- Plant Biology Major, Iowa State University, Ames, IA, USA
Numerous signal molecules move through the phloem to regulate development, including proteins, secondary metabolites, small RNAs and full-length transcripts. Several full-length mRNAs have been identified that move long distances in a shootward or rootward direction through the plant vasculature to modulate both floral and vegetative processes of growth. Here we discuss two recently discovered examples of long-distance transport of full-length mRNAs into roots and the potential target genes and pathways for these mobile signals. In both cases, the mobile RNAs regulate root growth. Previously, RNA movement assays demonstrated that transcripts of StBEL5, a transcription factor from the three-amino-loop-extension superclass, move through the phloem to stolon tips to enhance tuber formation in potato (Solanum tuberosum L.). StBEL5 mRNA originates in the leaf and its movement to stolons is induced by a short-day photoperiod. Movement of StBEL5 RNA to roots correlated with increased growth and the accumulation of several transcripts associated with hormone metabolism, including GA2-oxidase1, YUCCA1a and -c, several Aux/IAA types, and PIN1, -2, and -4 was observed. In another example, heterografting techniques were used to identify phloem-mobile Aux/IAA transcripts in Arabidopsis. Movement assays confirmed that these Aux/IAA transcripts are transported into the root system where they suppress lateral root formation. Phloem transport of both StBEL5 and Aux/IAA RNAs are linked to hormone metabolism and both target auxin synthesis genes or auxin signaling processes. The mechanisms of transport for these mobile RNAs, the impact they have on controlling root growth, and a potential transcriptional connection between the BEL1/KNOX complex and Aux/IAA genes are discussed.
Introduction
Mobile Signals to the Root
The phloem functions as a remarkably efficient conduit for transferring numerous signal molecules throughout the plant. Proteins, metabolites, small RNAs and messenger RNAs move readily through the sieve element system to regulate development and respond to environmental changes. Some of the best examples include the Flowering Locus T protein that mediates flowering (reviewed by Turck et al., 2008) and GAI and Knotted1-like transcripts that regulate SAM development and leaf architecture (Kim et al., 2001; Haywood et al., 2005). These signal molecules function by moving through the phloem in a shootward direction but there are also good examples of shoot-to-root signaling as well. These basipetally directed signals have been implicated in controlling development and in responding to nutritive stress. Well-documented cases range from leaf-derived signals for nodule formation to microRNAs that communicate nutritional imbalances.
Nodule formation in legumes is tightly controlled by an intricate root-to-shoot-to-root signaling loop termed autoregulation of nodulation (Ferguson et al., 2010). This long-distance signaling pathway includes peptide hormones, receptor kinases and small metabolites. During nodule formation, a specific peptide hormone is transported from roots to leaves triggering the production of a leaf-derived signal that moves down into roots to suppress further nodulation. As another example, by visualizing radiolabeled hormones in young seedlings, cytokinins have been implicated as signals that move basipetally via symplastic connections in the phloem into roots (Bishopp et al., 2011). This long-distance basipetal transport of cytokinin is critical in regulating polar auxin transport and maintaining the vascular pattern in the root meristem. Phloem-derived miRNAs have also been established as information molecules with the capacity to move down into roots. During phosphate deprivation, miR399 moves from shoots to roots to enhance inorganic phosphate (Pi) uptake and translocation (Lin et al., 2008). This miRNA targets the ubiquitin-conjugating E2 enzyme 24, designated PHO2, suppressing its activity (Aung et al., 2006). PHO2 functions to repress Pi uptake and a pho2 mutant over-accumulates Pi (Dong et al., 1998). Overall, the interaction between miR399 and PHO2 plays a crucial role of in the maintenance of Pi homeostasis. The phloem-mobile miR395 appears to operate in a similar fashion as a general component of the regulatory network of sulfate assimilation (Buhtz et al., 2010; Matthewman et al., 2012). Small RNAs involved in gene silencing also move from shoot to root across grafts and have the capacity to direct epigenetic modifications in the genome of recipient cells that can influence growth and development (Molnar et al., 2010; Bai et al., 2011; Melnyk et al., 2011).
Functional Phloem-Mobile RNAs
Either by phloem cell microdissection or analysis of phloem sap, the transcriptome of phloem is revealed to include thousands of full-length mRNAs with a wide range of potential functions (Omid et al., 2007; Deeken et al., 2008; Kehr and Buhtz, 2008). Despite the fact that so many mRNAs have been identified in phloem sap, the movement of only a few has been confirmed to be associated with a function. These include StBEL5 (Banerjee et al., 2006) and POTH1 (Mahajan et al., 2012) of potato, CmGAI of pumpkin (Haywood et al., 2005), PFP-LeT6 from tomato (Kim et al., 2001), and AUX/IAA (Notaguchi et al., 2012) and FLOWERING LOCUS T (Li et al., 2011; Lu et al., 2012) from Arabidopsis. Whereas, there are numerous examples of acropetal movement of mRNAs confirmed through grafting experiments (Kim et al., 2001; Haywood et al., 2005; Kanehira et al., 2010; Yang and Yu, 2010), there are only a few examples of full-length RNAs moving in a rootward direction. GAI transcripts of apple exhibited the capacity to move in both directions across a graft union, from scion to stock and from stock to scion (Xu et al., 2010). Two specific transcript types, AUX/IAA and StBEL5, have been shown to move into roots and affect a phenotype. Both are involved in auxin synthesis or signaling. The mechanisms of their movement and the potential regulatory networks they affect are the topics of this report.
AUX/IAA mRNAs Move Into Roots and Suppress Growth
AUX/IAA (Auxin/indoleacetic acid) proteins are important transcriptional regulators involved in auxin signaling (Tiwari et al., 2001). In general, they act as repressors by interacting with auxin response factors bound to auxin response elements of target genes that control numerous aspects of growth (Tiwari et al., 2001, 2004). Two types of AUX/IAA transcripts were first identified in phloem sap of melon (Cucumis melo). These melon RNAs were detected in the scion of heterografts of pumpkin/melon confirming that specific AUX/IAA RNAs were capable of long-distance trafficking (Omid et al., 2007). Using tobacco/Arabidopsis heterografts, a subsequent screen of the AUX/IAA family in Arabidopsis revealed two AUX/IAA RNAs, designated IAA18 and −28, capable of long-distance transport (Notaguchi et al., 2012). Using a GUS marker in transgenic lines, promoter activity of both of these genes was localized to leaf vascular tissue.
To assess the function of phloem-mobile IAA18 in root formation, the gain-of-function mutant, diaa18, was used in heterografts with WT stocks (Notaguchi et al., 2012). This dominant mutant, in which the protein is not targeted for degradation by the 26S proteasome, exhibits stable repressor activity in the auxin signaling cascade (Chapman and Estelle, 2009; Vanneste and Friml, 2009). In this model, because the IAA18 protein is not degraded in the diaa18 mutant, it functions to repress root growth even in the presence of auxin. When IAA18 is degraded, auxin response factors are released and auxin activity and lateral root growth are enhanced (Notaguchi et al., 2012). Previous work showed that diaa18 plants exhibited severe defects in lateral root formation, reflective of its role in root development (Rogg et al., 2001; Fukaki et al., 2002; Uehara et al., 2008; Péret et al., 2009; Notaguchi et al., 2012). Micrografting experiments showed that the capacity of diaa18 to repress root growth was transmitted from the scion to the rootstock (Notaguchi et al., 2012). This repression of root growth by the diaa18 scion occurred both with and without auxin treatments.
