- 1 Environmental Biotechnology Research Center, Korea Research Institute of Bioscience and Biotechnology (KRIBB), Daejeon, Korea
- 2 Division of Biosystems and Bioengineering, University of Science and Technology, Daejeon, Korea
- 3 Department of Biological Sciences, Chungnam National University, Daejeon, Korea
- 4 School of Biological Sciences and Genomics and Breeding Institute, Seoul National University, Seoul, Korea
Phospholipase A2 (PLA2) hydrolyzes phospholipids at the sn-2 position to yield lysophospholipids and free fatty acids. Of the four paralogs expressed in Arabidopsis, the cellular functions of PLA2α in planta are poorly understood. The present study shows that PLA2α possesses unique characteristics in terms of spatiotemporal subcellular localization, as compared with the other paralogs that remain in the ER and/or Golgi apparatus during secretory processes. Only PLA2α is secreted out to extracellular spaces, and its secretion to apoplasts is modulated according to the developmental stages of plant tissues. Observation of PLA2α-RFP transgenic plants suggests that PLA2α localizes mostly at the Golgi bodies in actively growing leaf tissues, but is gradually translocated to apoplasts as the leaves become mature. When Pseudomonas syringae pv. tomato DC3000 carrying the avirulent factor avrRpm1 infects the apoplasts of host plants, PLA2α rapidly translocates to the apoplasts where bacteria attempt to become established. PLA2α promoter::GUS assays show that PLA2α gene expression is controlled in a developmental stage- and tissue-specific manner. It would be interesting to investigate if PLA2α functions in plant defense responses at apoplasts where secreted PLA2α confronts with invading pathogens.
Introduction
Phospholipase A2 (PLA2) is widespread throughout nature and stereospecifically catalyzes the hydrolysis of phospholipids at sn-2 to produce lysophospholipids and free fatty acids, which are important mediators or precursors in signal transduction pathways in animal cells (Schaloske and Dennis, 2006; Burke and Dennis, 2009). There is evidence that plant PLA2s are also involved in diverse biological and physiological processes such as senescence, wound healing, elicitor and stress responses, defense against pathogens, and the induction of secondary metabolite accumulation (Wang, 2001, 2004; Ryu, 2004; Scherer et al., 2007; Seo et al., 2008; Kirik and Mudgett, 2009; Mansfeld, 2009; Froidure et al., 2010; Liao and Burns, 2010).
There are four PLA2 paralogs in Arabidopsis: PLA2α, PLA2β, PLA2γ, and PLA2δ. The paralogs PLA2γ and PLA2δ are expressed solely in pollen, localized in the endoplasmic reticulum (ER)/Golgi bodies and ER, respectively, and mediate pollen germination and tube growth (Kim et al., 2011). PLA2β is localized in the ER and expressed in different tissues such as young seedlings, elongating flower stems, and pollen, and it mediates cell elongation, shoot gravitropism, stomatal opening, and pollen development (Lee et al., 2003; Kim et al., 2011). Although PLA2α appears to be ubiquitous in diverse organs (Ryu et al., 2005; Mansfeld and Ulbrich-Hofmann, 2007; Kim et al., 2011), its temporal and spatial expression dynamics in different tissues and its subcellular translocation during different developmental stages are unknown.
In this study, we report that PLA2α in Arabidopsis moves from the ER/Golgi apparatus to the apoplasts as the leaves become mature, and that PLA2α gene expression is controlled in both a developmental stage- and organ-dependent manner. Several lines of evidence suggest that secretory proteins or proteins enhancing secretory pathways play important roles in plant defense responses (Wang et al., 2005; Kwon et al., 2008; Sup Yun et al., 2008). Thus, we examined if the secretion of PLA2α to apoplasts is modulated by pathogen infection. Interestingly, translocation of PLA2α to apoplasts was rapidly enhanced in response to the inoculation of Pseudomonas syringae pv. tomato DC3000 carrying avrRpm1 (Pst-avrRpm1). These observations suggest that PLA2α proteins secreted into apoplasts in response to bacterial infection may play a role in host defense responses.
Materials and Methods
Plant Materials and Reagents
Arabidopsis thaliana (Col-0) plants were grown in soil pots at 22°C, 60% relative humidity, with a 16-h photoperiod and a photon flux density of 110 μmol m-2 s-1.
GUS Staining
For histochemical localization studies, a PLA2α-promoter::GUS (ProPLA2α::GUS) construct was cloned by incorporating the PLA2α sequence upstream of the ATG start codon (from -1175 bp to +3 bp) into the HindIII and BamHI sites of the pBI101 vector. The resulting plasmids were inserted into Agrobacterium tumefaciens strain EHA105, which was transformed into Arabidopsis using the floral dip method as described previously (Bech1998told and Pelletier, 1998). Histochemical GUS assays to show tissue-specific PLA2α expression at different developmental stages were performed as previously described (Jefferson et al., 1987). Tissues from ProPLA2α::GUS-transformed plants were immersed in GUS solution [1 mM X-gluc, 100 mM sodium phosphate buffer (pH 7.0), 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, 10 mM EDTA, and 0.1% (v/v) Triton X-100] and incubated for 12 h at 37°C due to its weak staining. After GUS staining, 100% ethanol was used to remove the chlorophyll.
