- Department of Biochemistry, Biophysics, and Molecular Biology, Iowa State University, Ames, IA, USA
The hemicellulosic polysaccharide xyloglucan (XyG), found in the primary cell walls of most plant tissues, is important for structural organization of the cell wall and regulation of growth and development. Significant recent progress in structural characterization of XyGs from different plant species has shed light on the diversification of XyG during plant evolution. Also, identification of XyG biosynthetic enzymes and examination of their interactions suggests the involvement of a multiprotein complex in XyG biosynthesis. This mini-review presents an updated overview of the diversity of XyG structures in plant taxa and recent findings on XyG biosynthesis.
Structural Diversity of the Xyloglucans
Xyloglucan (XyG) is a hemicellulosic polysaccharide found in all land plants (Popper and Fry, 2003, 2004; Popper, 2008; Sarkar et al. 2009) in varying amounts; for example, XyG is a major hemicellulosic component of the primary cell wall of flowering plants (up to 25%), but a minor, sometimes barely detectable, constituent of grasses (less than 2%; Hsieh and Harris, 2009). XyGs have a β-1,4-glucan backbone that is highly branched, with characteristic repetitive patterns of α-Xyl residues linked to glucose at the O-6 position. The side chain xylosyl residues can be further substituted with different mono-, di-, or trisaccharides; the pattern of these substitutions produces the broad diversity of XyG structural motifs that are present in different plant species (Hoffman et al., 2005; Pena et al., 2008; Hsieh and Harris, 2009). A single-letter nomenclature was introduced by Fry et al. (1993), to describe the XyG backbone substitution pattern. For example, the letter G indicates an unbranched Glc residue, and X denotes the α-D-Xyl-(1 → 6)-β-D-Glc motif in the xylosylated glucan backbone. Xyl residues can carry a β-D-Gal (L motif), an α-L-Ara (S motif), or a β-D-Xyl (U motif; Hoffman et al., 2005). In turn, Gal residues in the L side chain can be linked to an α-L-Fuc residue (F motif; Hoffman et al., 2005), or an α-L-Gal (J motif; Hantus et al., 1997); also, the Ara residue in the S side chain can carry a β-L-Ara (T motif; York et al., 1996). In addition, Pena et al. (2008) found that avascular plants, such as mosses and liverworts, can form XyG with a 2,4-linked Xyl residue, where xylosyl can be substituted with a β-D-GalA and a β-D-Gal (P motif), with an α-L-Ara and a β-D-Gal (M motif), an α-L-Ara and the disaccharide β-D-Gal-(1 → 6)-β-D-GalA (N motif), or the disaccharide β-D-Gal-(1 → 6)-β-D-GalA and a β-D-Gal at the O-2 and O-4 positions, respectively.
Different patterns and types of XyG subunits can be found in different species and different tissues. Most vascular seed-bearing plants synthesize XXXG-type XyG (O’Neill and York, 2003; Hoffman et al., 2005), but grasses and some lamiids produce XXGG- and XXGGG-type XyG with fewer Xyls on the glucan backbone (Gibeaut et al., 2004; Hoffman et al., 2005). Typically, XXXG-type XyG comprises XXXG, XXFG, XXLG, and XLFG subunits, which are present in different proportions depending on the plant tissue and developmental stage (Pauly et al., 2001; Obel et al., 2009). In many flowering and non-flowering plants, the Fuc residue is linked to β-D-Gal at the O-2 position, while the XyG in Equisetum and Selaginella has the Fuc linked to an α-L-Ara residue at the O-2 position (E motif; Pena et al., 2008). It was proposed that fucosylated XyG was first synthesized in a common ancestor of hornworts and vascular plants (Pena et al., 2008). Gal residues in XXLG, XXFG, and XLFG can carry acetyl groups as was found in Arabidopsis and sycamore XyG (Kiefer et al., 1989; Gille et al., 2011).
