- Department of Biology, University of Copenhagen, Copenhagen, Denmark
Plant mitogen-activated protein kinase (MAPK) cascades generally transduce extracellular stimuli into cellular responses. These stimuli include the perception of pathogen-associated molecular patterns (PAMPs) by host transmembrane pattern recognition receptors which trigger MAPK-dependent innate immune responses. In the model Arabidopsis, molecular genetic evidence implicates a number of MAPK cascade components in PAMP signaling, and in responses to immunity-related phytohormones such as ethylene, jasmonate, and salicylate. In a few cases, cascade components have been directly linked to the transcription of target genes or to the regulation of phytohormone synthesis. Thus MAPKs are obvious targets for bacterial effector proteins and are likely guardees of resistance proteins, which mediate defense signaling in response to the action of effectors, or effector-triggered immunity. This mini-review discusses recent progress in this field with a focus on the Arabidopsis MAPKs MPK3, MPK4, MPK6, and MPK11 in their apparent pathways.
Introduction
Plants have evolved an effective basal defense system to detect and limit the growth of pathogens. Pathogens may be recognized by the host via the perception of conserved microbial structures termed pathogen-associated molecular patterns (PAMPs). PAMPs are recognized via transmembrane pattern recognition receptors (PRRs) that bind specific PAMPs and initiate intracellular immune responses (Zipfel, 2008). These PAMP-triggered immunity (PTI) responses include the generation of reactive oxygen species (ROS), extracellular alkalinization, and protein phosphorylation with associated gene regulation that ultimately restricts the growth of the microbial intruder (Gimenez-Ibanez and Rathjen, 2010).
Mitogen-activated protein kinase (MAPK) signaling plays central roles in such intracellular immunity pathways. In general, MAP kinase signaling is initiated by the stimulus-triggered activation of a MAP kinase kinase kinase (MAP3K; also called MEKK). MAP3K activation, which may be directly or indirectly effected by a PRR, in turn leads to the phosphorylation and activation of downstream MAP kinase kinases (MAP2K; also called MKK or MEK). Subsequently, the MAP2K phosphorylates the downstream MAPK sequentially leading to changes in its subcellular localization and/or phosphorylation of downstream substrates including transcription factors which alter patterns of gene expression (see Figure 1). General functions of MAPK cascades in plant biology have recently been reviewed elsewhere (Fiil et al., 2009; Rodriguez et al., 2010; Komis et al., 2011).
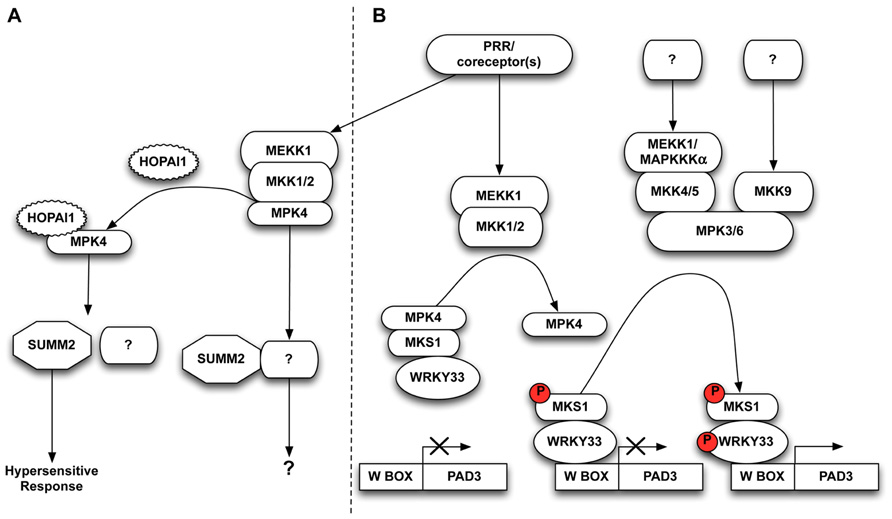
FIGURE 1. (A) MAPK signaling cascades are attractive targets for bacterial effectors. The P. syringae HopAI1 effector irreversibly inactivates MPK4 to prevent immune responses. The R protein SUMM2 may guard processes downstream of MPK4 independent from MKS1, and triggers a hypersensitive response in the event of loss or inactivation of MPK4. (B) PAMP perception by PRRs instigates a signaling cascade, often via co-receptors, which causes activation of MAP3K MEKK1 and two MAP2Ks MKK1 and MKK2. These phosphorylate and activate MPK4 which then phosphorylates its substrate MKS1, releasing MKS1 in complex with WRKY33. MPK3/MPK6 sequentially phosphorylate WRKY33 allowing it to promote PAD3 transcription, thus activating plant defense.
