- Energy Biosciences Institute, University of California, Berkeley, CA, USA
Prospective plant pathogens must overcome the physical barrier presented by the plant cell wall. In addition to being a preformed, passive barrier limiting access of pathogens to plant cells, the cell wall is actively remodeled and reinforced specifically at discrete sites of interaction with potentially pathogenic microbes. Active reinforcement of the cell wall through the deposition of cell wall appositions, referred to as papillae, is an early response to perception of numerous categories of pathogens including fungi and bacteria. Rapid deposition of papillae is generally correlated with resistance to fungal pathogens that attempt to penetrate plant cell walls for the establishment of feeding structures. Despite the ubiquity and apparent importance of this early defense response, relatively little is known about the underlying molecular mechanisms and cellular processes involved in the targeting and assembly of papillae. This review summarizes recent advances in our understanding of cell wall-associated defenses induced by pathogen perception as well as the impact of changes in cell wall polymers on interactions with pathogens and highlights significant unanswered questions driving future research in the area.
Introduction
Plants have evolved a multi-layered system of defenses to contend with the threat of infection by microbial pathogens. The current view of the plant immune system is that it can be conceptually divided into two modes of response. These modes are distinguished by the signals perceived to initiate the response and the nature or severity of the defense response exhibited, in particular, the presence or absence of the programmed cell death referred to as the hypersensitive response (HR). The first line of inducible defenses are initiated upon perception of conserved, microbe-derived elicitor molecules referred to as microbe- or pathogen-associated molecular patterns (MAMPs or PAMPs) or by host-derived elicitor molecules, referred to as damage-associated molecular patterns (DAMPs), which result from attempted infection by pathogens. This mode of plant immune response is referred to as pattern-triggered immunity (PTI; Jones and Dangl, 2006). The full spectrum of immune responses associated with PTI in plants is not currently understood, but discretely localized reinforcement of the cell wall through deposition of papillae at sites of pathogen detection appears to be a common component of the PTI response (Nicaise et al., 2009).
All prospective microbial phytopathogens must interact with the cell wall in some manner, and the nature of this interaction is typically determined by the lifestyle of the pathogen. Necrotrophic pathogens, which kill cells and feed on dead tissues, typically macerate plant tissues by secreting abundant hydrolytic enzymes that degrade cell wall polymers (Laluk and Mengiste, 2010). In contrast, biotrophic and hemibiotrophic pathogens must interact with living plant cells for all or part of their life cycles and typically employ more subtle strategies for interacting with the cell wall. Haustorium-forming pathogens, such as various fungal and oomycete mildews, must penetrate cell walls to establish haustorial feeding structures in close contact with the underlying host cell (Szabo and Bushnell, 2001). Bacterial phytopathogens typically manipulate plant cells by delivering effector proteins to host cells via secretion pili that must traverse the cell wall to access the underlying cell membrane and cytosol (Büttner and He, 2009). Deposition of papillae at sites of pathogen detection is thought to act as a physical barrier to limit access of pathogens to the underlying protoplast. Additionally, papillae are sites of accumulation of antimicrobial secondary metabolites (Bednarek et al., 2009; Clay et al., 2009). Rapid formation of papillae is correlated with enhanced resistance to fungal penetration, whereas, delayed papilla formation correlates with increased fungal penetration success (Bayles et al., 1990; Collins et al., 2003). Successful cell wall-associated defenses can halt invading pathogens at an early stage, before the establishment of disease, and can eliminate the need for more costly defense responses such as HR cell death. Therefore, it is of fundamental importance to understand the mechanisms through which cell wall-associated defenses are elaborated and to understand why these defenses fail to prevent infection by adapted pathogens. Here, we will focus on recent progress in our understanding of the role of the cell wall in plant–microbe interactions, concentrating primarily on interactions with biotrophic and hemibiotrophic pathogens.