The commelinid monocotyledons have predominantly non-fucosylated XXGn-type XyG. Thus, the Zingiberales and Commelinales have both XXGn and XXXG core motifs with few XXFG units, and no XLFG (Hsieh and Harris, 2009). In the Poales, the Poaceae have exclusively XXGn-type XyG without Fuc, but the other families contain either the mixed type XyG with XXXG and XXGn core motifs or only XXXG and XXFG motifs, but no XLFG (Hsieh and Harris, 2009). Frequently, in XXGG- or XXGGG-type XyG, one or two unbranched Glc residues have acetyl groups instead of the α-Xyl (Hoffman et al., 2005).
del Bem and Vincentz (2010) constructed an evolutionary model of the emergence of XyG-related genes in Viridiplantae, proposing a stepwise increase in XyG branching complexity starting from XyG-like molecules that contained only Glc and Xyl, which are found in streptophyte algae, to galactosylated motifs, which emerged in embryophytes, and finally to fucosylated XyGs, which emerged in the last common ancestors of spermatophytes. Although XyG has never been directly released from algae cell walls, some indirect evidence indicates that certain green algae contain XyG-like polysaccharide (VanSandt et al., 2007; Fry et al., 2008), additionally, XyG epitopes were immunologically detected in some members of evolutionarily advanced charophycean green algae orders (Sorensen et al., 2010, 2011). Since liverworts, which are believed to be the oldest extant land plant family (Qiu et al., 2006), have XXGG- and XXGGG-type XyG, it was proposed that XXXG-type motifs evolved later in hornworts and many vascular plants (Pena et al., 2008). Demonstration in hornworts of the presence of fucosylated XyG with similarities to seed-bearing plant XyG motifs allowed Pena et al. (2008) to propose that in some Lamiids and grasses, the genes encoding XyG-specific fucosyltransferases may have been eliminated completely, or are expressed only in specific cells or conditions.
Xyloglucan Biosynthesis
Xyloglucan biosynthesis requires glucan synthase to form the glucan backbone and requires multiple different types of glycosyl transferases to decorate the glucan chain to produce the broad diversity of XyG side chains found in various plants. Considering the high specificity of glycosyl transferases (Keegstra and Raikhel, 2001), the formation of each linkage is believed to require a distinct transferase; therefore, a combination of at least one (1,4)-β-glucan synthase, three (1,6)-α-xylosyltransferases, two (1,2)-β-galactosyltransferases, and one (1,2)-α-fucosyltransferase is needed to assemble a complete XLFG subunit.
The presence of other XyG motifs discovered in different taxa, and briefly described in the first section, implicates involvement of other types of glycosyltransferases in various plant species; these transferases may include XyG specific arabinosyltransferases, galacturonyltransferases, additional galactosyltransferases, and xylosyltransferases that would elongate diverse XyG side chains. For example, in Arabidopsis, XyG biosynthesis requires at least five types of enzymatic activities: UDP-Glc-dependent glucan synthase to synthesize the glucan chain, UDP-Xyl-dependent xylosyltransferases to transfer Xyl onto a specific Glc in the glucan chain, UDP-Gal-dependent galactosyltransferases to transfer Gal onto Xyl and elongate side chains attached to the second and third Glc in the XyG subunit, GDP-Fuc-dependent fucosyltransferase to transfer Fuc onto Gal in the side chain attached to the third Glc in the XyG subunit, and finally XyG specific acetyltransferase to attach the acetyl group to Gal.
Candidate genes encoding all these enzymes have been identified and partially characterized using a combination of biochemical and genetic approaches (Figure 1). For example, the amino acid sequence of fucosyltransferase (FUT1) purified from pea was used to identify Arabidopsis FUT1 from the GT37 gene family (Perrin et al., 1999), and heterologous expression demonstrated that Arabidopsis and pea FUT1 genes encode proteins with XyG fucosyltransferase activity (Perrin et al., 1999; Faik et al., 2000). Complete elimination of fucosylated XyG subunits in the Arabidopsis FUT1 T-DNA knock-out mutant suggests that XyG specific fucosyltransferase activity in Arabidopsis is encoded by a single gene (Keegstra and Cavalier, 2011).