RNA movement assays utilizing heterografts of WT and double mutants demonstrated that transcripts of both IAA18 and −28 moved from WT scions (Col-0) into both primary and lateral roots of stocks of the double mutant iaa18;iaa28 (Notaguchi et al., 2012). These results clearly establish the long-distance transport of both IAA types and a strong correlation of IAA18 movement with repression of root growth. Using a virus vector system and a Myc protein tag, Notaguchi et al. (2012) showed that, whereas IAA18 and −28 transcripts could move through the phloem, neither of their respective proteins has the capacity to enter the sieve tube system. These results suggest that these two proteins are unlikely to function as long-distance signaling agents in A. thaliana and are consistent with the established instability of these proteins (Rogg and Bartel, 2001).
StBEL5 is Transported Into Roots and Affects Growth
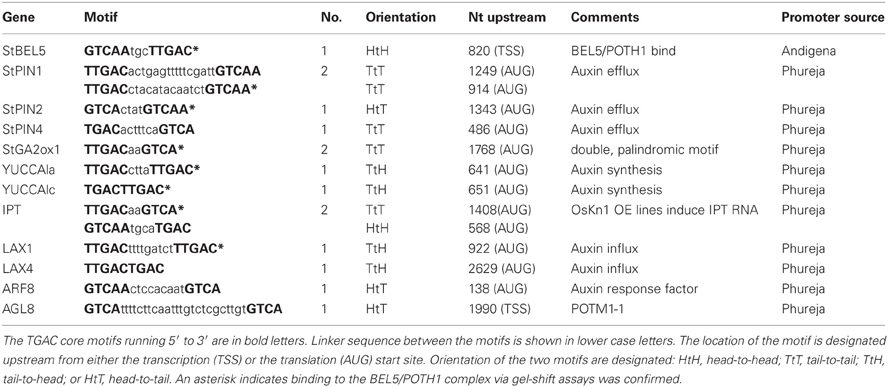
Previous work has clearly established the mRNA of StBEL5, a transcription factor of the three-amino-loop-extension superclass, as phloem mobile and enhancing the tuberization process in potato (Solanum tuberosum L.) by targeting genes that control growth (Banerjee et al., 2006). Working in tandem with the Knotted1-type transcription factor, POTH1, StBEL5 mediates vegetative development by regulating hormone levels (Chen et al., 2003). The BEL5/POTH1 complex binds specifically to a double TTGAC core motif present in target gene promoters (Chen et al., 2004).
RNA movement assays demonstrated that StBEL5 transcripts move through the phloem to stolon tips, the site of tuber induction. StBEL5 mRNA originates in the leaf, and its movement to stolons is induced by a short-day photoperiod. Recent work has established that StBEL5 also moves into roots and affects root growth. Movement into underground organs has been assayed in several studies in a transgenic line, designated GAS:BEL5, that transcribes StBEL5 in leaves only (Banerjee et al., 2006, 2009; Lin et al., 2013). Using the leaf-specific galactinol synthase (GAS) promoter (Ayre et al., 2003) to drive StBEL5 expression and a transgenic RNA-specific sequence tag, movement of StBEL5 RNA into other organs may be readily monitored by using qRT-PCR. In this system, any RNA driven by the GAS promoter and detected in organs other than the leaf is the result of long-distance transport. This promoter construct can essentially separate accumulation by transcription from accumulation by movement. These GAS:BEL5 lines are robust, slightly dwarf [probably due to the activation of GA2ox1 expression in GAS:BEL5 leaves (Lin et al., 2013)], and exhibit normal leaf morphology and enhanced root and tuber development. This phenotype was similar to 35S:BEL5 OE lines except for the shorter plant stature (Chen et al., 2003).
Using both movement assays in whole plants (Figure 1A) and heterografts of GAS:BEL5 scions and WT stocks (Figure 1B), movement of transgenic StBEL5 into roots was tested. The GAS promoter drives leaf-specific expression (Ayre et al., 2003) and in whole transgenic GAS:BEL5 plants, substantial amounts of transgenic StBEL5 RNA were transported into both primary and lateral roots (Figure 1A). To confirm this movement, heterografts of GAS:BEL5 scions and WT stocks were performed and RT-PCR assays were used to detect the StBEL5 transgenic RNA in the roots of WT stock material (Figure 1B). As a negative control GAS:GUS transgenic lines were grafted as scions onto WT stocks. Transgenic StBEL5 RNA was detected in lateral roots of WT stock from four separate GAS:BEL5/WT heterografts whereas, no GUS RNA was detected in lateral roots from WT stock from four separate GAS:GUS/WT heterografts (Figure 1B). In correlation with the long-distance transport of transgenic StBEL5 into roots, root growth was enhanced in the transgenic GAS:BEL5 lines in both soil-grown and in vitro plants by approximately 75% (Figure 2A). Root growth from these transgenic lines was more vigorous and robust than in WT lines (Figure 2B).
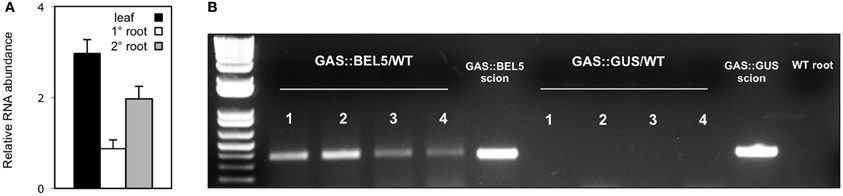
Figure 1. Movement of transgenic StBEL5 mRNA from leaf to root. Quantification of movement was performed on transgenic lines expressing full-length StBEL5 RNA driven by the galactinol synthase (GAS) promoter of melon (Cucumis melo) (A). This promoter is predominately expressed in the minor veins of leaf mesophyll but not in other parts of the plant (Ayre et al., 2003; Banerjee et al., 2009). Relative levels of StBEL5 RNA were quantified (A) from total RNA extracted from new leaves (■), and from either primary (□) or secondary ( ) root samples of a SD-grown GAS:BEL5 transgenic plants. One-step RT-PCR was performed using 200–250 ng of total RNA, a primer for the NOS terminator sequence specific to all transgenic RNAs and a gene-specific primer for the full-length StBEL5 transcript. These primers specifically amplify only transgenic BEL5 RNA. All PCR reactions were standardized and optimized to yield product in the linear range. Homogenous PCR products were quantified by using ImageJ software (Abramoff et al., 2004) and normalized by using 18S rRNA values. Standard errors of the means of three replicate samples are shown. For heterografts (B), micrografts were performed with replicates of either GAS:BEL5 scions on WT andigena stocks or GAS:GUS scions on WT andigena stocks. After 2 weeks in culture, grafts were moved to soil and grown under LDs for 3 weeks and then under SDs for 2 weeks before harvest of roots and leaves. After RNA extraction, RT-PCR with gene-specific primers was performed on RNA from WT lateral roots of both heterografts. A second PCR was performed with nested primers for both types. RNA from scion leaf samples was used as a positive control (scion samples). Two different gene-specific primers were used with a non-plant sequence tag specific for the transgenic StBEL5 RNA to discriminate from the native RNA. Four plants were assayed for both heterografts and are designated 1–4. Wild-type RNA from lateral roots of whole plants (S. tuberosum ssp. andigena) was used in the RT-PCR reactions with StBEL5 transgenic GSPs as a negative control (WT root). Similar negative results were obtained with RNA from WT leaves. (reprinted from Figure 1 of Lin et al., 2013; Copyright American Society of Plant Biologists, www.plantphysiol.org).
