. 281, 33511–33520.
Koo, A. J. K., Gao, X. L., Jones, A. D., and Howe, G. A. (2009). A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. Plant J. 59, 974–986.
LeClere, S., Tellez, R., Rampey, R. A., Matsuda, S. P. T., and Bartel, B. (2002). Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J. Biol. Chem. 277, 20446–20452.
Li, L., Li, C., Lee, G. I., and Howe, G. A. (2002). Distinct roles for jasmonate synthesis and action in the systemic wound response of tomato. Proc. Natl. Acad. Sci. U.S.A. 99, 6416–6421.
Li, L., Zhao, Y., McCaig, B. C., Wingerd, B. A., Wang, J., Whalon, M. E., Pichersky, E., and Howe, G. A. (2004). The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16, 126–143.
McConn, M., and Browse, J. (1996). The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8, 403–416.
Melotto, M., Mecey, C., Niu, Y., Chung, H. S., Katsir, L., Yao, J., Zeng, W., Thines, B., Staswick, P., Browse, J., Howe, G. A., and He, S. Y. (2008). A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 55, 979–988.
Mielke, K., Forner, S., Kramell, R., Conrad, U., and Hause, B. (2011). Cell-specific visualization of jasmonates in wounded tomato and Arabidopsis leaves using jasmonate-specific antibodies. New Phytol. 190, 1069–1080.
Miersch, O., Neumerkel, J., Dippe, M., Stenzel, I., and Wasternack, C. (2008). Hydroxylated jasmonates are commonly occurring metabolites of jasmonic acid and contribute to a partial switch-off in jasmonate signaling. New Phytol. 177, 114–127.
Mizutani, M., and Ohta, D. (2010). Diversification of P450 genes during land plant evolution. Annu. Rev. Plant Biol. 61, 291–315.
Moreno, J. E., Tao, Y., Chory, J., and Ballare, C. L. (2009). Ecological modulation of plant defense via phytochrome control of jasmonate sensitivity. Proc. Natl. Acad. Sci. U.S.A. 106, 4935–4940.
Nakamura, Y., Mithofer, A., Kombrink, E., Boland, W., Hamamoto, S., Uozumi, N., Tohma, K., and Ueda, M. (2011). 12-Hydroxyjasmonic acid glucoside is a COI1-JAZ-independent activator of leaf-closing movement in Samanea saman. Plant Physiol. 155, 1226–1236.
Nelson, D., and Werck-Reichhart, D. (2011). A P450-centric view of plant evolution. Plant J. 66, 194–211.
Pauwels, L., Morreel, K., De Witte, E., Lammertyn, F., Van Montagu, M., Boerjan, W., Inze, D., and Goossens, A. (2008). Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc. Natl. Acad. Sci. U.S.A. 105, 1380–1385.
Pieterse, C. M. J., Leon-Reyes, A., Van der Ent, S., and Van Wees, S. C. M. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316.
Pinot, F., and Beisson, F. (2011). Cytochrome P450 metabolizing fatty acids in plants: characterization, and physiological roles. FEBS J. 278, 195–205.
Ralph, S., Oddy, C., Cooper, D., Yueh, H., Jancsik, S., Kolosova, N., Philippe, R. N., Aeschliman, D., White, R., Huber, D., Ritland, C. E., Benoit, F., Rigby, T., Nantel, A., Butterfield, Y. S. N., Kirkpatrick, R., Chun, E., Liu, J., Palmquist, D., Wynhoven, B., Stott, J., Yang, G., Barber, S., Holt, R. A., Siddiqui, A., Jones, S. J. M., Marra, M. A., Ellis, B. E., Douglas, C. J., Ritland, K., and Bohlmann, J. (2006). Genomics of hybrid poplar (Populus trichocarpa x deltoides) interacting with forest tent caterpillars (Malacosoma disstria): normalized and full-length cDNA libraries, expressed sequence tags, and a cDNA microarray for the study of insect-induced defences in poplar. Mol. Ecol. 15, 1275–1297.
Reymond, P., Weber, H., Damond, M., and Farmer, E. E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12, 707–720.
Robson, F., Okamoto, H., Patrick, E., Harris, S. R., Wasternack, C., Brearley, C., and Turner, J. G. (2010). Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell 22, 1143–1160.
Sasaki, Y., Asamizu, E., Shibata, D., Nakamura, Y., Kaneko, T., Awai, K., Amagai, M., Kuwata, C., Tsugane, T., Masuda, T., Shimada, H., Takamiya, K., Ohta, H., and Tabata, S. (2001). Monitoring of methyl jasmonate-responsive genes in Arabidopsis by cDNA macroarray: self-activation of jasmonic acid biosynthesis and crosstalk with other phytohormone signaling pathways. DNA Res. 8, 153–161.
Sasaki-Sekimoto, Y., Taki, N., Obayashi, T., Aono, M., Matsumoto, F., Sakurai, N., Suzuki, H., Hirai, M. Y., Noji, M., Saito, K., Masuda, T., Takamiya, K., Shibata, D., and Ohta, H. (2005). Coordinated activation of metabolic pathways for antioxidants and defence compounds by jasmonates and their roles in stress tolerance in Arabidopsis. Plant J. 44, 653–668.
Schaller, A., and Stintzi, A. (2009). Enzymes in jasmonate biosynthesis – structure, function, regulation. Phytochemistry 70, 1532–1538.
Sheard, L. B., Tan, X., Mao, H. B., Withers, J., Ben-Nissan, G., Hinds, T. R., Kobayashi, Y., Hsu, F. F., Sharon, M., Browse, J., He, S. Y., Rizo, J., Howe, G. A., and Zheng, N. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468, 400–405.