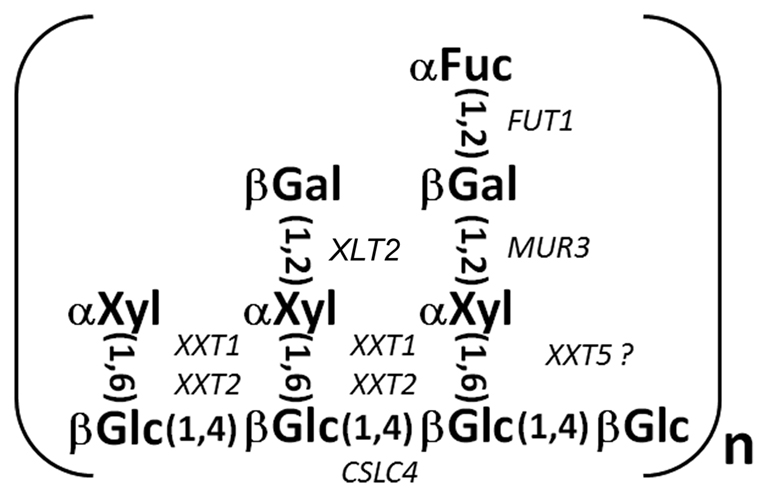
Figure 1. The structure of the XLFG XyG subunit. Glycosyl transferases known to form particular linkages are shown in corresponding positions. The catalytic activity of XXT5 and XLT2 has not been confirmed.
XyG galactosyltransferases have also been identified. Discovered in a screen for mutants with aberrant cell wall formation, Arabidopsis thaliana MUR3 (Reiter et al., 1997) encodes a XyG galactosyltransferase that adds galactose specifically to the third xylosyl residue, forming an XXLG subunit (Madson et al., 2003). More recently, a second XyG galactosyltransferase was discovered by RNA-Seq analysis of developing nasturtium seeds and then confirmed by mutation of the Arabidopsis ortholog (At5g62220). The Arabidopsis gene, named XyG L-side chain galactosyltransferase (XLT2) showed no redundancy with MUR3, and is required for galactosylation of the second xylose in the XyG subunit (Jensen et al., 2012).
A gene family (CAZy GT34Family), containing seven genes spread among three clades in Arabidopsis, was identified and predicted to encode XyG xylosyltransferases (Faik et al., 2002). Heterologous expression of two of those genes, XXT1 and XXT2, demonstrated that the encoded proteins have xylosyltransferase activity (Cavalier and Keegstra, 2006). The studies also demonstrated that XXT1 and XXT2 have the same substrate-acceptor specificity, catalyzing the substitution of two glycosidic residues in adjacent positions, and thereby generating a GGXXGG structure. Also, xxt1 xxt2 double mutant plants lack detectable XyG in their cell walls (Cavalier et al., 2008), confirming that XXT1 and XXT2 are XyG xylosyltransferases that are essential for XyG formation. Application of reverse genetics demonstrated that another member of the GT34 gene family, XXT5, is also involved in XyG biosynthesis, although its activity has not been demonstrated in vitro (Zabotina et al., 2008). The lack of detectable XyG in the xxt1 xxt2 double mutant plants challenges conventional models for the functional organization of the primary cell wall and also existing assumptions about linkage-specific enzyme relationships in polysaccharide biosynthesis. The xxt1 and xxt2 single mutant plants each have only a slight decrease in XyG content, but the xxt5 single mutant has a 50% reduction in XyG content and the XyG that is made in the xxt5 mutant plant shows an altered subunit composition (Zabotina et al., 2008). However, the knock-out of XXT5 does not eliminate xylosylation of any particular glucose in the XyG backbone, which suggests that the absence of XXT5 protein is compensated for, at least in part, by the presence of the other two xylosyltransferases. Thus, the ability of XXT1 and XXT2 to partially compensate for the lack of XXT5 in synthesizing fully xylosylated XyG subunits raises questions about their specificity with respect to which glucose in the glucan backbone is the target of their activity. The studies of two additional double mutants, xxt1 xxt5 and xxt2 xxt5, as well as a triple mutant line, xxt1 xxt2 xxt5, revealed further complexity in the functional relationship of XXT proteins. A combination of biochemical and immunological analyses (Zabotina et al., 2012) demonstrated that either XXT1 or XXT2 alone is sufficient to synthesize the complete set of XyG subunits, although in significantly lower amounts compared with plants that have XXT5 in addition to XXT1 and XXT2. Thus, either XXT1 or XXT2 is capable of adding all three of the xylosyl residues present in XyG; this confirms earlier in in vitro experiments (Cavalier and Keegstra, 2006). However in vivo, the efficiency of XXT1 or XXT2 when functioning alone is significantly lower than when the three proteins are present together. Also, although the XXT5 protein itself does not seem to be capable of adding xylosyl residues to the XyG backbone in vivo, the lack of this protein causes the most dramatic impact on XyG biosynthesis.
