- Plant Molecular Biology (Botany), Department Biology I, Ludwig Maximilians University, Munich, Germany
The concept of retrograde signaling posits that signals originating from chloroplasts or mitochondria modulate the expression of nuclear genes. A popular scenario assumes that signaling factors are generated in, and exported from the organelles, then traverse the cytosol, and act in the nucleus. In this scenario, which is probably over-simplistic, it is tacitly assumed that the signal is transferred by passive diffusion and consequently that changes in nuclear gene expression (NGE) directly reflect changes in the total cellular abundance of putative retrograde signaling factors. Here, this notion is critically discussed, in particular in light of an alternative scenario in which a signaling factor is actively exported from the organelle. In this scenario, NGE can be altered without altering the total concentration of the signaling molecule in the cell as a whole. Moreover, the active transport scenario would include an additional level of complexity, because the rate of the export of the signaling molecule has to be controlled by another signal, which might be considered as the real retrograde signal. Additional alternative scenarios for retrograde signaling pathways are presented, in which the signaling molecules generated in the organelle and the factors that trigger NGE are not necessarily identical. Finally, the diverse consequences of signal integration within the organelle or at the level of NGE are discussed. Overall, regulation of NGE at the nuclear level by independent retrograde signals appears to allow for more complex regulation of NGE than signal integration within the organelle.
What is a Retrograde Signal?
Changes in the developmental or metabolic state of chloroplasts or mitochondria can have profound effects on the rest of the plant cell and involve massive changes in the transcript profiles of nuclear genes. The concept of retrograde signaling therefore posits that signals originating in chloroplasts and/or mitochondria modulate nuclear gene expression (NGE). In the classical case, the retrograde signal is generated in the organelle(s), exported, and traverses the cytosol to act in the nucleus. Several such classical retrograde signals have been tentatively identified. However, messenger roles have also been proposed for factors which display some, but not all, of the characteristics attributed to classical retrograde signals.
The Spectrum of Putative Retrograde Signals – a Current Inventory
Several candidate signaling pathways and signaling molecules have been identified during the last three decades, and their number continues to increase. However, unambiguous experimental verification of a compound as a signaling molecule is an extremely difficult task, and in no case is the status of a candidate as a signaling molecule yet secure. Therefore, instead of presenting a detailed and exhaustive description of individual candidate signaling pathways/molecules (for recent reviews see Baier and Dietz, 2005; Beck, 2005; Nott et al., 2006; Pesaresi et al., 2007; Pogson et al., 2008; Kleine et al., 2009a; Chan et al., 2010; Pfannschmidt, 2010; Inaba et al., 2011), I wish to concentrate here on some basic aspects of retrograde signaling mechanisms and their impact on the character of the response.
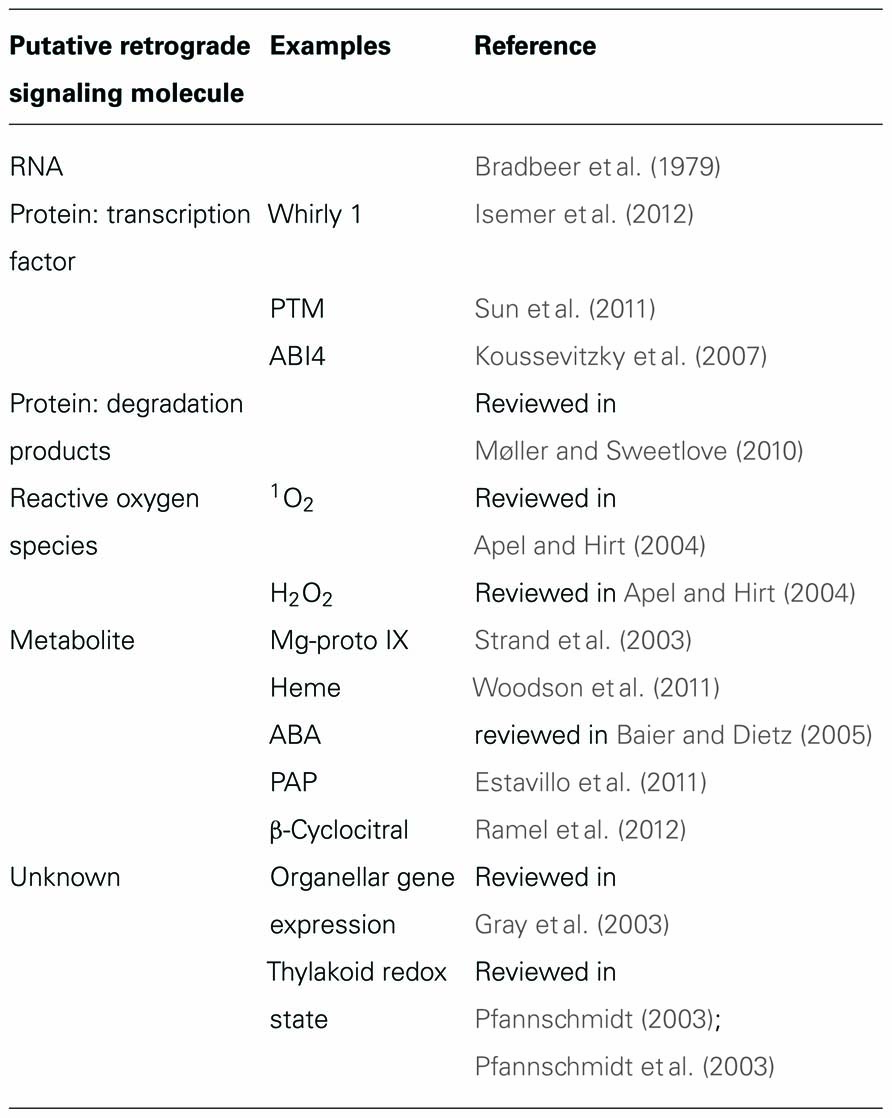
The pertinent retrograde signals are thought to derive from various sources, including the tetrapyrrole pathway, organellar gene expression (OGE), reactive oxygen species (ROS), or the redox state of the organelle (Table 1). Conversely, one can ask which of the principal types of biomolecules produced in organelles have the potential to act as a retrograde signal, i.e., encode sufficient information on the state of the organelle to trigger an appropriate response in the nucleus. Nucleic acids can efficiently store and transfer information, and their relocation from organelle to nucleus over evolutionary time is well established (reviewed in Kleine et al., 2009b). Hence, more than three decades ago, it was hypothesized that organelle-derived RNAs might regulate cytoplasmic protein synthesis (Bradbeer et al., 1979), but experimental support for this idea has not yet been forthcoming. ROS are by-products of several organellar processes and their accumulation is associated with changes in NGE (Apel and Hirt, 2004). However, their suitability as classical retrograde signals has been questioned, either because they are probably too short-lived to reach the nucleus [in the case of singlet oxygen (1O2)] or too unspecific (H2O2) to act as information carriers (reviewed in Møller and sweetlove, 2010). Although specific transporters for the export of chloroplast proteins, in particular transcription factors, have yet to be identified, evidence is accumulating that chloroplast-located transcription factors can be conveyed to the nucleus (Sun et al., 2011; Isemer et al., 2012). Nevertheless, metabolites still appear to be the most likely candidates for retrograde signaling molecules. This is because (i) the profile of metabolites exported from an organelle to the cytosol does indeed reflect the metabolic state of the organelle and (ii) numerous transporters exist that facilitate the controlled exchange of metabolites between organelles and the cytosol. Therefore, changes in the metabolic profile of the cytosol triggered by the organelle might well be used by the cell to adjust NGE, either by using a metabolite directly as a signaling molecule or by converting an appropriate metabolite into an active signaling molecule during its sojourn in the cytosol.