MAPK Cascades in PTI
A few PRRs have been documented to stimulate MAPK signaling upon perception of PAMPs. These include the flagellin receptor FLS2 (Felix et al., 1999; Gómez-Gómez and Boller, 2000), the bacterial elongation factor EF-Tu receptor EFR (Zipfel et al., 2006), and the chitin receptor CERK1 (Miya et al., 2007).
The Arabidopsis genome encodes 60 MAP3Ks, 10 MAP2Ks, and 20 MAPKs (Ichimura et al., 2002). This indicates that MAPK cascades may not simply consist of single MAP3Ks, MAP2Ks, and MAPKs connected together. Instead, it suggests that there is some level of redundancy, and that the spatial and temporal activities of different components may be strictly regulated to minimize wanton cross-talk. The three MAPKs MPK3, MPK4, and MPK6 are the most intensively studied plant MAPKs, and all three were implicated in defense signaling a decade ago (Petersen et al., 2000; Asai et al., 2002). MPK11, a close homolog to MPK4, has also recently been shown to be activated by PAMP treatment (Bethke et al., 2012).
MPK3, MPK4, and MPK6 are all activated by PAMPs such as flg22 (a conserved 22 amino acid flagellin peptide) and elf18 (elongation factor-Tu peptide; Felix et al., 1999; Zipfel et al., 2006). However, these three MAPK cascades are differently regulated already at the PRR level. For example, the two receptor kinases BAK1 and BKK1 genetically regulate PAMP signaling through their interactions with cognate PRRs (Roux et al., 2011; Schwessinger et al., 2011). The BAK1 mutant allele bak1-5 carries a Cys408Tyr substitution adjacent to its kinase catalytic loop. This impairs its flg22-regulated kinase activity and inhibits phosphorylation of MPK4. However, the catalytic complex formed between mutant BAK1 in bak1-5 and FLS2 is still able to induce phosphorylation of MPK3/MPK6 (Roux et al., 2011; Schwessinger et al., 2011). Interestingly, MPK3/MPK6 phosphorylation was impaired in only the double bak1-5 bkk1 background and not in the individual bak1-5 and bkk1 lines (Roux et al., 2011).
Asai et al. (2002) developed an elegant protoplast expression system in an attempt to identify signaling components downstream of FLS2. With this system they were able to show a complete MAPK cascade downstream of FLS2 consisting of the MAP3K MEKK1, two MAP2Ks (MKK4 and MKK5), and the MAPKs MPK3/MPK6. However, genetic evidence later showed that MEKK1 kinase activity was dispensable for MPK3/MPK6 activation, although mekk1 plants were impaired in MPK4 activation (Rodriguez et al., 2007). Interestingly, expressing a kinase dead version of MEKK1 in mekk1 plants completely restored the activation of MPK4 upon treatment with flg22, suggesting that MEKK1 may “simply” act as a scaffold protein (Rodriguez et al., 2007). Biochemical and genetic studies further revealed that the two MAP2Ks MKK1 and MKK2 interact with both MEKK1 and with MPK4, and that flg22-induced MPK4 activation is impaired in the double mkk1 mkk2 mutant. This indicates that MKK1 and MKK2 are partially redundant in MPK4 mediated downstream signaling (Gao et al., 2008; Qiu et al., 2008b).
MPK4 was originally reported as a negative regulator of plant immunity because the mpk4 mutant accumulates high levels of salicylic acid, constitutively expresses pathogenesis-related (PR) genes, and has a severely dwarfed growth phenotype (Petersen et al., 2000). This phenotype is very similar to that of the mekk1 single and mkk1 mkk2 double mutants, further supporting their functional relationships (Rodriguez et al., 2007; Gao et al., 2008; Qiu et al., 2008b).
MAPK Cascades in Effector-Triggered Immunity
In addition to PTI, plants also employ resistance (R) proteins as cytoplasmic receptors to directly or indirectly recognize specific pathogenic effector proteins injected into host cells as virulence factors. Effector-triggered immunity (ETI) and PTI share a number of responses, although ETI also includes varying levels of rapid, localized cell death in what is called the hypersensitive response. R protein-dependent recognition initiates immune responses in ETI. R proteins may recognize effector proteins either directly or indirectly by monitoring changes in the effector’s host target(s). This latter case gave rise to the guard hypothesis in which R proteins guard host guardees that are manipulated by pathogen effectors (Van Der Biezen and Jones, 1998).