The XXT1, XXT2, and XXT5 double and single mutant phenotypes indicate that these loci encode the major XyG xylosyltransferases. Two members of the Arabidopsis GT34 gene family (At4g37690 and At2g22900) are closely related to the galactomannan galactosyltransferases identified by Edwards et al. (1999), which suggests that they encode galactosyltransferases (Keegstra and Cavalier, 2011). Two other members of this family (At1g18690 and At1g74380) are expressed at very low levels in all tissues that were examined (http://www.weigelworld.org/resources/microarray/AtGenExpress/; Schmid et al., 2005) and are therefore unlikely to contribute significantly to XyG biosynthesis. This supports the hypothesis that XXT1, XXT2, and XXT5 are the main XyG xylosyltransferases that synthesize XyG, at least in the major plant tissues. Analysis of microarray data showed that these three XXT genes are expressed in all tissues that were analyzed, but have different levels of expression. In the majority of tissues, XXT2 has approximately twofold higher expression than XXT1 and XXT5, but XXT2 and XXT5 have comparable expression levels except in a few specialized tissues such as stamen and root pericycle, where XXT1 has the highest expression level of the three genes.
The members of the GT2 gene family, Arabidopsis cellulose synthase-like C genes (CSLC4, CSLC5, and CSLC6) are believed to be involved in XyG biosynthesis, being implicated in glucan backbone synthesis. Initially, CSLC4 was identified as a candidate gene for the (1,4)-β-glucan synthase in nasturtium and Arabidopsis (Cocuron et al., 2007). Heterologously expressed in Pichia pastoris cells, CSLC4 produces cellodextrins. Moreover, when XXT1 was co-expressed in the same cells, longer chains were detected, indicating that XXT1 can assist CSLC4 in glucan synthesis. Since yeast cells cannot produce UDP-xylose, xylosylation of the synthesized glucan oligomers was not observed. Later, reverse-genetic studies suggested that two other genes, CSLC5 and CSLC6, are also involved in XyG biosynthesis (Cavalier and Keegstra, 2010). Among these three genes, CSLC4 has higher levels of expression in all Arabidopsis tissues and also is expressed in the same tissues and with the same developmental timing as the XXTs, but expression of CSLC5 and CSLC6 is limited to specific tissues (Schmid et al., 2005).
In Arabidopsis XyG, acetyl groups are exclusively linked to galactosyl residues. Recently, the putative XyG specific acetyltransferases, AXY4 and AXY4L, were discovered in Arabidopsis using a forward genetic screen (Gille et al., 2011), although their catalytic activity and substrate specificity have yet to be confirmed. It is also unclear whether AXY4 attaches acetyl groups to all galactosyl residues or only to some of them, and whether this happens during XyG biosynthesis or after XyG is completely synthesized. When AXY4 was overexpressed in wild type Arabidopsis, cell wall XyG still contained unacetylated galactoses (Gille et al., 2011), suggesting the possible involvement of other AXY proteins. Whether AXY4 can interact with any of the proteins involved in XyG biosynthesis remains to be investigated as part of an exploration of the biological role of XyG acetylation.
Does a XyG Synthase Complex Exist?