What Sorts of Nuclear Genes are Regulated by Retrograde Signaling?
Frequently used marker genes for the analysis of retrograde signaling or the identification of putative signaling mutants are LHCB1, 2CPA, and APX2 (Oelmüller and Mohr, 1986; Karpinski et al., 1997; Heiber et al., 2007), which code for a photosynthetic antenna protein, a chloroplast peroxidase and a cytosolic ascorbate peroxidase, respectively. Transcriptomics-based studies in Arabidopsis thaliana have identified large sets of nuclear genes as possible targets of retrograde signaling by employing either putative signaling mutants and/or conditions thought to trigger retrograde signaling. Thus, nuclear genes coding for chloroplast proteins have been associated with GUN signaling (Strand et al., 2003) and a subsequent study showed that the ACGT motif, the core of both the light-responsive G box (CACGTG) and the abscisic acid (ABA) response element (ABRE), was over-represented in putative target genes of GUN signaling (Koussevitzky et al., 2007). Analysis of the time-dependent impact of redox signals showed rapid and dynamic changes in nuclear transcript accumulation, resulting in differential and specific expression patterns for genes associated with photosynthesis and metabolism (Bräutigam et al., 2009). Another study analyzed the overlap between the differential gene expression profiles of several mutants affected in photosynthetic acclimation. The identified 56 nuclear genes could represent either nuclear target genes of photosynthesis-relevant retrograde signaling or genes that are differentially expressed to compensate for the effects of suppression of retrograde signaling (Pesaresi et al., 2009). Interestingly, genes involved in many different processes, including stress responses, post-transcriptional, translational and post-translational regulation of gene expression, as well as metabolism, were represented in this set; however, few of them appeared to be directly involved in the photosynthetic process (Pesaresi et al., 2009). ROS-responsive genes were also identified and it was possible to discriminate between H2O2- and 1O2-responsive genes (op den Camp et al., 2003). Interestingly, nuclear genes coding for chloroplast proteins generally seem to be co-regulated to a certain degree irrespective of their biochemical function (Richly et al., 2003; Biehl et al., 2005; Leister et al., 2011), prompting the postulate that a transcriptional master switch might exist (Richly et al., 2003). Nuclear photosynthesis-associated genes, including genes coding for photosystem subunits, display a distinct regulation pattern, which is independent of the master switch, highlighting the unique role of photosynthesis as representing both stimulus and target of retrograde regulation (Biehl et al., 2005). The large-scale analysis of the transcript profiles of chloroplast- and mitochondrion-relevant genes in A. thaliana confirmed that the activity of gene sets involved in organellar energy production (OEP) or OGE in each of the organelles and in the nucleus is highly coordinated (Leister et al., 2011). Moreover, the same analysis indicated that dynamic inter- and intracompartmental transcriptional networks for OEP and OGE genes adjust the activity of organelles in response to the cellular energy state and environmental stresses (Leister et al., 2011).
Taken together, these studies suggest that large numbers of genes respond to retrograde signaling, but in most cases it remains unclear whether they are directly targeted by retrograde signaling or are regulated indirectly as part of broader adjustments that compensate for the original stimulus (in most cases mutations). The use of transient stimuli and kinetic analysis of transcriptional responses, as practiced by Bräutigam et al. (2009), seems the most appropriate way to address this question.
The Simplest Scenario: The Classical Retrograde Signal
As outlined above, classical signaling molecules are assumed to be generated in the organelle and make their way to the nucleus, where they modify NGE by either inducing or repressing the expression of specific genes (Figure 1A). Therefore, when gene expression patterns in retrograde signaling mutants are compared with the profile in wild-type (WT) plants, differential expression of the regulated genes will be observed (Figure 1B). Two principal modes of transfer of retrograde signaling molecules present themselves: transfer by passive diffusion or active transport involving specific transporters (Figure 1C). In principle, the latter mode of transfer would include already an additional level of complexity, because the rate of the export of the signaling molecule has to be controlled by another signal, which might be considered as the real retrograde signal (Figure 1C). Here, three putative classical retrograde signaling molecules are discussed in the context of these alternative modes of relocation: (i) the phosphonucleotide 3′-phosphoadenosine 5′-phosphate (PAP) and (ii) β-cyclocitral (β-CC), an oxidation product of β-carotene, both of which might be relocated by diffusion; and (iii) the tetrapyrrole Mg-protoporphyrin IX (Mg-proto IX).
