- 1 Department of Biology, Texas A&M University, College Station, TX, USA
- 2 Department of Biochemistry and Biophysics, Texas A&M University, College Station, TX, USA
Telomeres, the essential terminal regions of linear eukaryotic chromosomes, consist of G-rich DNA repeats bound by a plethora of associated proteins. While the general pathways of telomere maintenance are evolutionarily conserved, individual telomere complex components show remarkable variation between eukaryotic lineages and even within closely related species. The recent genome sequencing of the lycophyte Selaginella moellendorffii and the availability of an ever-increasing number of flowering plant genomes provides a unique opportunity to evaluate the molecular and functional evolution of telomere components from the early evolving non-seed plants to the more developmentally advanced angiosperms. Here we analyzed telomere sequence in S. moellendorffii and found it to consist of TTTAGGG repeats, typical of most plants. Telomere tracts in S. moellendorffii range from 1 to 5.5 kb, closely resembling Arabidopsis thaliana. We identified several S. moellendorffii genes encoding sequence homologs of proteins involved in telomere maintenance in other organisms, including CST complex components and the telomere-binding proteins, POT1 and the TRFL family. Notable sequence similarities and differences were uncovered among the telomere-related genes in some of the plant lineages. Taken together, the data indicate that comparative analysis of the telomere complex in early diverging land plants such as S. moellendorffii and green algae will yield important insights into the evolution of telomeres and their protein constituents.
Introduction
The ends of linear eukaryotic chromosomes terminate with a long stretch of simple tandem repeats of GT-rich telomeric DNA. These sequences, together with specific DNA binding proteins, comprise the telomeres. Telomeres are important for maintaining genome integrity, distinguishing natural DNA ends from double-strand (ds) breaks, and preventing illegitimate DNA repair. Proper maintenance of telomeric DNA length and structure is essential for normal cell viability (Zellinger and Riha, 2007).
Eukaryotic organisms across different taxa display remarkable variation in the length of telomere tracts. Unicellular ciliates and budding yeast typically harbor short telomeres that range from several dozen to several hundred base pairs, while humans and mice have telomeres in the range of 5–15 kb and 10–60 kb, respectively (Hug and Lingner, 2006). Plants also display dramatic variations in telomere length, with tracts spanning 2–5 kb in Arabidopsis thaliana to > 150 kb in tobacco (Richards and Ausubel, 1988; Fajkus et al., 1995; Shakirov and Shippen, 2004). In addition, telomere length varies not only from species to species, but even within different populations of the same species. Telomere length in different A. thaliana ecotypes (natural populations) varies as much as twofold (Shakirov and Shippen, 2004; Maillet et al., 2006), while some Zea mays recombinant inbred lines show up to 25-fold differences in telomere length (Burr et al., 1992).
Telomere binding proteins play essential roles in regulating telomere length by modulating telomerase access to chromosome ends. Numerous other proteins influence telomere length, including DNA damage response factors and DNA-modifying enzymes (Martinez and Blasco, 2011). Notably, a deletion screen of all non-essential genes in budding yeast identified ~200 candidates whose absence resulted in deregulated telomeres (Askree et al., 2004). While most of these genes likely affect telomere homeostasis indirectly, these genetic data underscore the dynamic and complex nature of telomere length regulation.
Plants and animals diverged over 1.5 bya (Yoon et al., 2004) and yet many aspects of telomere biology are conserved. For example, the most common telomere repeat sequence in plants is TTTAGGG, just one nucleotide longer than the 6-base sequence TTAGGG found in vertebrates (McKnight and Shippen, 2004). Many sequence and functional homologs of telomere-related genes in vertebrates and yeast have been identified in plants (Fitzgerald et al., 1999; Riha et al., 2002; Karamysheva et al., 2004; Shakirov et al., 2005; Song et al., 2008; Surovtseva et al., 2009). Indeed plants provide a unique opportunity to examine evolution of telomere composition, structure, and function due to the well-established evolutionary relationships within the plant kingdom. Here we exploit the recently sequenced genome of the lycophyte Selaginella moellendorffii (Banks et al., 2011) to characterize telomeric DNA and to identify genes with putative roles in telomere biology. Our analysis indicates that S. moellendorffii harbors short telomere tracts consisting of canonical TTTAGGG repeats. Furthermore, we find a full complement of the telomere-associated genes that have previously been described in other plants. Comparative studies of S. moellendorffi with other early diverging plants may be useful for studying the evolution of telomere proteins in plants.
Results and Discussion
Selaginella moellendorffii Telomeres
Sequence analysis of terminal chromosomal scaffolds indicates that S. moellendorffii telomeres, like those of most other plants, are composed of tandem arrays of (TTTAGGG)n repeats (Banks et al., 2011). To gauge the size of S. moellendorffii telomere tracts, we performed terminal restriction fragment (TRF) analysis using Tru11. The blot was hybridized with probe corresponding to four repeats of TTTAGGG. As shown in Figure 1A, S. moellendorffii telomere tracts migrated as a smear ranging from 1.5 to 5.5 kb, closely resembling telomere profile in many A. thaliana accessions (Richards and Ausubel, 1988; Shakirov and Shippen, 2004).