Identification of glycosyl transferases that can potentially fully decorate the XyG glucan backbone and synthesize the complete XyG structure (Figure 1) stimulated further investigations to understand their functional organization in the Golgi. The few available examples suggest that glycosyl transferases localized in the Golgi might function in multiprotein complexes (Atmodjo et al., 2011; Harholt et al., 2012). Indeed, recent data suggest that proteins involved in XyG biosynthesis are co-localized in multiprotein homo- and hetero-complexes and most likely interact through their catalytic domains. For example, BiFC assays using Arabidopsis protoplasts transiently expressing transferases fused with the C and N parts of YFP demonstrated approximate co-localization of XyG xylosyltransferases in two hetero-complexes, XXT2-XXT5 and XXT1-XXT2; XXT2 was also shown to form an XXT2-XXT2 homo-complex (Chou et al., 2012). Using the same approach, XXT5 and CSLC4 were shown to form a hetero-complex, XXT5-CSLC4. Results from in vitro pull-down experiments using recombinant proteins expressed without transmembrane domains confirmed the interactions between XXT2 and XXT5, and XXT1 and XXT2, suggesting that these proteins can physically interact through their catalytic domains (Chou et al., 2012).
A recent study (Davis et al., 2010) suggested that CSLC4 is positioned in the Golgi membrane with its active loop and both C- and N-termini protruding to the cytosolic side of the Golgi. BiFC assays showed that CSLC4 forms homo-complexes and switching the C and N YFP fragments fused to either the C or N-terminus of CSLC4 did not affect the intensity of reconstituted YFP fluorescence (Chou et al., 2012). This suggests that both termini are localized close to each other, while the CSLC4 active loops are positioned on the outer sides of the homo-complex, farther from each other.
Similarly, BiFC assays demonstrated that MUR3 and FUT1 also co-localize and interact with each other and with either XXT2 or CSLC4. These results have yet to be confirmed by an independent approach, but it is plausible to believe that during XyG formation, these proteins likely come into close proximity to XXTs to finalize the complete XyG structure. Whether all these proteins form a single multiprotein complex or a more dynamic structure that differs in different situations has yet to be investigated. For example, it has been suggested that the fucosyltransferase FUC1 functions independently from glucan elongation and xylosylation (Faik et al., 1997a,b). After using pea microsomal fractions to study XyG formation in vitro, the authors proposed that fucosylation of galactosylated heptasaccharide occurs after complete formation of the glucan backbone. Additionally, Chevalier et al. (2010), using Arabidopsis XXT1, MUR3, and FUT1 fused with GFP and expressed in N. tabacum BY-2 cells, showed that XXT1 and MUR3 localized predominantly in Golgi cis and medial cisternae, respectively, but FUT1 was detected in trans cisternae. This localization supports the hypothesis that the fucosyltransferase may be spatially independent. However, it is worth noting that tobacco XyG does not contain terminal fucose in its subunits; therefore Arabidopsis FUT1 may not have been able to establish the proper interactions to be correctly localized in N. tabacum Golgi. Further elucidation of the structure of the XyG synthase complex is needed to understand its functions, and the regulation of XyG biosynthesis. It is hypothesized (Gunl et al., 2011) that XyG synthesized in Golgi has, most likely, an XXXGXXFG repeating pattern and the diversity of XyG structure is created after its deposition into the cell wall. This regular structure would derive most efficiently from cooperative action of multiple XyG biosynthetic enzymes organized in a multiprotein complex in the Golgi, rather than from spatially and functionally independent enzymes.
Concluding Remarks
During the last decade, significant progress has been made in revealing the proteins involved in XyG biosynthesis, which, together with detailed structural studies, makes this important cell wall polysaccharide a good model for examining the mystery of polysaccharide evolution, formation, and modification. Accumulating evidence demonstrates that the evolution of polysaccharide structural complexity during land colonization produced key adaptations, including cell wall mechanical strengthening needed to support plants in the absence of water buoyancy and to protect them against biotic and abiotic environmental stresses. To provide this complexity, plants evolved complex and dynamic polysaccharide synthesizing machinery localized in the Golgi. Enzymes involved in polysaccharide biosynthesis are most likely organized in multiprotein complexes, the structure of which is probably dynamic, varying in different plant tissues and throughout plant development. The structural organization and regulation of XyG and other polysaccharide synthase complexes remain to be uncovered in the future.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was partially supported by the NSF grant (#1121163; December 2011–2015).
References
Atmodjo, M. A., Sakuragi, Y., Zhu, X., Burrell, A. J., Mohanty, S. S., Atwood, J. A. III, Orlando, R., Scheller, H. V., and Mohnen, D. (2011). Galacturonosyltransferase (GAUT)1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan:galacturonosyltransferase complex. Proc. Natl. Acad. Sci. U.S.A. 108, 20225–20230.