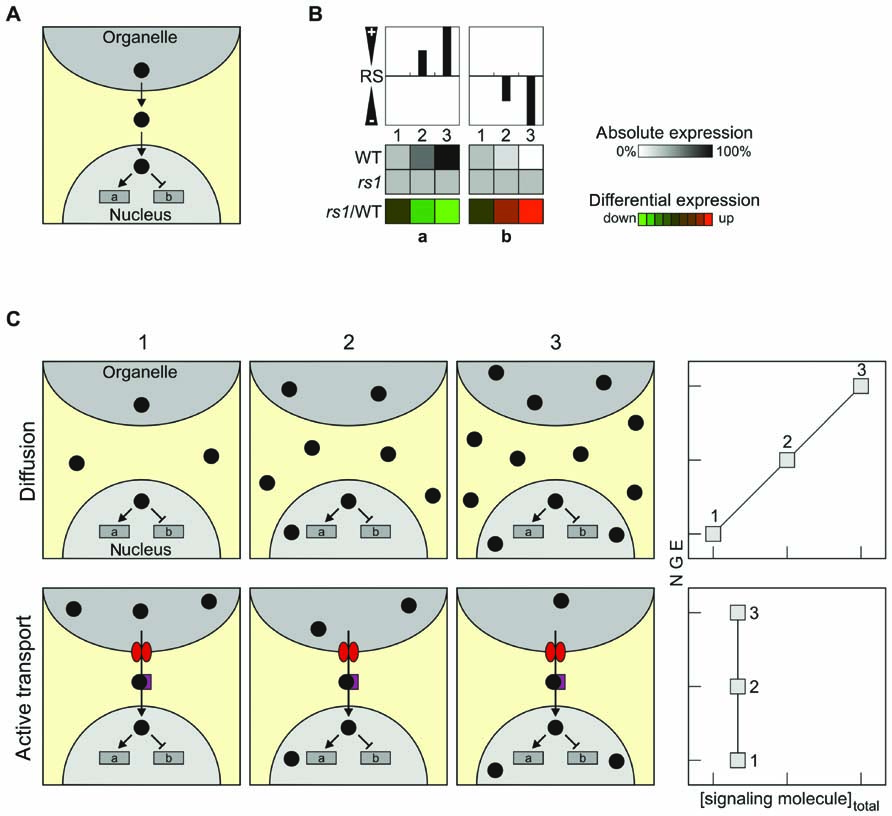
FIGURE 1. Characteristics and transport of classical retrograde signals. (A) Schematic overview of the mode of operation of the simplest scenario for retrograde signaling, the case of a classical retrograde signal. (B) Overview of how changes in the relative abundance of the retrograde signal (RS) in the nucleus might affect NGE. Relative levels of the RS increase from state 1 (0%), state 2 (50%) to state 3 (100%), resulting in either induction (gene a) or repression (gene b) of the expression of a nuclear reporter gene (indicated in grayscale). The repressive effect of the RS is symbolized in the top panel by negative values (“-”), the inducing effect by positive values (“+”). In a signaling mutant rs, no expression changes occur and therefore differential expression (rs1/WT) in terms of down-regulation (gene a) or up-regulation (gene b) is observed (shown in color scale). (C) Passive diffusion versus active transport of RS. If the signaling molecule is disseminated by diffusion (upper panels, “Diffusion”), the total concentration of the signaling molecule in all cellular components increases during the transition from state 1 to state 3, leading to changes in NGE. In this case, a linear correlation between the total concentration of the signaling molecule and NGE is expected. If the signaling molecule is actively transported into the nucleus (lower panel, “Active transport”), NGE can be altered by specifically changing the abundance of the signaling molecule in the nucleus – without altering its total concentration in the cell as a whole. In consequence, analyses of total cell extracts would fail to identify any correlation between the overall abundance of the signaling molecule and NGE. The tetrapyrrole Mg-protoporphyrin IX, for which total cellular levels fail to correlate with changes in NGE, might represent such a signal. Note that the “active transport scenario” would require regulation of the activity of the transport by another signal. In fact, in such a scenario the signal that up-regulates export might be considered as the real signal, and the transported compound as second messenger.
PAP and β-Cyclocitral – Two Novel Candidate Classical Signals
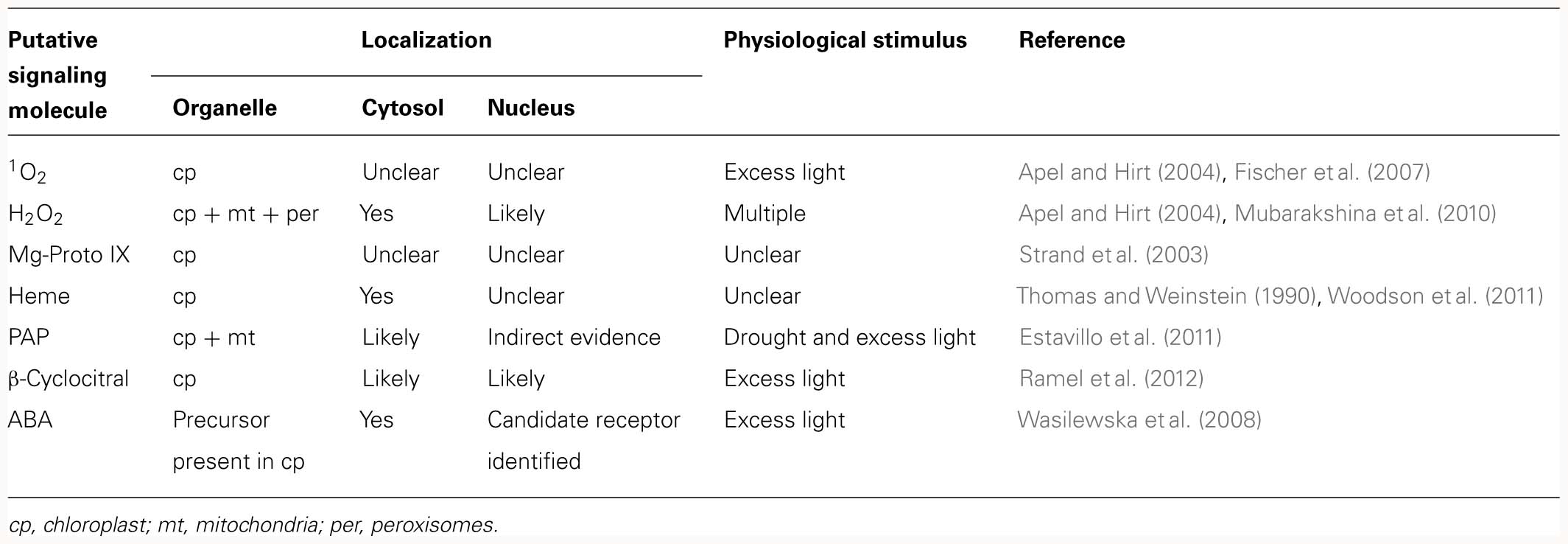
3′-Phosphoadenosine 5′-phosphate accumulates in A. thaliana in response to drought and high light (HL) stress (Estavillo et al., 2011). The enzyme SAL1, which is found in both mitochondria and chloroplasts, regulates PAP levels by dephosphorylating PAP to AMP; therefore, in sal1 mutants PAP accumulates 20-fold (Estavillo et al., 2011). Moreover, sal1 mutants display constitutive upregulation of many HL-regulated genes (Wilson et al., 2009). Except in the case of chloroplasts, where PAP could be detected by HPLC, attempts to identify PAP in subcellular fractions have failed for technical reasons (Estavillo et al., 2011). However, targeting of SAL1 to either the nucleus or chloroplasts in sal1 mutants reduces total PAP levels and decreases expression of the nuclear marker gene APX2, indirectly demonstrating that PAP must be able to move between cellular compartments (Table 2). Because total cellular levels of PAP and nuclear marker gene expression correlate well (Estavillo et al., 2011), a plausible mechanism for the effects of PAP on NGE is the “free diffusion” scenario (Figure 1C). In this scenario, the total concentration of the signaling molecule in the organelle must reach a certain level to enable diffusion into the nucleus to supply sufficient amounts to its putative site of action. Consequently, PAP seems to represent a classical signaling molecule which is synthesized in organelles of a given type, leaves that organelle and reaches the nucleus by passive diffusion. Moreover, the physiological relevance of PAP as a possible retrograde signal is strongly supported by the fact that PAP levels increase in response to at least two abiotic stresses.