The genetic characterization of the MEKK1/MKK1–MKK2/ MPK4 cascade as a negative regulatory pathway of defense responses was at odds with the activation of the pathway by PAMPs. Instead, it was possible that the severe phenotypes of the kinase knockout mutants were caused by activation of one or more R protein(s) guarding this kinase pathway. Indeed, in an elegant screen for suppressors of the mkk1 mkk2 double mutant, Zhang et al. (2012) identified the R protein SUMM2 (suppressor of mkk1 mkk2). The T-DNA insertion line summ2-8 completely suppressed the severe mkk1 mkk2 phenotype in respect to morphology, cell death, ROS levels and PR gene expression (Zhang et al., 2012). The analogous knockout phenotype of the upstream MAP3K mekk1 is also completely suppressed in the summ2-8 background. Interestingly, although the mpk4 mutant shares a similar phenotype with the knockouts of its upstream kinase partners, the mpk4 phenotype is not fully suppressed by the summ2-8 mutation, as double mpk4 summ2-8 mutants still retain residual cell death and low levels of ROS. This suggests that MPK4 is involved in other pathways independent of SUMM2, and that MPK4 may be guarded by additional R proteins (Zhang et al., 2012; Figure 1A).
The importance of MAPK signaling in immunity is emphasized by studies reporting bacterial effector proteins targeting MAPK cascades for downregulation (Zhang et al., 2007a,b, 2012; Cui et al., 2010). For example, the Pseudomonas syringae effector protein HopAI1 targets and irreversibly inactivates MPK3, MPK4, and MPK6, thereby suppressing immune responses which would otherwise inhibit bacterial growth (Zhang et al., 2007a, 2012). In addition, the P. syringae effector protein AvrB has been shown to interact with and induce the phosphorylation of MPK4, although it has not been shown if this phosphorylation occurs as a direct effect of AvrB action or via recognition of AvrB by the plant immune system (Cui et al., 2010).
In plants carrying functional SUMM2 alleles, immune responses are activated by bacterial effector proteins targeting the MPK4 pathway (Figure 1A). For example, inducible expression of the bacterial HopAI1 effector in wild-type plants gives rise to a defense phenotype similar to that seen in mekk1, mkk1 mkk2, and mpk4 mutants including elevated levels of ROS, PR gene expression, and cell death (Zhang et al., 2012). SUMM2 apparently does not interact directly with the kinase components of the MEKK1/MKK1–MKK2/MPK4 signaling cascade, suggesting that SUMM2 most likely guards a downstream target of MPK4 activity (Zhang et al., 2012). At present, the best studied in vivo substrate of MPK4 activity is MPK4 substrate 1 (MKS1) which forms a nuclear complex with MPK4 and the WRKY33 transcription factor (Andreasson et al., 2005; Qiu et al., 2008a). Phosphorylation of MKS1 follows MPK4 activation by flg22 perception and, once phosphorylated, MKS1 is released from complexes with MPK4, thereby releasing the WRKY33 transcription factor to bind to its cognate target genes (Qiu et al., 2008a). It has therefore been proposed that MPK4 and MKS1 sequester WRKY33 in the absence of pathogens, and free WRKY33 to induce resistance upon pathogen perception (Figure 1B, left).
As MKS1 is the only known direct target of MPK4, Zhang et al. (2012) tested whether MKS1 interacted with the R protein SUMM2 that seemingly guards MPK4 activity. However, no interaction between SUMM2 and MKS1 was detected. Since mks1 mutants have a wild-type growth phenotype, and the mpk4 phenotype is strongly suppressed in the mks1 background, SUMM2 may guard a process downstream of MPK4 that is independent of MKS1 (Petersen et al., 2010).
WRKY Transcription Factors
The plant-specific WRKY family is a large group of transcription factors which bind a conserved W-box sequence in the promoters of numerous genes including those encoding PR proteins. WRKY33 was found to induce the transcription of PHYTOALEXIN DEFICIENT 3 (PAD3) which encodes the cytochrome P450 monooxygenase 71B15 required for synthesis of the antimicrobial compound camalexin (Zhou et al., 1999; Qiu et al., 2008a; Figure 1B). The wrky33 mutant exhibits enhanced susceptibility toward necrotrophic pathogens such as Botrytis cinerea, while WRKY33 overexpression results in increased resistance due to enhanced PAD3 expression (Zheng et al., 2006).