Cavalier, D., and Keegstra, K. (2010). “Members of Arabidopsis cellulose synthase-like C (CSLC) family are involved in xyloglucan biosynthesis,” in Book of Abstracts of XII Cell Wall Meeting, Porto.
Cavalier, D. M., and Keegstra, K. (2006). Two xyloglucan xylosyltransferases catalyze the addition of multiple xylosyl residues to cellohexaose. J. Biol. Chem. 281, 34197–34207.
Cavalier, D. M., Lerouxel, O., Neumetzler, L., Yamauchi, K., Reinecke, A., Freshour, G., Zabotina, O. A., Hahn, M. G., Burgert, I., Pauly, M., Raikhel, N. V., and Keegstra, K. (2008). Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 20, 1519–1537.
Chevalier, L., Bernard, S., Ramdani, Y., Lamour, R., Bardor, M., Lerouge, P., Follet-Gueye, M.-L., and Driouich, A. (2010). Subcompartment localization of the side chain xyloglucan-synthesizing enzymes within Golgi stacks of tobacco suspension-cultured cells. Plant J. 64, 977–989.
Chou, Y.-H., Pogorelko, G., and Zabotina, O. A. (2012). Xyloglucan xylosyltransferases XXT1, XXT2, XXT5, and the glucan synthase CSLC4 form Golgi-localized multiprotein complexes. Plant Physiol. doi:10.1104/pp.112.199356
Cocuron, J. C., Lerouxel, O., Drakakaki, G., Alonso, A. P., Liepman, A. H., Keegstra, K., Raikhel, N., and Wilkerson, C. G. (2007). A gene from the cellulose synthase-like C family encodes a beta-1,4 glucan synthase. Proc. Natl. Acad. Sci. U.S.A. 104, 8550–8555.
Davis, J., Brandizzi, F., Liepman, A. H., and Keegstra, K. (2010). Arabidopsis mannan synthase CSLA9 and glucan synthase CSLC4 have opposite orientations in the Golgi membrane. Plant J. 64, 1028–1037.
del Bem, L., and Vincentz, M. (2010). Evolution of xyloglucan-related genes in green plants. BMC Evol. Biol. 10, 341. doi:10.1186/1471-2148-10-341
Edwards, M. E., Dickson, C. A., Chengappa, S., Sidebottom, C., Gidley, M. J., and Reid, J. S. G. (1999). Molecular characterisation of a membrane-bound galactosyltransferase of plant cell wall matrix polysaccharide biosynthesis. Plant J. 19, 691–697.
Faik, A., Bar-Peled, M., Derocher, A. E., Zeng, W., Perrin, R. M., Wilkerson, C., Raikhel, N. V., and Keegstra, K. (2000). Biochemical characterization and molecular cloning of an alpha-1,2-fucosyltransferase that catalyzes the last step of cell wall xyloglucan biosynthesis in pea. J. Biol. Chem. 275, 15082–15089.
Faik, A., Chileshe, C., Sterling, J., and Maclachlan, G. (1997a). Xyloglucan galactosyl- and fucosyltransferase activities from pea epicotyl microsomes. Plant Physiol. 114, 245–254.
Faik, A., Desveaux, D., and Maclachlan, G. (1997b). Xyloglucan galactosyl- and fucosyl-transferase activities in the cotyledons of developing nasturtium seeds. Plant Physiol. 114, 716–716.
Faik, A., Price, N. J., Raikhel, N. V., and Keegstra, K. (2002). An Arabidopsis gene encoding an alpha-xylosyltransferase involved in xyloglucan biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 99, 7797–7802.
Fry, S. C., Mohler, K. E., Nesselrode, B. H., and Frankova, L. (2008). Mixed-linlage bet-glucan: xyloglucan endotransglucosylase, a novel wall-remodelling enzyme from Equisetum (horsetails) and charophytic algae. Plant J. 55, 240–252.
Fry, S. C., York, W. S., Albersheim, P., Darvill, A., Hayashi, T., Joseleau, J. P., Kato, Y., Lorences, E. P., Maclachlan, G. A., McNeil, M., Mort, A. J., Reid, J. S. G., Seitz, H. U., Selvendran, R. R., Voragen, A. G. J., and White, A. R. (1993). An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Physiol. Plant. 89, 1–3.
