- Plant Biology Division, The Samuel Roberts Noble Foundation, Ardmore, OK, USA
The Arabidopsis cellulose synthase-like D (CSLD) 2 and 3 genes are known to function in root hair development. Here, we show that these genes also play a role in female gametophyte development because csld2 csld3 double mutants were observed to have low seed set that could be traced to defects in female transmission efficiency. Cell biological studies of csld2 csld3 ovules showed synergid cell degeneration during megagametogenesis and reduced pollen tube penetration during fertilization. Although CSLD2 and CSLD3 function redundantly in female gametophyte development, detailed analyses of root hair phenotypes of progeny from genetic crosses between csld2 and csld3, suggest that CSLD3 might play a more prominent role than CSLD2 in root hair development. Phylogenetic and gene duplication studies of CSLD2 and CSLD3 homologs in Arabidopsis lyrata, Populus, Medicago, maize, and Physcomitrella were further performed to investigate the course of evolution for these genes. Our analyses indicate that the ancestor of land plants possibly contained two copies of CSLD genes, one of which developed into the CSLD5 lineage in flowering plants, and the other formed the CSLD1/2/3/4 clade. In addition, CSLD2 and CSLD3 likely originated from a recent genome-wide duplication event explaining their redundancy. Moreover, sliding-window dN/dS analysis showed that most of the coding regions of CSLD2 and CSLD3 have been under strong purifying selection pressure. However, the region that encodes the N-terminus of CSLD3 has been under relatively relaxed selection pressure as indicated by its high dN/dS value, suggesting that CSLD3 might have gained additional functions through more frequent non-synonymous sequence changes at the N-terminus, which could partly explain the more prominent role of CSLD3 during root hair development compared to CSLD2.
Introduction
Cellulose, the major polysaccharide component of plant cell walls, consists of unbranched polymers of β-1,4-linked glucan residues that are connected by extensive hydrogen bonds to form cellulose microfibrils (Taylor, 2008). Cellulose is synthesized by a group of glycosyltransferases called cellulose synthases. In Arabidopsis, 10 cellulose synthase A (CesA) genes encode cellulose synthases that assemble into plasma membrane resident multimeric protein complexes, which catalyze the formation of individual glucan chains in developing primary and secondary walls (Endler and Persson, 2011). Although cellulose microfibrils form the core of the plant cell wall, a suite of other polysaccharides with a β-1,4-linked backbone collectively called hemicelluloses is essential for reinforcing its structural integrity. Hemicellulose is synthesized by Golgi-localized glycosyltransferases and is then secreted to the plasma membrane where they cross-link cellulose microfibrils to increase cell wall tensile strength (Scheller and Ulvskov, 2010). A third polysaccharide component of the cell wall is pectin. Compared to cellulose and hemicellulose, pectin is more structurally complex, and consists of a family of galacturonic-rich polysaccharides that require more than 60 transferase enzymes for its biosynthesis (Mohnen, 2008).
Some of the most significant advances toward understanding the importance of cell wall biosynthetic enzymes in plant development have come from genetic studies in Arabidopsis. For example, it was shown by mutant analyses that CESA1, CESA3, and CESA6 comprise the primary cell wall cellulose synthase complex and that this complex is essential for pollen development. One member of the primary cell wall cellulose synthase complex (CESA6) was partially redundant with other CESA isoforms such as CESA2 and CESA9 (Persson et al., 2007). In a parallel set of studies, it was demonstrated by bimolecular fluorescence complementation that CESA1, CESA3, and CESA6 indeed interact in vivo and through mutant analyses it was further shown that CESA5 and CESA2 were functionally redundant with CESA6. This led to the proposal that several CESA isoforms compete for the third position on the primary wall cellulose synthase complex that might be significant for fine tuning microfibril deposition during plant development (Desprez et al., 2007). Because the CESA2 protein can homodimerize in vitro, it was also proposed that CESA2 might form a complex with itself and contribute to cellulose synthase rosette assembly that regulates cell expansion during hypocotyl growth (Chu et al., 2007). Moreover, forward genetic screens have led to the isolation of the irx (for irregular xylem) set of mutants that were instrumental in identifying CESA4, CESA7, and CESA8 as components of the cellulose synthase complex responsible for cellulose biosynthesis in developing secondary cell walls (Taylor et al., 2003; Szyjanowicz et al., 2004).
Complete sequencing of the Arabidopsis genome led to the discovery of a large family of cellulose synthase-like (CSL) genes that encode proteins sharing various degrees of sequence similarity to CESA. The Arabidopsis CSL gene family can be divided into six groups namely CSLA, CSLB, CSLC, CSLD, CSLE, and CSLG (Richmond and Somerville, 2000). Two additional groups (CSLF and CSLH) have been found exclusively in grasses (Lerouxel et al., 2006) and a third group (CSLJ) is present in grasses and some dicot genomes (Dwivany et al., 2009; Yin et al., 2009). These CSL genes were predicted to encode processive glycosyl transferases that might be involved in the biosynthesis of the hemicellulose backbone of the cell wall (Richmond and Somerville, 2000). Indeed, activity for some CSLs in the synthesis of non-cellulosic polysaccharides has been reported. For instance, heterologous expression of recombinant Arabidopsis CSLA proteins in insect cells produced β-linked mannan polymers indicating that CSLAs encode mannan synthases (Liepman et al., 2005). These studies were reinforced by the observation that triple csla2 csla3 csla9 mutants lacked glucomannans and overexpression of CSLA9 enhanced glucomannan levels in Arabidopsis stems (Goubet et al., 2009). A Golgi-localized β-mannan synthase in guar seeds that is closely related to the CLSA genes in Arabidopsis and rice was also identified using heterologous expression in soybean somatic embryos (Dhugga et al., 2004) whereas expression of nasturtium and Arabidopsis CSLC in yeast provided evidence that CSLCs are β-1,4 glucan synthases that are involved in xyloglucan biosynthesis (Cocuron et al., 2007). In this regard, activities of some non-cellulosic polysaccharide synthesizing enzymes in grasses have been demonstrated using Arabidopsis as a heterologous system. Arabidopsis has been a useful model for elucidating function of grass cell wall synthesizing enzymes because it lacks some of the mixed polysaccharide backbones typically found in grasses. For example, the grass cell wall contains (1,3;1,4)-β-D-glucan, which is not present in Arabidopsis. Arabidopsis plants expressing rice CSLF (OsCSLF) and barley (HvCSLH) genes were able to produce (1,3;1,4)-β-D-glucans in their cell walls providing direct gain of function evidence that CSLF and CSLH are involved in (1,3;1,4)-β-D-glucan biosynthesis (Burton et al., 2006; Doblin et al., 2009).
Like CESA, the significance of CSL genes for various aspects of plant development has been demonstrated through mutant studies. For instance, it was found that Arabidopsis csla7 mutants are defective in pollen tube growth and embryogenesis indicating that mannan hemicellulosic polysaccharides that are synthesized by CSLAs are important for cell wall function needed to support these developmental processes (Goubet et al., 2003, 2009). In rice, mutants with altered leaf morphology and plant architecture were disrupted in OsCSLD1 and OsCSLD4 (Li et al., 2009; Hu et al., 2010; Wu et al., 2010; Luan et al., 2011). OsCSLD1 was also shown to be necessary for root hair development (Kim et al., 2007). More recently, the narrow-organ and warty leaf phenotype of the maize csld1 mutant was attributed to defects in cell division and expansion (Hunter et al., 2012). The Arabidopsis genome contains six CSLD genes and mutations to five of these genes have been reported to cause distinct phenotypes. csld5 mutants, for example, have stem growth defects (Bernal et al., 2007) whereas csld2 and csld3 have root hairs that rupture at the tip (Favery et al., 2001; Wang et al., 2001; Bernal et al., 2008; Galway et al., 2011; Park et al., 2011). Furthermore, CSLD1 and CSLD4 were shown to be essential for maintaining growth of pollen tubes for efficient transmission of male gametes (Bernal et al., 2008; Wang et al., 2011) indicating that the latter four CSLD genes encode proteins that synthesize important components of the cell wall of tip growing cells. However, recent double and triple mutant studies indicate that CSLD2 and CSLD3 may function cooperatively with CSLD5 in plant developmental processes besides tip growth (Yin et al., 2011).
In this paper, we present new data showing an additional function for CSLD2 and CSLD3. We observed that csld2 csld3 double mutants had reduced seed set, which we found was due to defects in female transmission. Cell biological analyses of the ovules of csld2 csld3 indicate that synergid cells degenerated during megagametogenesis resulting in the compromised ability of pollen tubes to efficiently fertilize ovules. Interestingly, while the above results suggest that CSLD2 and CSLD3 function redundantly during female gametophyte development, they might have evolved divergent roles in root hair development with CSLD3 playing a more prominent role. These conclusions are supported by phylogenetic and gene duplication studies showing that CSLD3 may have undergone more relaxed selection resulting in its having a divergent role in root hair growth compared to CSLD2.
Materials and Methods
Genetic Crosses and Analysis of Genetic Transmission
All of the Arabidopsis lines used in this study are of the Col-0 ecotype. Homozygous knockout lines of csld2 (SALK_119808, a T-DNA mutant of At5g16910) and csld3 (SALK_112105, a T-DNA mutant of At3g03050) were obtained from the Arabidopsis Biological Resource Center, and then backcrossed to the wild-type at least two times. The segregation of the progeny derived from the crosses was examined based on their root hair phenotypes (Bernal et al., 2008), and genotypes of these progeny were confirmed by polymerase chain reaction (PCR) following the standard protocol from the Salk Institute Genomic Analysis Laboratory1. For csld2, D2-LP (CACAAATGGCTGGCTCTAAAG) and D2-RP (AAAAAGGAACCCAAATGTTGG), for csld3, D3-LP (GGGAGATCAGATTTCCCAGTC) and D3-RP (TGTTCACCTCTTGATGATTTGG) were used as gene specific primers. The LBb1.3 (ATTTTGCCGATTTCGGAAC) primer from the left border of T-DNA was used either with D2-RP or D3-RP for the csld2 or csld3 T-DNA insertion mutants, respectively.
To determine gametophytic transmission of the csld2 and csld3 mutant alleles, reciprocal crosses were performed between wild-type and plants with only one functional copy of CSLD3 (i.e., csld2/csld2 CSLD3/csld3). The F1 seeds were harvested and pooled separately for the progeny that was generated by the crosses between the male mutant (i.e., csld2/csld2 CSLD3/csld3) and the female wild-type or of male wild-type to the female mutant. After sterilization and germination on plates, DNA was extracted. Progeny was genotyped for the presence of csld2 and csld3 T-DNA insertions by PCR. χ2 Values are indicated. P values were calculated using GraphPad QuickCalcs (available online at http://www.GraphPad.com/).