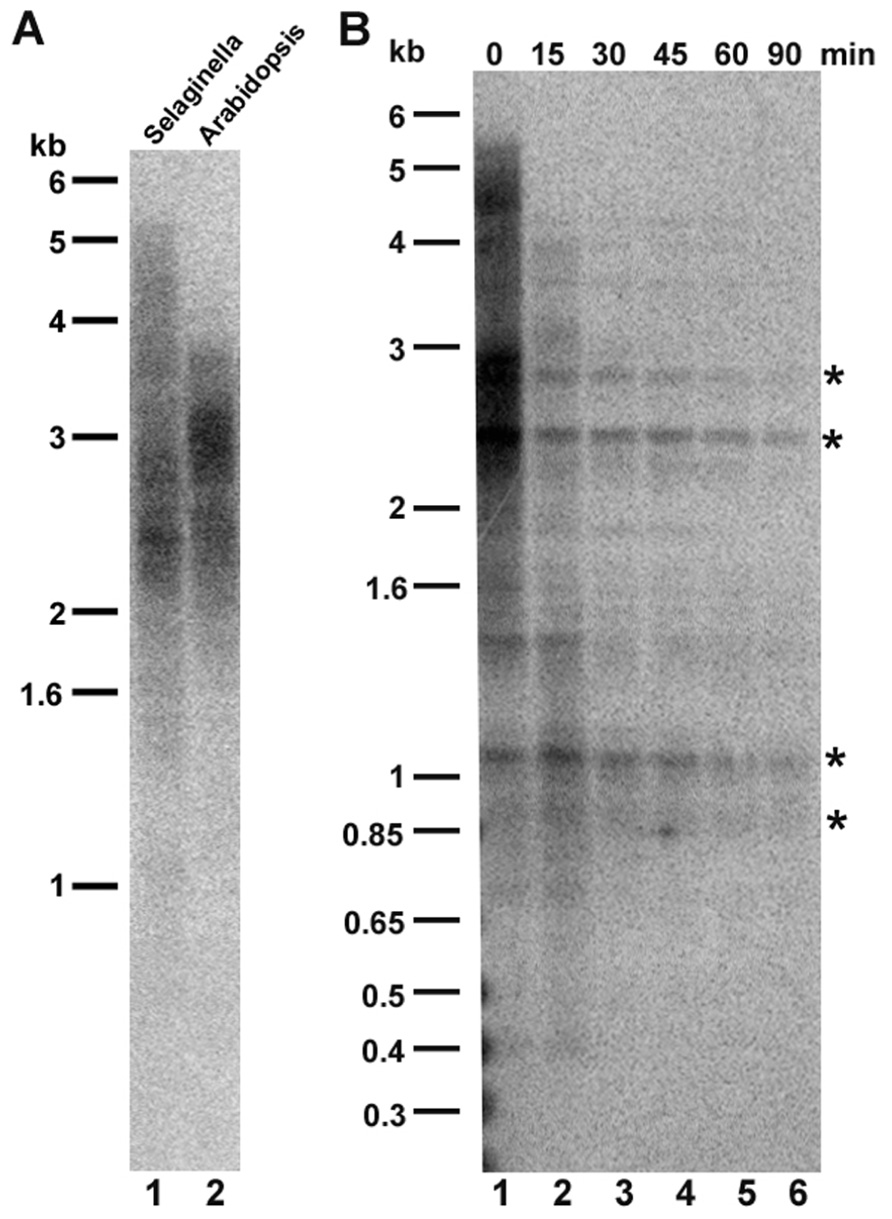
FIGURE 1. Telomere length analysis in Selaginella moellendorffii. (A) Comparative terminal restriction fragment (TRF) analysis of S. moellendorffii (lane 1) and A. thaliana (lane 2) telomeres. Molecular weight markers are shown on the left. (B) Bal31 digestion of S. moellendorffii telomeric DNA. Lane 1, Tru1I digestion of genomic DNA without prior Baltreatment (0 min). Lanes 2–6, Tru1I digestion of genomic DNA with Bal31 tr31eatment for 15, 30, 45, 60, and 90 min, respectively. Asterisks indicate cross-hybridizing interstitial telomeric DNA bands, which are not sensitive to Bal31 digestion for up to 90 min.
We verified that sequences detected by TRF analysis correspond to chromosome ends using the non-specific Bal31 exonuclease. Bal31 preferentially degrades DNA ends versus more internal genomic regions. DNA was pre-incubated with Bal31 prior to digestion with Tru1I, and a Southern blot was performed. After 15 min of Bal31 digestion, the hybridization products migrated faster on the gel and showed reduced intensity (Figure 1B, lane 2). With continued Bal31 incubation, the telomeric signal disappeared completely (Figure 1B, lanes 3–6). In contrast, several cross-hybridizing bands, corresponding to interstitial telomeric DNA were insensitive to Bal31 digestion for up to 90 min, supporting the conclusion that the Bal31-sensitive hybridization signal corresponds to terminal telomeric DNA. Thus, S. moellendorffii telomeres are comprised of 1.5–5.5 kb tracts of TTTAGG repeats.