Subcellular Localization of PLA2α-RFP in Arabidopsis
To investigate the dynamics of PLA2α subcellular localization, transgenic Arabidopsis plants carrying Pro35S::PLA2α-RFP were generated (Lee et al., 2010). Leaf tissues from the PLA2α-RFP transgenic Arabidopsis plants were viewed at different developmental stages using a laser scanning confocal microscope (Meta system, Zeiss) after incubation in water or 1 N KNO3 for 5 min to trigger plasmolysis. RFP-fluorescence was excited at 543 nm and the emitted fluorescence was collected with a band-pass filter at 560–615 nm.
Co-Localization Assay of PLA2α-RFP and ST-GFP
To investigate whether PLA2α-RFP proteins are co-localized with a Golgi marker ST-GFP, transgenic Arabidopsis plants expressing both PLA2α-RFP and ST-GFP were generated (Lee et al., 2010). Close-to-mature leaves of 3-week-old transgenic plants were observed with a laser scanning confocal microscope (Meta system, Zeiss). RFP and GFP fluorescence was detected using at 543/560–615 nm and 488/505–530 nm excitation/emission filter sets, respectively.
Bacterial Inoculation of Plants
Pseudomonas syringae pv. tomato DC3000 carrying avrRpm1 (Pst-avrRpm1) were obtained from Y. J. Kim (Korea University, Seoul, Korea). Plants were inoculated by spreading a bacterial suspension (1 × 108 CFU ml-1 in 0.015% v/v Silwet L-77 and 10 mM MgCl2) onto the adaxial leaf surfaces of Arabidopsis carrying Pro35S::PLA2α-RFP. Plants designated as NT were given no treatment, whereas mock plants were treated with 0.015% v/v Silwet L-77 and 10 mM MgCl2. All the data presented in this study were obtained from at least three independent replicates.
Results
Histochemical Analysis of GUS Activity of the PLA2α Promoter
Although RT-PCR analysis shows that PLA2α transcripts are present in different parts of Arabidopsis tissues (Kim et al., 2011), there is little information regarding PLA2α gene expression at different developmental stages. To elucidate the cell type-specific expression patterns of the PLA2α gene, transgenic Arabidopsis lines were generated that expressed the beta-glucuronidase (GUS) reporter gene under the control of the PLA2α promoter (Figure 1). GUS activity was detected in the cotyledons, the shoot apex, the hypocotyl, and the vascular tissues of 7-day-old germinated seedlings (Figure 1A). Strong GUS activity was detected in the shoot apex in 14-day-old seedlings and 3-week-old plants, and was preferentially expressed in young leaves rather than old leaves (Figures 1A–C). No GUS activity was detected in the roots at this stage. In 5-week-old plants, GUS expression was found in the cauline leaves, sepals, styles, and pedicel of reproductive tissues (Figure 1D). The apical end of