Growth Conditions and Evaluation of Root Hair Phenotypes
Arabidopsis seeds were surface sterilized and planted as detailed in Dyachok et al. (2009). To evaluate root hair phenotypes, 4- or 5-day-old seedlings were examined with an inverted Nikon Eclipse TE300 compound microscope and photographed with a Nikon DXM1200 digital camera (Nikon Corporation, Melville, NY, USA). Root hair length from digital images was measured using ImageJ ver. 1.42q software2.
Complementation of csld2 csld3 Double Mutant
The full length genomic DNA of CSLD3 was amplified using primers D3-KpnI-F (CTGGTACCTGAAAGTCTCTGAAGAACAACG) and D3-XbaI-R (AGTCTAGAATCTGTTGCCATTAGAAATCTC). The resulting 5.8 kb DNA fragment that contained 1.6 kb of the 5′ upstream from the start codon and 450 bp of the downstream sequence from the stop codon of CSLD3 was subcloned into a pCAMBIA2300. Sequencing analysis detected no PCR errors in the construct. The csld2 csld3 mutant seedlings were transformed by using Agrobacterium tumefaciens strain LBA4404 as previously described (Clough and Bent, 1998) and 14 independent lines were analyzed. The presence of the transgene was confirmed by PCR genotyping using the primers of D2-RP, D3-RP, and LBb1.3 as described earlier, and D3-IN (GAAAAGCCGTGTGCCAGAAG), D3-UTR (ACATTTCTGAGCCATTATTC), CAMBIA (TCACGACGTTGTAAAACGAC).
Histochemical Staining for β-Glucuronidase (GUS) Activity in Pollen Tubes
Histochemical staining of the flowers to monitor GUS activity in pollen was performed as previously described (Jefferson et al., 1987). The LAT52:GUS pollen (Johnson et al., 2004) was transferred onto wild-type and csld2 csld3 mutant stigma. Sixteen hours after pollination, flowers were vacuum-infiltrated for 15 min in a GUS substrate solution of 100 mM sodium phosphate, pH 7.2, 0.2% (v/v) Triton X-100, 2 mM potassium ferricyanide, 2 mM potassium ferrocyanide, and 2 mM 5-bromo-4-chloro-3-indolyl-β-D-glucuronide, and then incubated for an additional 12 h at 37°C. Samples were then transferred into a mixture of 1:1 ethanol:acetic acid to remove chlorophyll pigmentation. Finally, the tissues were cleared by Hoyer’s solution (7.5 g Arabic gum, 100 g chloral hydrate, 5 ml glycerol, and 60 ml deionized water) prior to photography.
Evaluation of Synergid Cells in Wild-Type and Mutant Ovules
Processing of ovules for microscopic analysis was as previously described (Christensen et al., 1997) with minor modifications. Briefly, wild-type and csld2 csld3 mutants flowers were emasculated at the 12b stage (Smyth et al., 1990), and then incubated in a growth chamber (22–23°C, 16 h light and 8 h dark cycle) for 2–3 days to ensure that the pistils proceeded into the FG7 stage in the absence of fertilization (Christensen et al., 1997). The pistils were then cut and fixed in a solution of 4% glutaraldehyde in cacodylate (pH 6.9) for 2 h at room temperature. Following fixation, the tissues were dehydrated in a graded ethanol series followed by clearing in a mixture of 2:1 benzyl benzoate:benzyl alcohol. The pistils were mounted in immersion oil and sealed with coverslips using nail polish.
A Leica TCS SP2 AOBS confocal laser scanning microscope (Leica Microsystems, Exton, PA, USA) was used to examine the female gametophytes within the pistils. The 595 nm laser line of the HeNe laser was used to observe ovule autofluorescence. Fifteen optical sections at 1.34 μm interval were collected with a 63× water immersion objective. Optical sections were compiled into a single image using the maximum projection command of the Leica confocal software.
CSLD Gene and Protein Sequence Retrieval
Sequences of Arabidopsis thaliana CSLD genes were obtained from TAIR-www.arabidopsis.org excluding CSLD6 and its orthologs because CSLD6 in Arabidopsis appears to be a pseudogene (Verhertbruggen et al., 2011). Sequences from Arabidopsis lyrata, Populus trichocarpa, Medicago truncatula, maize, and Physcomitrella patens were retrieved using Arabidopsis thaliana CSLD protein sequences to search against the respective proteome database in Phytozome V8.0 at JGI (Goodstein et al., 2011). The resulting protein and cDNA sequences in Medicago were further curated to find proper intron locations based on the GT/AG rule by comparing with other CSLD proteins (Breathnach et al., 1978). A few Medicago sequences were not included in the phylogenetic analysis because these sequences showed clear sequencing errors or alternative intron starting/ending sites could not be located (i.e., Medtr1g047090).
Sequence and Evolutionary Analyses
The full length protein sequences from all species in this study were aligned by the MAFFT Version 6.0 program with minor manual adjustments (Katoh et al., 2009). Phylogenetic analysis by the Bayesian statistical method (BEAST) was completed using BEAST v1.7.1 software package (Drummond et al., 2012). To further evaluate the evolution of CSLD family, the Maximum-Parsimony (MP) method was performed using MEGA 4.0 with 1000 replicates (Tamura et al., 2007). The Bayesian posterior support values were found to be comparable at most nodes with the MP bootstrap values. The changes of dN/dS ratios between taxa were estimated by MEGA 4.0 (Tamura et al., 2007) and K-Estimator 6.0 (Comeron, 1999). The ancestral sequence of Arabidopsis thaliana CSLD2 and CSLD3 was predicted by Diverge 2.0 (Gu and Vander, 2002). To check for patterns of segmental duplication, the DotPlot function of the PipMaker program at http://pipmaker.bx.psu.edu/pipmaker was used (Schwartz et al., 2000). The gene structure was displayed by the GSDS program at http://gsds.cbi.pku.edu.cn/ (Guo et al., 2007).
Results
CSLD2 CSLD3 Double Mutants are Defective in Female Genetic Transmission
The Arabidopsis CSLD2 and CSLD3 genes are highly expressed in root tissues, and they have been shown to play important roles in root hair development (Favery et al., 2001; Wang et al., 2001; Bernal et al., 2008; Galway et al., 2011; Park et al., 2011). Moreover, they were shown to be expressed in a variety of other tissues (Zimmermann et al., 2004; Bernal et al., 2008), suggesting that these two genes may participate in the development of organs or tissues other than root hairs. However, the lack of distinct phenotypes in other plant organs might be due to functional redundancy between these two homologous CSLD genes. We specifically selected the non-functional mutant alleles of csld2 (SALK_16910) and csld3 (SALK_03050) for our studies as described previously (Bernal et al., 2008), in contrast to the partially functional alleles of csld3 (Favery et al., 2001; Wang et al., 2001; Galway et al., 2011).
Interestingly, we found that siliques of csld2 csld3 double mutants contained a significant number of aborted seeds, while those of wild-type or single csld2 or csld3 mutants did not (Figure 1). To determine whether seed abortion in csld2 csld3 double mutants was due to abnormal genetic transmission, 142 seedlings of the F2 progeny were genotyped by PCR (Table 1). We found that the number of double homozygote (csld2/csld2 csld3/csld3) and double heterozygote (CSLD2/csld2 CSLD3/csld3) individuals in the F2 population was less than expected suggesting inefficient genetic transmission during csld2 csld3 reproduction.

Figure 1. csld2 csld3 double mutants have siliques with fewer seeds. Whole-mount images of the mature siliques cleared with 0.2N NaOH and 1% SDS solution. (A) Siliques from wild-type, csld2 and csld3 single mutants, and the csld2 csld3 double mutant. (B) Siliques from the wild-type, csld2/csld2 CSLD3/csld3 individuals and siliques resulting from reciprocal crosses between wild-type and csld2/csld2 CSLD3/csld3. Bars = 5 mm.
Like csld3 single mutants, csld2 csld3 double mutants lacked root hairs due to very early rupturing of the tip (Bernal et al., 2008). To determine whether the lack of root hairs and low seed set phenotype of the csld2 csld3 double mutant was indeed due to the combined reduced expression of CSLD2 and CSLD3, csld2 csld3 was transformed with a wild-type CSLD3 gene under the control of its native promoter (Figure A1 in Appendix). The root hairs of the resulting transgenic plants were noticeably longer and swollen at their bases, and ruptured at their tips much later than csld3 single or csld2 csld3 double mutants. Such root hairs defects were reminiscent of the root hair phenotypes of csld2 single mutants (Figures A2A–D in Appendix; Bernal et al., 2008). Moreover, the csld2 csld3 complemented lines also showed normal seed set similar to csld2 single mutants and wild-type providing definitive proof that reduced seed set in csld2 csld3 double mutant was due to the disruption of both CSLD2 and CSLD3 (Figures A2E–G in Appendix).
To determine whether the reduced seed set of csld2 csld3 double mutants was due to male or female transmission defects, we performed reciprocal crosses between individuals with only one copy of CSLD3 (csld2/csld2 CSLD3/csld3) and wild-type plants (CSLD2/CSLD2 CSLD3/CSLD3). As outlined in Table 2, if genetic transmission is normal, the F1 population would be expected to have 50% double heterozygotes (CSLD2/csld2 CSLD3/csld3) and 50% as single heterozygotes of CSLD2 (CSLD2/csld2 CSLD3/CSLD3). When csld2/csld2 CSLD3/csld3 pollen was used to fertilize wild-type plants, the distribution of CSLD2/csld2 CSLD3/csld3 and CSLD2/csld2 CSLD3/CSLD3 progeny were observed at the expected ratio of 1:1 (Table 2). However, when wild-type pollen was used to fertilize csld2/csld2 CSLD3/csld3 plants, the number of CSLD2/csld2 CSLD3/csld3 individuals declined significantly suggesting that csld2 csld3 transmission through female gametophytes is the primary cause of reduced seed set in csld2 csld3 double mutants. All of the ovules of csld2 csld3 double mutants that developed into seeds germinated normally and developed into normal plants (data not shown). No deformed seeds in siliques of the double mutant were found (Figure 2C). These results indicate that reduced seed set in csld2 csld3 plants can be attributed to events prior to embryogenesis and such defects are likely occurring in female reproductive structures.