MPK3 and MPK6 activities also induce the production of camalexin. Transient overexpression of the constitutively active, phospho-mimic mutant forms of MKK4/MKK5 (MKK4DD and MKK5DD), which are the upstream MAP2Ks of MPK3/MPK6, has been reported to induce transcription of both PAD2, which encodes γ-glutamylcysteine synthetase functioning in glutathione biosynthesis, and PAD3. Both PAD2 and PAD3 are necessary for camalexin production (Parisy et al., 2007; Ren et al., 2008). Pathogen-induced camalexin accumulation is partially comprised in mpk3 but not notably in mpk6 mutants, yet camalexin accumulation in mpk3 mpk6 double mutants is almost completely abolished (Ren et al., 2008). While this implicates MPK3/MPK6 in camalexin synthesis, caution should be applied in evaluating results obtained from the mpk3 mpk6 double mutant as it is arrested at the cotyledon stage and is unable to initiate true leaves (Wang et al., 2007). Upstream of MPK3/MPK6 in camalexin induction, MKK4 and MKK5 are activated by the MAP3Ks MEKK1 and MAPKKKα in response to fungal pathogens (Ren et al., 2008). Yet another MAP2K, MKK9, whose upstream MAP3K(s) remains unidentified, is also involved in MPK3/MPK6 activation, as plants expressing phospho-mimic MKK9DD produce even more camalexin than plants expressing MKK4DD or MKK5DD (Xu et al., 2008).
To delineate the link between MPK3/MPK6 activation and camalexin accumulation, Mao et al. (2011) elegantly introduced the phospho-mimic mutant NtMEK2DD, an MKK4 and/or MKK5 ortholog from Nicotiana tabacum, into an array of different wrky mutants in a search for essential transcription factors involved in MPK3/MPK6 mediated camalexin induction. Interestingly, NtMEKK2DD was able to induce camalexin accumulation in all tested mutant lines except wrky33. In addition, WRKY33 proved to be a substrate of MKP3/MPK6 activity, and overexpression of non-phosphorylatable forms of WRKY33 could not fully complement the inability of wrky33 mutants to express PAD3 and accumulate camalexin (Mao et al., 2011; Figure 1B, right).
WRKY33-induced PAD3 expression therefore appears to involve both MPK4- and MPK3/MPK6-mediated signaling (Andreasson et al., 2005; Qiu et al., 2008a; Mao et al., 2011). Mao et al. (2011) proposed a model in which PAD3-mediated camalexin induction occurs differentially depending on the type of pathogen causing the immune response. In this model, bacterial pathogens induce an MPK4 mediated response while fungal pathogens initiate an MPK3/MPK6 mediated response. This hypothesis is based on overexpression of the constitutively active MKK4/MKK5 ortholog NtMEKK2DD, rendering MPK3/MPK6 hyperactive and able to induce PAD3 expression (Mao et al., 2011). In support of this hypothesis, the mpk3 mpk6 double mutant is comprised in B. cinerea-induced PAD3 induction (Ren et al., 2008). Nonetheless, and as noted above, some care should be taken with experiments based on mpk3 mpk6 double mutants given their developmental lethality (Wang et al., 2007).
An alternative model may therefore be proposed which combines the MPK4 and MPK3/MPK6 pathways into a dual control of PAD3 regulation in response to pathogen perception (Figure 1B). In such a model, WRKY33 is sequestered in a nuclear complex comprising at least MPK4 and MKS1 in unchallenged plants, and is released following PAMP perception (Qiu et al., 2008a). Phosphorylation is dispensable for WRKY33 to bind its cognate W-box cis-elements, although it does promote transcriptional activation (Mao et al., 2011). This is illustrated by the fact that PAD3 expression is induced in mpk4 plants (Qiu et al., 2008a), perhaps due to the basal activity of free non-phosphorylated WRKY33 or by free WRKY33 activated by basal MPK3 and/or MPK6 activity. In this scenario, once WRKY33 is released from its nuclear complex with MPK4 and MKS1, it is phosphorylated and hence activated by MPK3/MPK6, thereby inducing camalexin levels through PAD3 expression. The elevated PAD3 expression induced from NtMEKK2DD hyper-activated MPK3/MPK6 (Mao et al., 2011) is not in conflict with this model, as it is likely that hyperactive MPK3/MPK6 are able to phosphorylate residual free WRKY33, thus bypassing other possible feedback mechanisms in PAD3 expression.