the pedicel is particularly dark-stained, apparently due to its thickened cell tissues based on a comparison with the GUS staining of the control transgenic plants harboring the 35S promoter fused with the GUS gene (Figure 1I). In plants transformed with ProPLA2α::GUS, GUS expression was also detected in the developing siliques (Figures 1E–H) and in the main roots of flowering plants (Figure 1J). Taken together, these data indicate that PLA2α gene expression is controlled in a unique developmental stage- and tissue-specific manner.
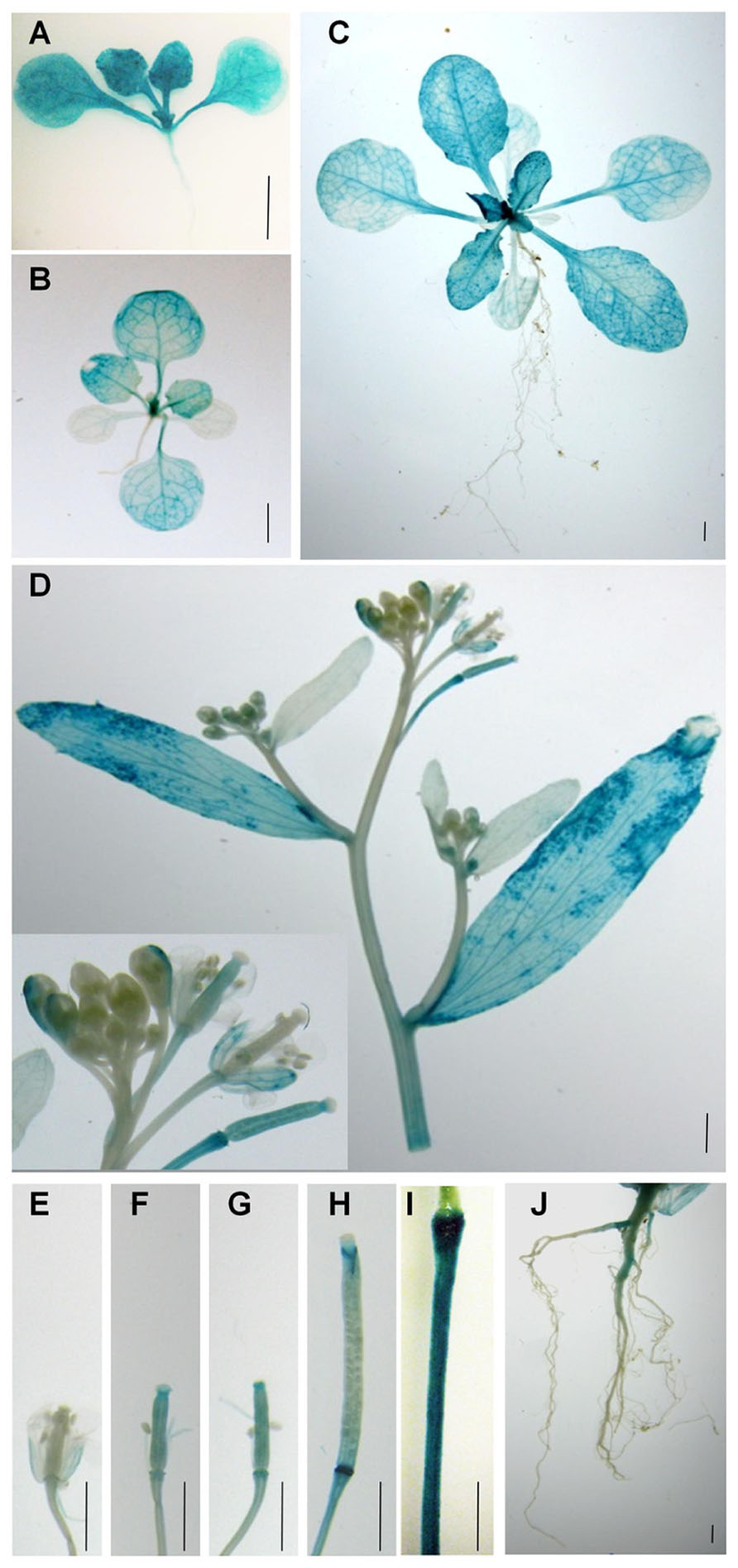
FIGURE 1. Spatial and temporal expression of PLA2α. Spatiotemporal expression patterns of the PLA2α gene in transgenic Arabidopsis plants harboring the PLA2α promoter fused with the GUS gene. Promoter activity was visualized by histochemical GUS staining. (A) Seven-day-old plant. (B) Fourteen-day-old plant. (C) Three-week-old plant. (D) Flower cluster, cauline leaf, and stem of a 5-week-old plant. (E–H) Carpels and developing siliques of a 5-week-old plant. (I) Pedicel of the control transgenic plants harboring the 35S promoter fused with the GUS gene. (J) Root of a 6-week-old plant. Bars = 2 mm.
Subcellular Localization of PLA2α
Lee et al. (2010) reported that fluorescence signals for PLA2α-fusion proteins were observed at the Golgi apparatus of root hair cells. However, Froidure et al. (2010) showed time-dependent localization of PLA2α using a transient expression system incorporating N. tabacum. The YFP reporter fused with PLA2α was detected in cytoplasmic vesicles around the nucleus 36 h after agroinfiltration to tobacco leaves, and was detected at the extracellular spaces outside the cells at a later time point (48 h after agroinfiltration). To resolve these inconsistencies, we investigated in more detail the subcellular localization of PLA2α by analyzing the fluorescence of fusion proteins in transgenic plants carrying Pro35S::PLA2α-RFP. The leaves of 4-week-old PLA2α-RFP transgenic seedlings were viewed using a laser scanning confocal microscope. The results showed that the subcellular localization of PLA2α was dependent on the developmental stages of leaf tissue. PLA2α-RFP fusion proteins were present primarily at the Golgi apparatus in pre-mature young leaves (Figure 2A), whereas in mature leaves, they were detected primarily in the apoplasts (Figure 2B). Even after cell plasmolysis was induced by treatment with 1 N KNO3 for 5 min, the PLA2α-RFP signal remained in the extracellular spaces or diffused into the gap between the cell wall and the plasma membrane that is induced by plasmolysis (Figure 2C). These results indicate that PLA2α is indeed localized in the apoplasts of mature leaves.