Cell Wall Appositions
Observations of cell wall-associated defense responses to attempted pathogen invasion were first made over a century ago (Smith, 1900). In the intervening years, numerous studies have focused on characterization of papillae through ultrastructural observations or histochemical and immunohistochemical analyses. These studies have revealed significant details about the structure and composition of papillae and related structures such as collars at the neck regions of fungal haustoria and haustorial encasements (Figure 1). The specific biochemical constituents of papillae seem to vary somewhat between plant species, but classes of compounds commonly found associated with papillae include: callose; phenolics including lignin; and phenolic conjugates such as phenolic–polyamines; reactive oxygen species (ROS); peroxidases; cell wall structural proteins such as arabinogalactan proteins and hydroxyproline-rich glycoproteins; and cell wall polymers including pectin and xyloglucans (Aist, 1976; Zeyen et al., 2002).
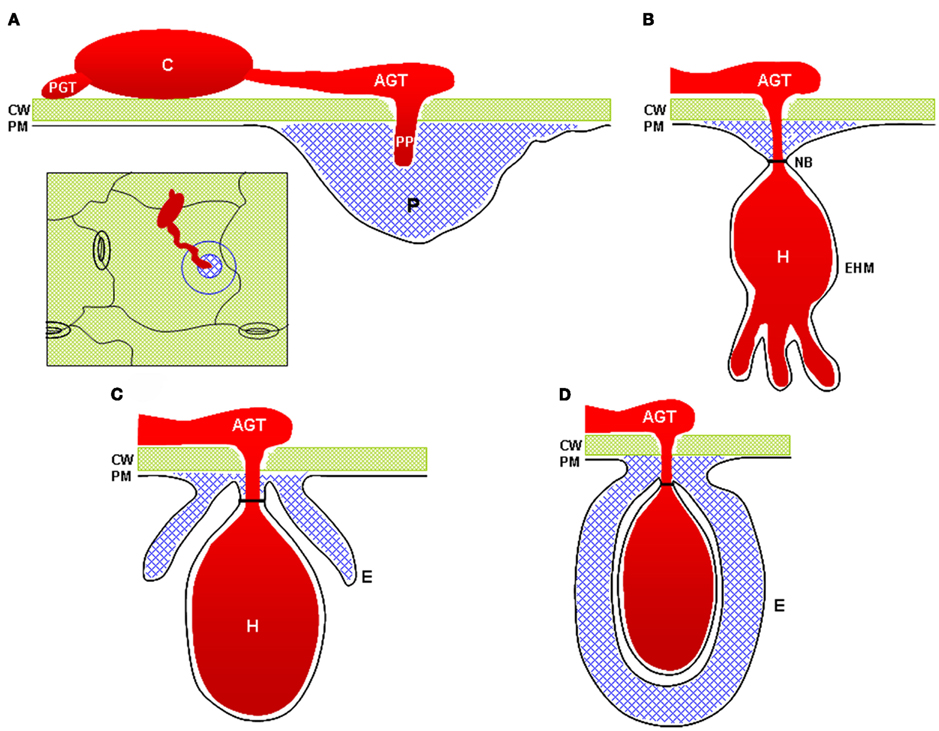
Figure 1. Cell wall-associated structures commonly observed at sites of interaction with powdery mildews and other fungal pathogens. (A) A fungal penetration attempt halted by deposition of a cell wall apposition (blue). Inset image illustrates a top-down view of the penetration site as typically visualized by light microscopy. (B) A successful penetration event in which the fungus has formed a haustorial feeding structure. The cell wall apposition materials form a neck-band or collar around the neck of the haustorium. (C) A haustorium partially surrounded by a haustorial encasement. Encasements contain materials similar to those found in cell wall appositions. (D) A fully encased haustorium. CW, cell wall; PM, plasma membrane; C, conidiospore; PGT, primary germ tube (note that not all powdery mildew species develop PGTs); AGT, appressorial germ tube; PP, penetration peg; H, haustorium; EHM, extra-haustorial membrane; NB, haustorial neck-band; P, papilla (e.g., cell wall apposition); E, haustorial encasement.