In this model, MPK4 and MPK3/MPK6 function together as a binary switch conferring dual level regulation. Clarification of the mode of action in which MPK4 and MPK3/MPK6 function clearly needs further elucidation and should include experiments using catalytically inactive and/or inactivatable MPK4 (Petersen et al., 2000; Brodersen et al., 2006). Application of fungal PAMPs to plants expressing catalytically inactive MPK4 might indicate whether phosphorylation of free WRKY33 by endogenous MPK3/MPK6 is enough to induce expression of PAD3.
MAPK in General Stress Signaling
The refined work of Popescu et al. (2009) identified a MAP2K–MAPK phosphorylation network covering 570 MAPK substrates by combinatorially pairing active MAP2Ks with MAPKs, and then subjecting them to a protein microarray phosphorylation assay. Interestingly, the substrates identified were enriched for transcription factors involved in stress responses. Notably, MPK6 phosphorylated 32% of the identified targets, of which 40% overlapped with MPK3 targets (Popescu et al., 2009). This is in agreement with earlier data, similarly obtained from a protein microarray study (Feilner et al., 2005). Equally noteworthy is the finding that MPK3 also shared 50% of its targets with MPK4, revealing intensive synergy in MAPK signaling (Popescu et al., 2009).
In addition to MAPK cascades, ROS also play a pivotal role in stress signaling (Rodriguez et al., 2010). OXI1, a serine/threonine kinase induced by general ROS-generating stimuli, is required for full activation of MPK3/MPK6 after treatment with H2O2 (Rentel et al., 2004). Although OXI1 is characterized as an upstream regulator of MPK3/MPK6 activation, MPK3/MPK6 have been shown to phosphorylate OXI1 in vitro. This suggests that there is a feedback loop, but in vivo data supporting such a loop has not been shown (Forzani et al., 2011).
In addition to MAPK cascade signaling, PAMP perception also induces Ca2+ dependent kinases (CDPKs) by regulating Ca2+ influx channels (Ma et al., 2009; Kwaaitaal et al., 2011). Recent findings indicate that Ca2+ ATPases regulate Ca2+ efflux and function to regulate innate immune defenses (Zhu et al., 2010). Of particular interest is the Ca2+ ATPase ACA8 which was shown to interact with FLS2, and which may well regulate CDPK signaling through flg22 perception (Frei dit Frey et al., 2012).
MPK8 activity has been shown to negatively regulate the expression of OXI1 in order to maintain ROS homeostasis. Remarkably, activation of MPK8 is not limited to the upstream MAP2K MKK3, as the Ca2+ binding protein calmodulin (CaM) is able to bind and activate MPK8 in an Ca2+-dependent manner (Takahashi et al., 2011). CaM-mediated MPK8 activation is interesting because it bypasses the traditional, sequential activation of MAPKs and also unequivocally links MAPK activation with the ROS burst and ion flux during stress signaling. In addition, CaM also mediates MAPK downregulation. MAP kinase phosphatase 1 (MKP1), which interacts with MPK3, MPK4, and MPK6 (Ulm et al., 2002), binds CaM in a Ca2+-dependent manner and stimulates MKP1 phosphatase activity (Lee et al., 2008). The associations between CDPKs and MAPK cascades have recently been review elsewhere (Wurzinger et al., 2011).
Much progress has been made in understanding how MAPK signaling functions in plant immunity. In Arabidopsis, 3 of the 60 identified MAP3Ks are involved in defense, namely MEKK1 (Asai et al., 2002), EDR1 (Frye et al., 2001), and MEKKα (del Pozo et al., 2004; Ren et al., 2008). In addition, at least 6 of the 10 identified MAP2Ks (MKK1, MKK2, MKK4, MKK5, MKK7, and MKK9) are involved in defense signaling (Asai et al., 2002; Djamei et al., 2007; Dóczi et al., 2007; Zhang et al., 2007b; Yoo et al., 2008). This situation requires tight regulation of the spatial and temporal kinase activities in order to impose specificity upon downstream signaling. To shed light on this regulation, high-throughput methods such as those used by Popescu et al. (2009) are particularly valuable and help to outline MAPK signaling cascades. While this progress may be lauded, further work needs to focus on identifying direct, in vivo kinase substrates and their respective phosphorylation sites. This may bring us closer to bridging the apparent gap between PRRs and MAPK cascades, and to understanding how specificity is achieved among MAPK pathways both spatially and temporally.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants to John Mundy from the Danish Research Council for Technology and Production (23-03-0076) and the Strategic Research Council (09-067148) and to Milena Roux from the Natural Science Council (11-116368).