Oxidation products of β-carotene, like β-CC, have been suggested to act as “downstream” messengers or proxies for rising 1O2 levels (Ramel et al., 2012; see below). β-CC is volatile; therefore, it should be capable of traversing the cytosol to regulate gene expression in the nucleus. β-CC is generated under physiological conditions, following exposure to excess light which promotes generation of 1O2, which in turn oxidizes β-carotene. Moreover, treatment of plants with exogenous β-CC induces changes in the expression of many 1O2-responsive genes. Therefore, it is tempting to speculate that β-CC represents a classical retrograde signal which is disseminated by diffusion. Because this HL-induced signaling pathway may involve at least two signaling molecules, 1O2 and β-CC, this instance of retrograde signaling is discussed further in the next section.
Is Mg-Proto IX a Classical Retrograde Signaling Molecule or Part of a More Complex Pathway?
When it became clear that most gun mutants were affected in chloroplast tetrapyrrole biosynthesis (Mochizuki et al., 2001), the obvious next step was to test whether an intermediate in tetrapyrrole biosynthesis might be responsible for changes in NGE. Therefore, the possibility that the concentrations of various tetrapyrroles might be correlated with the expression of nuclear marker genes in A. thaliana was analyzed in several studies with conflicting results (Mochizuki et al., 2001, 2008; Strand et al., 2003; Moulin et al., 2008). Strand et al. (2003) claimed to have found a direct correlation between changes in the total cellular level of Mg-proto IX and NGE, although an earlier study by the same group had come to a different conclusion (Mochizuki et al., 2001). Later investigations employing more precise and reproducible tests, as well as additional genotypes, confirmed that no correlation exists between total Mg-proto IX levels and NGE (Mochizuki et al., 2008; Moulin et al., 2008). However, lack of correlation between changes in the total concentration of Mg-proto IX in plant leaves and alterations in the expression profiles of certain nuclear marker genes does not suffice to rule out the possibility that Mg-proto IX acts as a classical plastid signal. The finding simply argues against diffusion as a transport mechanism for the signal. It does not exclude the possibility that the signaling molecule is actively transported from its source to the nucleus. If the signaling molecule is actively transported into the nucleus, its concentration there could still increase (with corresponding effects on NGE), while the total cellular concentration of the signaling molecule remained unchanged (Figure 1C). Whether or not this scenario holds for Mg-proto IX remains to be clarified; as a phototoxic molecule, it seems rather unlikely to act as a signaling molecule (Mochizuki et al., 2010). Heme is a somewhat better candidate retrograde signaling molecule deriving from the tetrapyrrole biosynthesis pathway (Woodson et al., 2011), but levels of total heme and LHCB1 mRNA also fail to correlate (Voigt et al., 2010; Woodson et al., 2011).
Nevertheless, a correlation between perturbations in chloroplast tetrapyrrole biosynthesis and NGE changes undoubtedly exists, and future research has to clarify what retrograde signaling mechanisms mediate this relationship.
More Complex Scenarios of Retrograde Signaling
The classical scenario for a retrograde signaling molecule, in which the molecule that is generated in the organelle itself relays information to the nucleus, might be true for the phosphonucleotide PAP during drought and HL stress and for β-CC after HL stress (Table 2), but need not be valid for other retrograde signals. Thus, ABA, which has been proposed as a putative plastid signal, is generated in the cytosol but derives from a precursor (xanthoxin) which is synthesized in the chloroplast (reviewed in Baier and Dietz, 2005; Figure 2A). Alternatively, the original metabolite triggering retrograde signaling might not even leave the organelle (Figure 2B). For instance, singlet oxygen (1O2) is generated in chloroplasts and regulates the expression of a set of nuclear genes, although it is thought to be largely restricted to plastids with little, if any, leakage to the cytosol (Apel and Hirt, 2004; Fischer et al., 2007). In consequence, an unidentified second messenger should exist which relays the information from the plastid to the nucleus, and the two chloroplast proteins Executer 1 and 2 (reviewed in Kim et al., 2008), as well as ABA (Kim et al., 2009), seem to be involved in this retrograde signaling pathway. More recently, β-CC was suggested to act as a “downstream” messenger of 1O2 (Ramel et al., 2012), possibly representing a classical retrograde signal which traverses the cytosol to regulate gene expression in the nucleus (see above).