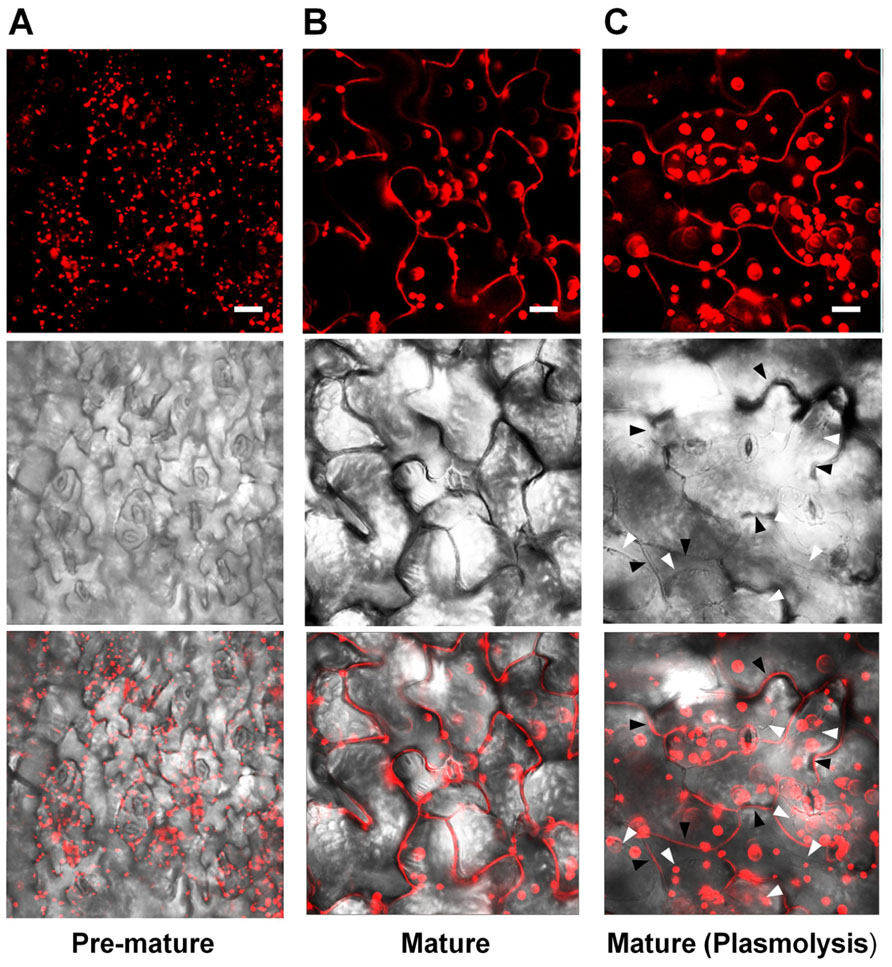
FIGURE 2. Subcellular localization of PLA2α at different leaf ages in PLA2α-RFP transgenic Arabidopsis plants. (A,B) Epidermal cells in pre-mature leaf tissues (A) and mature leaf tissues (B) from Pro35SPLA2α-RFP transgenic Arabidopsis plants (4-week-old) were viewed using confocal microscopy. (C) Mature leaf tissues were incubated in 1 N KNO3 to induce plasmolysis before being viewed with confocal microscopy. The slightly decreased clarity of images in (C) appears to result from the diffusion of PLA2α-RFP into the expanded apoplasts due to plasmolysis. White arrows in (C) indicate the plasmolyzed plasma membrane, whereas black arrows indicate extracellular spaces. Fluorescent (top), bright field (middle), and merged images (bottom) are presented. Bars = 20 μm.
Co-Localization of PLA2α with a Golgi Marker
As secretion of proteins to apoplasts is known to occur through ER and Golgi bodies, PLA2α-RFP signals were mostly detected at the Golgi bodies in pre-mature young leaves (Figure 2A). However, the fluorescent spots become gradually bigger as the leaves become mature, leading us to suspect that they may be other cellular organelles. To investigate if the big PLA2α fluorescent spots are real Golgi bodies, we performed co-localization assay of PLA2α with a Golgi body marker, sialyltransferase (ST). Close-to-mature leaves of transgenic Arabidopsis plants expressing both PLA2α-RFP and ST-GFP were observed with a laser scanning confocal microscope. As shown in Figure 3, big spots of PLA2α-RFP signals are mostly overlapped with the spots of a Golgi body marker (ST-GFP), confirming that PLA2α is localized in Golgi apparatus before secretion to the apoplasts. We found that the apparent big size spots result from aggregation of several Golgi bodies and strong brightness of RFP-fluorescence. Aggregation of Golgi bodies appears to be gradually enhanced as the leaves become mature.
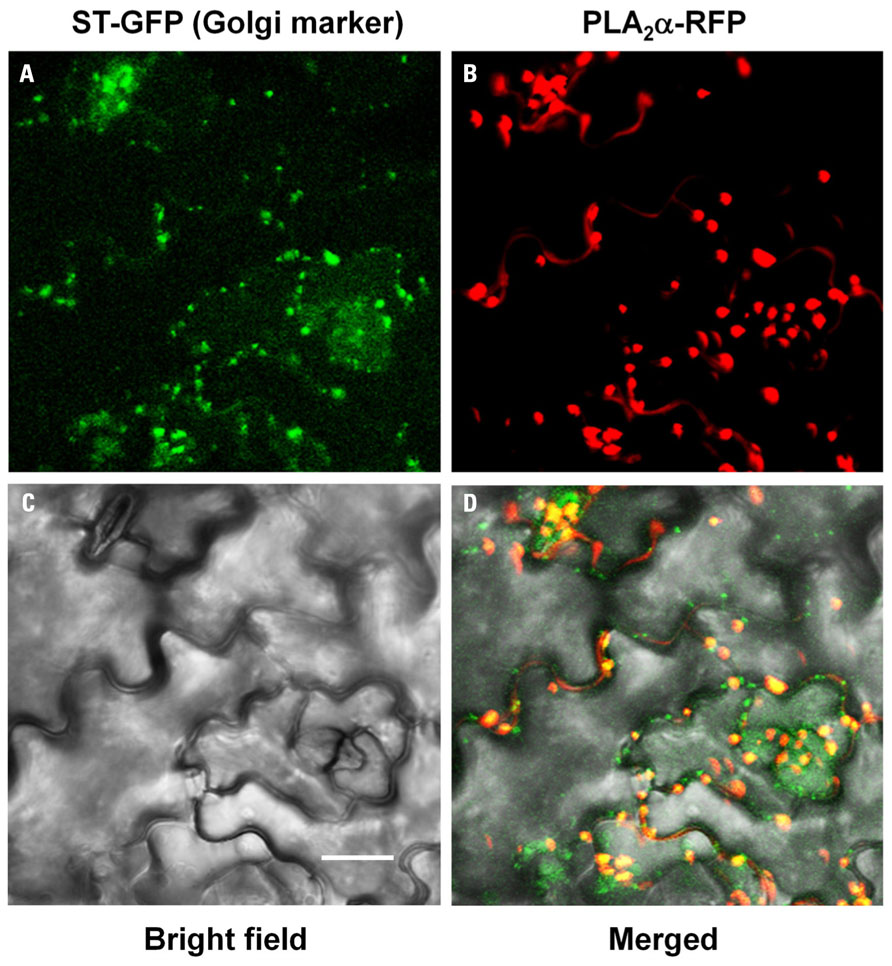