References
Andreasson, E., Jenkins, T., Brodersen, P., Thorgrimsen, S., Petersen, N. H., Zhu, S., Qiu, J.-L., Micheelsen, P., Rocher, A., Petersen, M., Newman, M. A., Bjørn Nielsen, H., Hirt, H., Somssich, I., Mattsson, O., and Mundy, J. (2005). The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 24, 2579–2589.
Asai, T., Tena, G., Plotnikova, J., Willmann, M. R., Chiu, W. L., Gomez-Gomez, L., Boller, T., Ausubel, F. M., and Sheen, J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983.
Bethke, G., Pecher, P., Eschen-Lippold, L., Tsuda, K., Katagiri, F., Glazebrook, J., Scheel, D., and Lee, J. (2012). Activation of the Arabidopsis thaliana mitogen-activated protein kinase MPK11 by the flagellin-derived elicitor peptide, flg22. Mol. Plant Microbe Interact. 25, 471–480.
Brodersen, P., Petersen, M., Nielsen, H. B., Zhu, S., Newman, M., Shokat, K. M., Rietz, S., Parker, J., and Mundy, J. (2006). Arabidopsis MAP kinase 4 regulates salicylic acid- and jasmonic acid/ethylene-dependent responses via EDS1 and PAD4. Plant J. 47, 532–546.
Cui, H., Wang, Y., Xue, L., Chu, J., Yan, C., Fu, J., Chen, M., Innes, R. W., and Zhou, J.-M. (2010). Pseudomonas syringae effector protein AvrB perturbs Arabidopsis hormone signaling by activating MAP kinase 4. Cell Host Microbe 7, 164–175.
del Pozo, O., Pedley, K. F., and Martin, G. B. (2004). MAPKKK[alpha] is a positive regulator of cell death associated with both plant immunity and disease. EMBO J. 23, 3072–3082.
Djamei, A., Pitzschke, A., Nakagami, H., Rajh, I., and Hirt, H. (2007). trojan horse strategy in Agrobacterium transformation: abusing MAPK defense signaling. Science 318, 453–456.
Dóczi, R., Brader, G., Pettkó-Szandtner, A., Rajh, I., Djamei, A., Pitzschke, A., Teige, M., and Hirt, H. (2007). The Arabidopsis mitogen-activated protein kinase kinase MKK3 is upstream of group C mitogen-activated protein kinases and participates in pathogen signaling. Plant Cell 19, 3266–3279.
Feilner, T., Hultschig, C., Lee, J., Meyer, S., Immink, R. G. H., Koenig, A., Possling, A., Seitz, H., Beveridge, A., Scheel, D., Cahill, D. J., Lehrach, H., Kreutzberger, J., and Kersten, B. (2005). High throughput identification of potential Arabidopsis mitogen-activated protein kinases substrates. Mol. Cell. Proteomics 4, 1558–1568.
Felix, G., Duran, J. D., Volko, S., and Boller, T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18, 265–276.
Fiil, B. K., Petersen, K., Petersen, M., and Mundy, J. (2009). Gene regulation by MAP kinase cascades. Curr. Opin. Plant Biol. 12, 615–621.
Forzani, C., Carreri, A., de la Fuente van Bentem, S., Lecourieux, D., Lecourieux, F., and Hirt, H. (2011). The Arabidopsis protein kinase Pto-interacting 1-4 is a common target of the oxidative signal-inducible 1 and mitogen-activated protein kinases. FEBS J. 278, 1126–1136.
Frei dit Frey, N., Mbengue, M., Kwaaitaal, M., Nitsch, L., Altenbach, D., Haweker, H., Lozano-Duran, R., Njo, M. F., Beeckman, T., Huettel, B., Borst, J. W., Panstruga, R., and Robatzek, S. (2012). Plasma membrane calcium ATPases are important components of receptor-mediated signaling in plant immune responses and development. Plant Physiol. 159, 798–809.
Frye, C. A., Tang, D., and Innes, R. W. (2001). Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc. Natl. Acad. Sci. U.S.A. 98, 373–378.
Gao, M., Liu, J., Bi, D., Zhang, Z., Cheng, F., Chen, S., and Zhang, Y. (2008). MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 18, 1190–1198.