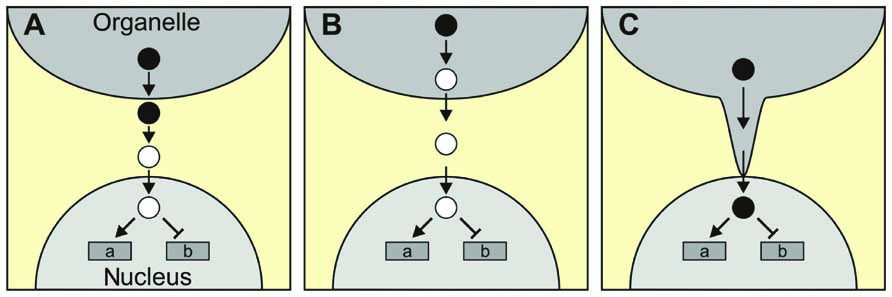
FIGURE 2. More complex scenarios for retrograde signaling. (A) The signaling molecule generated in and exported from the organelle (filled circles), and the signaling molecule that enters the nucleus (open circles) might not be identical. ABA, which is synthesized in the cytosol from a chloroplast precursor, is a possible example. (B) The signaling molecule generated in the organelle might not even leave the organelle. 1O2 serves here as an example, as it could, in principle, generate volatile oxidation products of carotenoids that serve as “downstream” messengers. (C) Direct delivery of the signaling molecule to the nucleus via stromules.
So far, proteins that specifically transport signaling molecules from the organelle to the nucleus have not been identified. In an extreme case, the signaling molecules could be delivered directly to its target if organelle and nucleus were physically connected. In fact, stromules might represent such connection points between plastids and the nucleus (Figure 2C). Stromules are stroma-filled tubules that extend from the surface of plastids, are extremely variable in length and are highly dynamic structures (reviewed in Hanson and Sattarzadeh, 2008). Interestingly, stromules are induced by stress treatments, including drought and salt stress, and application of ABA (Gray et al., 2012) – conditions which are also thought to be associated with retrograde signaling.
Integration of Multiple Signals
The existence of multiple candidate signaling molecules, including those discussed above, ABA, 1O2 (and its tentative downstream signal β-CC), Mg-Proto IX and PAP, suggests that it is very likely that more than one retrograde signaling pathway exists. Moreover, the generation of signaling molecules in the organelle will itself be affected by multiple factors. Two model cases that take account of this complexity are discussed here. In scenario 1, each of two retrograde signals (RS1 and RS2) can act independently to activate or repress NGE. In scenario 2, only one retrograde signal (RS) operates, but its formation is controlled by two organellar factors (O1 and O2) that can act synergistically or in opposite senses (Figure 3A). To simulate the effects on NGE of the two retrograde signals in scenario 1 and the two organellar factors in scenario 2, three discrete doses for each retrograde signal or organellar factor were considered: 0, 50, and 100%. If both retrograde signals or organellar factors are assumed to act in the same sense, i.e., both stimulate (or repress) gene expression or generation of the retrograde signal, the two scenarios result in identical effects on NGE in WT plants (Figures 3B,C). Furthermore, where mutants for the two retrograde signaling pathways or organellar factors are available, the differential expression patterns determined in the two scenarios will be identical. Therefore, it will not be possible to discriminate between the two scenarios on the basis of expression data from WT and mutant plants defective in signaling under conditions in which both retrograde signaling factors or both organellar signaling factors act additively.
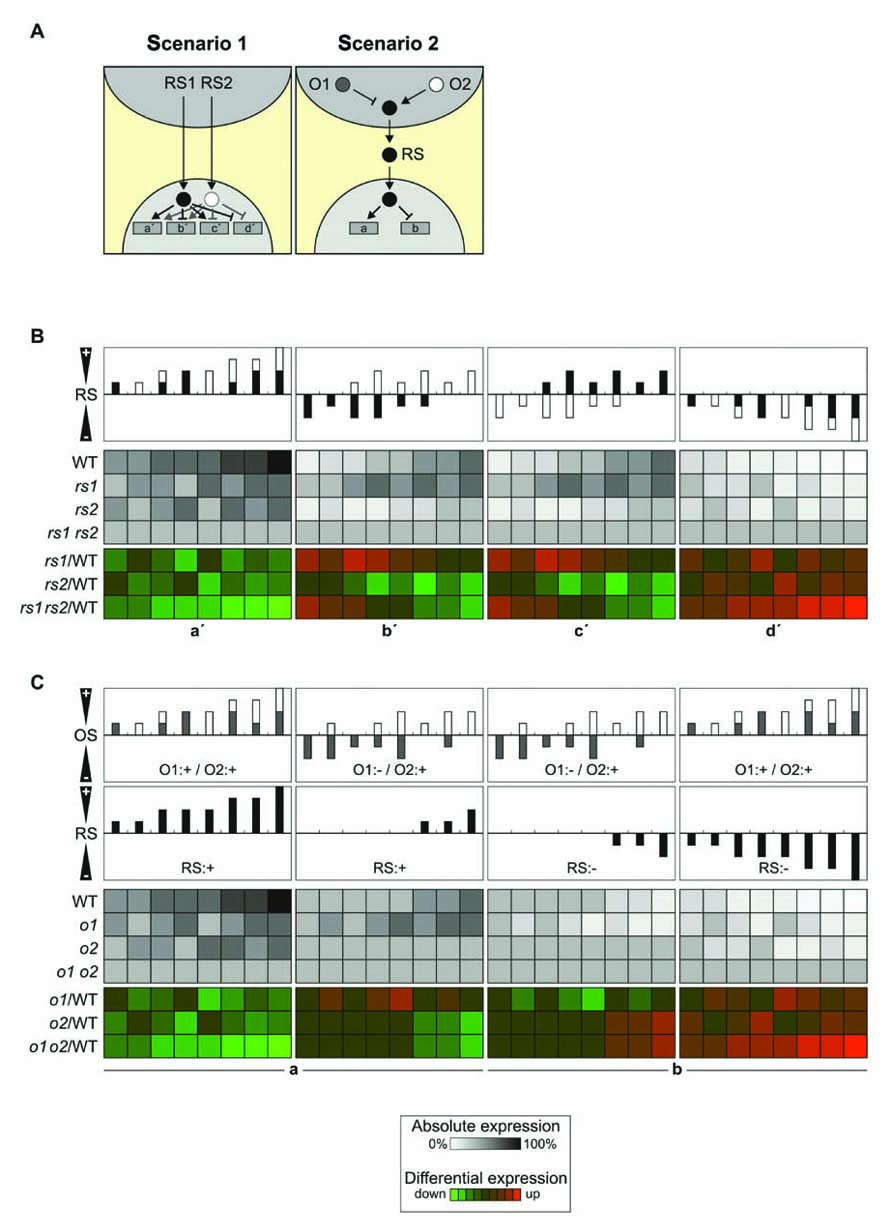
FIGURE 3. Signal integration. (A) Schematic overview of the integration of two retrograde signals (RS1 and RS2) in the nucleus (left panel) or in the organelle (right panel). O1 and O2 refer to signals acting within the organelle. Activating signals are indicated by arrows, repressing signals by blunt-ended lines. Left panel (two retrograde signals, integration in the nucleus): gene a′, induced by RS1 and RS2; gene b′, repressed by RS1 and induced by RS2; gene c′, induced by RS1 and repressed by RS2; gene d′, repressed by RS1 and RS2. Right panel (two organellar signals, integration in the organelle): RS induces gene a and represses gene b as in Figure 1A. (B) Signal integration at the gene level. In this model each of the two retrograde signals can accumulate to 0, 50, or 100% [depicted as black (RS1) or white (RS2) bars], analogous to the three states discussed in Figures 1B,C, resulting in nine different combinations, of which eight are shown in the top panels (the combination RS1: 0%/RS2: 0% is not shown). The effects on NGE are shown in WT and corresponding single (rs1 and rs2) and double (rs1 rs2) signaling mutants in grayscale. In color scale, effects on differential gene expression (mutant versus WT) are shown. In the left/right panel, the expected impact of additive inducing/repressing effects of the retrograde signals on genes a′ and d′ is shown. In the middle panel (b′/c′), the two retrograde signals act antagonistically and the outcome for the expression of genes b′ and c′ is very similar. (C) As in (B), the two organellar signals can act additively with the same polarity (O1:+/O2:+) or act antagonistically (O1:-/O2:+; O1:+/O2:-; upper panel). The net effect of OS1 and OS2 on RS is shown in the second uppermost panel and can either induce gene expression (the two left panels, gene a) or repress gene expression (the two right panels, gene b). The resulting effects on NGE are again shown in grayscale for absolute expression and in color scale for differential expression (mutant versus WT).