Gibeaut, D. M., Pauly, M., Bacic, T., and Fincher, G. B. (2004). Changes in cell wall polysaccharides in developing barley (Hordeum vulgare) coleoptiles. Planta 221, 729–738.
Gille, S., de Souza, A., Xiong, G., Benz, M., Cheng, K., Schultink, A., Reca, I. B., and Pauly, M. (2011). O-acetylation of arabidopsis hemicellulose xyloglucan requires AXY4 or AXY4L, proteins with a TBL and DUF231 domain. Plant Cell 23, 4041–4053.
Gunl, M., Neumetzler, L., Kraemer, F., de Souza, A., Schultink, A., Pena, M., York, W. S., and Pauly, M. (2011). AXY8 encodes an alpha-fucosidase, underscoring the importance of apoplastic metabolism on the fine structure of Arabidopsis cell wall polysaccharides. Plant Cell 23, 4025–4040.
Hantus, J., Pauly, M., Darvill, A. G., Albersheim, P., and York, W. S. (1997). Structural characterization of novel L-galactose-containing oligosaccharide subunits of jojoba seed xyloglucans. Carbohydr. Res. 304, 11–20.
Harholt, J., Jensen, J. K., Verhertbruggen, Y., Sogaard, C., Bernard, S., Nafisi, M., Poulsen, C. P., Geshi, N., Sakuragi, Y., Driouich, A., Knox, J. P., and Scheller, H. V. (2012). ARAD proteins associated with pectic Arabinan biosynthesis form complexes when transiently overexpressed in planta. Planta. doi:10.1007/s00425-012-1592-1593
Hoffman, M., Jia, Z. H., Pena, M. J., Cash, M., Harper, A., Blackburn, A. R., Darvill, A., and York, W. S. (2005). Structural analysis of xyloglucans in the primary cell walls of plants in the subclass Asteridae. Carbohydr. Res. 340, 1826–1840.
Hsieh, Y. S. Y., and Harris, P. J. (2009). Xyloglucans of monocotyledons have diverse structures. Mol. Plant 2, 943–965.
Jensen, J. K., Schultink, A., Keegstra, K., Wilkerson, C. G., and Pauly, M. (2012). RNA-Seq analysis of developing nasturtium seeds (Tropaeolum majus): identification and characterization of an additional, galactosyltransferase involved in xyloglucan biosynthesis. Mol. Plant. doi:10.1093/mp/sss032
Keegstra, K., and Cavalier, D. (2011). “Glycosyltransferases of the GT34 and GT37 families,” in Annual Plant Reviews, Vol. 41, Plant Polysaccharides: Biosynthesis and Bioengineering, ed. P. Ulskov (Oxford: Blackwell Publishing), 235–250.
Keegstra, K., and Raikhel, N. (2001). Plant glycosyltransferases. Curr. Opin. Plant Biol. 4, 219–224.
Kiefer, L. L., York, W. S., Darvill, A. G., and Albersheim, P. (1989). Structure of plant-cell walls.27. xyloglucan isolated from suspension-cultured sycamore cell-walls is o-acetylated. Phytochemistry 28, 2105–2107.
Madson, M., Dunand, C., Li, X., Verma, R., Vanzin, G. F., Caplan, J., Shoue, D. A., Carpita, N. C., and Reiter, W. D. (2003). The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 15, 1662–1670.
Obel, N., Erben, V., Schwarz, T., Kuhnel, S., Fodor, A., and Pauly, M. (2009). Microanalysis of plant cell wall polysaccharides. Mol. Plant 2, 922–932.
O’Neill, M. A., and York, W. S. (2003). “The composition and structure of plants primary cell walls,” in The Plant Cell Wall, ed. J. K. C. Rose (Oxford: Blackwell), 1–54.
Pauly, M., Qin, Q., Greene, H., Albersheim, P., Darvill, A., and York, W. S. (2001). Changes in the structure of xyloglucan during cell elongation. Planta 212, 842–850.