Gimenez-Ibanez, S., and Rathjen, J. P. (2010). The case for the defense: plants versus Pseudomonas syringae. Microbes Infect. 12, 428–437.
Gómez-Gómez, L., and Boller, T. (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011.
Ichimura, K., Shinozaki, K., Tena, G., Sheen, J., Henry, Y., Champion, A., Kreis, M., Zhang, S., Hirt, H., Wilson, C., Heberle-Bors, E., Ellis, B. E., Morris, P. C., Innes, R. W., Ecker, J. R., Scheel, D., Klessig, D. F., Machida, Y., Mundy, J., Ohashi, Y., and Walker, J. C. (2002). Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 7, 301–308.
Komis, G., Illés, P., Beck, M., and Šamaj, J. (2011). Microtubules and mitogen-activated protein kinase signalling. Curr. Opin. Plant Biol. 14, 650–657.
Kwaaitaal, M., Huisman, R., Maintz, J., Reinstädler, A., and Panstruga, R. (2011). Ionotropic glutamate receptor (iGluR)-like channels mediate MAMP-induced calcium influx in Arabidopsis thaliana. Biochem. J. 440, 355–365.
Lee, K., Song, E. H., Kim, H. S., Yoo, J. H., Han, H. J., Jung, M. S., Lee, S. M., Kim, K. E., Kim, M. C., Cho, M. J., and Chung, W. S. (2008). Regulation of MAPK phosphatase 1 (AtMKP1) by calmodulin in Arabidopsis. J. Biol. Chem. 283, 23581–23588.
Ma, W., Qi, Z., Smigel, A., Walker, R. K., Verma, R., and Berkowitz, G. A. (2009). Ca2+, cAMP, and transduction of non-self perception during plant immune responses. Proc. Natl. Acad. Sci. U.S.A. 106, 20995–21000.
Mao, G., Meng, X., Liu, Y., Zheng, Z., Chen, Z., and Zhang, S. (2011). Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23, 1639–1653.
Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., Shirasu, K., Narusaka, Y., Kawakami, N., Kaku, H., and Shibuya, N. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 19613–19618.
Parisy, V., Poinssot, B., Owsianowski, L., Buchala, A., Glazebrook, J., and Mauch, F. (2007). Identification of PAD2 as a γ-glutamylcysteine synthetase highlights the importance of glutathione in disease resistance of Arabidopsis. Plant J. 49, 159–172.
Petersen, K., Qiu, J.-L., Lütje, J., Fiil, B. K., Hansen, S., Mundy, J., and Petersen, M. (2010). Arabidopsis MKS1 is involved in basal immunity and requires an intact N-terminal domain for proper function. PLoS ONE 5, e14364. doi: 10.1371/journal.pone.0014364
Petersen, M., Brodersen, P., Naested, H., Andreasson, E., Lindhart, U., Johansen, B., Nielsen, H. B., Lacy, M., Austin, M. J., Parker, J. E., Sharma, S. B., Klessig, D. F., Martienssen, R., Mattsson, O., Jensen, A. B., and Mundy, J. (2000). Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 103, 1111–1120.
Popescu, S. C., Popescu, G. V., Bachan, S., Zhang, Z., Gerstein, M., Snyder, M., and Dinesh-Kumar, S. P. (2009). MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 23, 80–92.
Qiu, J.-L., Fiil, B. K., Petersen, K., Nielsen, H. B., Botanga, C. J., Thorgrimsen, S., Palma, K., Suarez-Rodriguez, M. C., Sandbech-Clausen, S., Lichota, J., Brodersen, P., Grasser, K. D., Mattsson, O., Glazebrook, J., Mundy, J., and Petersen, M. (2008a). Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27, 2214–2221.
Qiu, J.-L., Zhou, L., Yun, B.-W., Nielsen, H. B., Fiil, B. K., Petersen, K., MacKinlay, J., Loake, G. J., Mundy, J., and Morris, P. C. (2008b). Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol. 148, 212–222.
Ren, D., Liu, Y., Yang, K.-Y., Han, L., Mao, G., Glazebrook, J., and Zhang, S. (2008). A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 105, 5638–5643.
Rentel, M. C., Lecourieux, D., Ouaked, F., Usher, S. L., Petersen, L., Okamoto, H., Knight, H., Peck, S. C., Grierson, C. S., Hirt, H., and Knight, M. R. (2004). OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis. Nature 427, 858–861.