In contrast, under conditions where the two retrograde signals or organellar factors act in opposite senses, differences in differential expression between signaling mutants and WT plants will emerge. In the case of two retrograde signals whose effects are integrated in the nucleus, differential up-regulation for one signaling mutant but down-regulation for the other signaling mutant will be observed (Figure 3B). On the other hand, where signal integration occurs already in the organelle, opposite regulation of the two organellar signals will essentially neutralize their effects on the generation of the retrograde signal (Figure 3C). In the model presented here, five out of eight conditions fail to produce a retrograde signal in the WT, and opposite differential regulation in signaling mutants is found only in one condition.
Overall, regulation of NGE by two independent retrograde signals appears to allow for more complex regulation of NGE, and this should be detectable by opposite regulation of NGE in corresponding signaling mutants under appropriate conditions. Of course, the crucial question is whether such complex regulatory scenarios are actually realized and whether sufficient expression data are available to decide whether signal integration occurs in the organelle or the nucleus. In fact, two instances of synergistic effects of two signaling mutants on NGE have been described so far. The first example concerns the gun mutants and the second involves mutants defective in OGE. The gun phenotype was originally defined as derepressed LHCB1 expression after treatment with norflurazon (NF). Then the double mutants gun1 gun4 and gun1 gun5 were shown to exhibit higher levels of LHCB1 mRNA after NF treatment than did the individual single mutants, indicating that GUN1 affects a separate pathway from GUN4 and GUN5 (Mochizuki et al., 2001). However, the same group showed in a later publication that the synergy between gun1 and gun5 does not extend to the gun1–9 allele (Koussevitzky et al., 2007). Nevertheless, the available data are not sufficient to decide whether signal integration occurs in the chloroplast or at the nuclear level.
Changes in OGE affect the expression of nuclear genes, in particular genes coding for photosynthetic proteins (reviewed in Pesaresi et al., 2007). Pesaresi et al. (2006) showed that simultaneous perturbation of OGE in both mitochondria and chloroplasts down-regulates nuclear photosynthetic genes much more drastically than do treatments that affect OGE in only the one or the other. For this study, mutations affecting mitochondrial or chloroplast ribosomes, as well as mutations affecting a dual-targeted tRNA synthetase were analyzed. For all nuclear photosynthesis genes studied, the double mutant prpl11 mrpl11, in which ribosomal function was impaired in both types of organelles, always produced fewer transcripts than either of the single mutants prpl11 and mrpl11, indicating that both plastid OGE and mitochondrial OGE stimulate NGE under the growth conditions employed in the study. In this case also, it is not possible to discriminate between signal integration in the organelle and the nucleus based on the expression profiles. However, because the two mutations affect separate organelles it is tempting to speculate that signal integration occurs in the cytosol or in the nucleus as outlined in Figure 3B.
Concluding Remarks: Solid Criteria to Define a Retrograde Signal
How can a candidate signaling molecule be unambiguously confirmed to be a real signal? In light of the different retrograde signaling scenarios outlined above, it is not possible to draw up a universal list of characteristics and experiments that could unequivocally decide whether a putative retrograde signal is a true retrograde signal. Nevertheless, a number of conclusions can be drawn from the considerations set out above. A classical retrograde signal should be detectable in all relevant compartments (organelle, cytosol and nucleus), assuming that passage through the cytosol is not rendered superfluous by the use of stromules. So far, no putative classical signal has been unambiguously detected in all relevant compartments (Table 2). Secondly, for a signal which follows the free diffusion scenario a correlation between overall signal level and NGE should exist, opening the way for systematic approaches involving LC–MS or GC–MS analyses. Finally, how can one show that a signaling molecule is not only required but also sufficient to trigger NGE? Feeding experiments in which PAP (the candidate most likely to conform to the “free diffusion” scenario) was supplied via the roots have so far failed to confirm a signaling role for it (Estavillo et al., 2011), whereas Mg-proto IX was reported to repress LHCB expression when fed to protoplasts for 5 h at relatively high concentrations (Strand et al., 2003). These two examples highlight the dilemma of feeding experiments. On the one hand, a negative result cannot conclusively disqualify a candidate retrograde signal, unless one can monitor the accumulation of the added molecule at its destination. Conversely, a positive result is only valid if the feeding assays truly mimic physiological conditions. Nevertheless, feeding experiments and in vitro assays with subcellular fractions (in which target nuclear genes are directly exposed to the signaling molecules), together with the transient activation of retrograde signaling pathways by inducible gene expression systems, are certainly needed to clarify the role of putative signaling molecules.