Shyu, C., Figueroa, P., DePew, C. L., Cooke, T. F., Sheard, L.B., Moreno, J., Katsir, L., Zheng, N., Browse, J., and Howe, G.A. (2012). JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell. (in press).
Staswick, P. E., and Tiryaki, I. (2004). The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16, 2117–2127.
Staswick, P. E., Yuen, G. Y., and Lehman, C. C. (1998). Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J. 15, 747–754.
Stenzel, I., Hause, B., Miersch, O., Kurz, T., Maucher, H., Weichert, H., Ziegler, J., Feussner, I., and Wasternack, C. (2003). Jasmonate biosynthesis and the allene oxide cyclase family of Arabidopsis thaliana. Plant Mol. Biol. 51, 895–911.
Stitz, M., Gase, K., Baldwin, I. T., and Gaquerel, E. (2011). Ectopic expression of AtJMT in Nicotiana attenuata: creating a metabolic sink has tissue-specific consequences for the jasmonate metabolic network and silences downstream gene expression. Plant Physiol. 157, 341–354.
Stumpe, M., Gobel, C., Faltin, B., Beike, A. K., Hause, B., Himmelsbach, K., Bode, J., Kramell, R., Wasternack, C., Frank, W., Reski, R., and Feussner, I. (2010). The moss Physcomitrella patens contains cyclopentenones but no jasmonates: mutations in allene oxide cyclase lead to reduced fertility and altered sporophyte morphology. New Phytol. 188, 740–749.
Suza, W. P., Rowe, M. L., Hamberg, M., and Staswick, P. E. (2010). A tomato enzyme synthesizes (+)-7-iso-jasmonoyl-L-isoleucine in wounded leaves. Planta 231, 717–728.
Suza, W. P., and Staswick, P. E. (2008). The role of JAR1 in jasmonoyl-L-isoleucine production in Arabidopsis wound response. Planta 227, 1221–1232.
Swiatek, A., Van Dongen, W., Esmans, E. L., and Van Onckelen, H. (2004). Metabolic fate of jasmonates in tobacco bright yellow-2 cells. Plant Physiol. 135, 161–172.
Tamogami, S., Ralkwal, R., and Agrawal, G. K. (2008). Interplant communication: airborne methyl jasmonate is essentially converted into JA and JA-Ile activating jasmonate signaling pathway and VOCs emission. Biochem. Biophys. Res. Commun. 376, 723–727.
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., Nomura, K., He, S. Y., Howe, G. A., and Browse, J. (2007). JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448, 661–665.
VanDoorn, A., Bonaventure, G., Schmidt, D. D., and Baldwin, I. T. (2011). Regulation of jasmonate metabolism and inactivation of systemic signaling in Solanum nigrum: COI1 and JAR4 play overlapping yet distinct roles. New Phytol. 190, 640–652.
Wang, L., Halitschke, R., Kang, J. H., Berg, A., Harnisch, F., and Baldwin, I. T. (2007). Independently silencing two JAR family members impairs levels of trypsin proteinase inhibitors but not nicotine. Planta 226, 159–167.
Wasternack, C. (2007). Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 100, 681–697.
Woodward, A. W., and Bartel, B. (2005). Auxin: regulation, action, and interaction. Ann. Bot. 95, 707–735.
Wu, J. S., and Baldwin, I. T. (2010). New insights into plant responses to the attack from insect herbivores. Annu. Rev. Genet. 44, 1–24.
Wu, J. S., Wang, L., and Baldwin, I. T. (2008). Methyl jasmonate-elicited herbivore resistance: does MeJA function as a signal without being hydrolyzed to JA? Planta 227, 1161–1168.
Xie, D. X., Feys, B. F., James, S., Nieto-Rostro, M., and Turner, J. G. (1998). COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094.
Yan, J. B., Zhang, C., Gu, M., Bai, Z. Y., Zhang, W. G., Qi, T. C., Cheng, Z. W., Peng, W., Luo, H. B., Nan, F. J., Wang, Z., and Xie, D. X. (2009). The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21, 2220–2236.
Yan, Y., Stolz, S., Chetelat, A., Reymond, P., Pagni, M., Dubugnon, L., and Farmer, E. E. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19, 2470–2483.
Yoshihara, T., Omer, E. A., Koshino, H., Sakamura, S., Kikuta, Y., and Koda, Y. (1989). Structure of a tuber-inducing stimulus from potato leaves. Agric. Biol. Chem. 53, 2835–2837.
Zhang, Y., and Turner, J. (2008). Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS ONE 3, e3699. doi:10.1371/journal.pone.0003699
Zhu, Z., An, F., Feng, Y., Li, P., Xue, L., A, M., Jiang, Z., Kim, J. M., To, T. K., Li, W., Zhang, X., Yu, Q., Dong, Z., Chen, W. Q., Seki, M., Zhou, J. M., and Guo, H. (2011). Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 12539–12544.
Keywords: oxylipin metabolism, lipid signaling, cytochrome P450, Arabidopsis, plant hormone, plant defense, jasmonate, omega oxidation
Citation: Koo AJK and Howe GA (2012) Catabolism and deactivation of the lipid-derived hormone jasmonoyl-isoleucine. Front. Plant Sci. 3:19. doi: 10.3389/fpls.2012.00019
Received: 21 December 2011;
Accepted: 18 January 2012;
Published online: 07 February 2012.
Edited by:
Kent D. Chapman, University of North Texas, USAReviewed by:
Paul Staswick, University of Nebraska – Lincoln, USASrinivasa Rao Uppalapati, The Samuel Roberts Noble Foundation, USA
Copyright: © 2011 Koo and Howe. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Gregg A. Howe, Department of Energy-Plant Research Laboratory, Michigan State University, 122 Plant Biology Building, East Lansing, MI 48824-1312, USA. e-mail: howeg@msu.edu