The β-1,3-glucan polymer callose is an abundant and ubiquitous component of cell wall appositions. However, the role of callose in papillae is not clear. The Arabidopsis thaliana callose synthase responsible for wound- and pathogen-induced callose deposition was identified as being encoded by PMR4/GSL5 (Jacobs et al., 2003; Nishimura et al., 2003). Loss of function pmr4 mutants retained the ability to deposit papillae at sites of attempted powdery mildew penetration and only a minor increase in penetration frequency by the barley powdery mildew Blumeria graminis f.sp. hordei was observed, suggesting that callose does not play a dramatic role as a structural barrier in papillae, at least in the context of powdery mildew penetration. In contrast, RNAi knockdown of a callose synthase in lemon (Citrus limon) leaves resulted in enhanced susceptibility to the bacterial pathogen Xanthomonas citri (Enrique et al., 2011). X. campestris pv campestris has been shown to block callose deposition in Nicotiana benthamiana and Arabidopsis through the production of the exopolysaccharide xanthan (Yun et al., 2006). These results suggest that callose deposition in papillae may play a role in limiting access of Xanthomonas to host cells. However, whether callose acts as a direct physical barrier is still not clear as it is not known whether the absence of callose has additional effects on papilla structure or other defenses in lemon leaves. Unexpectedly, pmr4 mutants displayed enhanced resistance to normally virulent biotrophic pathogens including Golovinomyces cichoracearum, G. orontii, and Hyaloperonospora arabidopsidis (Vogel and Somerville, 2000; Jacobs et al., 2003). Enhanced resistance was dependent on an intact salicylic acid (SA) defense signaling pathway and common SA-induced genes were found to be upregulated in pmr4 and hyperinduced in pmr4 upon powdery mildew inoculation. These results imply that callose or the PMR4 protein itself negatively regulates SA synthesis or signaling. One possible interpretation of these unexpected results is that callose serves as a protective containment barrier to shield the plant cell from toxic metabolites that accumulate in papillae and haustorial encasements. Such a hypothesis is supported by the observations that callose-containing cell wall appositions occur at sites of plasmodesmata in cells neighboring those undergoing HR cell death and that cells that have undergone HR typically become encased by callose (Jacobs et al., 2003; An et al., 2006). Alternatively, callose deposition in papillae and haustorial encasements may limit diffusion of pathogen-derived elicitors, thus reducing the level of activation of the SA-dependent defense pathway.
The mechanisms of regulation and targeting for PMR4/GSL5 callose deposition are largely unknown. Recently, the barley ADP-ribosylation factor (ARF) GTPase ARFA1b/1c was found to be important for callose deposition at powdery mildew penetration sites (Böhlenius et al., 2010). RNAi knockdown or expression of a dominant negative ARFA1b/1c variant abolished callose accumulation at penetration sites and resulted in increased fungal penetration success. ARFA1b/1c was found to localize to an endosomal multi-vesicular body compartment that accumulated at fungal penetration sites prior to the accumulation of callose. The authors propose a model whereby cell surface callose synthase enzymes are activated throughout the cell and the resulting extracellular callose is internalized into multi-vesicular bodies and delivered to penetration sites in an ARFA1b/1c dependent process. Interestingly, the Arabidopsis ARF-GEF (guanine nucleotide exchange factor) MIN7 is required for normal levels of callose deposition in response to the Pseudomonas syringae pv. tomato (Pst) ΔCEL mutant, suggesting that an ARF-dependent process may also play a role in callose deposition at sites of pathogen detection in Arabidopsis (Nomura et al., 2006).