Telomere-Related Genes in S. moellendorffii
POT1 Proteins
Single-strand (ss) telomere-binding proteins represent a key component of the telomere cap. Such proteins control telomerase access to the telomere and ensure chromosome end protection (de Lange, 2009). Overall, ss telomere binding proteins share limited sequence similarity, but they all bear signature N-terminal oligonucleotide/oligosacchaaride folds (OB-folds). One key ss telomere binding protein is Protection of telomeres (POT1; Baumann and Cech, 2001). In the moss Physcomitrella patens, a single-copy POT1 gene encodes a typical DNA binding protein that efficiently binds ss telomeric substrates in vitro (Shakirov et al., 2010). Furthermore, similar to its mammalian and fission yeast counterparts, P. patens POT1 is involved in telomere end protection (capping). While PpPOT1-deficient moss can survive long-term in culture, the mutant strain is sterile and shows end-to-end chromosome fusions, indicating that the overall telomere protective function of POT1 is conserved bet-ween early diverging land plants and other eukaryotes (Shakirov et al., 2010).
Despite conservation of POT1 function in the earliest land plant lineages, several lines of biochemical and genetic evidence indicate that the functions of POT1 in vascular plants (starting with S. moellendorffii) may have changed substantially. First, biochemical analysis of POT1 proteins from 13 plants representing major evolutionary branches of plants has indicated that the ability to bind telomeric DNA has been lost for most plant POT1 proteins, including POT1 from S. moellendorffii (Shakirov et al., 2009a,b). In fact, besides the P. patens POT1 protein and its ortholog from the green alga Ostreococcus lucimarinus, only two other POT1 proteins (from Asparagus officinalis and Z. mays) out of a total of 16 surveyed have retained the capacity to bind telomeric DNA in vitro (Shakirov et al., 2009b). However, both A. officinalis and Z. mays are unusual plants with respect to telomere biology. A. officinalis possesses unconventional telomere repeats TTAGGG instead of the canonical TTTAGGG, while Z. mays belongs to the only plant family surveyed other than Brassicaceae that harbors duplicated POT1 genes (Shakirov et al., 2009a,b). Thus, the ability of POT1 proteins from A. officinalis and Z. mays to bind telomeric DNA may have been conserved due to unusual changes in organismal telomere biology (A. officinalis) or protein sub-functionalization (Z. mays). Alternatively, the ability of A. officinalis and Z. mays POT1 proteins to bind telomeric DNA may have evolved independently through parallel evolution.
The second line of evidence supporting unusually fast evolution of POT1 functions in vascular plants comes from the studies of A. thaliana. Unlike the situation in humans and most other organisms, A. thaliana and other members of the Brassicaceae family possess two full-length POT1 proteins (Shakirov et al., 2005, 2009b). Genetic and biochemical studies indicate that AtPOT1a is a positive regulator of telomere length working in the context of telomerase holoenzyme (Surovtseva et al., 2007). In contrast, POT1b is implicated in chromosome end protection (Shakirov et al., 2005). Notably, AtPOT1a has a high binding specificity for the RNA subunit of telomerase (Cifuentes-Rojas et al., 2011), an unexpected mode of action for an OB-fold containing protein originally evolved to bind DNA. Unlike A. thaliana, but similar to the situation in P. patens, the S. moellendorffii genome encodes only a single POT1 protein. As expected from phylogenetic positions of their corresponding species, S. moellendorffii POT1 shares more amino acid similarity with P. patens POT1 (60%), than with A. thaliana POT1 proteins (46% to AtPOT1a and 47% to AtPOT1b; Shakirov et al., 2009b). Despite the overall higher amino acid conservation between P. patens and S. moellendorffii POT1 proteins, the loss of telomeric DNA binding capacity (Shakirov et al., 2009b) clearly suggests that the functional role of POT1 in S. moellendorffii telomere biology may in fact be more analogous to the situation in A. thaliana than in P. patens. Determining whether P. patens POT1 binds telomerase RNA must await the identification of this molecule in moss.
TRFL Proteins
The second class of telomeric DNA binding proteins associates with ds telomeric DNA. This family of telomere repeat binding factors (TRF) shares a conserved Myb-related DNA-binding domain in the C-terminus and a central dimerization domain (Bilaud et al., 1996). Mammals and other vertebrates encode two ds telomere binding proteins, TRF1 and TRF2, with distinct functions in telomere homeostasis. TRF1 is thought to act primarily in telomere length control, while TRF2 is required for chromosome end protection through participation in T-loop formation (Broccoli et al., 1997; Griffith et al., 1999).