FIGURE 3. Co-localization of PLA2α-RFP with ST-GFP, a Golgi body marker. Close-to-mature leaves of transgenic Arabidopsis plants expressing both PLA2α-RFP and ST-GFP were observed with a laser scanning confocal microscope. PLA2α-RFP signals are mostly overlapped with ST-GFP, a Golgi body marker. ST-GFP (A), PLA2α-RFP (B), bright field (C), and merged image (D) are presented. Bar = 20 μm.
PLA2α Translocates to Apoplasts in Response to the Inoculation of Avirulent Bacteria
As leaves become mature, PLA2α is secreted into the apoplast, where it generates its lipid products, lysophospholipids and free fatty acids. The lipid products have been suggested to function as bio-active molecules that mediate a variety of cellular processes. Apoplasts are an important site for the interaction of plant cell defense mechanisms with invading bacteria, which attempt to become established in the apoplasts. If PLA2α positively participates in defense responses to pathogen attack, we hypothesized that its translocation to the apoplasts would be enhanced when pathogens are inoculated. As speculated, the translocation of PLA2α to the apoplasts was enhanced at 3 h post-inoculation of avirulent bacteria, Pst-avrRpm1, in young leaves (Figure 4C) and in close-to-mature leaves (Figure 5C), as compared to non-treated controls (NT) and 0.015% Silwet/10 mM MgCl2-treated mocks (Figures 4A,B and 5A,B).
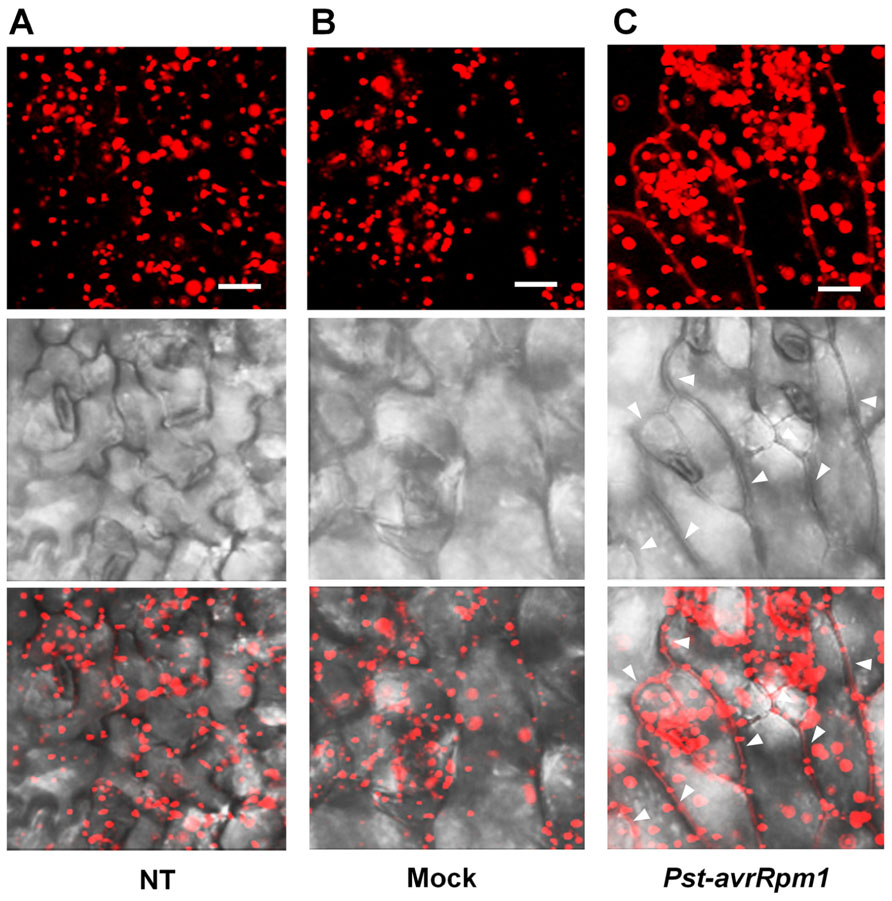
FIGURE 4. Translocation of PLA2α to apoplasts was enhanced by the inoculation of bacteria, Pst-avrRpm1, in pre-mature young leaves. (A–C) Images showing increased fluorescence intensity and vesicle sizes followed by the translocation of PLA2α to apoplasts at 3 h post-inoculation of Pst-avrRpm1 (C) compared to the no-treatment control (A) and 0.015% Silwet/10 mM MgCl2-treated mock (B) in pre-mature young leaves where PLA2α is normally localized primarily in Golgi bodies. Fluorescent (top), bright field (middle), and merged images (bottom) are presented. Bars = 20 μm.