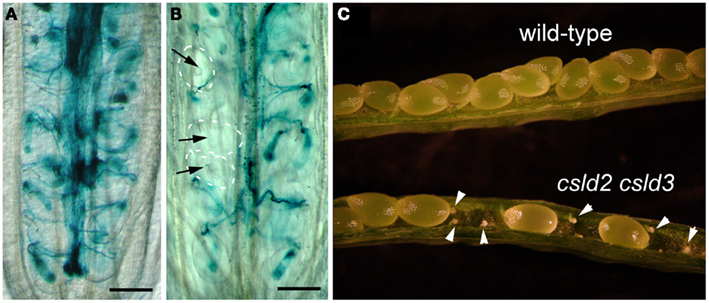
Figure 2. Defective seed set in csld2 csld3 double mutants is the result of a failure in fertilization. (A,B) Whole-mount images of GUS stained ovules. LAT52:GUS pollen was used to pollinate wild-type (A) and the csld2 csld3 double mutant (B) stigmas. Note that pollen tubes have not penetrated some ovules of csld2 csld3 mutants (black arrows). On the other hand, all of the wild-type ovules show pollen penetration. (C) Dissected view of mature siliques. Note that siliques of the csld2 csld3 double mutant contained unfertilized and desiccated ovules (white arrowheads), while the wild-type produces a complete set of seeds. Bars = 100 μm (A,B).
Synergid Cells of CSLD2 CSLD3 Degenerate during Female Gametophyte Development
To investigate the nature of the defect in female genetic transmission, we fertilized csld2 csld3 double mutants and wild-type with pollen from a wild-type plant expressing the pollen tube marker, LAT52:GUS (Johnson et al., 2004). We found successful penetration of pollen tubes in most of the ovules of wild-type. In contrast, pollen failed to penetrate some of the ovules of csld2 csld3 double mutants (Figures 2A,B).
The observation that pollen tubes failed to penetrate some ovules of csld2 csld3 double mutants led us to hypothesize that the developmental and structural integrity of the female gametophyte might be compromised. To test this hypothesis, we emasculated csld2 csld3 flowers at stage 12 (Smyth et al., 1990) and waited for 48–72 h to ensure that the female gametophytes were sufficiently developed, and then we examined ovule structure by confocal microscopy. Christensen et al. (1997) demonstrated that flowers at the FG7 stage, a stage when medial stamens become stalked (Smyth et al., 1990), the female gametophyte consists of four cells: one central cell, one egg cell, and two synergid cells. They documented that synergid cell degeneration occurs only after pollen tubes penetrated the ovule. In our experience, the ability to clearly image synergid cells in isolated Arabidopsis ovules with the confocal microscope depended on their correct orientation when mounted on glass slides. This prevented us from correlating the percentage of non-degenerated synergid cells in csld2 csld3 mutants with seed set. However, in ovules where synergid cells were clearly visible, 100% of wild-type ovules had non-degenerated synergid cells (Figure 3A). On the other hand, from a total of 100 ovules examined, 30% of csld2 csld3 ovules contained degenerated synergid cells (Figures 3B,C). This result suggests that the loss of CSLD2 and CSLD3 triggered the synergid cells to degenerate during megagametogenesis, and this degeneration could be the cause of the partial infertility of the double mutant.
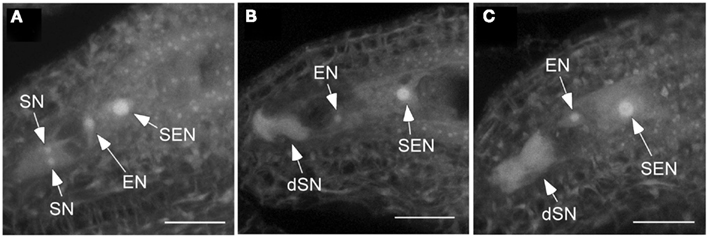
Figure 3. Synergid cells of csld2 csld3 degenerate before fertilization. (A) Secondary endosperm nucleus (SEN), egg cell nucleus (EN), and the two synergid cell nuclei (SN) in the wild-type. (B,C) Show images at the same stage of female gametophyte development (FG7) in csld2 csld3 double mutant. Note that csld2 csld3 has degenerated synergid nucleus (dSN) as evident by increased autofluorescence and absence of a distinct synergid cell nuclei. Thirty percent of ovules of csld2 csld3 contained degenerated synergid cells. Images shown are a projection of fifteen 1.34 μm confocal optical sections. Bars = 20 μm.
CSLD2 and CSLD3 Show Dosage-Dependent Effects during Root Hair Development
In a previous study, we showed that single csld2 mutants had shorter root hairs compared to wild-type due to rupturing of the root hair tip (Bernal et al., 2008). Surprisingly, we found that double heterozygous plants (CSLD2 csld2/CSLD3 csld3) at the F1 generation, showed ruptured root hair tips, indicating that a complete set of both CSLD genes is required for normal root hair development (Figures 4A,B). This notion is supported by the observation that the extent of root hair rupturing depended on the number of functional copies of CSLD2 or CSLD3. For example, CSLD2/csld2 CSLD3/CSLD3 and CSLD2/CSLD2 CSLD3/csld3 had a higher percentage of shorter root hairs than the wild-type. Furthermore, csld2 single mutants with two copies of CSLD3 (csld2/csld2 CSLD3/CSLD3) had a larger percentage of root hairs that were longer compared to csld2/csld2 CSLD3/csld3 (Figure 4C). Altogether, these results suggest that CSLD2 and CSLD3 have dosage-dependent effects on root hair development.
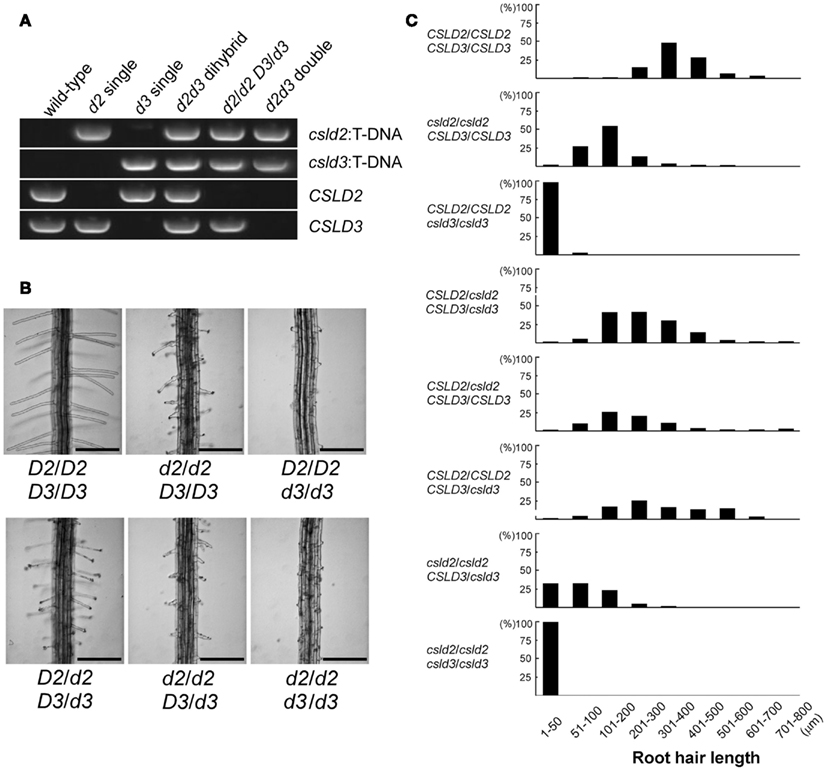
Figure 4. Dosage-dependent effects of CSLD2 (D2) and CSLD3 (D3) on root hair development. (A) T-DNA PCR verifying the genotype of the mutants. (B) Representative images of root hairs. Note that the root hair phenotype differs among individuals depending on the number of functional copies of the CSLD genes. Bars = 250 μm. (C) Quantification of root hair length from individuals with different copies of the CSLD2 or CSLD3 genes. One hundred and fifty to 200 mature root hairs were measured from 4- to 5-day-old seedlings.
CSLD2 and CSLD3 Exert Different Impacts on Root Hair Growth
As noted above, double heterozygous plants (CSLD2/csld2 CSLD3/csld3) already exhibited ruptured root hairs that mirrored the phenotype of csld2 (Figure 4B). However, as reported previously, root hairs of csld2 would typically swell at the base after tip rupturing but then are able to resume tip growth if tip rupturing is mild (Bernal et al., 2008). Because of this cycle of mild tip rupturing and tip growth resumption, root hairs of csld2 single mutants were characterized by uneven swelling along their length, and in some cases root hairs developed two growing tips before eventually rupturing (Figure 5A). In contrast, double heterozygous plants did not show the unusual root hair growth behavior exhibited by csld2 single mutants. Instead, root hairs of CSLD2/csld2 CSLD3/csld3 would first elongate like wild-type but eventually rupture later during root hair development (Figure 5B). Because root hairs of CSLD2/csld2 CSLD3/csld3 ruptured later, they were longer than those of csld2 single mutants. Likewise, CSLD2/CSLD2 CSLD3/csld3 plants had longer root hairs than CSLD2/csld2 CSLD3/CSLD3 plants (Figure 4C).
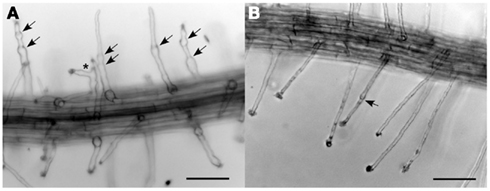
Figure 5. Tip rupture and re-growth of csld2 single mutants (A) and csld2 csld3 dihybrids (B). The arrows indicate the points of tip growth rupture and growth resumption. The asterisk indicates a branched root hair. Bars = 100 μm.
On the other hand, having only a single copy of CSLD3 gene (csld2/csld2 CSLD3/csld3), was sufficient for many root hairs to elongate to lengths of 200 μm or more, whereas having two copies of CSLD2 (CSLD2/CSLD2 csld3/csld3) was not sufficient for root hairs to proceed beyond initiation (Figure 4C). Taken together, these results suggest that CSLD3 is critical during both the onset of root hair initiation and sustained root hair elongation whereas CSLD2 is more important for sustained root hair elongation.
Expression of CSLD3 is Modulated by a Larger Array of Transcriptional Regulatory Elements than CSLD2
On the basis of the above studies, CSLD2 and CSLD3 might have overlapping functions during both root hair and female gametophyte development. However, because csld3 and csld2 mutants are different with regard to the severity of their root hair phenotype (Figure 4; Bernal et al., 2008), it is possible that functional differences might exist between the two genes. To understand the partial conservation and divergence between these two closely related CSLD homologs, the upstream regions of CSLD2 and CLSD3 were examined and compared. cis-Regulatory elements of CSLD2 and CSLD3 were analyzed by the AtcisDB program at AGRIS (Arabidopsis Gene Regulatory Information Server; Davuluri et al., 2003; Yilmaz et al., 2011). We found that the upstream region of CSLD3 contained 64 predicted binding sites for various transcriptional regulators while that of CSLD2 contained only 35 sites. Twenty three of these binding sites were shared by both genes, suggesting that the regulation of transcription between CSLD2 and CSLD3 might be similar during certain developmental stages. However, because 40 of the predicted binding sites are only present in the upstream region of CSLD3, it appears that expression of CSLD3 is controlled by a more diversified group of regulators. This implies that CSLD3 may play a more divergent role than CSLD2.