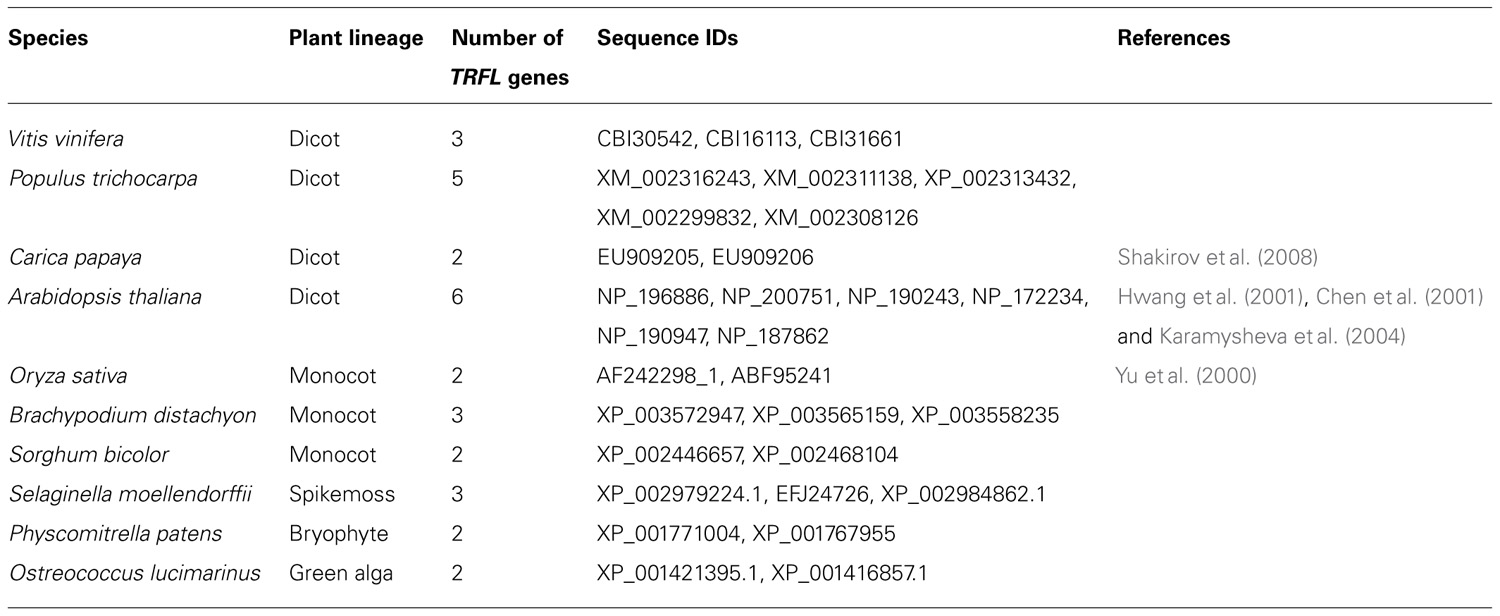
Unlike vertebrates, plants possess two related families of TRF-like (TRFL) proteins, class I and class II (Chen et al., 2001; Hwang et al., 2001; Karamysheva et al., 2004). In A. thaliana, there are 12 TRFL proteins, 6 in class I and 6 in class II. Members of class II do not bind ds TTTAGGG repeats in vitro. In contrast, all six members of class I specifically bind ds telomeric DNA (Karamysheva et al., 2004). This interaction is dependent on the presence of a unique plant-specific Myb-extension motif, located at the extreme C-terminus of the TRFL protein (Figure 2). Overall, plants display remarkable variation in the number of class I TRFL genes. In dicot species, TRFL gene amplification appears to be a common theme, with three genes in grapes (Vitis vinifera), five genes in poplar (Populus trichocarpa), and six genes in A. thaliana (Table 1). In contrast, sequenced genomes of monocots and non-flowering plants, including S. moellendorffii, harbor 2 or 3 TRFL genes. While the precise role of individual TRFL proteins in plants remains unclear, amplification of TRFL gene family may provide a route for sub- and neo-functionalization with the potential for more dynamic control of telomere length or telomerase activity.

FIGURE 2. Multiple alignment of C-terminal regions of plant group I TRFL proteins. Position of conserved plant-specific MYB-extension motif is indicated. Abbreviations: At, Arabidopsis thaliana; Cp, Carica papaya; Pt, Populus trichocarpa; Vv, Vitis vinifera; Os, Oryza sativa; Sb, Sorghum bicolor; Bd, Brachypodium distachyon; Pp, Physcomitrella patens; Sm, Selaginella moellendorffii; Ol, Ostreococcus lucimarinus. Accession IDs: AtTBP1, NP_196886; AtTRP1, NP_200751; AtTRFL1, NP_190243; AtTRFL2, NP_172234; AtTRFL4, NP_190947; AtTRFL9, NP_187862; CpTRFL1, EU909205; CpTRFL2, EU909206; PtTRFL1, XM_002316243; PtTRFL2, XM_002311138; PtTRFL3, XP_002313432; PtTRFL4, XM_002299832; PtTRFL5, XM_002308126; VvTRFL1, CBI30542; VvTRFL2, CBI16113; VvTRFL3, CBI31661; OsTBP1, AF242298_1; OsTBP2, ABF95241; SbTRFL1, XP_002446657; SbTRFL2, XP_002468104; BdTRFL1, XP_003572947; BdTRFL2, XP_003565159; BdTRFL3, XP_003558235; PpTRFL1, XP_001771004; PpTRFL2, XP_001767955; SmTRFL2, XP_002979224.1; SmTRFL3, EFJ24726; SmTRFL1, XP_002984862.1; OlTRFL1, XP_001421395.1; OlTRFL2, XP_001416857.1. Protein alignment was generated using MEGA5 software (Tamura et al., 2011) and visualized in the BOXSHADE format.
We also examined the evolutionary relationship of the available class I full-length plant TRFL proteins, using three A. thaliana class II proteins as the outgroup (Figure 3). As expected, class II proteins form a separate clade distinct from class I, consistent with the lack of the C-terminal Myb-extension motif. The evolutionary relationship of class I proteins correlates with the phylogenetic position of the corresponding plant species. Notably, the two TRFL proteins from green alga form a sister clade to all TRFL proteins from land plants, suggesting significant sequence divergence in this ancestral lineage.