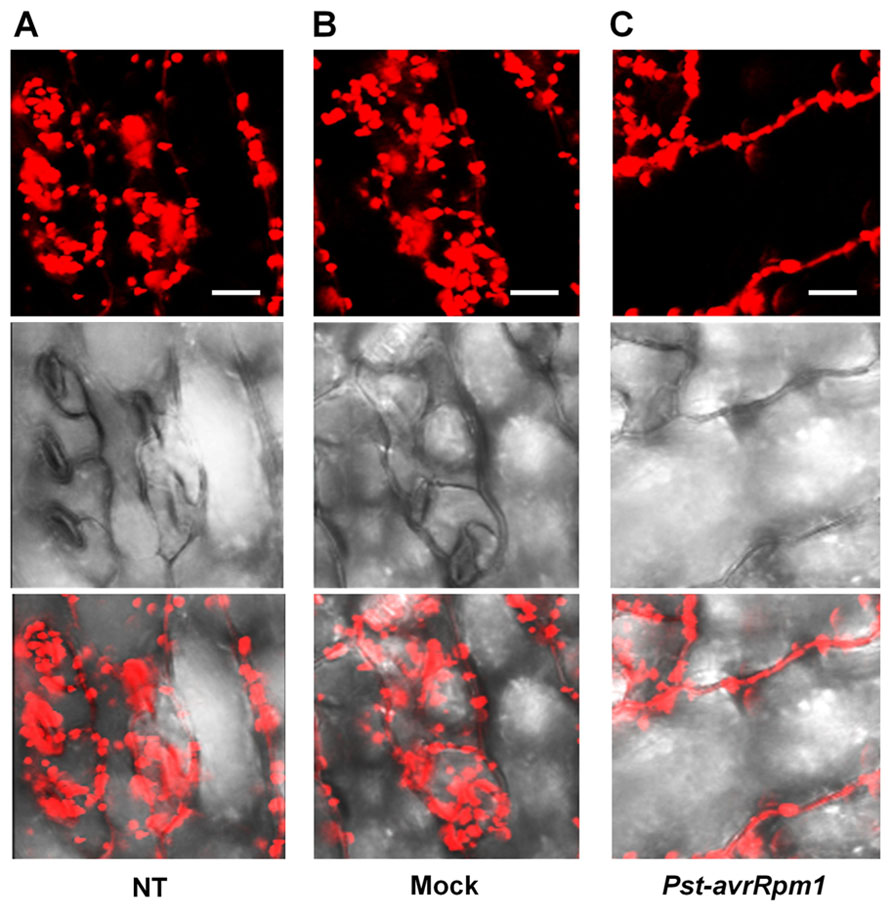
FIGURE 5. Enhanced translocation of PLA2α to apoplasts in response to the inoculation of Pst-avrRpm1 in close-to-mature leaves. (A–C) Significant enhancement of PLA2α translocation to apoplasts was observed at 3 h post-inoculation of Pst-avrRpm1 (C) compared to the no-treatment control (A) and 0.015% Silwet/10 mM MgCl2-treated mock (B) in close-to-mature leaves where PLA2α is already detected at a low level in the apoplasts. Fluorescent (top), bright field (middle), and merged images (bottom) are presented. Bars = 20 μm.
Discussion
PLA2α is expressed in a tissue- and developmental stage-specific manner in Arabidopsis plant tissues. Relatively strong activities of the PLA2α promoter were observed in actively growing seedlings and young leaves. Expression decreased slightly as leaves became mature. This expression pattern of PLA2α is different from that of PLA2β, which is expressed at a very low level in the mature leaves (Lee et al., 2003). This pattern of expression is also observed in the cauline leaves of the inflorescence stems, which display strong expression of PLA2α but low expression of PLA2β. At the young seedling and reproductive organ developmental stages, both PLA2 paralogs display similar expression patterns; strong expression in actively growing tissues and reproductive organs such as sepals, pedicels, and styles of open flowers, but low expression in petals, stigmas, and ovaries. Expression of both PLA2 paralogs was detected in developing siliques but not in maturing seeds. However, PLA2α was not expressed in pollen tissues, in contrast to the strong expression of PLA2β. In the root, PLA2α was expressed at the late stages of growth, whereas PLA2β was expressed in roots from seedling stages (Lee et al., 2003). These results suggest that PLA2α and PLA2β may play a role in plant growth and development in harmony with holding their own cellular roles at the different cellular localization and expressing tissues.