Sequence and Evolutionary Analyses of CSLD Genes
To further elucidate the similarity and divergence between CSLD2 and CSLD3 genes, sequence comparison, and evolutionary analysis of CSLD2/3 and their close homologs in other plant species were performed. All Arabidopsis CSLD sequences were found to be very similar. In particular, the Arabidopsis CSLD2 and CSLD3 were identified to be the most closely related homologous pair (Bernal et al., 2008) and they showed very similar intron/exon organization (Figure 6 and Figure A1 in Appendix). Due to the high sequence homology of the CSLD genes, we investigated their evolutionary relationship. CSLD and selected CESA protein sequences were collected from several species including Arabidopsis thaliana, A. lyrata, poplar, Medicago, maize, and Physcomitrella (Figure 6). These sequences were used to construct a phylogenetic tree while using selected CESA genes as an out group but excluding CSLD6 and its orthologs because CSLD6 appears to be a pseudogene by virtue of having a truncated transcript and extremely low expression in most tissues (Bernal et al., 2008; Verhertbruggen et al., 2011).
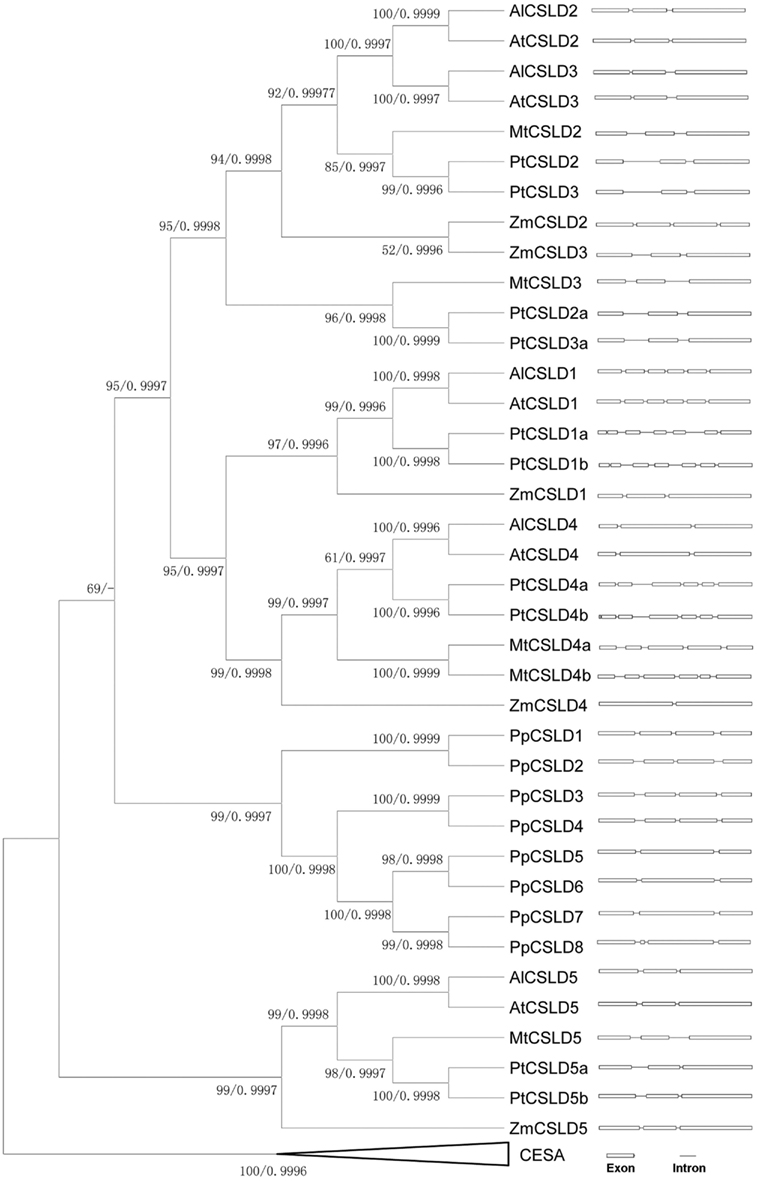
Figure 6. Phylogenetic tree and intron-exon distribution of CSLDs from Arabidopsis thaliana, A. lyrata, Populus, Medicago, maize, and Physcomitrella. Bootstrap support values using MP method and Bayesian posterior values were labeled at each node respectively.
Topology of this tree indicates that before the separation of vascular plants and non-vascular plants, there might have been two copies of CSLD ancestral genes. It appears that each of these two ancestral genes evolved separately, forming two branches that led to the development of the CSLD5 branch and other CSLD gene branch. It also appears that the CSLD5 gene became lost in non-vascular plants and was only retained in flowering plants, suggesting that its function may be different from its paralogs in Arabidopsis.
Moreover, since the split of vascular and non-vascular plants, CSLD genes in the CLSD1/2/3/4 branch had experienced multiple duplication events, resulting in the formation of multiple copies of the CSLD genes. The ancestor of flowering plants could have experienced more than one duplication event, forming multiple copies of the CSLD genes that evolved independently. Furthermore, it seems that the duplication event that generated CSLD2 and CSLD3 could have happened after the separation of the rosid II clade (Arabidopsis) from the rosid I clade (poplar and Medicago) 83∼107 Million Years Ago (MYA; Wang et al., 2009), but before the split of Arabidopsis thaliana and A. lyrata about 5 MYA (Kuittinen et al., 2004). It has been proposed that Arabidopsis thaliana could have experienced three genome-wide duplication events with the most recent one (the third genome-wide duplication) happening 24–40 MYA (Simillion et al., 2002; Blanc et al., 2003; Jansson and Douglas, 2007). This suggested that the formation of CSLD2 and CSLD3 might have occurred during the third genome-wide duplication event in Arabidopsis. Such conclusions are supported by two other phylogenetic methods namely the Neighbor-Joining and Maximum-likelihood methods (data not shown).
To further explore the differences among CSLD genes, their intron arrangements were compared. As shown in Figure 6, exons of most CSLD gene are separated by two introns. Taking a closer look at the exon/intron distribution in the CSLD2/3 subfamilies, introns of CSLD genes in rosid II lineage appear to be much shorter than those in the rosid I lineage, indicating that they probably had experienced different courses of evolution since their separation. The high similarity in intron arrangement between CSLD2 and CSLD3 in Arabidopsis thaliana additionally suggests that these two genes may have been produced more recently.
To support the above hypotheses, dot-matrix analyses were done by comparing the 50-kb genomic sequences upstream and downstream of the target Arabidopsis CSLD genes. A comparison between CSLD2 and CSLD3 and adjacent regions showed a pattern of segmental duplication (Figure 7A). It was also found that the synonymous nucleotide change (dS) value can be used as an estimate of the duplication age (Kong et al., 2007) and the dS value between the CSLD2 and CSLD3 pair is 0.81, equal to the mean dS value of the chromosomal blocks that resulted from the third genome-wide duplication (Blanc et al., 2003). Therefore, it can be concluded that CSLD2 and CSLD3 resulted from the most recent genome-wide duplication event in Arabidopsis. Since the formation of CSLD2 and CSLD3 was more recent, they could be more functionally redundant, but could also have developed novel functions during the course of recent evolution.
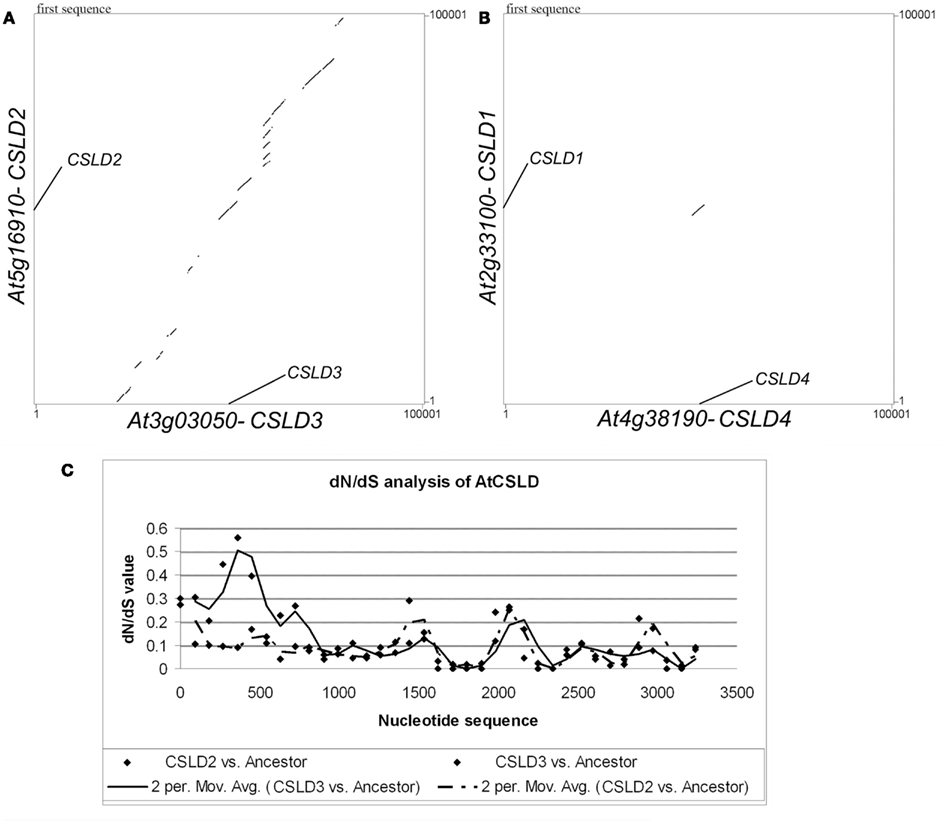
Figure 7. Sequence analyses of CSLD genes. (A) Dot-matrix analysis comparing the flanking genomic regions (50 kb on both sides) of CSLD2 and CSLD3. (B) dN/dS sliding-window (window size 180, movement 90) analysis of CSLD2 (dash line) and CSLD3 (solid line) with their predicted common ancestor.