In addition to callose, phenolic polymers are incorporated into papillae. Phenolics are thought to contribute to the physical barrier through cross linking to form a hardened wall that cannot be easily degraded by enzymes employed by pathogens (Zeyen et al., 2002). Accumulation of a specific phenolic–polyamine conjugate correlated with resistance in mlo barley and was found to have direct antifungal activity, suggesting that phenolic polymers may have multiple defense functions in papillae (von Röpenack et al., 1998). Recently, Arabidopsis mutants impaired in specific isoforms of cinnamyl alcohol dehydrogenase (CAD), enzymes that catalyze the final step of mono-lignol biosynthesis, were found to be more susceptible to Pst (Tronchet et al., 2010). Increased levels of multiplication and more severe disease symptoms were observed on cad-b1 single mutants and cad-C/cad-D double mutants inoculated with either virulent Pst DC3000 or avirulent DC3000 (AvrPphB). The mutants exhibited altered expression profiles for SA biosynthesis and response genes after bacterial inoculation, complicating interpretation of the results. It is not yet clear if phenolic polymers or lignin have a direct effect on antibacterial defenses as a structural barrier and/or through antimicrobial activity or if the effects are indirectly exerted through altered SA biosynthesis or response.
Plant cells respond to invasion by haustorium-forming pathogens through the deposition of several related cell wall-associated structures including papillae, haustorial encasements, and haustorial collars or neckbands (Zeyen et al., 2002; Micali et al., 2011; Figure 1). Ultrastructural observations have suggested that haustorial encasements and collars are extensions of or derived from papillae (Zeyen et al., 2002). More recent immunocytochemical characterization of haustorial encasements supports the notion that these structures are extensions of papillae. Immunofluorescence and immunogold labeling detected callose, arabinogalactan proteins, rhamnogalacturonan I, a β-linked galactose-containing protein, and xyloglucan in Arabidopsis encasements completely or partially surrounding G. orontii haustoria, but failed to detect high- or low-ester homogalacturonans or extensins (Micali et al., 2011). The same epitopes were detected in papillae, reinforcing the notion that these structures are related. In addition to cell wall materials, papillae, and encasements also contain membrane lipid materials and multi-vesicular bodies that may be involved in delivery of materials for construction of the papilla (Meyer et al., 2009). Haustorial encasements appear to be defensive structures rather than playing a role in haustorium accommodation as they are not observed around haustoria in compatible interactions, suggesting that adapted fungi have the ability to suppress formation of encasements. Encasements were observed around ∼20% of G. orontii haustoria in Arabidopsis, but were not observed around G. cichoracearum haustoria, suggesting that G. cichoracearum can suppress this defense response more effectively than G. orontii (Koh et al., 2005; Meyer et al., 2009). Understanding how haustorium-forming pathogens suppress the formation of encasements and defeat papilla-associated cell wall defenses will be a significant challenge for future research.
Plasma Membrane-Cell Wall Adhesion
Plant cells, like those of other multicellular organisms, maintain a connection to and communication with their extracellular environment. Mammalian cells connect to and communicate with their extracellular matrix in part through plasma membrane (PM)-localized integrin proteins that recognize components of the extracellular matrix via an Arg-Gly-Asp (RGD) protein motif (Gee et al., 2008). In plants, a similar connection between the PM and cell wall dependent on RGD or similar motifs appears to exist (Canut et al., 1998). RGD-mediated PM-cell wall adhesion appears to play a role in resistance to pathogens. PM-wall adhesion can be disrupted by treatment with peptides containing the RGD motif (Canut et al., 1998). Such treatment causes the disappearance of PM attachment sites, referred to as Hechtian strands, that become visible upon plasmolysis and subsequently changes the appearance of the plasmolyzed protoplasts from a concave morphology to convex. Pretreatment with RGD peptides caused a decrease in callose and H2O2 accumulation at penetration sites in both the cowpea rust (Uromyces