) root samples of a SD-grown GAS:BEL5 transgenic plants. One-step RT-PCR was performed using 200–250 ng of total RNA, a primer for the NOS terminator sequence specific to all transgenic RNAs and a gene-specific primer for the full-length StBEL5 transcript. These primers specifically amplify only transgenic BEL5 RNA. All PCR reactions were standardized and optimized to yield product in the linear range. Homogenous PCR products were quantified by using ImageJ software (Abramoff et al., 2004) and normalized by using 18S rRNA values. Standard errors of the means of three replicate samples are shown. For heterografts (B), micrografts were performed with replicates of either GAS:BEL5 scions on WT andigena stocks or GAS:GUS scions on WT andigena stocks. After 2 weeks in culture, grafts were moved to soil and grown under LDs for 3 weeks and then under SDs for 2 weeks before harvest of roots and leaves. After RNA extraction, RT-PCR with gene-specific primers was performed on RNA from WT lateral roots of both heterografts. A second PCR was performed with nested primers for both types. RNA from scion leaf samples was used as a positive control (scion samples). Two different gene-specific primers were used with a non-plant sequence tag specific for the transgenic StBEL5 RNA to discriminate from the native RNA. Four plants were assayed for both heterografts and are designated 1–4. Wild-type RNA from lateral roots of whole plants (S. tuberosum ssp. andigena) was used in the RT-PCR reactions with StBEL5 transgenic GSPs as a negative control (WT root). Similar negative results were obtained with RNA from WT leaves. (reprinted from Figure 1 of Lin et al., 2013; Copyright American Society of Plant Biologists, www.plantphysiol.org).
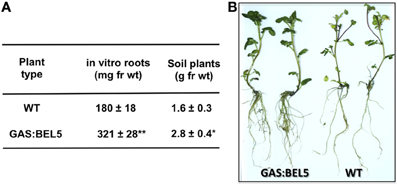
Figure 2. Root development of transgenic lines of Solanum tuberosum ssp. andigena grown in vitro and in soil. For root fresh weight harvests (A), in vitro plantlets were grown for 4 weeks at 27°C under 16 h light, 8 h dark. Roots from in vitro transgenic lines were generally longer and more robust than WT controls (B). Soil plants were grown in pots in a growth chamber under long days (16 h light, 8 h dark) at 24°C days and 18°C nights and harvested after 7 weeks. The SE of the mean of several plants is shown (A). One asterisk indicates a significant difference (p < 0.05) and two asterisks, a significant difference (p < 0.01) using a Student's t test. (Reprinted from Figure 2 of Lin et al., 2013; Copyright American Society of Plant Biologists, www.plantphysiol.org).
Auto-Regulation of StBEL5 in Roots
Examination of the upstream sequence of the StBEL5 promoter revealed the presence of the BEL/Knox tandem TTGAC motif (Chen et al., 2004) with the two core motifs on opposite strands of the DNA (Lin et al., 2013). This discovery suggested the possibility that the StBEL5 gene could be auto-regulated. To test this possibility, two promoter constructs of StBEL5, both approximately 2.0 kb in length, were designed and fused to GUS, one with both motifs intact (proBEL5) and another with one of the motifs deleted, designated mut-proBEL5 (Figure 3). In transgenic lines driving GUS expression with the WT proBEL5 construct, activity was detected in both primary and lateral roots and in the phloem and cortex of primary roots (Lin et al., 2013). In the transgenic lines with the mut-proBEL5 construct, GUS activity was greatly reduced in lateral roots and in the phloem and cortex of primary roots. Overall, these results suggest that auto-regulation of the StBEL5 gene is occurring in roots and that this expression is localized to cortical and phloem cells. Consistent with these observations, levels of endogenous StBEL5 RNA are increased 2.4-fold in lateral roots of GAS:BEL5 plants relative to StBEL5 in WT roots (Lin et al., 2013). Figure 1A shows the relative level of transgenic StBEL5 RNA that is transported into these lateral roots.
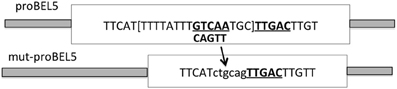
Figure 3. Schematic of the modification of the wild-type StBEL5 promoter sequence. To create the mutated BEL5 promoter used in the transgenic lines reported here, one of the tandem TTGAC cis-elements (underlined and bold) that make up the binding motif for StBEL5 and its Knotted1-like partner, POTH1 (Chen et al., 2004), was deleted. To facilitate cloning, this 5-base motif plus the TGC linker and eight other bp (all in brackets) were removed and replaced by the ctgcag sequence. The intact wild-type double motif sequence begins 820 nt upstream from the start of the StBEL5 5′ untranslated region (Chatterjee et al., 2007). (Reprinted from Figure 6 of Lin et al., 2013; Copyright American Society of Plant Biologists, www.plantphysiol.org).
Results
Long-Distance Transport of StBEL5 Into Roots Targets Gene Expression
Because of the role the StBEL5/KNOX complex plays in targeting genes involved in hormone synthesis and signaling pathways and with recent results confirming StBEL5′s effect on the induction of GA2-oxidase1, YUCCA1a, and isopentenyl transferase (IPT) in roots (Lin et al., 2013), a search for tandem TGAC motifs present in upstream sequence of several hormone genes was undertaken. Sequences meeting the criteria of a double motif present within 2.0 kb of the start codon (LAX4 was the only exception) included three PIN genes (PIN1, −2, and −4), two YUCCA1 genes (1a and 1c), two LIKE-AUX1 genes (LAX1 and −4), one auxin response factor (ARF8), and AGL8, which is the potato MADS box gene, POTM1-1, shown previously by Rosin et al. (2003) to be involved in cyokinin regulation (Table 1). These elements contained linkers between the core motifs ranging from 2 to 24 nt. The motifs of YUCCA1c and LAX4 contained no linker. All four 5′ to 3′ DNA strand orientations for the two core motifs are represented by this group: head-to-head, tail-to-tail, head-to-tail, and tail-to-head (Table 1).
To assay RNA levels of target induction by mobile transgenic StBEL5, RNA samples were taken from lateral roots of the same soil-grown plants used in Figure 1A and compared to target RNAs from WT roots. In this phloem-transport induction system, GAS:BEL5 transgenic lines express StBEL5 only in leaves so that any BEL5 RNA detected elsewhere (e.g., roots) represents long-distance transport. qRT-PCR was performed on eight of the potato genes of Table 1 plus StBEL5, YUCCA1a, and GA2-oxidase1 as controls. StBEL5 induces expression of YUCCA1a, GA2oxidase1 and its own gene in lateral roots (Lin et al., 2013). Six of the candidate genes exhibited induction in response to StBEL5 accumulation (Figures 4A,B). Levels of LAX4 and ARF8 exhibited no significant increase. Both PIN2 and YUCCA1c transcript levels increased by approximately 5-fold.