Pena, M. J., Darvill, A. G., Eberhard, S., York, W. S., and O’Neill, M. A. (2008). Moss and liverwort xyloglucans contain galacturonic acid and are structurally distinct from the xyloglucans synthesized by hornworts and vascular plants. Glycobiology 18, 891–904.
Perrin, R. M., Derocher, A. E., Bar-Peled, M., Zeng, W., Norambuena, L., Orellana, A., Raikhel, N. V., and Keegstra, K. (1999). Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science 284, 1976–1979.
Popper, Z. A. (2008). Evolution and diversity of green plant cell walls. Curr. Opin. Plant Biol. 11, 1–7.
Popper, Z. A., and Fry, S. C. (2003). Primary cell wall composition of bryophytes and charophytes. Ann. Bot. 91, 1–12.
Popper, Z. A., and Fry, S. C. (2004). Primary cell wall composition of pteridophytes and spermatophytes. New Phytol. 64, 165–174.
Qiu, Y.-L., Li, L., Wang, B., Chen, Z., Knoop, V., Groth-Malonek, M., Dombrovska, O., Lee, J., Kent, L., Rest, J., Estabrook, G. F., Hendry, T. A., Taylor, D. W., Testa, C. M., Ambros, M., Crandall-Stotler, B., Duff, R. J., Stech, M., Frey, W., Quandt, D., and Davis, C. C. (2006). The deepest divergences in land plants inferred from phylogenomic evidence. Proc. Natl. Acad. Sci. U.S.A. 103, 15511–15516.
Reiter, W. D., Chapple, C., and Somerville, C. R. (1997). Mutants of Arabidopsis thaliana with altered cell wall polysaccharide composition. Plant J. 12, 335–345.
Sarkar, P., Bosneaga, E., and Auer, M. (2009). Plant cell walls throughout evolution: towards a molecular understanding of their design principles. J. Exp. Bot. 60, 3615–3635.
Schmid, M., Davison, T. S., Henz, S. R., Pape, U. J., Demar, M., Vingron, M., Scholkopf, B., Weigel, D., and Lohmann, J. U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37, 501–506.
Sorensen, I., Domozych, D., and Willats, W. G. T. (2010). How have plant cell walls evolved? Plant Physiol. 153, 366–372.
Sorensen, I., Pettolino, F. A., Bacic, A., Ralph, J., Lu, F., O’Neill, M. A., Fei, Z., Rose, J. K. C., Domozych, D. S., and Willats, W. G. T. (2011). The charophycean green algae provide insights into the early origins of plant cell walls. Plant J. 68, 201–211.
Van Sandt, V. S., Stieperaere, H., Guisez, Y., Verbelen, J. P., and Vissenberg, K. (2007). XET activity is found near sites of growth and cell elongation in bryphytes and some green algae: new insights into the evolution of primary cell wall elongation. Ann. Bot. 99, 39–51.
York, W. S., Kolli, V. S. K., Orlando, R., Albersheim, P., and Darvill, A. G. (1996). The structure of arabinoxyloglucans produced by solanaceous plants. Carbohydr. Res. 285, 99–128.
Zabotina, O. A., Avci, U., Cavalier, D., Pattathil, S., Chou, Y.-H., Eberhard, S., Danhof, L., Keegstra, K., and Hahn, M. G. (2012). Mutations in multiple XXT genes of Arabidopsis reveal the complexity of xyloglucan biosynthesis. Plant Physiol. doi:10.1104/pp.112.198119
Keywords: xyloglucan structure, biosynthesis, glycosyltransferases, multiprotein complex
Citation: Zabotina OA (2012) Xyloglucan and its biosynthesis. Front. Plant Sci. 3:134. doi: 10.3389/fpls.2012.00134
Received: 15 March 2012; Accepted: 05 June 2012
Published online: 25 June 2012.
Edited by:
Seth DeBolt, University of Kentucky, USAReviewed by:
Jocelyn Kenneth Campbell Rose, Cornell University, USABreeanna Urbanowicz, University of Georgia, USA
Copyright: © 2012 Zabotina. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Olga A. Zabotina, Department Biochemistry, Biophysics, and Molecular Biology, Iowa State University, 3212 Molecular Biology Building, Ames, IA 50011, USA. e-mail: zabotina@iastate.edu