vignae)-pea (Pisum sativum) and G. cichoracearum-cowpea (Vigna unguiculata) interactions and increased the frequency of penetration by both pathogens (Mellersh and Heath, 2001). A localized decrease in PM-wall adhesion was observed at penetration sites in the cowpea rust–cowpea interaction and the appearance of cell wall defense responses was correlated with the maintenance of PM-wall adhesion. These results suggest that some pathogens may disrupt PM-wall adhesion to promote infection and suppress cell wall-associated defense responses. This notion has been further substantiated by the discovery of an RGD-containing effector protein, IPI-O, from the oomycete pathogen Phytophthora infestans (Senchou et al., 2004). Treatment of Arabidopsis suspension-cultured cells with recombinant IPI-O disrupted PM-wall adhesion and disruption was dependent on an intact RGD motif. IPI-O was found to bind the Arabidopsis lectin receptor kinase LecRK-I.9 (Gouget et al., 2005). Arabidopsis lecRK-I.9 mutants were susceptible to a normally incompatible isolate of P. brassicae whereas lines overexpressing LecRK-I.9 became resistant to a normally virulent P. brassicae isolate (Bouwmeester et al., 2011). PM-wall adhesion was also partially compromised in lecRK-I.9 mutant plants. Similarly, transgenic expression of the IPI-O effector in planta resulted in susceptibility to incompatible P. brassicae and disruption of PM-wall adhesion. Interestingly, lecRK-I.9 and 35S::IPI-O plants were also impaired in callose deposition induced by the Pst DC3000 hrcC mutant and by the flagellin-derived flg22 peptide, further supporting the correlation between disruption of PM-wall adhesion and abrogation of cell wall-associated defense responses. It will be interesting to see whether interfering with PM-wall adhesion is a widespread pathogenesis strategy among phytopathogens. Powdery mildews do not appear to utilize this strategy, as PM-wall adhesion was found to increase slightly in both compatible and incompatible plant–powdery mildew interactions (Mellersh and Heath, 2001).
Recently, the Arabidopsis NDR1 protein was found to share structural similarity to mammalian integrins and to contain a solvent exposed Asn-Gly-Asp (NGD) motif (Knepper et al., 2011). NDR1 is a PM-localized protein that is required for HR induction mediated by numerous resistance genes of the coiled coil–nucleotide binding site-leucine-rich repeat class (Aarts et al., 1998). Despite the importance of NDR1 in resistance protein signaling, its cellular function has remained enigmatic. Knepper and colleagues demonstrated that ndr1 mutants are impaired in PM-wall adhesion and that NGD-containing peptides can disrupt adhesion, suggesting that the NGD motif is functionally equivalent to RGD. Responsiveness of ndr1 mutants to flg22 peptide was reduced. Additionally, electrolyte leakage was increased in ndr1 plants inoculated with avirulent Pst DC3000 (AvrRpt2), suggesting that NDR1 impacts fluid loss from cells. However, treatment with NGD- or RGD-containing peptides did not alter the HR in response to avirulent Pst, indicating that the participation of NDR1 in promoting PM-wall adhesion does not account for its function in resistance protein signaling.
Impacts of Cell Wall Alterations on Plant–Microbe Interactions
In addition to pathogen resistance conferred by active reinforcement of plant cell walls, it is expected that differences in cell wall composition between plant species may account, at least in part, for limitations of pathogen host range (Vorwerk et al., 2004). Due to the complexity of the cell wall and the fact that many mutants with alterations in cell wall polymer composition display significant pleiotropic effects, it has proven difficult to directly address this hypothesis. However, analysis of a number of mutants has revealed correlations between altered cell wall compositions and altered susceptibility to pathogens. The Arabidopsis powdery mildew resistant mutants pmr5 and pmr6 both display similar patterns of cell wall alteration when analyzed by Fourier transform infrared (FTIR) spectroscopy (Vogel et al., 2002, 2004). The FTIR spectra of these mutants suggested an increase in pectin content and decrease in pectin methyl esterification or O-acetylation. The resistance of pmr5 and pmr6 appears specific to powdery mildews as these mutants were susceptible to Pst and H. arabidopsidis. PMR6 was found to encode a putative pectate lyase, consistent with the apparent