Arabidopsis PLA2 genes encode proteins with N-terminal signal peptides, which are predicted to be secreted via ER and Golgi bodies to apoplasts and/or vacuoles. PLA2β has a KTEL sequence at its C-terminus, which is similar to the canonical ER-retention signal KDEL, and was shown to be localized in the ER (Seo et al., 2008). The PLA2γ and PLA2δ isoforms, which are solely expressed in pollen, are localized in the ER and/or Golgi (Kim et al., 2011). PLA2β, PLA2γ, and PLA2δ, which share high sequence homologies with each other, are expressed during pollen growth and development and play critical roles during pollen germination and tube growth (Kim et al., 2011). In contrast to PLA2γ and PLA2δ, PLA2β is expressed in tissues such as actively growing leaves and elongating stems, and regulates shoot cell elongation and stem gravitropism, likely as a downstream component of auxin signaling (Lee et al., 2003). In addition, PLA2β is expressed in guard cells in response to light and modulates light-induced stomatal opening (Seo et al., 2008).
PLA2α localizes primarily at Golgi bodies in actively growing young leaves but translocates to apoplasts as the leaves become mature. In root tissues, PLA2α localizes at Golgi bodies (Lee et al., 2010). Localization of PLA2α in Golgi bodies was confirmed by co-localization assay with a Golgi body marker. Studies in animal cells indicate that lysophospholipids, which are generated by PLA2, modulate retrograde trafficking and the cisternal structure of the Golgi complex by modifying membrane tubule formation (de Figueiredo et al., 1998; Brown et al., 2003). As in animal cells, PLA2 at Golgi bodies in plant cells may play an important role in the intracellular trafficking of proteins. Membrane fusion, formation, and intracellular trafficking at the Golgi bodies are prominent processes during active growth stages of plant tissues. Consistent with this hypothesis, Golgi-localized PLA2α in root hairs appeared to act in the trafficking of PIN proteins (Lee et al., 2010). In actively growing young leaf tissues, PLA2α localized at Golgi bodies may facilitate growth and development by mediating vesicular trafficking.
Once leaf tissues are mature, PLA2α translocates to the apoplasts by way of Golgi bodies. Among the four Arabidopsis PLA2 paralogs, only PLA2α translocates to the apoplasts of mature leaves. The reason for PLA2α movement from ER and Golgi bodies to apoplasts in the mature leaves is unknown. It could be speculated that as leaves are mature the demand for PLA2α activity diminishes in ER and Golgi bodies since the demand for vesicular trafficking for active growth decreases. If so, PLA2α may rather translocalize to the apoplasts, probably, in order to fulfill some other mission in the apoplasts of the mature leaves. Translocation of PLA2α is supported by its unique enzyme characteristics. The optimal pH range for PLA2α activity is quite broad compared with other PLA2 paralogs, from pH 6 to 11 (Lee et al., 2005), so that PLA2α may fully be functional not only in the ER and Golgi bodies but also in the apoplasts. In contrast, PLA2β, which is localized in the ER, has a narrow range of optima from pH 6 to 7, whereas PLA2γ and PLA2δ, which are localized in the ER/Golgi bodies and ER of pollens, respectively, have optima at pH 7–9 and pH 8–9, respectively (Lee et al., 2005).
Structurally, apoplasts are formed by a continuation of the cell walls of adjacent cells and the associated extracellular spaces. The apoplast is important for a plant's interaction with the environment and microbes. The apoplast is also a site for cell-to-cell communication. Therefore, we examined the dynamics of PLA2α secretion to apoplasts in response to bacterial attack. When Pst-avrRpm1 infects host plants, the pathogens cannot enter into the cytoplasm but remain in the apoplasts and attempt to become established. Our results indicate that PLA2α rapidly translocates to apoplasts in response to pathogen invasion. These data prompt us to speculate that PLA2α may play a certain role in the apoplasts, where host cells confront invading pathogens. Thus, it would be of interest to investigate the role of PLA2α in a variety of cellular processes, including host defense responses to pathogen attacks.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank H. J. Kim for helping us to write the manuscript. This work was supported by a 2nd Bio-Green grant (PJ009029) from Rural Development Administration, by a grant (IPET 109106-3); by a grant from the KRIBB Initiative Program; and by a Joint Research Support Project of University of Science and Technology. Krishna Kumar was supported by National Institute for International Education of Korea.
References
Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82, 259–266.
Brown, W. J., Chambers, K., and Doody, A. (2003). Phospholipase A2 (PLA2) enzymes in membrane trafficking: mediators of membrane shape and function. Traffic 4, 214–221.
Burke, J. E., and Dennis, E. A. (2009). Phospholipase A2 structure/function, mechanism, and signaling. J. Lipid Res. 50, S237–S242.
de Figueiredo, P., Drecktrah, D., Katzenellenbogen, J. A., Strang, M., and Brown, W. J. (1998). Evidence that phospholipase A2 activity is required for Golgi complex and trans Golgi network membrane tubulation. Proc. Natl. Acad. Sci. U.S.A. 95, 8642–8647.
Froidure, S., Canonne, J., Daniel, X., Jauneau, A., Briere, C., Roby, D., and Rivas, S. (2010). AtsPLA2-alpha nuclear relocalization by the Arabidopsis transcription factor AtMYB30 leads to repression of the plant defense response. Proc. Natl. Acad. Sci. U.S.A. 107, 15281–15286.