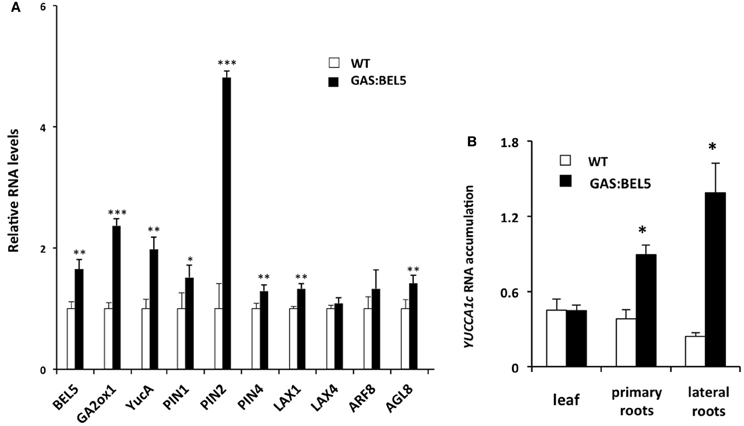
Figure 4. RNA accumulation of several genes in secondary roots of GAS:BEL5 plants (A). In these transgenic lines (■), transgenic StBEL5 transcripts accumulate in roots (Lin et al., 2013). Quantitative real-time RT-PCR with gene-specific primers was used to calculate the relative amounts of each RNA (A). StBEL5 was included as a positive control. Each sample was measured in triplicate and normalized against StActin8 RNA. The fold change in expression was calculated as the comparative threshold cycle method value relative to the mean values obtained from the WT samples (□). YUCCA1c induction in primary and lateral roots of GAS:BEL5 plants (B). Quantification of mRNA in these samples was performed as described by Lin et al. (2013). Standard deviations of the means of three biological replicates are shown with one, two, and three asterisks indicating significant differences (p < 0.05, p < 0.01, p < 0.001, respectively) using a Student's t-test. YucA, YUCCA1a.
Because the GAS:BEL5 line expresses StBEL5 in the leaves, the possibility exists that these target RNAs are up-regulated in leaves and may either move down to roots or activate pathways that lead to their induction in roots. Previous work with this system showed that several genes activated by StBEL5 in roots or stolons were not induced in leaves. These included YUCCA1a and -c, ISOPENTENYL TRANSFERASE, and StBEL5 and −22 (Lin et al., 2013, Figure 4B). One notable exception is GA2ox1 which is induced in both leaves and roots (Lin et al., 2013). As previously discussed, this increase in leaves may very likely explain the slight dwarf phenotype exhibited by GAS:BEL5 transgenic lines. An assay for leaf RNA of the four induced auxin genes, StPIN1, −2, −4, and StLAX1, in the same GAS:BEL5 line used for root induction was performed (Figure 5A). There was no induction of StLAX1 in leaves but the three PIN genes showed slight increases of their transcript levels in leaves that corresponded very closely to their induction levels in roots (Figure 5B). The one exception was StPIN2 which exhibited a 1.8-fold increase in leaves but a 4.8-fold increase in roots. This very high level of root-specific accumulation was also reported for the tomato ortholog of PIN2 (Pattison and Catala, 2012). The correlation in transcript levels in both leaves and roots for StPIN1 and −4 would suggest a similar transcriptional relationship. It is conceivable that StPIN2 transcripts are transported to roots but to date there is no report of any phloem-mobile PIN mRNAs.
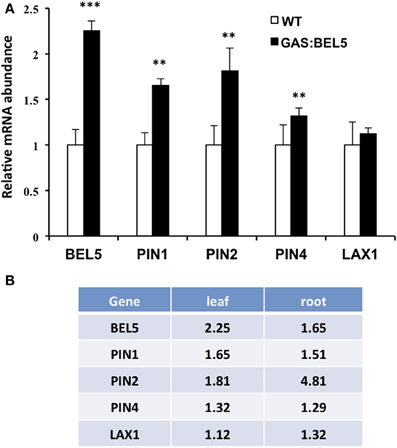
Figure 5. Accumulation of RNA for select target genes in leaves of GAS:BEL5 plants (A) and a comparison of values in both leaves and roots (B). In these transgenic lines (■), transgenic StBEL5 transcripts are transcribed in leaves and move to roots (Lin et al., 2013). Quantitative real-time RT-PCR with gene-specific primers was used to calculate the relative amounts of each RNA. StBEL5 was included as a positive control. Each sample was measured in triplicate and normalized against StActin8 RNA. The fold change in expression was calculated as the comparative threshold cycle method value relative to the mean values obtained from the WT samples (□). The WT value is equivalent to 1.0. Standard deviations of the means of three biological replicates are shown with two and three asterisks indicating significant differences (p < 0.01 and p < 0.001, respectively) using a Student's t-test.
To determine if the StBEL5/POTH1 complex interacts with the double elements identified in upstream sequence of the induced genes, gel-shift assays were undertaken on select targets from Table 1 and Figure 4 (Figure 6). The four cis-elements tested for binding represented three of the four strand orientations: tail-to-head for YUCCA1c, tail-to-tail for StIPT and StPIN1, and head-to-tail for StPIN2. The linker region between the TTGAC core motifs of these four elements ranges from no linkers for YUCCA1c to a 13-nt linker for the StPIN1 motif. StIPT and StPIN2 contain linkers of 2 and 4 nt, respectively. Despite the diversity in strand orientation and linker length, binding of proteins to these elements was consistently strongest for the BEL5/POTH1 complex and the StBEL5 protein alone (Figure 6). No interaction was observed with the glutatione S-transferase (GST) protein alone.
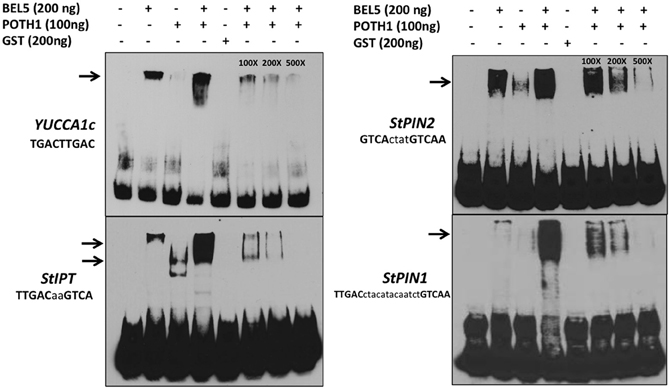
Figure 6. Gel-shift assays of various tandem TGAC core motifs (bold, upper case nucleotides) in four putative target genes of StBEL5 and POTH1 with a range of linker sequence (lower case nucleotides) between motifs. Included here are YUCCA1c, StIPT (isopentenyl transferase), and StPIN1 and −2. The StBEL5 and POTH1 proteins were expressed and purified with a carboxyl-terminal glutatione S-transferase (GST) fusion tag. Each DNA bait was tested for binding with StBEL5-GST, POTH1-GST or GST alone or with StBEL5-GST and POTH1-GST together. Ten fm of synthesized DNA probes 50 nt in length labeled with biotin were used in the binding reaction. The amounts of StBEL5 and POTH1 proteins used in these assays were adjusted to achieve equivalent molarity. Unlabeled DNA bait at 100X, 200X, and 500X concentrations relative to the labeled probe was used in the competition assays. Arrows indicate approximate location of shifted bands.