Lastly, to further understand the functional conservation and divergence between CSLD2 and CSLD3, an investigation of the evolutionary pressure on this pair by a dN/dS sliding-window analysis was performed to compare the CDS sequences of CSLD2 and CSLD3 with the sequence of their estimated common ancestor. As shown in Figure 7B, it is clear that the evolution of CSLD2 and CSLD3 was under purifying selection, especially, the regions that encode the C-terminus of these proteins. However, the regions that encode the N-terminus of both proteins appeared to be under relaxed selection pressure as shown by mild dN/dS values (Figure 7B). These mild values suggested more frequent non-synonymous substitutions, which could potentially allow functional divergence. Additionally, the region encoding the N-terminus of CSLD3 showed much higher dN/dS values. This would suggest that CSLD3 may have gained additional functions during evolution compared to CSLD2, which could be recognized by its more predominant function in root hair development.
Discussion
The possibility that CSLDs synthesize a β-1,4 glucan polysaccharide that assembles into cellulose-like microfibrils at the apex of growing root hairs was elegantly demonstrated by Park et al. (2011). They showed that replacing CSLD3 catalytic activity with the CESA6 catalytic domain could rescue the root hair defects of the csld3 mutant while retaining its apical plasma membrane localization. Given their broad expression patterns, CSLD function is likely not limited to tip growing cells (Zimmermann et al., 2004; Bernal et al., 2008). For instance, there is recent genetic evidence showing that CSLD5 in cooperation with CSLD2 and CSLD3 is pivotal for Arabidopsis development by impacting diffuse growing cells of the stem (Yin et al., 2011). Here, double mutant studies uncovered a role for CSLD2 and CSLD3 in female gametophyte development, consistent with previous genetic subtraction and cell-type specific transcript profiling experiments demonstrating that CSLD2 and CSLD3 are expressed in various cells and tissues within the ovule (Johnston et al., 2007; Wuest et al., 2010). Interestingly, a gene trap insertional mutagenesis screen identified an Arabidopsis mutant called astlik, which had multiple insertions and deletions within the genome including a deletion in the CSLD3 gene. Astlik had siliques containing a mixture of normal seeds and infertile ovules that was reminiscent of the csld2 csld3 phenotype reported here. However, it was not shown conclusively, whether the CSLD3 gene was responsible for the partial infertility of the astlik mutant because of the complex genomic rearrangement induced by the gene trap mutagenesis strategy (Brukhin et al., 2011). Nonetheless, the fact that the partial infertility phenotype of csld2 csld3 can be rescued with the CSLD3 genomic DNA driven by its own promoter indicates that both CSLD2 and CSLD3 function redundantly in female gametophyte development.
Light microscopic studies indicate that the defects in female transmission we observed in csld2 csld3 double mutants could be explained in part by altered function of the synergid cells. During female gametophyte development in Arabidopsis, the megaspore undergoes three rounds of mitosis to produce an eight-nucleated cell. After nuclear migration and cellularization, the megagametophyte forms a seven-celled structure consisting of three antipodal cells, two synergid cells, one central cell, and one egg cell (Schneitz et al., 1995; Christensen et al., 1997; Drews et al., 1998). During fertilization, the pollen tube penetrates one of the two synergid cells. This synergid cell undergoes cell death (Christensen et al., 1997; Punwani and Drews, 2008), which then induces the pollen tube to stop growing, and discharge one sperm to the egg cell and one sperm to the central cell to initiate double fertilization (Faure et al., 2002; Berger et al., 2008). In Arabidopsis, synergid cell degeneration is only triggered several minutes after the pollen tube makes contact (Sandaklie-Nikolova et al., 2007). On the other hand, synergid cell death in Nicotiana has been reported to occur before pollination indicating that synergid degeneration in some plant species is an important component of female gametophyte development (Huang and Russell, 1992). Studies of mutants that fail to undergo synergid cell death after pollen tube contact have implicated a mitochondria-localized DnaJ chaperonin and a FERONIA receptor-like kinase in the synergid cell death program in Arabidopsis (Christensen et al., 2002; Rotman et al., 2003). Here, we found that several ovules in csld2 csld3 double mutants had apparently dead synergid cells even before fertilization (Figure 3). Because synergid cells are known to secrete attractants that guide pollen tubes to the female gametophyte, their death in ovules of csld2 csld3 could be detrimental to pollen attraction (Higashiyama et al., 2001; Kasahara et al., 2005; Okuda et al., 2009). This notion is supported by the observation that several ovules in the csld2 csld3 pistils were not penetrated by pollen tubes and therefore explains the large percentage of undeveloped ovules in csld2 csld3 siliques (Figure 2).
It is unclear how the lack of CSLD2 and CSLD3 contribute to synergid cell death. Previous studies have shown that synergid cells have unique structural features that could likely be impacted by the absence of the specific cell wall polysaccharide synthesized by CSLD2 and CSLD3. Most notable is the synergid cell wall adjacent to the micropylar pole of the embryo sac. This region of the synergid cell called the filiform apparatus is typically thickened and contains numerous finger-like projections that extend into the synergid cytoplasm (Weterings and Russell, 2004). Several functions have been proposed for the filiform apparatus one of which is to facilitate the secretion of a pollen tube attractant (Punwani and Drews, 2008). Although we have not examined the ultrastructure of csld2 csld3 synergid cells, other female gametophytic mutants with defects in the filiform apparatus exhibit female transmission abnormalities that mirror csld2 csld3. For example, mutants in the synergid-expressed MYB98 transcription factor, which is a member of the R2R3-MYB gene family, had desiccated ovules, reduced transmission through the female gametophyte, and defective pollen tube guidance (Kasahara et al., 2005). Interestingly, downstream targets of MYB98 include filiform apparatus-localized secreted proteins with predicted signaling roles (Punwani et al., 2007). The defective cell walls of csld2 csld3 synergids could compromise the timely secretion of signal peptides needed for pollen tube attraction. Detailed studies of pollen tube growth patterns are needed to determine the specific stage of pollen tube guidance that is affected by the polysaccharide product of CSLDs.
Recent complementation studies showed that expressing a 35S: YFP-CSLD2 construct can fully restore root hairs of csld3 to the wild-type phenotype (Yin et al., 2011). This indicates that like in the female gametophyte, CSLD2 and CSLD3 might function redundantly during root hair development. However, the fact that csld3 single mutants had more severe root hair defects than csld2 (Bernal et al., 2008), suggests only partial redundancy with CSLD3 having a more prominent role than CSLD2 in root hair growth. This notion is reinforced by the observation that seedlings with only a single copy of CSLD3 (csld2/csld2 CSLD3/csld3) had a high percentage of root hairs that were able to elongate to lengths of up 200 μm or more (Figure 4C). This raises the possibility that CSLD2 and CSLD3 deposit their respective polysaccharide products in different regions of the root hair cell. Early ultrastructural studies have shown that the inner cell wall surface of elongating root hairs at the extreme apex contain cellulose microfibrils that are deposited randomly, forming a thin primary wall layer, while the sub-apex develops a secondary cell wall layer at the inner surface with cellulose microfibrils deposited longitudinally. These longitudinal bundles of microfibrils were shown to extend to the base of the root hair (Sassen et al., 1985). Such a pattern of wall organization could be significant for root hair growth because randomly organized microfibrils at the tip can be more readily extended by the action of expansins without disruption of the covalent bonds (McQueen-Mason and Cosgrove, 1995; Cho and Cosgrove, 2002). It is tempting to speculate that CSLD3 might contribute more to cell wall structure at the tip apex, which is consistent with its localization at the extreme root hair apex (Park et al., 2011). CSLD2 on the other hand could have a more prominent role in the synthesis of parallel microfibril bundles that thicken and mechanically strengthen the basal cell wall of the root hair. If this is the case, it could explain why csld2 root hairs swell at the base and are able to resume tip growth after rupturing, whereas csld3 root hairs are never able to elongate beyond initiation (Bernal et al., 2008). This could also explain why root hairs of dihybrid seedlings (CSLD2/csld2 CSLD3/csld3) grow with a confined diameter and eventually rupture at the tip before maturation. Previously, we showed by promoter-reporter fusions that CSLD3 expression in seedling roots extended from trichoblasts to rapidly elongating root hairs while CSLD2 expression was more prominent in more mature root hairs (Bernal et al., 2008). Thus, the dosage-dependent effects of CSLD2 and CSLD3 on root hair development could also be explained by differences in their expression levels in seedling roots.
In addition to having high coding sequence homology, redundancy in CSLD2 and CSLD3 function can also be inferred from bioinformatic studies of their upstream regions. For instance, according to AGRIS, upstream regions of CSLD2 and CSLD3 were predicted to contain a variety of transcriptional regulatory binding sites with 23 of these sites common to both CSLD2 and CSLD3. These include binding motifs for the WRKY, MYB, homeobox, and Auxin Response Factor (ARF) transcription factors. Some of the functions of the aforementioned transcription factors are consistent with the phenotypes reported here. For example, it was shown that WRKY75 plays a role during root development and specifies root hair number (Devaiah et al., 2007; Rushton et al., 2010) while an Endoplasmic Reticulum (ER)-tethered R2R3-MYB transcription factor was recently implicated in controlling root hair length in part through auxin signaling pathways (Slabaugh et al., 2011). In addition, overexpressing a rice WRKY protein resulted in sterile plants that were likely caused by defective female reproductive organs (Ramamoorthy et al., 2008). Moreover, as noted earlier, the Arabidopsis MYB98 protein was found to be required for the guidance of pollen tubes and the formation of the filiform apparatus in synergid cells (Punwani et al., 2007).
The common binding motifs for transcriptional regulatory elements in the upstream regions of CSLD2 and CSLD3 could partly explain their redundancy. However, the fact that CSLD3 contains 41 additional binding motifs in its upstream region compared to CSLD2 indicates that CSLD3 expression could be controlled by a larger group of transcription factors. This might explain why csld2 and csld3 single mutants exhibit root hair defects with different levels of severity. For example, the upstream region of CSLD3 contains three sites for basic helix-loop-helix (bHLH)-type transcription factors not present in CSLD2. The presence of bHLH sites has significant implications for root hair development because GLABRA3 (GL3), a bHLH transcription factor, forms a complex with WEREWOLF (WER) and a WD40 protein, to positively regulate GL2, which in turn inhibits the generation of root hairs, and promotes cells to differentiate into non-hair cells (Ishida et al., 2007, 2008; Seo et al., 2011). More recently, large scale comparative transcriptional profiling of root epidermal cell mutants, including csld3, and auxin/ethylene treatment uncovered three distinct types of bHLH proteins that participate in root hair development in a stage-specific manner (Bruex et al., 2012). Given that the CSLD3 upstream region contains three predicted bHLH binding sites, it is likely that bHLH transcription factors could be involved in regulating CSLD3 expression during early stages of root hair development. It is possible that this additional level of regulation would enable CSLD3 to have a more pronounced impact on root hair formation than CSLD2. The more prominent role of CSLD3 in root hair development compared to CSLD2 is also supported by our previous experiments showing that induction of CSLD3 expression by low temperature treatment could reverse the defective root hairs of csld2 single mutants whereas cold induction of CSLD2 expression caused only partial rescue of csld3 defects (Bernal et al., 2008).