alteration of pectin content and/or structure in the mutant (Vogel et al., 2002). PMR5 encodes a protein of unknown function that belongs to a large gene family encoding proteins that contain a plant-specific DUF231 domain (Vogel et al., 2004). Interestingly, DUF231 family proteins were found to contain sequence similarity to the N-terminal domain of Cryptococcus neoformans Cas1p, a protein that is involved in polysaccharide acetylation (Janbon et al., 2001; Anantharaman and Aravind, 2010). The C-terminus of Cas1p shares sequence similarity with a small family of Arabidopsis proteins that include REDUCED WALL ACETYLATION2 (RWA2). Arabidopsis rwa2 mutants were found to have reduced acetylation of both pectic and non-pectic wall polysaccharides (Manabe et al., 2011). Interestingly, rwa2 showed increased resistance to the necrotrophic fungal pathogen Botrytis cinerea, but no alteration in susceptibility to the powdery mildew G. cichoracearum. Localization of RWA2 to the endoplasmic reticulum and the fact that the reduction in wall acetylation was not specific to a particular wall polysaccharide prompted Manabe and colleagues to hypothesize that RWA2 acts upstream of polysaccharide acetyl transfer. These authors further speculate that DUF231 proteins may be involved in specific polysaccharide acetylation, potentially as transferase enzymes. This notion is supported by the identification of the Arabidopsis axy4 mutant which specifically lacks xyloglucan O-acetylation and is impaired in a DUF231 protein (Gille et al., 2011). It remains to be determined if PMR5 alters pectin acetylation either directly as a transferase enzyme or indirectly in some other manner and, if this is the case, how an alteration in pectin acetylation results in powdery mildew resistance. Vogel et al. (2004) hypothesized that alteration in pectin modification may result in the release of defense elicitor-active fragments upon degradation by powdery mildew hydrolytic enzymes. Ongoing research on pectin modification by RWA2 and DUF231 proteins may soon shed light on this hypothesis.
Perspective
The cell wall represents a first line of defense for plant cells against infection by microbial pathogens. Despite over a century of study, many questions about the role of the cell wall in pathogen defense remain to be answered: what are the major factors underlying success or failure of wall-associated defenses in a given plant–microbe interaction? What are the rate-limiting steps that affect timing of papilla deposition and can these be rationally altered to enhance resistance? How significant a factor are differences in wall composition in determining pathogen host range? How does PM-wall adhesion contribute to defense activation? Answering these questions may provide tools to promote rational engineering of disease resistance through enhancement of wall-associated defense responses and should further our understanding of the dynamic nature of the plant cell wall.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author thanks Dr. Clarice Souza and Dr. Shauna Somerville for critical reading of the manuscript. Funding was provided in part by a NIH postdoctoral fellowship (F32-GM0834393) and by NSF (Award # 0519898 and 0929226 to Dr. Shauna Somerville). The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
References
Aarts, N., Metz, M., Holub, E., Staskawicz, B. J., Daniels, M. J., and Parker, J. E. (1998). Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 95, 10306–10311.
Aist, J. R. (1976). Papillae and related wound plugs of plant cells. Annu. Rev. Phtyopathol. 14, 145–163.
An, Q., Ehlers, K., Kogel, K-H., van Bel, A. J. E., and Hückelhoven, R. (2006). Multivesicular compartments proliferate in susceptible and resistant MLA12-barley leaves in response to infection by the biotrophic powdery mildew fungus. New Phytol. 172, 563–576.
Anantharaman, V., and Aravind, L. (2010). Novel eukaryotic enzymes modifying cell surface biopolymers. Biol. Direct 5, 1.
Bayles, C. J., Ghemawat, M. S., and Aist, J. R. (1990). Inhibition by 2-deoxy-D-glucose of callose formation, papilla deposition, and resistance to powdery mildew in an mlo barley mutant. Physiol. Mol. Plant Pathol. 36, 63–72.
Bednarek, P., Pi’slewska-Bednarek, M., Svatoš, A., Schneider, B., Doubský, J., Mansurova, M., Humphry, M., Consonni, C., Panstruga, R., Sanchez-Vallet, A., Molina, A., and Schulze-Lefert, P. (2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 232, 101–106.