Induction of AUX/IAA in Roots of GAS:BEL5 Plants
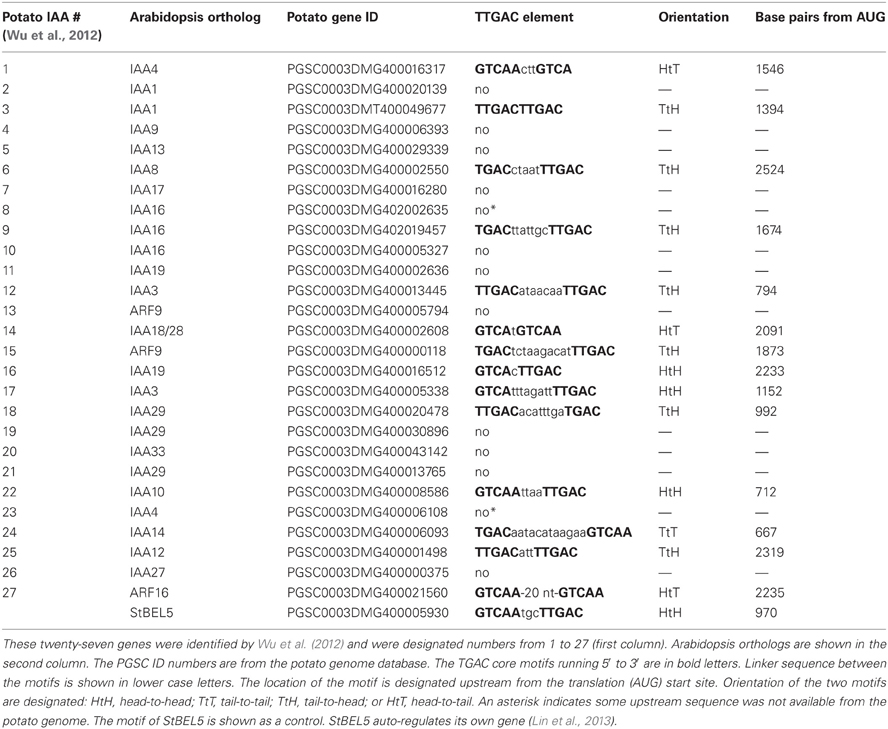
Using ChIP-seq, targets of KNOTTED1 in maize included many genes involved with gibberellic acid, brassinosteroid, cytokinin, and auxin synthesis and signaling pathways (Bolduc et al., 2012). Among the target hormonal genes that bound to KNOTTED1, differential gene expression occurred preferentially for auxin-related genes, including transcription factors involved in auxin signaling like AUX-IAA and ARFs. KNOTTED1 binds to eighteen AUX/IAA and twenty ARF genes, almost half of the AUX-IAA and ARF genes identified in the maize genome (Bolduc et al., 2012; Supplemental Table 7). The ChIP-seq analysis further demonstrated that KNOX transcription factors have degenerate binding sites and that specificity may be acquired through an interaction with binding partners like the BEL1-type transcription factors. In potato, the BEL1/KNOX interaction has been well-documented (Chen et al., 2004; Lin et al., 2013). Of the twenty-seven potato AUX/IAA genes, fourteen contain the tandem TGAC core motif representing all four orientations in their upstream sequences (Table 2). These observations coupled with the results of Bolduc et al. (2012) suggest the intriguing possibility that a connection exists between the BEL1/KNOX transcriptional complex and AUX/IAA gene activity. To test this hypothesis, induction of select StAUX/IAA genes was assayed in the roots of the GAS:BEL5 line. Four StIAA genes, IAA3, −14, −22, and −24, that contained the double TTGAC motif (Table 2) also exhibited induction in correlation with an increase in StBEL5 transcripts, whereas two genes without double TTGAC motifs in their upstream sequence, StIAA4 and −5, exhibited no such increase (Figure 7). The group of genes showing positive induction represented all four motif orientations (tail-to-head, head-to-tail, head-to-head, and tail-to-tail, respectively) with linker sequence between TGAC cores ranging from 0 (StIAA3) to 12 (StIAA24) nt. Induction levels of the four StIAA genes exhibited a negative correlation with the length of the linker sequence suggesting that linker length may influence rate of transcriptional activity. The strongest induction was with StIAA3 and levels of induction decreased steadily as the linker length increased from 0 to 1, 4, and 12 nt (Figure 7, Table 2).
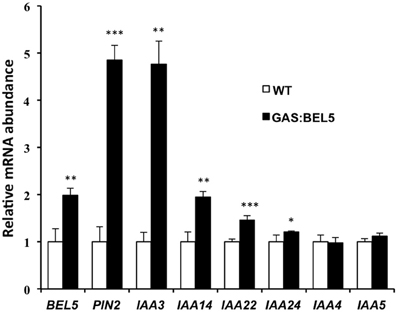
Figure 7. RNA accumulation of several StAUX/IAA genes in secondary roots of GAS:BEL5 plants. In these transgenic lines (■), transgenic StBEL5 transcripts accumulate in roots (Lin et al., 2013). Quantitative real-time RT-PCR with gene-specific primers was used to calculate the relative amounts of each RNA. Each sample was measured in triplicate and normalized against StActin8 RNA. The fold change in expression was calculated as the comparative threshold cycle method value relative to the mean values obtained from the WT samples (□). Standard deviations of the means of three biological replicates are shown with one, two, and three asterisks indicating significant differences (p < 0.05, p < 0.01, p < 0.001, respectively) using a Student's t-test. StBEL5 and StPIN2 were included as positive controls. StIAA4 and -5 do not contain a double TTGAC motif in their upstream sequence (Table 2).
Discussion
IAA18 and StBEL5 Regulate Transcription
Two unique full-length messenger RNAs that originate in leaf vascular tissue, transverse graft unions to move into roots and impact growth. Both are transcription regulators. IAA18, a putative transcription repressor, dimerizes with auxin response factors (ARFs) to suppress their effect in enhancing auxin activity. In Arabidopsis, there are twenty-nine AUX/IAA genes and in rice and maize there are thirty-one (Liscum and Reed, 2002; Jain et al., 2006; Wang et al., 2010). Most AUX/IAA proteins contain four highly conserved domains that facilitate dimerization to target proteins and degradation of the AUX/IAA proteins via the ubiquitin-proteasome pathway (Dharmasiri et al., 2005). Under low auxin concentration, ARFs are inhibited by their interaction with domains III and IV of the AUX/IAAs (Ulmasov et al., 1997; Hagen and Guilfoyle, 2002). Elevated auxin levels releases ARFs from the repressor heterodimer by promoting the degradation of AUX/IAA proteins (Tiwari et al., 2001; Berleth et al., 2004; Dharmasiri et al., 2005). AUX/IAA proteins function in regulating lateral root formation, apical dominance, apical hook curvature, petiole epinasty, leaf architecture, and fruit development. In the model presented by Notaguchi et al. (2012), shoot-derived IAA transcripts are transported into the root tip and contribute to the regulation of lateral root formation. They propose that this regulation is accomplished by an interaction between IAA18 and cytokinin as negative repressors, and auxin as the positive regulator. Their results, however, do not rule out the possible involvement of other signaling agents, including other mobile AUX/IAA transcripts.
Whereas IAA18 functions as a transcription regulator through its interaction with ARFs, the mobile RNA, StBEL5, encodes a transcription factor that acts directly in tandem with a KNOTTED1-like partner to bind to conserved motifs in upstream sequence of numerous target genes that impact hormone synthesis and transport. StBEL5 is regulated in several ways. Its transcription is activated by low levels of white light and short days then facilitate long-distance transport of its mRNA (Banerjee et al., 2006; Chatterjee et al., 2007). RNA chaperone proteins bind to StBEL5 RNA to potentially mediate movement, stabilize the RNA, and repress translation (Cho et al., 2012). Established as a chaperone for mobile RNAs in pumpkin, a polypyrimidine tract-binding (PTB) protein of potato binds to the 3′ untranslated region of StBEL5 (Mahajan et al., 2012). Interaction with a PTB protein can mediate movement and repress translation until it is activated at functional sites in specific cells of roots and stolons. Similar to other BEL1-like transcription factors, StBEL5 is functional with an interactive KNOX partner that is co-expressed and co-localized (Bhatt et al., 2004; Cole et al., 2006).