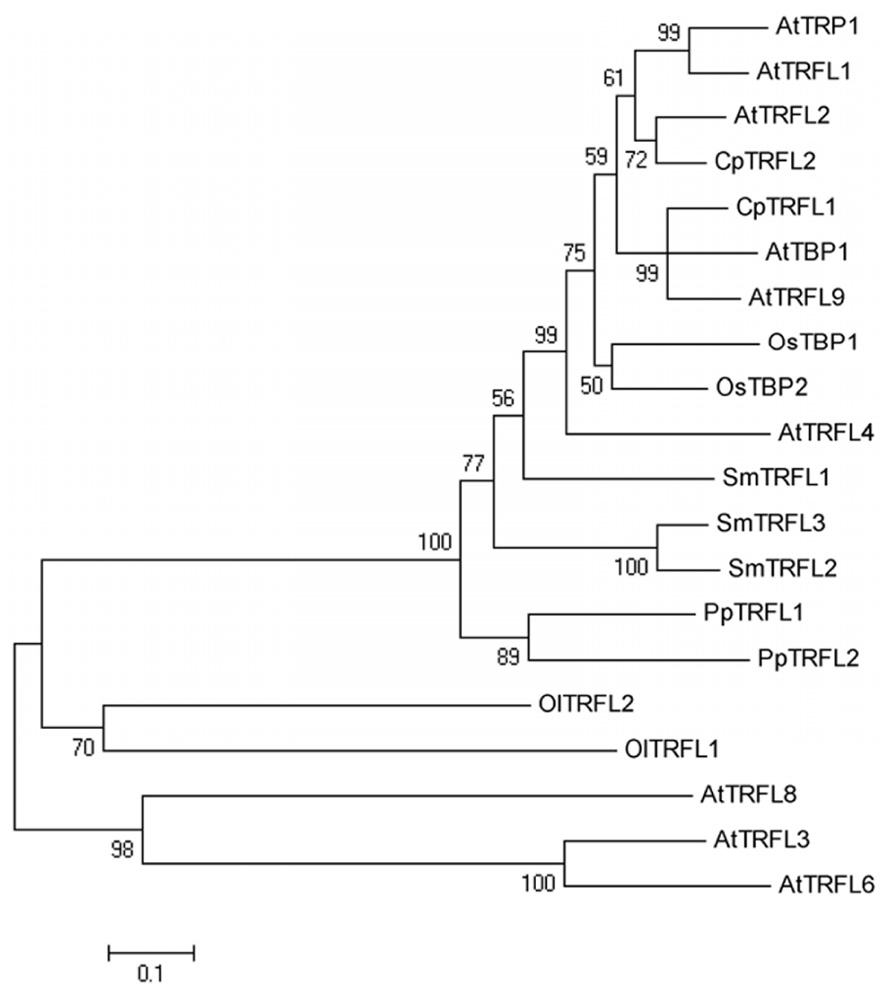
FIGURE 3. Evolutionary relationships of plant group I TRFL proteins. The evolutionary history of 17 currently available full-length TRFL proteins was inferred using the Neighbor-Joining method (Saitou and Nei, 1987). A. thaliana Group II TRFL proteins lacking the Group I-specific Myb-extension motif (AtTRFL3, AtTRFL6 and AtTRFL8) were used as outgroup (Karamysheva et al., 2004). The bootstrap consensus tree was inferred from 1000 replicates, and the percentage of replicate trees in which the associated taxa clustered together in the bootstrap test are shown next to the branches. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965). All positions containing gaps and missing data were eliminated. There were a total of 162 positions in the final dataset. Evolutionary analyses were conducted in MEGA5 (Tamura et al., 2011).
CST Components
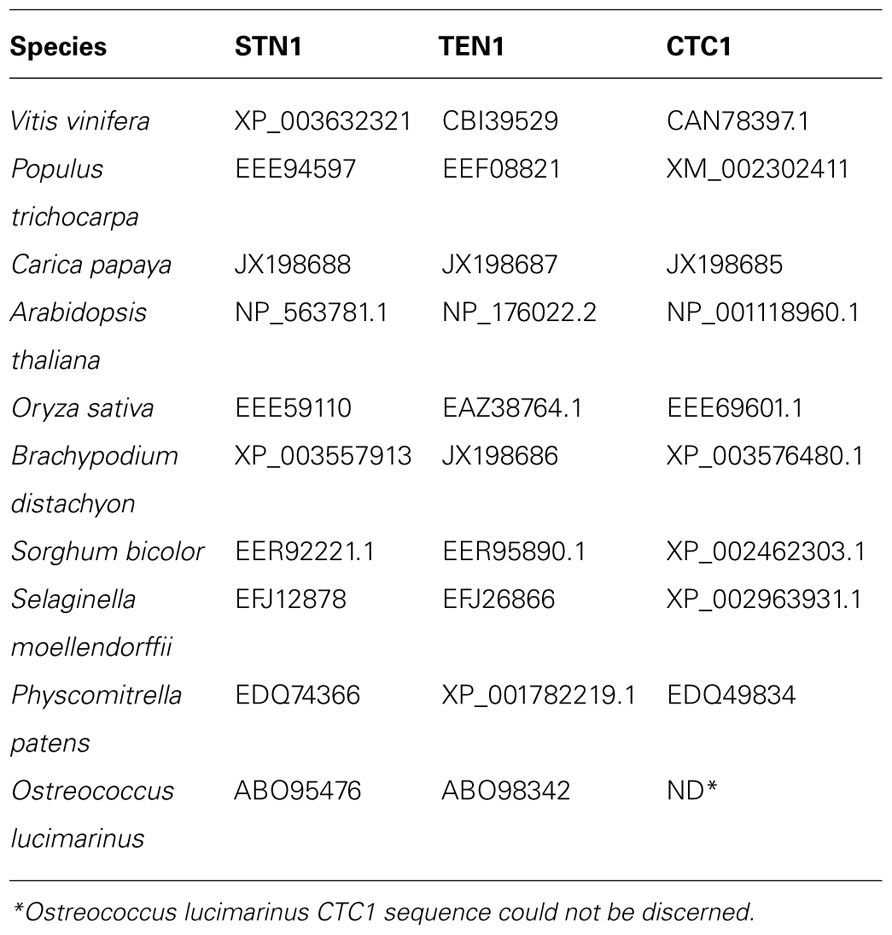
A third group of evolutionarily conserved plant telomere proteins is a trimeric complex composed of CTC1, STN1, and TEN1, termed CST (Price et al., 2010). In the budding yeast Saccharomyces cerevisiae, a similar complex, composed of Cdc13, Stn1, and Ten1 proteins, was described over 20 years ago, but only recently has this complex come to light in multicellular eukaryotes (Wellinger, 2009). Individual CST components are highly divergent, with only 10–20% amino acid sequence identity between corresponding proteins from different eukaryotic lineages (Price et al., 2010). A. thaliana mutants deficient in STN1 and CTC1 are characterized by severe defects in telomere maintenance, massive end-to-end chromosome fusions, and elevated rates of telomere recombination (Song et al., 2008; Surovtseva et al., 2009). As in vertebrates (Casteel et al., 2009), the A. thaliana CTC1 subunit of CST physically interacts with the catalytic subunit of DNA polymerase α (Price et al., 2010), thus linking the CST complex to the telomere replication pathway.
To gain a better understanding of the evolution of CST complex in plants, we looked for the presence of genes encoding CST complex subunits in selected plant species with completely sequenced genomes. Single-copy STN1 and TEN1 genes were found in all organisms surveyed (Table 2). In addition, with the exception of the green alga O. lucimarinus, a single copy of CTC1 gene was also identified in all plant species analyzed, including S. moellendorffii (Table 2). The apparent absence of a clear CTC1 homolog in O. lucimarinus is intriguing. Strikingly, none of the CTC1 orthologs can be readily identified in several species of genera Chlamydomonas, Micromonas, and Chlorella, which belong to evolutionarily distinct lineages of green algae. These data indicate that either CTC1 sequence has diverged beyond recognition or that in green algae this protein has been functionally replaced by an unrelated polypeptide. This observation is in line with the observed sequence divergence of green algae TRFL proteins and suggests that many components of the telomere complex in green algae diverged substantially from their counterparts in land plants.