Böhlenius, H., Mørch, S. M., Godfrey, D., Nielsen, M. E., and Thordal-Christensen, H. (2010). The multivesicular body-localized GTPase ARFA1b/1c is important for callose deposition and ROR2 syntaxin-dependent preinvasive basal defense in barley. Plant Cell 22, 3831–3844.
Bouwmeester, K., de Sain, M., Weide, R., Gouget, A., Klamer, S., Canut, H., and Govers, F. (2011). The lectin receptor kinase LecRK-I.9 is a novel Phytophthora resistance component and a potential host target for an RXLR effector. PLoS Pathog. 7, e1001327. doi: 10.1371/journal.ppat.1001327
Büttner, D., and He, S. Y. (2009). Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 150, 1656–1664.
Canut, H., Carrasco, A., Galaud, J., Cassan, C., Bouyssou, H., Vita, N., Ferrera, P., and Pont-Lezica, R. (1998). High affinity RGD-binding sites at the plasma membrane of Arabidopsis thaliana link the cell wall. Plant J. 16, 63–71.
Clay, N. K., Adio, A. M., Denoux, C., Jander, G., and Ausubel, F. M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323, 95–100.
Collins, N. C., Thordal-Christensen, H., Lipka, V., Bau, S., Kombrink, E., Qiu, J. L., Huckelhoven, R., Stein, M., Freialdenhoven, A., Somerville, S. C., and Schulze-Lefert, P. (2003). SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425, 973–977.
Enrique, R., Siciliano, F., Favaro, M. A., Gerhardt, N., Roeschlin, R., Rigano, L., Sendin, L., Castagnaro, A., Vojnov, A., and Marano, M. R. (2011). Novel demonstration of RNAi in citrus reveals importance of citrus callose synthase in defence against Xanthomonas citri subsp. citri. Plant Biotechnol. J. 9, 394–407.
Gee, E. P., Ingber, D. E., and Stultz, C. M. (2008). Fibronectin unfolding revisited: modeling cell traction-mediated unfolding of the tenth type-III repeat. PLoS ONE 3, e2373. doi: 10.1371/journal.pone.0002373
Gille, S., de Souza, A., Xiong, G., Benz, M., Cheng, K., Schultink, A., Reca, I. B., and Pauly, M. (2011). O-acetylation of Arabidopsis hemicellulose xyloglucan requires AXY4 or AXY4L, proteins with a TBL and DUF231 domain. Plant Cell 23, 4041–4053.
Gouget, A., Senchou, V., Govers, F., Sanson, A., Barre, A., Rougé, P., Pont-Lezica, R., and Canut, H. (2005). Lectin receptor kinases participate in protein-protein interactions to mediate plasma membrane-cell wall adhesions in Arabidopsis. Plant Physiol. 140, 81–90.
Jacobs, A. K., Lipka, V., Burton, R. A., Panstruga, R., Strizhov, N., Schulze-Lefert, P., and Fincher, G. B. (2003). An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell 15, 2503–2513.
Janbon, G., Himmelreich, U., Moyrand, F., Improvisi, L., and Dromer, F. (2001). Cas1p is a membrane protein necessary for the O-acetylation of the Cryptococcus neoformans capsular polysaccharide. Mol. Microbiol. 42, 453–467.
Knepper, C., Savory, E. A., and Day, B. (2011). Arabidopsis NDR1 is an integrin-like protein with a role in fluid loss and plasma membrane-cell wall adhesion. Plant Physiol. 156, 286–300.
Koh, S., Andre, A., Edwards, H., Ehrhardt, D., and Somerville, S. (2005). Arabidopsis thaliana subcellular responses to compatible Erisyphe cichoracearum infections. Plant J. 44, 516–529.
Laluk, K., and Mengiste, T. (2010). Necrotroph attacks on plants: wanton destruction or covert extortion? Arabidopsis Book 8, e0136.