A Network of Controls
In a remarkable feedback process, auxin moves through the phloem into roots (Chhun et al., 2007) and controls degradation of AUX/IAA proteins, proteins encoded by phloem-mobile transcripts (Notaguchi et al., 2012). Now we add a third component, mobile transcripts of StBEL5, a transcription factor that targets and induces both biosynthetic (YUCCA1) and transport (PIN, LAX) genes in the IAA pathway. These layers of regulation suggest a complex and finely tuned network of control that has very likely evolved across species with fundamental conserved components. There are twenty-seven non-redundant AUX/IAA genes identified in potato (Wu et al., 2012; Table 2) but as yet, none have been confirmed as phloem-mobile. If any potato AUX/IAA transcripts are mobile, then one would expect their levels in phloem cells to be relatively high. Both IAA18 and StBEL5 are transcribed in phloem cells and subsequently loaded into sieve elements (Banerjee et al., 2006; Notaguchi et al., 2012). Based on RNA-Seq data, six of the twenty-seven AUX/IAA RNAs of potato, including the ortholog of IAA18, exhibit very high abundance levels in phloem cells comparable to two known mobile RNAs (Table 3). This data is consistent with the possibility that select AUX/IAA RNAs of potato may also be phloem mobile.
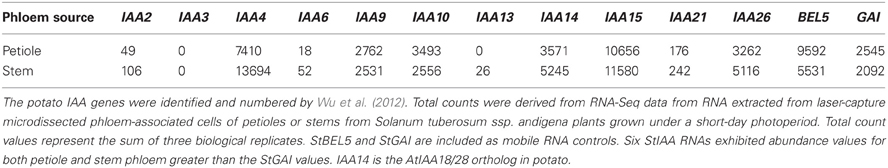
Table 3. Abundance of AUX/IAA transcripts of potato from phloem-associated cells of petioles and stems.
Movement and localization of signals and elicitors are key issues when considering auxin's role in meristem development, vascular tissue development and the genesis of lateral roots. To add one more layer of control, StBEL5 auto-regulates its own expression in roots and stolons (Lin et al., 2013). StBEL5 transcription is localized to root phloem and accumulation of its mobile, transgenic RNA in roots was correlated with increased stele diameter of primary roots, anomalies in the organization of the vascular core, and increased lateral root formation (Lin et al., 2013). All three of these observations can be explained by changes in hormone activity of cytokinin, gibberellic acid, and auxin (Dettmer et al., 2009; Gou et al., 2010; De Smet, 2012). Induction assays showed that StBEL5 targets genes are involved in the metabolism of all three (Figures 4 and 7; Lin et al., 2013). Both zeatin and isopentenyl types of cytokinins increased in over-expression lines of StBEL5 in potato (Chen et al., 2003) and over-expression of BEL1-like genes produced plants that were dwarf due to decreases in active gibberellin levels (Dong et al., 2000; Müller et al., 2001). In similar fashion, a BEL1-like transcription factor was required for regulating both cytokinin and auxin signaling pathways, including PIN1 activity, to establish the correct pattern of ovule development in Arabidopsis (Bencivenga et al., 2012).
A Transcriptional Connection?
ChIP-seq analysis revealed numerous target genes of KNOTTED1 in maize involved with hormone synthesis and signaling and demonstrated that KN1 plays a key role in a regulatory network that influences myriad aspects of development (Bolduc et al., 2012). The maize ga2ox1-like binding motif with two TGAC core motifs (Bolduc and Hake, 2009) was identified as an important element present within the KN1-bound regions. Several AUX-IAA and ARF genes were identified as targets as well. Because of the strong interaction of the StBEL5/POTH1 complex with TGAC core elements (Figure 6, Chen et al., 2004), it is plausible that BEL1-like transcription factors are also involved in many of the interactions documented in this ChIP-seq analysis. For example, at least three genes identified in the KNOTTED1 screen, GA2ox1, IPT, and PIN1, contain elements that bind to StBEL5/POTH1 and are induced by StBEL5 (Figures 4 and 6, Lin et al., 2013).
Although the strongest relative in vitro binding was observed with KNOTTED1 protein alone with tandem TGAC core motifs on the same strand of DNA with a 3-bp linker gap (Bolduc et al., 2012, Supplemental Table 4), we have consistently observed that the most robust gel shifts involved the POTH1/BEL5 heterodimer with motifs on both the same and opposite strands and with linkers between core motifs ranging from zero to 13 bp (Table 1; Figure 6; Chen et al., 2004; Lin et al., 2013). This discrepancy in binding affinity in response to the strand orientation of tandem motifs or linker length between motifs may be a function of the spatial dynamics of the KNOX/KNOX or POTH1/BEL5 dimers. Even though the homeodomains of KNOTTED1- and BEL1-like transcription factors are almost identical, molecular weight differences are significant. StBEL5 has a molecular weight of 76 kDa, whereas the KNOTTED1-types (including POTH1) are approximately 37–40 kDa. A BEL1/KNOX transcriptional complex may very well impart a degree of specificity and spatial flexibility not provided by a KNOTTED1 homodimer.
In summary, these results confirm that AUX/IAA RNAs like IAA18 and StBEL5 and its transcriptional partners are involved in a complex developmental network that regulates hormone activity in roots through the long-distance transport of their mRNAs. Despite the apparent complexity of this system, however, it is almost certain that other long-distance signaling agents, including phloem-mobile RNAs, are also functional in regulating root morphology and are awaiting discovery.
Materials and Methods
Plant Material
Soil-grown plants were maintained in a growth chamber under either a long-day (16 h light at 22°C, 8 h dark at 18°C) or a short-day (8 h light at 22°C, 16 h dark at 18°C for 12 days) photoperiod with a fluence rate of 400 μmol m−2 s−1. Leaves and roots from GAS:BEL5 plants were harvested from soil-grown plants at the 12- to 13-leaf stage, frozen in liquid nitrogen and stored at -80°C prior to RNA extraction. Primary and secondary root types could be distinguished by their morphology. Primary roots are thicker and often exhibit a light purple color, whereas secondary roots are bright white.
Real Time qRT-PCR
Total RNA was extracted from all the plant tissues using the RNeasy Plant Mini Kit (Qiagen, USA) according to manufacturer's instructions. RNA samples with 260/280 ratio from 1.9 to 2.1 and 260/230 ratio from 2.0 to 2.5 were used for qRT-PCR analysis. qRT-PCR analysis was performed with qScript™ One-Step SYBR Green qRT-PCR Kit (Quanta Biosciences) following manufacturer's protocol. Briefly, 50 ng aliquots of total RNA template was subjected to each qRT-PCR reaction in a final volume of 15 μl containing 7.5 μl One-step SYBR Green Master Mix and 0.3 μl of qScript One-step Reverse Transcriptase along with target specific primers (200 nM). All reactions were performed in triplicate using Illumina Eco qPCR machine (Illumina, USA) with fast qPCR cycling parameters (cDNA synthesis: 50°C, 5 min; Taq activation: 95°C, 2 min; PCR cycling (40 cycles): 95°C, 3 s/60°C, 30 s). StACT8 (accession number GQ339765) was used as an endogenous control for normalization of the total RNA template in a reaction. The relative gene quantification (comparative threshold cycle) method (Livak and Schmittgen, 2001) was used to calculate the expression levels of different target genes. Primers ranged from 98 to 160 bp and were mostly designed spanning the introns in order to detect any genomic DNA contamination. Specificity of primers was determined by melting curve analyses and agarose gel (3%) electrophoresis performed following the qRT-PCR experiments. A standard curve was generated based on 6-point (10-fold) serial dilutions of cDNA to calculate the gene specific PCR efficiency. PCR efficiencies of primers ranged from 97 to 110%.