Finally, the reconstruction of signaling networks and of the synergistic and antagonistic integration of diverse retrograde inputs will be necessary for the knowledge-based modification of retrograde signaling. Studies, like the one of gene regulatory networks involving AP2/EREB transcription factors (Dietz et al., 2010), might serve as prototypes for systematic genome analysis of retrograde signaling, demonstrating that the examples provided here (see Figure 3) certainly represent simplifications of a much more complex phenomenon.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to the Deutsche Forschungsgemeinschaft for funding (grant FOR 804) and Paul Hardy for critical reading of the manuscript.
References
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399.
Baier, M., and Dietz, K. J. (2005). Chloroplasts as source and target of cellular redox regulation: a discussion on chloroplast redox signals in the context of plant physiology. J. Exp. Bot. 56, 1449–1462.
Biehl, A., Richly, E., Noutsos, C., Salamini, F., and Leister, D. (2005). Analysis of 101 nuclear transcriptomes reveals 23 distinct regulons and their relationship to metabolism, chromosomal gene distribution and co-ordination of nuclear and plastid gene expression. Gene 344, 33–41.
Bradbeer, J. W., Atkinson, Y. E., Börner, T., and Hagemann, R. (1979). Cytoplasmic synthesis of plastid polypeptides may be controlled by plastid-synthesized RNA. Nature 279, 816–817.
Bräutigam, K., Dietzel, L., Kleine, T., Ströher, E., Wormuth, D., Dietz, K. J., Radke, D., Wirtz, M., Hell, R., Dörmann, P., Nunes-Nesi, A., Schauer, N., Fernie, A. R., Oliver, S. N., Geigenberger, P., Leister, D., and Pfannschmidt, T. (2009). Dynamic plastid redox signals integrate gene expression and metabolism to induce distinct metabolic states in photosynthetic acclimation in Arabidopsis. Plant Cell 21, 2715–2732.
Chan, K. X., Crisp, P. A., Estavillo, G. M., and Pogson, B. J. (2010). Chloroplast-to-nucleus communication: current knowledge, experimental strategies and relationship to drought stress signaling. Plant Signal. Behav. 5, 1575–1582.
Dietz, K. J., Vogel, M. O., and Viehhauser, A. (2010). AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma 245, 3–14.
Estavillo, G. M., Crisp, P. A., Pornsiriwong, W., Wirtz, M., Collinge, D., Carrie, C., Giraud, E., Whelan, J., David, P., Javot, H., Brearley, C., Hell, R., Marin, E., and Pogson, B. J. (2011). Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23, 3992–4012.
Fischer, B. B., Krieger-Liszkay, A., Hideg, E., Snyrychova, I., Wiesendanger, M., and Eggen, R. I. (2007). Role of singlet oxygen in chloroplast to nucleus retrograde signaling in Chlamydomonas reinhardtii. FEBS Lett. 581, 5555–5560.
Gray, J. C., Hansen, M. R., Shaw, D. J., Graham, K., Dale, R., Smallman, P., Natesan, S. K., and Newell, C. A. (2012). Plastid stromules are induced by stress treatments acting through abscisic acid. Plant J. 69, 387–398.
Gray, J. C., Sullivan, J. A., Wang, J. H., Jerome, C. A., and Maclean, D. (2003). Coordination of plastid and nuclear gene expression. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358, 135–144; discussion 144–135.
Hanson, M. R., and Sattarzadeh, A. (2008). Dynamic morphology of plastids and stromules in angiosperm plants. Plant Cell Environ. 31, 646–657.
Heiber, I., Ströher, E., Raatz, B., Busse, I., Kahmann, U., Bevan, M. W., Dietz, K. J., and Baier, M. (2007). The redox imbalanced mutants of Arabidopsis differentiate signaling pathways for redox regulation of chloroplast antioxidant enzymes. Plant Physiol. 143, 1774–1788.
Inaba, T., Yazu, F., Ito-Inaba, Y., Kakizaki, T., and Nakayama, K. (2011). Retrograde signaling pathway from plastid to nucleus. Int. Rev. Cell Mol. Biol. 290, 167–204.
Isemer, R., Mulisch, M., Schäfer, A., Kirchner, S., Koop, H. U., and Krupinska, K. (2012). Recombinant Whirly1 translocates from transplastomic chloroplasts to the nucleus. FEBS Lett. 586, 85–88.
Karpinski, S., Escobar, C., Karpinska, B., Creissen, G., and Mullineaux, P. M. (1997). Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell 9, 627–640.
Kim, C., Lee, K. P., Baruah, A., Nater, M., Gobel, C., Feussner, I., and Apel, K. (2009). 1O2-mediated retrograde signaling during late embryogenesis predetermines plastid differentiation in seedlings by recruiting abscisic acid. Proc. Natl. Acad. Sci. U.S.A. 106, 9920–9924.
Kim, C., Meskauskiene, R., Apel, K., and Laloi, C. (2008). No single way to understand singlet oxygen signalling in plants. EMBO Rep. 9, 435–439.
Kleine, T., Voigt, C., and Leister, D. (2009a). Plastid signalling to the nucleus: messengers still lost in the mists? Trends Genet. 25, 185–192.
Kleine, T., Maier, U. G., and Leister, D. (2009b). DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu. Rev. Plant Biol. 60, 115–138.
Koussevitzky, S., Nott, A., Mockler, T. C., Hong, F., Sachetto-Martins, G., Surpin, M., Lim, J., Mittler, R., and Chory, J. (2007). Signals from chloroplasts converge to regulate nuclear gene expression. Science 316, 715–719.