Manabe, Y., Nafisi, M., Verhertbruggen, Y., Orfila, C., Gille, S., Rautengarten, C., Cherk, C., Marcus, S. E., Somerville, S., Pauly, M., Knox, J. P., Sakuragi, Y., and Scheller, H. V. (2011). Loss-of-function mutation of REDUCED WALL ACETYLATION2 in Arabidopsis leads to reduced cell wall acetylation and increased resistance to Botrytis cinerea. Plant Physiol. 155, 1068–1078.
Mellersh, D. G., and Heath, M. C. (2001). Plasma membrane-cell wall adhesion is required for expression of plant defense responses during fungal penetration. Plant Cell 13, 413–424.
Meyer, D., Pajonk, S., Micali, C., O’Connell, R., and Schulze-Lefert, P. (2009). Extracellular transport and integration of plant secretory proteins into pathogen-induced cell wall compartments. Plant J. 57, 986–999.
Micali, C. O., Neumann, U., Grunewald, D., Panstruga, R., and O’Connell, R. (2011). Biogenesis of a specialized plant-fungal interface during host cell internalization of Golovinomyces orontii haustoria. Cell. Microbiol. 13, 210–226.
Nicaise, V., Roux, M., and Zipfel, C. (2009). Recent advances in PAMP-triggered immunity against bacteria: pattern recognition receptors watch over and raise the alarm. Plant Physiol. 150, 1638–1647.
Nishimura, M. T., Stein, M., Hou, B-H., Vogel, J. P., Edwards, H., and Somerville, S. C. (2003). Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301, 969–972.
Nomura, K., DebRoy, S., Lee, Y. H., Pumpin, N., Jones, J., and He, S. Y. (2006). A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313, 220–223.
Senchou, V., Weide, R., Carrasco, A., Bouyssou, H., Pont-Lezica, R., Govers, F., and Canut, H. (2004). High affinity recognition of a Phytophthora protein by Arabidopsis via an RGD motif. Cell. Mol. Life Sci. 61, 502–509.
Szabo, L. J., and Bushnell, W. R. (2001). Hidden robbers: the role of fungal haustoria in parasitism of plants. Proc. Natl. Acad. Sci. U.S.A. 98, 7654–7765.
Tronchet, M., Balagué, C., Kroj, T., Jouanin, L., and Roby, D. (2010). Cinnamyl alcohol dehydrogenases-C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis. Mol. Plant Pathol. 11, 83–92.
Vogel, J., and Somerville, S. (2000). Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proc. Natl. Acad. Sci. U.S.A. 97, 1897–1902.
Vogel, J. P., Raab, T. K., Schiff, C., and Somerville, S. C. (2002). PMR6, a pectate-lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 14, 2095–2106.
Vogel, J. P., Raab, T. K., Somerville, C. R., and Somerville, S. C. (2004). Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J. 40, 968–978.
von Röpenack, E., Parr, A., and Schulze-Lefert, P. (1998). Structural analysis and dynamics of soluble and cell wall-bound phenolics in a broad spectrum resistance to the powdery mildew fungus in barley. J. Biol. Chem. 273, 9013–9022.
Vorwerk, S., Somerville, S., and Somerville, C. (2004). The role of plant cell wall polysaccharide composition in disease resistance. Trends Plant Sci. 9, 203–209.
Keywords: papilla, cell wall apposition, disease resistance, pathogen, immunity, cell wall adhesion, callose
Citation: Underwood W (2012) The plant cell wall: a dynamic barrier against pathogen invasion. Front. Plant Sci. 3:85. doi: 10.3389/fpls.2012.00085
Received: 15 March 2012; Accepted: 16 April 2012;
Published online: 07 May 2012.
Edited by:
Seth DeBolt, University of Kentucky, USAReviewed by:
Brad Day, Michigan State University, USACarloalberto Petti, University of Kentucky Lexington, USA
Copyright: © 2012 Underwood. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: William Underwood, Energy Biosciences Institute, University of California, 130 Calvin Lab, MC5230, Berkeley, CA 94720, USA. e-mail: williamu@berkeley.edu