Protein Expression and Purification
Glutathione S-transferase (GST) fusion constructs of StBEL5 and POTH1 described by Chen et al. (2004) were used for preparation of GST-tagged StBEL5 and POTH1. For GST-POTH1 expression, pGEX-POTH1 was transformed into BL21 Star (DE3) E. coli cells (Invitrogen, CA). Cells were grown at 37°C until the OD600 reached 0.6, induced with 1.0 mM IPTG and cultured at 30°C for 5 h. For expression of GST-StBEL5, pGEX-StBEL5 was transformed into ArticExpress E. coli cells (Agilent, CA). Cells were grown at 37°C until the OD600 reached 0.6, induced with 1.0 mM IPTG and cultured at 4°C for 48 h. The GST-tagged protein purification was performed using the Pierce GST Spin Purification kit.
Gel-Shift Assays
Oligos with 3′ biotin labeling were synthesized by the DNA Sequencing and Synthesis Facility, Iowa State University, Ames, IA. Double-stranded DNA was prepared by hybridization of complementary synthetic oligonucleotides. Gel-shift assays were performed using the LightShift Chemiluminescent EMSA Kit from Thermo Scientific according to the manufacturer's protocol provided in the kit with the following modifications. Twenty ul DNA-binding reactions were set up on ice containing 20 mM HEPES (pH 7.5), 10% glicerol (V/V), 0.5% Triton X-100 (V/V), 0.5 mM EDTA (pH 8.0), 50 mM KCl, 2 mM MgCl2, 20 ng/μl BSA, 1 mM DTT, 50 ng/ul of poly(dI-dC) as a non-specific competitor. Ten fmol of labeled DNA was used for all targets. 200 ng of StBEL5, 100 ng of POTH1 and 200 ng of GST proteins were used as indicated in Figure 6. The binding system was incubated on ice for 60 min before electrophoresis. For the competitive assay, unlabeled double stranded DNA fragments were incubated with the recombinant protein on ice for 30 min before addition of the labeled probe.
RNA-Seq Analysis
RNA-Seq was performed on RNA extracted from laser-capture microdissected (LCM) phloem cells of potato (S. tuberosum ssp. andigena) plants grown under short-day conditions (Yu et al., 2007). RNA was isolated using the PicoPure™ RNA Isolation kit (Arcturus). RNA amplification using 70–260 pg of LCM RNA samples from three biological replicates from both stem and petiole sections was performed. The Ovation RNA-Seq kit (NuGEN) was used for cDNA synthesis. One ug of cDNA from each replicate sample was subjected to 100-cycle paired- or single-end sequence cluster generation on an Illumina HiSeq 2000 instrument at the DNA Facility, Iowa State University. The reads from sequencing were saved in fastq format and aligned to the potato genome (PGSC_DM_v3.4_gene.fasta & PGSC_DM_v3.4_gene.gff from http://solanaceae.plantbiology.msu.edu/pgsc_download.shtml) with Genomic Short-read Nucleotide Alignment Program (GSNAP). False Discovery rate (FDR) was set at 0.05. Read numbers were recorded for each replicate and analyzed for abundance. The longest peptide sequence was selected from the genome database for all the genes in both the stem and petiole lists (PGSC_DM_v3.4_pep_representative.fasta). Functions of these genes were analyzed with Blast2Go (http://www.blast2go.com/b2glaunch/start-blast2go).
Primers and Oligos Used for qRT-PCR and Gel-Shift Assays
Primers used for qRT-PCR
BEL5F: 5′-CTGCAACAGCTAGGAATGATG-3′
BEL5R: 5′-ATGATTTTGTCTGAATCCTTTGGG-3′
GA2ox1F: 5′- AGGCACAGAGTGATCGCAGAT-3′
GA2ox1R: 5′- TGGTGGCCCTCCAAAGTAAA-3′
YUCCA1aF: 5′-CATTATCACAAATAAAAACCGGAAA-3′
YUCCA1aR: 5′-TGCCATCTAAAAATCTTGCACC-3′
PIN1F: 5′- GCACCAAATCCTGGCATGT-3′
PIN1R: 5′- AGCTGTATTCTTGTGTGCTTTGGT-3′
PIN2F: 5′-GCAAGTTTGATTGGACTCATTTG-3′
PIN2R: 5′-TGAGATCGAACCTTTTACAATCG-3′
PIN4F: 5′- GTTTCATTGCGGCGGATTC-3′
PIN4R: 5′- CCCATAGCGAAAGAACAACC-3′
LAX1F: 5′- GCGCCTATCCACCACTAAGAAG-3′
LAX1R: 5′- GAAGTATTGCAACAACCCCATGTA-3′
LAX4F: 5′- GCCACCGTGCACAACTAGG-3′
LAX4R: 5′- CACTAACACATTTTGGGTTACATGC-3′
ARF8F: 5′- CAGCCTAAGCGGCATCTTCT-3′
ARF8R: 5′- AGCCTTTTGGCGCTAACAAA-3′
AGL8F: 5′- AGCAAAACAACCAGCTTTCCAA-3′
AGL8R: 5′- TGATCCCACTGATTTTGCTGTG-3′
YUCCA1cF: 5′-ACAAATACAAAGAGGTGTGTATTCGT-3′
YUCCA1cR: 5′-CGTAGTCATTGTATCCGTCCTGA-3′
ACTF: 5′-TGATTGGTATGGAAGCTGCAG-3′
ACTR: 5′-CCACTGAGCACAATGTTACCG-3′
StIAA3 Fw: CTGATCTTCGATCAATTTCATGG
StIAA3 Rv: GACCTATTGCTGCCTTGTGCTA
StIAA4 Fw: CCAGCATTACTATTAGGCGAGG
StIAA4 Rv: CCATGTCGTAATCAGGTAAAGC
StIAA5 Fw: GACTACTGAGGCCAAAGGACTTG
StIAA5 Rv: TGTGGTCTCATTTGATCATTTGC
StIAA14 Fw: TGATGTGAATGAGCTAACGAGATG
StIAA14 Rv: GCTTCTGCAACTACACTTGAACAA
StIAA22 Fw: TAGGTCAGACAAAGAATCAACTT
StIAA22 Rv: CCATCCACTAAAATTTCCTTCTA
StIAA24 Fw: AAGCCAATTGATGGTGTGCA
StIAA24 Rv: ATACAAGCACATGAAAACAACAA
Sequences of oligos used for gel shifts
YUCCA1cS: 5′-AAAAAATTACAAATAAATGACTTGACTAATGTTGTTATTAATCTCCACA-3′
YUCCA1cA: 5′-TGTGGAGATTAATAACAACATTAGTCAAGTCATTTATTTGTAATTTTTT-3′
IPTS: 5′-TTTTTTTTTGGTTTTAAGTTTGACAAGTCAGGTCTAATTTGACATCCTT-3′
IPTA: 5′-AGGATGTCAAATTAGACCTGACTTGTCAAACTTAAAACCAAAAAAAAA-3′
PIN1Fw: 5′-GTCTGTGTATGATTTTGACCTACATACAATCTGTCAACTAATGTGTATGA-3′
PIN1Rv: 5′-TCATACACATTAGTTGACAGATTGTATGTAGGTCAAAATCATACACAGAC-3′
PIN2Fw: 5′-AAATGTGAAAGTCACTATGTCAATCATTATTT-3′
PIN2Rv: 5′-AAATAATGATTGACATAGTGACTTTCACATTT-3′
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Thanks to Mei Yu for her help in optimizing the gel-shift assay protocol and to Sung Ki Cho and Coralie Lashbrook for their contributions to the RNA-Seq project. Research on StBEL5 described in this report was supported by the National Research Initiative grant no. 2008-02806 from the USDA National Institute of Food and Agriculture and National Science Foundation-Plant Genome Research Program award no. 082065.