Leister, D., Wang, X., Haberer, G., Mayer, K. F., and Kleine, T. (2011). Intracompartmental and intercompartmental transcriptional networks coordinate the expression of genes for organellar functions. Plant Physiol. 157, 386–404.
Mochizuki, N., Brusslan, J. A., Larkin, R., Nagatani, A., and Chory, J. (2001). Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. U.S.A. 98, 2053–2058.
Mochizuki, N., Tanaka, R., Grimm, B., Masuda, T., Moulin, M., Smith, A. G., Tanaka, A., and Terry, M. J. (2010). The cell biology of tetrapyrroles: a life and death struggle. Trends Plant Sci. 15, 488–498.
Mochizuki, N., Tanaka, R., Tanaka, A., Masuda, T., and Nagatani, A. (2008). The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 105, 15184–15189.
Møller, I. M., and Sweetlove, L. J. (2010). ROS signalling – specificity is required. Trends Plant Sci. 15, 370–374.
Moulin, M., McCormac, A. C., Terry, M. J., and Smith, A. G. (2008). Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signaling is not due to Mg-protoporphyrin IX accumulation. Proc. Natl. Acad. Sci. U.S.A. 105, 15178–15183.
Mubarakshina, M. M., Ivanov, B. N., Naydov, I. A., Hillier, W., Badger, M. R., and Krieger-Liszkay, A. (2010). Production and diffusion of chloroplastic H2O2 and its implication to signalling. J. Exp. Bot. 61, 3577–3587.
Nott, A., Jung, H. S., Koussevitzky, S., and Chory, J. (2006). Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 57, 739–759.
Oelmüller, R., and Mohr, H. (1986). Photooxidative destruction of chloroplasts and its consequences for expression of nuclear genes. Planta 167, 106–113.
op den Camp, R. G., Przybyla, D., Ochsenbein, C., Laloi, C., Kim, C., Danon, A., Wagner, D., Hideg, E., Gobel, C., Feussner, I., Nater, M., and Apel, K. (2003). Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15, 2320–2332.
Pesaresi, P., Hertle, A., Pribil, M., Kleine, T., Wagner, R., Strissel, H., Ihnatowicz, A., Bonardi, V., Scharfenberg, M., Schneider, A., Pfannschmidt, T., and Leister, D. (2009). Arabidopsis STN7 kinase provides a link between short- and long-term photosynthetic acclimation. Plant Cell 21, 2402–2423.
Pesaresi, P., Masiero, S., Eubel, H., Braun, H. P., Bhushan, S., Glaser, E., Salamini, F., and Leister, D. (2006). Nuclear photosynthetic gene expression is synergistically modulated by rates of protein synthesis in chloroplasts and mitochondria. Plant Cell 18, 970–991.
Pesaresi, P., Schneider, A., Kleine, T., and Leister, D. (2007). Interorganellar communication. Curr. Opin. Plant Biol. 10, 600–606.
Pfannschmidt, T. (2003). Chloroplast redox signals: how photosynthesis controls its own genes. Trends Plant Sci. 8, 33–41.
Pfannschmidt, T. (2010). Plastidial retrograde signalling – a true “plastid factor” or just metabolite signatures? Trends Plant Sci. 15, 427–435.
Pfannschmidt, T., Schütze, K., Fey, V., Sherameti, I., and Oelmüller, R. (2003). Chloroplast redox control of nuclear gene expression – a new class of plastid signals in interorganellar communication. Antioxid. Redox Signal. 5, 95–101.
Pogson, B. J., Woo, N. S., Forster, B., and Small, I. D. (2008). Plastid signalling to the nucleus and beyond. Trends Plant Sci. 13, 602–609.
Ramel, F., Birtic, S., Ginies, C., Soubigou-Taconnat, L., Triantaphylides, C., and Havaux, M. (2012). Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc. Natl. Acad. Sci. U.S.A. 109, 5535–5540.
Richly, E., Dietzmann, A., Biehl, A., Kurth, J., Laloi, C., Apel, K., Salamini, F., and Leister, D. (2003). Covariations in the nuclear chloroplast transcriptome reveal a regulatory master-switch. EMBO Rep. 4, 491–498.
Strand, A., Asami, T., Alonso, J., Ecker, J. R., and Chory, J. (2003). Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421, 79–83.
Sun, X., Feng, P., Xu, X., Guo, H., Ma, J., Chi, W., Lin, R., Lu, C., and Zhang, L. (2011). A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nat. Commun. 2, 477.
Thomas, J., and Weinstein, J. D. (1990). Measurement of heme efflux and heme content in isolated developing chloroplasts. Plant Physiol. 94, 1414–1423.
Voigt, C., Oster, U., Börnke, F., Jahns, P., Dietz, K. J., Leister, D., and Kleine, T. (2010). In-depth analysis of the distinctive effects of norflurazon implies that tetrapyrrole biosynthesis, organellar gene expression and ABA cooperate in the GUN-type of plastid signalling. Physiol. Plant. 138, 503–519.
Wasilewska, A., Vlad, F., Sirichandra, C., Redko, Y., Jammes, F., Valon, C., Frei Dit Frey, N., and Leung, J. (2008). An update on abscisic acid signaling in plants and more. Mol. Plant 1, 198–217.
Wilson, P. B., Estavillo, G. M., Field, K. J., Pornsiriwong, W., Carroll, A. J., Howell, K. A., Woo, N. S., Lake, J. A., Smith, S. M., Millar, H. A., Von Caemmerer, S., and Pogson, B. J. (2009). The nucleotidase/phosphatase SAL1 is a negative regulator of drought tolerance in Arabidopsis. Plant J. 58, 299–317.
Keywords: retrograde signaling, plastid signaling, nuclear gene expression, signal integration
Citation: Leister D (2012) Retrograde signaling in plants: from simple to complex scenarios. Front. Plant Sci. 3:135. doi:10.3389/fpls.2012.00135
Received: 25 May 2012; Accepted: 05 June 2012;
Published online: 19 June 2012.
Edited by:
Tatjana Kleine, Ludwig-Maximilians-Universität München, GermanyCopyright: © 2012 Leister. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Dario Leister, Plant Molecular Biology (Botany), Department Biology I, Ludwig Maximilians University Munich, Großhaderner Str. 2, D-82152 Planegg-Martinsried, Germany. e-mail: leister@lmu.de