The partial redundancy and partial divergence between CSLD2 and CSLD3 during root hair development provide a good example of functional evolution of duplicated gene pairs. Duplicated genes may have experienced rapid subfunctionalization after their formation, accompanied by prolonged and substantial neofunctionalization (He and Zhang, 2005). CSLD2 and CSLD3 seem to function jointly to execute their ancestor’s function in root hair and female gametophyte development, which is known as subfunctionalization. However, CSLD3’s more prominent role in root hairs indicates that it might have been neofunctionalized during evolution starting from the duplication event. A similar scenario was previously observed in Arabidopsis as shown by studies of the kinesin 14 (KIN14) genes during reproductive development (Quan et al., 2008). KIN14a and KIN14b genes share extensive functional similarity during male and female reproductive development. However, KIN14a was shown to play a more prominent role in male meiotic spindle organization than KIN14b because homozygote of the KIN14a mutation displayed abnormal chromosome segregation and severe sterility, whereas a homozygous mutant of KIN14b looked like wild-type plants (Quan et al., 2008). KIN14a and KIN14b were also found to have resulted from the third genome-wide duplication event in Arabidopsis, suggesting that this partial functional redundancy and partial divergence between duplicate genes may be common for many genes that originated during the same period. Moreover, the partial divergence of functions could be explained by considerable neofunctionalization on one of the duplicate pair. Even though neofunctionalization suggested in this study does not appear to be significant, more detailed characterization of both genes, particularly with regard to differences in their biochemical properties, may reveal additional divergence between this pair of genes, and allow a deeper understanding of the evolution of this duplicated gene pair.
In summary, we showed that the double mutation of Arabidopsis CSLD2 and CSLD3 causes partial infertility, which can be traced to synergid cell death before the pollen tube penetrates the ovules. Single mutants did not show any female gametophyte defects indicating a redundant role of CSLD2 and CSLD3 in megagametogenesis. On the other hand, the genetic and phenotypic analyses of root hair development of double mutants demonstrated that CSLD2 and CSLD3 have overlapping but not fully redundant functions. An attempt to explain their functional divergence and redundancy from a phylogenetic point of view indicated that CSLD2 and CSLD3 might have evolved from a recent genome-wide duplication event and that they have undergone purifying selection pressure. The two genes share high sequence similarity with each other, but have notable variance in sequences particularly within the N-terminal region, which eventually allowed CSLD3 to acquire more prominent function in root hair development.
Author Contributions
Cheol-Min Yoo conducted cell biological and genetic studies of the csld mutants. Li Quan performed the phylogenetic and bioinformatic analyses of CSLD genes. Cheol-Min Yoo, Li Quan, and Elison B. Blancaflor wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Samuel Roberts Noble Foundation. We thank Dr. Ravishankar Palanivelu (University of Arizona) for sharing the Lat52:GUS lines.
Footnotes
References
Berger, F., Hamamura, Y., Ingouff, M., and Higashiyama, T. (2008). Double fertilization – caught in the act. Trends Plant Sci. 13, 437–443.
Bernal, A. J., Jensen, J. K., Harholt, J., Sorensen, S., Moller, I., Blaukopf, C., Johansen, B., de Lotto, R., Pauly, M., Scheller, H. V., and Willats, W. G. T. (2007). Disruption of ATCSLD5 results in reduced growth, reduced xylan and homogalacturonan synthase activity and altered xylan occurrence in Arabidopsis. Plant J. 52, 791–802.
Bernal, A. J., Yoo, C-M., Mutwil, M., Jensen, J. K., Hou, G., Blaukopf, C., Sorensen, I., Blancaflor, E. B., Scheller, H. V., and Willats, W. G. T. (2008). Functional analysis of the cellulose synthase-like genes CSLD1, CSLD2, and CSLD4 in tip-growing Arabidopsis cells. Plant Physiol. 148, 1238–1253.
Blanc, G., Hokamp, K., and Wolfe, K. H. (2003). A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 13, 137–144.
Breathnach, R., Benoist, C., O’Hare, K., Gannon, F., and Chambon, P. (1978). Ovalbumin gene: evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries. Proc. Natl. Acad. Sci. U.S.A. 75, 4853–4857.
Bruex, A., Kainkaryam, R. M., Wieckowski, Y., Kang, Y. H., Bernhardt, C., Xia, Y., Zheng, X., Wang, J. Y., Lee, M. M., Benfey, P., Woolf, P. J., and Schiefelbein, J. (2012). A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet. 8, e1002446. doi:10.1371/journal.pgen.1002446
Brukhin, V. B., Jaciubek, M., Carpio, A. B., Kuzmina, V., and Grossniklaus, U. (2011). Female gametophytic mutants of Arabidopsis thaliana identified in a gene trap insertional mutagenesis screen. Int. J. Dev. Biol. 55, 73–84.
Burton, R. A., Wilson, S. M., Hrmova, M., Harvey, A. J., Shirley, N. J., Stone, B. A., Newbigin, E. J., Bacic, A., and Fincher, G. B. (2006). Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-beta-D-glucans. Science 311, 1940–1942.
Cho, H.-T., and Cosgrove, D. J. (2002). Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell 14, 3237–3253.
Christensen, C. A., Gorsich, S. W., Brown, R. H., Jones, L. G., Brown, J., Shaw, J. M., and Drews, G. N. (2002). Mitochondrial GFA2 is required for synergid cell death in Arabidopsis. Plant Cell 14, 2215–2232.
Christensen, C. A., King, E. J., Jordan, J. R., and Drews, G. N. (1997). Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex. Plant Reprod. 10, 49–64.
Chu, Z., Chen, H., Zhang, Y., Zhang, Z., Zheng, N., Yin, B., Yan, H., Zhu, L., Zhao, X., Yuan, M., Zhang, X., and Xie, Q. (2007). Knockout of the AtCESA2 gene affects microtubule orientation and causes abnormal cell expansion in Arabidopsis. Plant Physiol. 143, 213–224.
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743.
Cocuron, J.-C., Lerouxel, O., Drakakaki, G., Alonso, A. P., Liepman, A. H., Keegstra, K., Raikhel, N., and Wilkerson, C. G. (2007). A gene from the cellulose synthase-like C family encodes a β-1,4 glucan synthase. Proc. Natl. Acad. Sci. U.S.A. 104, 8550–8555.
Comeron, J. M. (1999). K-estimator: calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics 15, 763–764.
Davuluri, R. V., Sun, H., Palaniswamy, S. K., Matthews, N., Molina, C., Kurtz, M., and Grotewold, E. (2003). AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics 4, 25. doi:10.1186/1471-2105-4-25
Desprez, T., Juraniec, M., Crowell, E. F., Jouy, H., Pochylova, Z., Parcy, F., Hofte, H., Gonneau, M., and Vernhettes, S. (2007). Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 104, 15572–15577.
Devaiah, B. N., Karthikeyan, A. S., and Raghothama, K. G. (2007). WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 143, 1789–1801.
Dhugga, K. S., Barreiro, R., Whitten, B., Stecca, K., Hazebroek, J., Randhawa, G. S., Dolan, M., Kinney, A. J., Tomes, D., Nichols, S., and Anderson, P. (2004). Guar seed β-mannan synthase is a member of the cellulose synthase super gene family. Science 303, 363–366.
Doblin, M. S., Pettolino, F. A., Wilson, S. M., Campbell, R., Burton, R. A., Fincher, G. B., Newbigin, E., and Bacic, A. (2009). A barley cellulose synthase-like CSLH gene mediates (1,3;1,4)-β-D-glucan synthesis in transgenic Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 5996–6001.
Drews, G. N., Lee, D., and Christensen, C. A. (1998). Genetic analysis of female gametophyte development and function. Plant Cell 10, 5–17.
Drummond, A. J., Suchard, M. A., Xie, D., and Rambaut, A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. doi:10.1093/molbev/mss075
Dwivany, F. M., Yulia, D., Burton, R. A., Shirley, N. J., Wilson, S. M., Fincher, G. B., Bacic, A., Newbigin, E., and Doblin, M. S. (2009). The cellulose-synthase like C (CSLC) family of barley includes members that are integral membrane proteins targeted to the plasma membrane. Mol. Plant 2, 1025–1039.
Dyachok, J., Yoo, C.-M., Palanichelvam, K., and Blancaflor, E. B. (2009). Sample preparation for fluorescence imaging of the cytoskeleton in fixed and living plant roots. Methods Mol. Biol. 586, 157–169.
Endler, A., and Persson, S. (2011). Cellulose synthases and synthesis in Arabidopsis. Mol. Plant 4, 199–211.
Faure, J. E., Rotman, N., Fortune, P., and Dumas, C. (2002). Fertilization in Arabidopsis thaliana wild type: developmental stages and time course. Plant J. 30, 481–488.
Favery, B., Ryan, E., Foreman, J., Linstead, P., Boudonck, K., Steer, M., Shaw, P., and Dolan, L. (2001). KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev. 15, 79–89.
Galway, M. E., Eng, R. C., Schiefelbein, J. W., and Wasteneys, G. O. (2011). Root hair-specific disruption of cellulose and xyloglucan in AtCSLD3 mutants, and factors affecting the post-rupture resumption of mutant root hair growth. Planta 233, 985–999.
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., Mitros, T., Dirks, W., Hellsten, U., Putnam, N., and Rokhsar, D. S. (2011). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186.
Goubet, F., Barton, C. J., Mortimner, J. C., Yu, X., Zhang, Z., Miles, G. P., Richens, J., Liepman, A. H., Seffen, K., and Dupree, P. (2009). Cell wall glucomannan in Arabidopsis is synthesized by CSLA glycosyltransferase, and influences the progression of embryogenesis. Plant J. 60, 527–538.
Goubet, F., Misrahi, A., Park, S. K., Zhang, Z. N., Twell, D., and Dupree, P. (2003). AtCSLA7, a cellulose synthase-like putative glycosyltransferase, is important for pollen tube growth and embryogenesis in Arabidopsis. Plant Physiol. 131, 547–557.
Gu, X., and Vander, V. K. (2002). DIVERGE: phylogeny-based analysis for functional-structural divergence of a protein family. Bioinformatics 18, 500–501.
Guo, A. Y., Zhu, Q. H., Chen, X., and Luo, J. C. (2007). GSDS: a gene structure display server. Yi Chuan 29, 1023–1026.
He, X., and Zhang, J. (2005). Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 169, 1157–1164.
Higashiyama, T., Yabe, S., Sasaki, N., Nishimura, Y., Miyagishima, S., Kuroiwa, H., and Kuroiwa, T. (2001). Pollen tube attraction by the synergid cell. Science 293, 1480–1483.