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS fusions: β-glucuronidase a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907.
Kim, H. J., Ok, S. H., Bahn, S. C., Jang, J., Oh, S. A., Park, S. K., Twell, D., Ryu, S. B., and Shin, J. S. (2011). Endoplasmic reticulum- and Golgi-localized phospholipase A2 plays critical roles in Arabidopsis pollen development and germination. Plant Cell 23, 94–110.
Kirik, A., and Mudgett, M. B. (2009). SOBER1 phospholipase activity suppresses phosphatidic acid accumulation and plant immunity in response to bacterial effector AvrBsT. Proc. Natl. Acad. Sci. U.S.A. 106, 20532–20537.
Kwon, C., Bednarek, P., and Schulze-Lefert, P. (2008). Secretory pathways in plant immune responses. Plant Physiol. 147, 1575–1583.
Lee, H. Y., Bahn, S. C., Kang, Y. M., Lee, K. H., Kim, H. J., Noh, E. K., Palta, J. P., Shin, J. S., and Ryu, S. B. (2003). Secretory low molecular weight phospholipase A2 plays important roles in cell elongation and shoot gravitropism in Arabidopsis. Plant Cell 15, 1990–2002.
Lee, H. Y., Bahn, S. C., Shin, J. S., Hwang, I., Back, K., Doelling, J. H., and Ryu, S. B. (2005). Multiple forms of secretory phospholipase A2 in plants. Prog. Lipid Res. 44, 52–67.
Lee, O., Kim, S., Kim, H., Hong, J., Ryu, S., Lee, S., Ganguly, A., and Cho, H. (2010). Phospholipase A2 is required for PIN-FORMED protein trafficking to the plasma membrane in the Arabidopsis root. Plant Cell 22, 1812–1825.
Liao, H. L., and Burns, J. K. (2010). Light controls phospholipase A2alpha and beta gene expression in Citrus sinensis. J. Exp. Bot. 61, 2469–2478.
Mansfeld, J. (2009). Plant phospholipases A2: perspectives on biotechnological applications. Biotechnol. Lett. 31, 1373–1380.
Mansfeld, J., and Ulbrich-Hofmann, R. (2007). Secretory phospholipase A2-α from Arabidopsis thaliana: functional parameters and substrate preference. Chem. Phys. Lipids 150, 156–166.
Ryu, S. B. (2004). Phospholipid-derived signaling mediated by phospholipase A in plants. Trends Plant Sci. 9, 229–235.
Ryu, S. B., Lee, H. Y., Doelling, J. H., and Palta, J. P. (2005). Characterization of a cDNA encoding Arabidopsis secretory phospholipase A2-alpha, an enzyme that generates bioactive lysophospholipids and free fatty acids. Biochim. Biophys. Acta 1736, 144–151.
Schaloske, R., and Dennis, E. (2006). The phospholipase A2 superfamily and its group numbering system. Biochim. Biophys. Acta 1761, 1246–1259.
Scherer, G. F., Zahn, M., Callis, J., and Jones, A. M. (2007). A role for phospholipase A in auxin-regulated gene expression. FEBS Lett. 581, 4205–4211.
Seo, J., Lee, H. Y., Choi, H., Choi, Y., Lee, Y., Kim, Y. W., Ryu, S. B., and Lee, Y. (2008). Phospholipase A2-b mediates light-induced stomatal opening in Arabidopsis. J. Exp. Bot. 59, 3587–3594.
Sup Yun, H., Panstruga, R., Schulze-Lefert, P., and Kwon, C. (2008). Ready to fire: secretion in plant immunity. Plant Signal. Behav. 3, 505–508.
Wang, D., Weaver, N. D., Kesarwani, M., and Dong, X. (2005). Induction of protein secretory pathway is required for systemic acquired resistance. Science 308, 1036–1040.
Keywords: phospholipase A2, translocation, apoplast, bacterial infection, subcellular localization
Citation: Jung J, Kumar K, Lee HY, Park Y-I, Cho H-T and Ryu SB (2012) Translocation of phospholipase A2α to apoplasts is modulated by developmental stages and bacterial infection in Arabidopsis. Front. Plant Sci. 3:126. doi:10.3389/fpls.2012.00126
Received: 22 December 2011; Accepted: 25 May 2012;
Published online: 18 June 2012.
Edited by:
Xuemin Wang, University of Missouri-St Louis and Donald Danforth Plant Science Center, USAReviewed by:
Stephan Pollmann, Universidad Politécnica de Madrid, SpainIngo Heilmann, Martin-Luther-University Halle-Wittenberg, Germany
Ying Gu, Pennsylvania State University, USA
Copyright: © 2012 Jung, Kumar, Lee, Park, Cho and Ryu. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Stephen Beungtae Ryu, Environmental Biotechnology Research Center, Korea Research Institute of Bioscience and Biotechnology (KRIBB), 125 Gwahak-ro, Yuseong-gu, Daejeon 306-809, Korea. e-mail: sbryu@kribb.re.kr
 Jihye Jung1,2
Jihye Jung1,2