Interestingly, it has been argued that the budding yeast S. cerevisiae replaced CTC1 with Cdc13 (Mitton-Fry et al., 2002), The two proteins share little sequence similarity, but possess structurally similar OB-fold DNA binding domains and interact with well-conserved protein binding partners (STN1 and TEN1). Our genome analysis indicates that single-copy genes encoding CST complex subunits are present in all land plants analyzed, from the earlier evolved non-seed plant lineages, represented by P. patens and S. moellendorffii, to the more developmentally advanced flowering plants. Thus, the important functions of CST complex in chromosome end protection and/or telomere replication are likely to be conserved throughout evolution of land plants.
Conclusion and Outlook
Lycophytes occupy a unique phylogenetic position in the evolution of land plants, as they are ancient representatives of vascular plants and sister to Euphyllophites (which include flowering plants). Since most components of the telomere maintenance machinery have previously been analyzed only in Angiosperms, we examined the telomere repeat array and sequence homologs of telomere-related factors in S. moellendorffii. As in A. thaliana and P. patents, telomere tracts are short and consist of the canonical TTTAGGG repeat. Furthermore, similar to the situation in Angiosperms and other land plants, the S. moellendorffii genome harbors homologs of POT1, TRFL, STN1, CTC1, and TEN1 genes. Finally, our study revealed marked sequence divergence in telomere components of green algae relative to S. moellendorffii, arguing that future comparative studies among these organisms may provide important insight into the evolution of the telomere complex in plants.
Materials and Methods
Telomere Length Analysis and Bal31 Digestion
Selaginella moellendorffii DNA was extracted as described by Cocciolone and Cone (1993). To detect telomeric DNA repeats, genomic DNA was digested with Tru1I (Fermentas; recognition sequence TTAA) and subjected to Southern blotting with 32P-labeled (TTTAGGG)4 as a probe (Fitzgerald et al., 1999). Radioactive signals were scanned by a STORM PhosphorImager (Molecular Dynamics), and the data were analyzed by IMAGEQUANT software (Molecular Dynamics). For the Bal31 exonuclease assay, 100 µg of S. moellendorffii genomic DNA was incubated with 50 units of Bal31 (New England Biolabs) or with H2O (0 min time point) in 1× Bal31 reaction buffer at 30°C.Equal amounts of sample were removed at 15 or 30 min intervals for 90 min. Reactions were stopped by the addition of 20 mM EGTA and heating to 65°C for 15 min. DNA in each sample was precipitated with isopropanol and ammonium acetate, followed by Tru1I digestion. Digested DNA was separated on 0.8% agarose, blotted onto a nitrocellulose membrane and subjected to hybridization as described above.