Hu, J., Zhu, L., Zeng, D. L., Gao, Z. Y., Guo, L. B., Fang, Y. X., Zhang, G. H., Dong, G. J., Yan, M. X., Liu, J., and Qian, Q. (2010). Identification and characterization of NARROW AND ROLLED LEAF 1, a novel gene regulating leaf morphology and plant architecture in rice. Plant Mol. Biol. 73, 283–292.
Huang, B.-Q., and Russell, S. D. (1992). Female germ unit: organization, isolation, and function. Int. Rev. Cytol. 140, 233–292.
Hunter, C. T., Kirienko, D. H., Sylvester, A. W., Peter, G. F., McCarty, D. R., and Koch, K. E. (2012). Cellulose synthase-like D1 is integral to normal cell division, expansion, and leaf development in maize. Plant Physiol. 158, 708–724.
Ishida, T., Hattori, S., Sano, R., Inoue, K., Shirano, Y., Hayashi, H., Shibata, D., Sato, S., Kato, E. T., Tabata, S., Okada, K., and Wada, T. (2007). Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell 19, 2531–2543.
Ishida, T., Kutata, T., Okada, K., and Wada, T. (2008). A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 59, 365–386.
Jansson, S., and Douglas, C. J. (2007). Populus: a model system for plant biology. Annu. Rev. Plant Biol. 58, 435–458.
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS fusion beta glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO 6, 3901–3908.
Johnson, M. A., von Besser, K., Zhou, Q., Smith, E., Aux, G., Patton, D., Levin, J. Z., and Preuss, D. (2004). Arabidopsis hapless mutations define essential gametophytic functions. Genetics 168, 971–982.
Johnston, A. J., Meier, P., Gheyselinck, J., Wuest, S. E. J., Federer, M., Schlagenhauf, E., Becker, J. D., and Grossniklaus, U. (2007). Genetic subtraction profiling identifies genes essential for Arabidopsis reproduction and reveals interaction between the female gametophyte and the maternal sporophyte. Genome Biol. 8, R204.
Kasahara, R. D., Portereiko, M. F., Sandaklie-Nikolova, L., Rabiger, D. S., and Drews, G. N. (2005). MYB98 is required for pollen tube guidance and synergid cell differentiation in Arabidopsis. Plant Cell 17, 2981–2992.
Katoh, K., Asimenos, G., and Toh, H. (2009). Multiple alignment of DNA sequences with MAFFT. Methods Mol. Biol. 537, 39–64.
Kim, C. M., Park, S. H., Je, B. I., Park, S. H., Park, S. J., Piao, H. L., Eun, M. Y., Dolan, L., and Han, C.-D. (2007). OsCSLD1, a cellulose synthase-like D1 gene, is required for root hair morphogenesis in rice. Plant Physiol. 143, 1220–1230.
Kong, H., Landherr, L. L., Frohlich, M. W., Leebens-Mack, J., Ma, H., and De Pamphilis, C. W. (2007). Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant J. 50, 873–885.
Kuittinen, H., de Haan, A. A., Vogl, C., Oikarinen, S., Leppälä, J., Koch, M., Mitchell-Olds, T., Langley, C. H., and Savolainen, O. (2004). Comparing the linkage maps of the close relatives Arabidopsis lyrata and A. thaliana. Genetics 168, 1575–1584.
Lerouxel, O., Cavalier, D. M., Liepman, A. H., and Keegstra, K. (2006). Biosynthesis of plant cell wall polysaccharides – a complex process. Curr. Opin. Plant Biol. 9, 621–630.
Li, M., Xiong, G., Li, R., Cui, J., Tang, D., Zhang, B., Pauly, M., Cheng, Z., and Zhou, Y. (2009). Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth. Plant J. 60, 1055–1069.
Liepman, A. H., Wilkerson, C. G., and Keegstra, K. (2005). Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc. Natl. Acad. Sci. U.S.A. 102, 2221–2226.
Luan, W., Liu, Y., Zhang, F., Song, Y., Wang, Z., Peng, Y., and Sun, Z. (2011). OsCD1 encodes a putative member of the cellulose synthase-like D sub-family and is essential for rice plant architecture and growth. Plant Biotechnol. J. 9, 513–524.
McQueen-Mason, S., and Cosgrove, D. J. (1995). Expansin mode of action on cell walls: analysis of wall hydrolysis, stress relaxation, and binding. Plant Physiol. 107, 87–100.
Okuda, S., Tsutsui, H., Shiina, K., Sprunck, S., Takeuchi, H., Yui, R., Kasahara, R. D., Hamamura, Y., Mizukami, A., Susaki, D., Kawano, N., Sakakibara, T., Namiki, S., Itoh, K., Otsuka, K., Matsuzaki, M., Nozaki, H., Kuroiwa, T., Nakano, A., Kanaoka, M. M., Dresselhaus, T., Sasaki, N., and Higashiyama, T. (2009). Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature 458, 357–361.
Park, S., Szumlanski, A. L., Gu, F., Guo, F., and Nielsen, E. (2011). A role for CSLD3 during cell-wall synthesis in apical plasma membranes of tip-growing root-hair cells. Nat. Cell Biol. 13, 973–980.
Persson, S., Paredez, A., Carroll, A., Palsdottir, H., Doblin, M., Poindexter, P., Khitrov, N., Auer, M., and Somerville, C. R. (2007). Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 15566–15571.
Punwani, J. A., and Drews, G. N. (2008). Development and function of the synergid cell. Sex. Plant Reprod. 21, 7–15.
Punwani, J. A., Rabiger, D. S., and Drews, G. N. (2007). MYB98 positively regulates a battery of synergid-expressed genes encoding filiform apparatus localized proteins. Plant Cell 19, 2557–2568.
Quan, L., Xiao, R., Li, W., Oh, S.-A., Kong, H., Ambrose, J. C., Malcos, J. L., Cyr, R., Twell, D., and Ma, H. (2008). Functional divergence of the duplicated AtKIN14a and AtKIN14b genes: critical roles in Arabidopsis meiosis and gametophyte development. Plant J. 53, 1013–1026.
Ramamoorthy, R., Jiang, S. Y., Kumar, N., Venkatesh, P. N., and Ramachandran, S. (2008). A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol. 49, 865–879.
Richmond, T. A., and Somerville, C. R. (2000). The cellulose synthase superfamily. Plant Physiol. 124, 495–498.
Rotman, N., Rozier, F., Boavida, L., Dumas, C., Berger, F., and Faure, J. E. (2003). Female control of male gamete delivery during fertilization in Arabidopsis thaliana. Curr. Biol. 13, 432–436.
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258.
Sandaklie-Nikolova, L., Palanivelu, R., King, E. J., Copenhaver, G. P., and Drews, G. N. (2007). Synergid cell death in Arabidopsis is triggered following direct interaction with the pollen tube. Plant Physiol. 144, 1753–1762.
Sassen, M. M. A., Traas, J. A., and Wolters-Arts, A. M. C. (1985). Deposition of cellulose microfibrils in cell walls of root hairs. Euro. J. Cell Biol. 37, 21–26.
Schneitz, K., Huelskamp, M., and Pruitt, R. E. (1995). Wild-type ovule development in Arabidopsis thaliana: a light microscope study of cleared whole-mount tissue. Plant J. 7, 731–749.
Schwartz, S., Zhang, Z., Frazer, K. A., Smit, A., Riemer, C., Bouck, J., Gibbs, R., Hardison, R., and Miller, W. (2000). PipMaker-a web server for aligning two genomic DNA sequences. Genome Res. 10, 577–586.
Seo, E., Yu, J., Ryu, K. H., Lee, M. M., and Lee, I. (2011). WEREWOLF, a regulator of root hair pattern formation, controls flowering time through the regulation of FT mRNA stability. Plant Physiol. 156, 1867–1877.
Simillion, C., Vandepoele, K., Van Montagu, M. C. E., Zabeau, M., and Van de Peer, Y. (2002). The hidden duplication past of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 99, 13627–13632.
Slabaugh, E., Held, M., and Brandizzi, F. (2011). Control of root hair development in Arabidopsis thaliana by an endoplasmic reticulum anchored member of the R2R3-MYB transcription factor family. Plant J. 67, 395–405.
Smyth, D. R., Bowman, J. L., and Meyerowitz, E. M. (1990). Early flower development in Arabidopsis. Plant Cell 2, 755–767.
Szyjanowicz, P. M. J., McKinnon, I., Taylor, N. G., Gardiner, J., Jarvis, M. C., and Turner, S. R. (2004). The irregular xylem 2 mutant is an allele of korrigan that affects the secondary cell wall of Arabidopsis thaliana. Plant J. 37, 730–740.
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0 . Mol. Biol. Evol. 24, 1596–1599.
Taylor, N. G. (2008). Cellulose biosynthesis and deposition in higher plants. New Phytol. 178, 239–252.
Taylor, N. G., Howells, R. M., Huttly, A. K., Vickers, K., and Turner, S. R. (2003). Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. U.S.A. 100, 1450–1455.
Verhertbruggen, Y., Yin, L., Oikawa, A., and Scheller, H. (2011). Mannan synthase activity in the CSLD family. Plant Signal. Behav. 6, 1620–1623.
Wang, H., Moore, M. J., Soltis, P. S., Bell, C. D., Brockington, S. F., Alexandre, R., Davis, C. C., Latvis, M., Manchester, S. R., and Soltis, D. E. (2009). Rosid radiation and the rapid rise of angiosperm-dominated forests. Proc. Natl. Acad. Sci. U.S.A. 106, 3853–3858.
Wang, W., Wang, L., Chen, C., Xiong, G., Tan, X.-Y., Yang, K.-Z., Wang, Z.-C., Zhou, Y., Ye, D., and Chen, L.-Q. (2011). Arabidopsis CSLD1 and CSLD4 are required for cellulose deposition and normal growth of pollen tubes. J. Exp. Bot. 62, 5161–5177.
Wang, X., Cnops, G., Vanderhaeghen, R., De Block, S., Van Montagu, M., and Van Lijsebettens, M. (2001). AtCSLD3, a cellulose synthase-like gene important for root hair growth in Arabidopsis. Plant Physiol. 126, 575–586.
Weterings, K., and Russell, S. D. (2004). Experimental analysis of the fertilization process. Plant Cell 16, S107–S118.
Wu, C., Fu, Y., Hu, G., Si, H., Cheng, S., and Liu, W. (2010). Isolation and characterization of a rice mutant with narrow and rolled leaves. Planta 232, 313–324.
Wuest, S. E., Vijverberg, K., Schmidt, A., Weiss, M., Gheyselinck, J., Lohr, M., Wellmer, F., Rahnenfuehrer, J., von Mering, C., and Grossniklaus, U. (2010). Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr. Biol. 20, 506–512.