Rodriguez, M. C., Adams-Phillips, L., Liu, Y., Wang, H., Su, S. H., Jester, P. J., Zhang, S., Bent, A. F., and Krysan, P. J. (2007). MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 143, 661–669.
Rodriguez, M. C., Petersen, M., and Mundy, J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61, 621–649.
Roux, M., Schwessinger, B., Albrecht, C., Chinchilla, D., Jones, A., Holton, N., Malinovsky, F. G., Tör, M., De Vries, S., and Zipfel, C. (2011). The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23, 2440–2455.
Schwessinger, B., Roux, M., Kadota, Y., Ntoukakis, V., Sklenar, J., Jones, A., and Zipfel, C. (2011). Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 7, e1002046. doi: 10.1371/journal.pgen.1002046
Takahashi, F., Mizoguchi, T., Yoshida, R., Ichimura, K., and Shinozaki, K. (2011). Calmodulin-dependent activation of MAP kinase for ROS homeostasis in Arabidopsis. Mol. Cell 41, 649–660.
Ulm, R., Ichimura, K., Mizoguchi, T., Peck, S. C., Zhu, T., Wang, X., Shinozaki, K., and Paszkowski, J. (2002). Distinct regulation of salinity and genotoxic stress responses by Arabidopsis MAP kinase phosphatase 1. EMBO J. 21, 6483–6493.
Van Der Biezen, E. A., and Jones, J. D. G. (1998). Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci. 23, 454–456.
Wang, H., Ngwenyama, N., Liu, Y., Walker, J. C., and Zhang, S. (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19, 63–73.
Wurzinger, B., Mair, A., Pfister, B., and Teige, M. (2011). Cross-talk of calcium-dependent protein kinase and MAP kinase signaling. Plant Signal. Behav. 6, 8–12.
Xu, J., Li, Y., Wang, Y., Liu, H., Lei, L., Yang, H., Liu, G., and Ren, D. (2008). Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J. Biol. Chem. 283, 26996–27006.
Yoo, S.-D., Cho, Y.-H., Tena, G., Xiong, Y., and Sheen, J. (2008). Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451, 789–795.
Zhang, J., Shao, F., Li, Y., Cui, H., Chen, L., Li, H., Zou, Y., Long, C., Lan, L., Chai, J., Chen, S., Tang, X., and Zhou, J. M. (2007a). A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1, 175–185.
Zhang, X., Dai, Y., Xiong, Y., DeFraia, C., Li, J., Dong, X., and Mou, Z. (2007b). Overexpression of Arabidopsis MAP kinase kinase 7 leads to activation of plant basal and systemic acquired resistance. Plant J. 52, 1066–1079.
Zhang, Z., Wu, Y., Gao, M., Zhang, J., Kong, Q., Liu, Y., Ba, H., Zhou, J., and Zhang, Y. (2012). Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11, 253–263.
Zheng, Z., Qamar, S. A., Chen, Z., and Mengiste, T. (2006). Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48, 592–605.
Zhou, N., Tootle, T. L., and Glazebrook, J. (1999). Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell 11, 2419–2428.
Zhu, X., Caplan, J., Mamillapalli, P., Czymmek, K., and Dinesh-Kumar, S. P. (2010). Function of endoplasmic reticulum calcium ATPase in innate immunity-mediated programmed cell death. EMBO J. 29, 1007–1018.
Zipfel, C. (2008). Pattern-recognition receptors in plant innate immunity. Curr. Opin. Immunol. 20, 10–16.
Keywords: calcium signaling, hypersensitive response, MAP kinase cascade, MAP kinase substrates, pathogen effectors, pattern recognition receptors, reactive oxygen species, resistance proteins
Citation: Rasmussen MW, Roux M, Petersen M and Mundy J (2012) MAP kinase cascades in Arabidopsis innate immunity. Front. Plant Sci. 3:169. doi:10.3389/fpls.2012.00169
Received: 02 May 2012; Paper pending published: 24 May 2012;
Accepted: 09 July 2012; Published online: 24 July 2012.
Edited by:
Alex Jones, The Sainsbury Laboratory, UKReviewed by:
Birgit Kersten, Johann Heinrich von Thünen Institute, GermanyJesus V. Jorrin Novo, University of Cordoba, Spain
Copyright: © 2012 Rasmussen, Roux, Petersen and Mundy. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution, and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: John Mundy, Department of Biology, University of Copenhagen, Ole Maaløes Vej 5, 2200 Copenhagen, Denmark. e-mail:bXVuZHlAYmlvLmt1LmRr