Blast Searches and Gene Predictions
BLAST searches were performed at the Phytozome v8 portal1 using the tblastn option and amino acid sequence of A. thaliana telomere proteins as a query. O. lucimarinus BLAST was performed at the corresponding genome portal2 using the similar pproach. GenBank accession numbers of all proteins are given in Tables 1 and 2.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jody Banks for coordinating collaborative work on the Selaginella moellendorffii genome, and Tom McKnight for helpful discussions concerning telomere protein evolution. This work was supported by a grant from NIH (GM065383) to Dorothy E. Shippen.
Footnotes
References
Askree, S. H., Yehuda, T., Smolikov, S., Gurevich, R., Hawk, J., Coker, C., Krauskopf, A., Kupiec, M., and McEachern, M. J. (2004). A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc. Natl. Acad. Sci. U.S.A. 101, 8658–8663.
Banks, J. A., Nishiyama, T., Hasebe, M., Bowman, J. L., Gribskov, M., dePamphilis, C., Albert, V. A., Aono, N., Aoyama, T., Ambrose, B. A., Ashton, N. W., Axtell, M. J., Barker, E., Barker, M. S., Bennetzen, J. L., Bonawitz, N. D., Chapple, C., Cheng, C., Correa, L. G., Dacre, M., DeBarry, J., Dreyer, I., Elias, M., Engstrom, E. M., Estelle, M., Feng, L., Finet, C., Floyd, S. K., Frommer, W. B., Fujita, T., Gramzow, L., Gutensohn, M., Harholt, J., Hattori, M., Heyl, A., Hirai, T., Hiwatashi, Y., Ishikawa, M., Iwata, M., Karol, K. G., Koehler, B., Kolukisaoglu, U., Kubo, M., Kurata, T., Lalonde, S., Li, K., Li, Y., Litt, A., Lyons, E., Manning, G., Maruyama, T., Michael, T. P., Mikami, K., Miyazaki, S., Morinaga, S., Murata, T., Mueller-Roeber, B., Nelson, D. R., Obara, M., Oguri, Y., Olmstead, R. G., Onodera, N., Petersen, B. L., Pils, B., Prigge, M., Rensing, S. A., Riaño-Pachóon, D. M., Roberts, A. W., Sato, Y., Scheller, H. V., Schulz, B., Schulz, C., Shakirov, E. V., Shibagaki, N., Shinohara, N., Shippen, D. E., Sørensen, I., Sotooka, R., Sugimoto, N., Sugita, M., Sumikawa, N., Tanurdzic, M., Theissen, G., Ulvskov, P., Wakazuki, S., Weng, J. K., Willats, W. W., Wipf, D., Wolf, P. G., Yang, L., Zimmer, A. D., Zhu, Q., Mitros, T., Hellsten, U., Loqué, D., Otillar, R., Salamov, A., Schmutz, J., Shapiro, H., Lindquist, E., Lucas, S., Rokhsar, D., and Grigoriev, I. V. (2011). The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332, 960–963.
Baumann, P., and Cech, T. R. (2001). Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292, 1171–1175.
Bilaud, T., Koering, C. E., Binet-Brasselet, E., Ancelin, K., Pollice, A., Gasser, S. M., and Gilson, E. (1996). The telobox, a Myb-related telomeric DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res. 24, 1294–1303.
Broccoli, D., Smogorzewska, A., Chong, L., and de Lange, T. (1997). Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 17, 231–235.
Burr, B., Burr, F. A., Matz, E. C., and Romero-Severson, J. (1992). Pinning down loose ends: mapping telomeres and factors affecting their length. Plant Cell 4, 953–960.
Casteel, D. E., Zhuang, S., Zeng, Y., Perrino, F. W., Boss, G. R., Goulian, M., and Pilz, R. B. (2009). A DNA polymerase-alpha primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J. Biol. Chem. 284, 5807–5818.
Chen, C. M., Wang, C. T., and Ho, C. H. (2001). A plant gene encoding a Myb-like protein that binds telomeric GGTTAG repeats in vitro. J. Biol. Chem. 276, 16511–16519.
Cifuentes-Rojas, C., Kannan, K., Tseng, L., and Shippen, D. E. (2011). Two RNA subunits and POT1a are components of Arabidopsis telomerase. Proc. Natl. Acad. Sci. U.S.A. 108, 73–78.
Cocciolone, S. M., and Cone, K. C. (1993). Pl-Bh, an anthocyanin regulatory gene of maize that leads to variegated pigmentation. Genetics 135, 575–588.
Fajkus, J., Kovarik, A., Kralovics, R., and Bezdek, M. (1995). Organization of telomeric and subtelomeric chromatin in the higher plant Nicotiana tabacum. Mol. Gen. Genet. 247, 633–638.
Fitzgerald, M. S., Riha, K., Gao, F., Ren, S., McKnight, T. D., and Shippen, D. E. (1999). Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc. Natl. Acad. Sci. U.S.A. 96, 14813–14818.
Griffith, J. D., Comeau, L., Rosenfield, S., Stansel, R. M., Bianchi, A., Moss, H., and de Lange, T. (1999). Mammalian telomeres end in a large duplex loop. Cell 97, 503–514.
Hwang, M. G., Chung, I. K., Kang, B. G., and Cho, M. H. (2001). Sequence-specific binding property of Arabidopsis thaliana telomeric DNA binding protein 1 (AtTBP1). FEBS Lett. 503, 35–40.