Yilmaz, A., Mejia-Guerra, M. K., Kurz, K., Liang, X., Welch, L., and Grotewold, E. (2011). AGRIS: the Arabidopsis gene regulatory information server, an update. Nucleic Acids Res. 39, D1118–D1122.
Yin, L., Verhertbruggen, Y., Oikawa, A., Manisseri, C., Knierim, B., Prak, L., Jensen, J. K., Knox, J. P., Auer, M., Willats, W. G. T., and Scheller, H. V. (2011). The cooperative activities of CSLD2, CSLD3, and CSLD5 are required for normal Arabidopsis development. Mol. Plant 4, 1024–1037.
Yin, Y. B., Huang, J. L., Xu, Y., Yin, Y. B., Huang, J. L., and Xu, Y. (2009). The cellulose synthase superfamily in fully sequenced plants and algae. BMC Plant Biol. 9, 99. doi:10.1186/1471-2229-9-99
Appendix
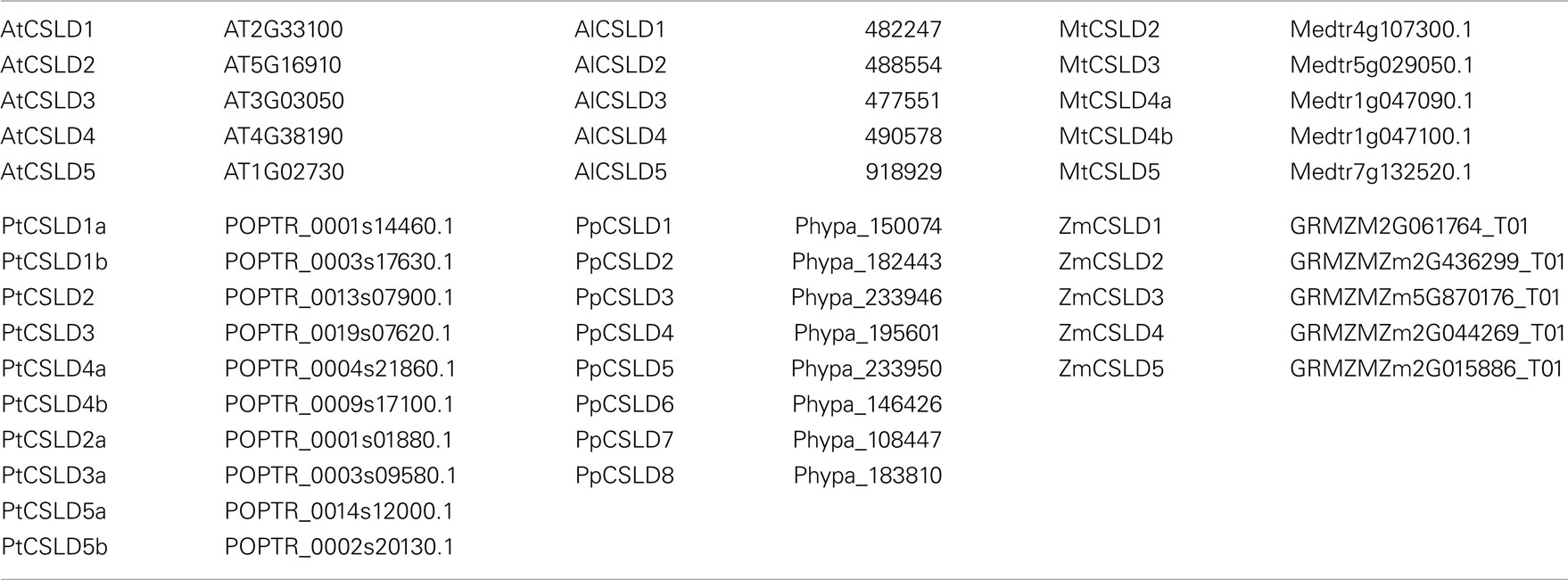
Table A1. Nomenclature of CSLD genes and their corresponding accession numbers in TAIR and Phytozome.
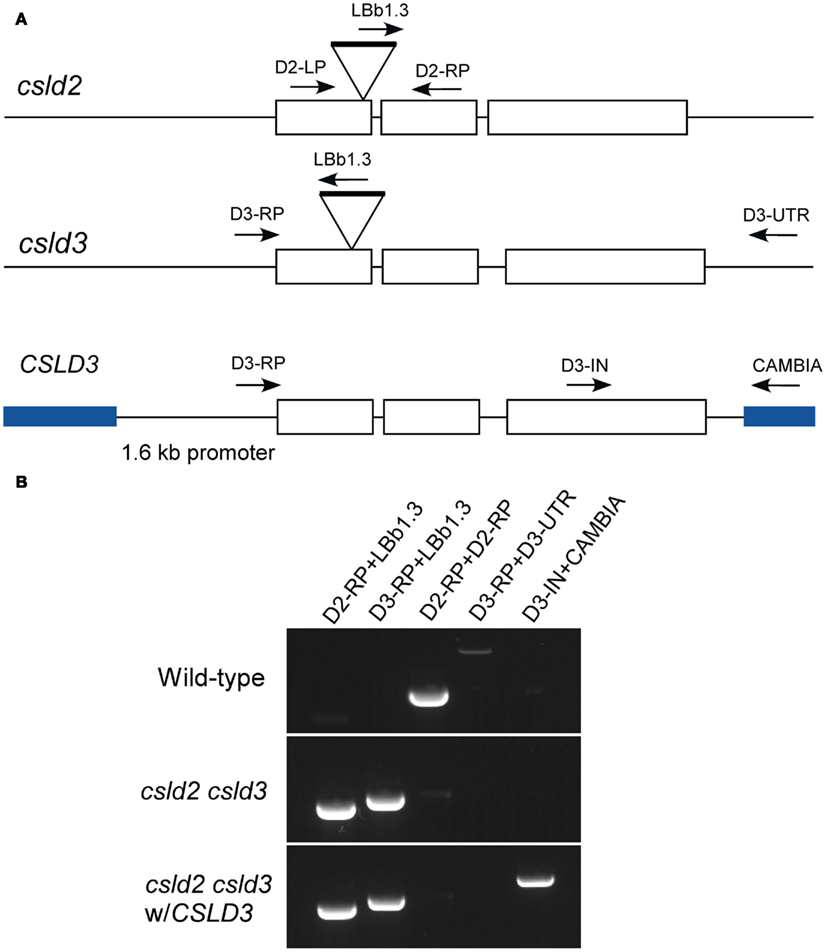
Figure A1. Generation of csld2 csld3 complemented lines. (A) Genome organization of CSLD2 and CSLD3. Boxes indicate exons, and lines indicate introns. csld2 and csld3 are the T-DNA mutants, which have hits on their first exon. A wild-type full length CSLD3 was subcloned into a pCAMBIA2300 as indicated by blue lines. The arrows indicate the position of the primers used for genotyping. (B) Genotyping by PCR using different sets of primers indicated in (A). The complemented line does not have the native CSLD3 but has the transgenic CSLD3 in the background of csld2 csld3 double homozygote.
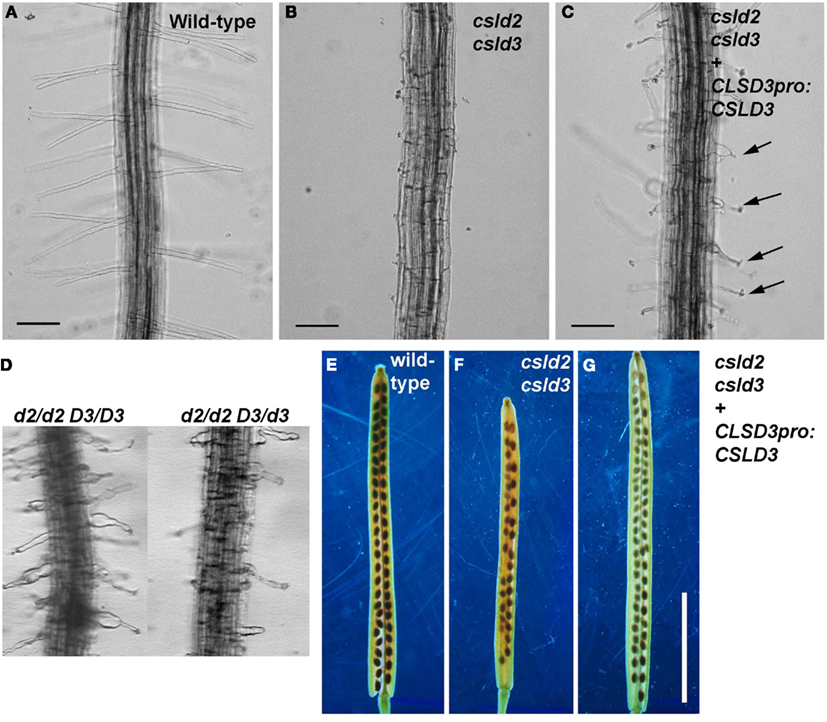
Figure A2. Complementation of csld2 csld3 double mutants. (A,E) Are wild-type, (B,F) are csld2 csld3, and (C,G) are the complemented line (csld2 csld3 w/CSLD3) as validated by the genotyping experiments shown in Figure A1B. (D) Root hairs of csld2 single mutants (d2/d2 D3/D3) and seedlings with one copy of CSLD3 (d2/d2 D3/d3). The complemented csld2 csld3 line showed CSLD2-like single mutant root hair phenotypes [arrows in (C)] and wild-type-like siliques (G). Bars = 100 μm [for (A–D)]. Bars = 5 mm [for (E–G)].
Keywords: Arabidopsis, cellulose synthase, female gametophyte, gene duplication, phylogenetics, root hairs
Citation: Yoo C-M, Quan L and Blancaflor EB (2012) Divergence and redundancy in CSLD2 and CSLD3 function during Arabidopsis thaliana root hair and female gametophyte development. Front. Plant Sci. 3:111. doi: 10.3389/fpls.2012.00111
Received: 29 February 2012; Accepted: 08 May 2012;
Published online: 29 May 2012.
Edited by:
Erik Nielsen, University of Michigan, USAReviewed by:
Viktor Zarsky, Charles University, Czech RepublicMary Lai Preuss, Webster University, USA
Moira Galway, St. Francis Xavier University, Canada
Copyright: © 2012 Yoo, Quan and Blancaflor. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Elison B. Blancaflor, Plant Biology Division, The Samuel Roberts Noble Foundation, 2510 Sam Noble Parkway, Ardmore, OK, USA. e-mail: eblancaflor@noble.org
†Cheol-Min Yoo and Li Quan have contributed equally to this work.