Karamysheva, Z. N., Surovtseva, Y. V., Vespa, L., Shakirov, E. V., and Shippen, D. E. (2004). A C-terminal Myb extension domain defines a novel family of double-strand telomeric DNA-binding proteins in Arabidopsis. J. Biol. Chem. 279, 47799–47807.
Maillet, G., White, C. I., and Gallego, M. E. (2006). Telomere-length regulation in inter-ecotype crosses of Arabidopsis. Plant Mol. Biol. 62, 859–866.
Martinez, P., and Blasco, M. A. (2011). Telomeric and extra-telomeric roles for telomerase and the telomere-binding proteins. Nat. Rev. Cancer 11, 161–176.
Mitton-Fry, R. M., Anderson, E. M., Hughes, T. R., Lundblad, V., and Wuttke, D. S. (2002). Conserved structure for single-stranded telomeric DNA recognition. Science 296, 145–147.
Price, C. M., Boltz, K. A., Chaiken, M. F., Stewart, J. A., Beilstein, M. A., and Shippen, D. E. (2010). Evolution of CST function in telomere maintenance. Cell Cycle 9, 3157–3165.
Richards, E. J., and Ausubel, F. M. (1988). Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell 53, 127–136.
Riha, K., Watson, J. M., Parkey, J., and Shippen, D. E. (2002). Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. Embo J. 21, 2819–2826.
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Shakirov, E. V., McKnight, T. D., and Shippen, D. E. (2009a) POT1-independent single-strand telomeric DNA binding activities in Brassicaceae. Plant J. 58, 1004–1015.
Shakirov, E. V., Song, X., Joseph, J. A., and Shippen, D. E. (2009b). POT1 proteins in green algae and land plants: DNA-binding properties and evidence of co-evolution with telomeric DNA. Nucleic Acids Res. 37, 7455–7467.
Shakirov, E. V., Perroud, P. F., Nelson, A. D., Cannell, M. E., Quatrano, R. S., and Shippen, D. E. (2010). Protection of Telomeres 1 is required for telomere integrity in the moss Physcomitrella patens. Plant Cell 22, 1838–1848.
Shakirov, E.V., Salzberg, S.L. and Shippen, D.E. (2008) Analysis of Carica papaya telomeres and telomere-associated proteins: insights into the evolution of telomere maintenance in Brassicales. Tropical Plant Biol. 1, 202–215.
Shakirov, E. V., and Shippen, D. E. (2004). Length regulation and dynamics of individual telomere tracts in wild-type Arabidopsis. Plant Cell 16, 1959–1967.
Shakirov, E. V., Surovtseva, Y. V., Osbun, N., and Shippen, D. E. (2005). The Arabidopsis Pot1 and Pot2 proteins function in telomere length homeostasis and chromosome end protection. Mol. Cell. Biol. 25, 7725–7733.
Song, X., Leehy, K., Warrington, R. T., Lamb, J. C., Surovtseva, Y. V., and Shippen, D. E. (2008). STN1 protects chromosome ends in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 105, 19815–19820.
Surovtseva, Y. V., Churikov, D., Boltz, K. A., Song, X., Lamb, J. C., Warrington, R., Leehy, K., Heacock, M., Price, C. M., and Shippen, D. E. (2009). Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol. Cell 36, 207–218.
Surovtseva, Y. V., Shakirov, E. V., Vespa, L., Osbun, N., Song, X., and Shippen, D. E. (2007). Arabidopsis POT1 associates with the telomerase RNP and is required for telomere maintenance. EMBO J. 26, 3653–3661.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739.
Wellinger, R. J. (2009). The CST complex and telomere maintenance: the exception becomes the rule. Mol. Cell 36, 168–169.
Yoon, H. S., Hackett, J. D., Ciniglia, C., Pinto, G., and Bhattacharya, D. (2004). A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 21, 809–818.
Yu, E. Y., Kim, S.E., Kim, J.H., Ko, J.H., Cho, M.H. and Chung, I.K. (2000) Sequence-specific DNA recognition by the Myb-like domain of plant telomeric protein RTBP1. J Biol Chem. 275, 24208–24214.
Zellinger, B., and Riha, K. (2007). Composition of plant telomeres. Biochim. Biophys. Acta 1769, 399–409.
Zuckerkandl, E., and Pauling, L. (1965). Molecules as documents of evolutionary history. J. Theor. Biol. 8, 357–366.
Addendum
The occurrence of Arabidopsis-type telomeres was previously shown by FISH in Selaginella martensii (J. Fuchs and I. Schubert “Arabidopsis-type telomere sequences on chromosome termini of Selaginella martensii Spring (Pteridophyta)” Biol. Zent. Bl. 115, 260–265, 1996).
Keywords: telomere, Selaginella, POT1, TRFL1, CST complex
Citation: Shakirov EV and Shippen DE (2012) Selaginella moellendorffii telomeres: conserved and unique features in an ancient land plant lineage. Front. Plant Sci. 3:161. doi: 10.3389/fpls.2012.00161
Received: 16 May 2012; Accepted: 29 June 2012;
Published online: 19 July 2012.
Edited by:
Amy Litt, The New York Botanical Garden, USAReviewed by:
Stephan Wenkel, University of Tuebingen, GermanyJill Christine Preston, University of Vermont, USA
Copyright: © 2012 Shakirov and Shippen. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Dorothy E. Shippen, Department of Biochemistry and Biophysics, Texas A&M University, 2128 TAMU, College Station, TX 77843-2128, USA. e-mail: dshippen@tamu.edu