References
Abramoff, M. D., Magelhaes, P. J., and Ram, S. J. (2004). Image processing with ImageJ. Biophot. Int. 11, 36–42.
Aung, K., Lin, S. I., Wu, C. C., Huang, Y. T., Su, C. L., and Chiou, T. J. (2006). pho2, a phosphate over accumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol. 141, 1000–1011. doi: 10.1104/pp.106.078063
Ayre, B. G., Blair, J. E., and Turgeon, R. (2003). Functional and phylogenetic analyses of a conserved regulatory program in the phloem of minor veins. Plant Physiol. 133, 1229–1239. doi: 10.1104/pp.103.027714
Bai, S., Kasai, A., Yamada, K., Li, T., and Harada, T. (2011). A mobile signal transported over a long distance induces systemic transcriptional gene silencing in a grafted partner. J. Expt. Bot. 62, 4561–4570.
Banerjee, A. K., Chatterjee, M., Yu, Y., Suh, S. G., Miller, W. A., and Hannapel, D. J. (2006). Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway. Plant Cell 18, 3443–3457. doi: 10.1105/tpc.106.042473
Banerjee, A. K., Lin, T., and Hannapel, D. J. (2009). Untranslated regions of a mobile transcript mediate RNA metabolism. Plant Physiol. 151, 1831–1843. doi: 10.1104/pp.109.144428
Bencivenga, S., Simonini, S., Benková, E., and Colombo, L. (2012). The transcription factors BEL1 and SPL are required for cytokinin and auxin signaling during ovule development in Arabidopsis. Plant Cell 24, 2886–2897. doi: 10.1105/tpc.112.100164
Berleth, T., Krogan, N. T., and Scarpella, E. (2004). Auxin signals—turning genes on and turning cells around. Curr. Opin. Plant Biol. 7, 553–563.
Bhatt, A. M., Etchells, J. P., Canales, C., Lagodienko, A., and Dickinson, H. (2004). VAAMANA–a BEL1-like homeodomain protein, interacts with KNOX proteins BP and STM and regulates inflorescence stem growth in Arabidopsis. Gene 328, 103–111. doi: 10.1016/j.gene.2003.12.033
Bishopp, A., Lehesranta, S., Vatén, A., Help, H., El-Showk, S., Scheres, B., et al. (2011). Phloem-transported cytokinin regulates polar auxin transport and maintains vascular pattern in the root meristem. Curr. Biol. 21, 927–932.
Bolduc, N., and Hake, S. (2009). The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell 21, 1647–1658. doi: 10.1105/tpc.109.068221
Bolduc, N., Yilmaz, A., Mejia-Guerra, M. K., Morohashi, K., O'Connor, D., Grotewold, E., et al. (2012). Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Develop. 26, 1685–1690.
Buhtz, A., Pieritz, J., Springer, F., and Kehr, J. (2010). Phloem small RNAs, nutrient stress responses, and systemic mobility. BMC Plant Biol. 10:64. doi: 10.1186/1471-2229-10-64
Chapman, E. J., and Estelle, M. (2009). Mechanism of auxin-regulated gene expression in plants. Ann. Rev. Genet. 43, 265–285. doi: 10.1146/annurev-genet-102108-134148
Chatterjee, M., Banerjee, A. K., and Hannapel, D. J. (2007). A BELL1-like gene of potato is light activated and wound inducible. Plant Physiol. 145, 1435–1443. doi: 10.1104/pp.107.105924
Chen, H., Banerjee, A. K., and Hannapel, D. J. (2004). The tandem complex of BEL and KNOX partners is required for transcriptional repression of ga20ox1. Plant J. 38, 276–284. doi: 10.1111/j.1365-313X.2004.02048.x
Chen, H., Rosin, F. M., Prat, S., and Hannapel, D. J. (2003). Interacting transcription factors from the TALE superclass regulate tuber formation. Plant Physiol. 132, 1391–1404. doi: 10.1104/pp.103.022434
Chhun, T., Uno, Y., Taketa, S., Azuma, T., Ichii, M., Okamoto, T., et al. (2007). Saturated humidity accelerates lateral root development in rice (Oryza sativa L.) seedlings by increasing phloem-based auxin transport. J. Exp. Bot. 58, 1695–1704.
Cho, S. K., Kang, I. H., Carr, T., and Hannapel, D. J. (2012). Using the yeast three-hybrid system to identify proteins that interact with a phloem-mobile mRNA. Front. Plant Physiol. 3:189. doi: 10.3389/fpls.2012.00189
Cole, M., Nolte, C., and Werr, W. (2006). Nuclear import of the transcription factor SHOOT MERISTEMLESS depends on heterodimerization with BLH proteins expressed in discrete sub-domains of the shoot apical meristem of Arabidopsis thaliana. Nucleic Acids Res. 34, 1281–1292. doi: 10.1093/nar/gkl016
Deeken, R., Ache, P., Kajahn, I., Klinkenberg, J., Bringmann, G., and Hedrich, R. (2008). Identification of Arabidopsis thaliana phloem RNAs provides a search criterion for phloem-based transcripts hidden in complex datasets of microarray experiments. Plant J. 55, 746–759. doi: 10.1111/j.1365-313X.2008.03555.x
De Smet, I. (2012). Lateral root initiation: one step at a time. New Phytol. 193, 867–873. doi: 10.1111/j.1469-8137.2011.03996.x
Dettmer, J., Elo, A., and Helariutta, Y. (2009). Hormone interactions during vascular development. Plant Mol. Biol. 69, 347–360.
Dharmasiri, N., Dharmasiri, S., and Estelle, M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445. doi: 10.1038/nature03543
Dong, B., Rengel, Z., and Delhaize, E. (1998). Uptake and translocation of phosphate by pho2 mutant and wild-type seedlings of Arabidopsis thaliana. Planta 205, 251–256. doi: 10.1007/s004250050318
Dong, Y. H., Yao, J. L., Atkinson, R. G., Putterill, J. J., Morris, B. A., and Gardner, R. C. (2000). MDH1: an apple homeobox gene belonging to the BEL1 family. Plant Mol. Biol. 42, 623–633.
Ferguson, B. J., Indrasumunar, A., Hayashi, S., Lin, M.-H., Lin, Y.-H., Reid, D. E., and Gresshoff, P. M. (2010). Molecular analysis of legume nodule development and autoregulation. J. Integ. Plant Biol. 52, 61–76.
Fukaki, H., Tameda, S., Masuda, H., and Tasaka, M. (2002). Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY- ROOT/IAA14 gene of Arabidopsis. Plant J. 29, 153–168. doi: 10.1046/j.0960-7412.2001.01201.x
Gou, J., Strauss, S. H., Tsai, C. J., Fang, K., Chen, Y., Jiang, X., et al. (2010). Gibberellins regulate lateral root formation in Populus through interactions with auxin and other hormones. Plant Cell 22, 623–639. doi: 10.1105/tpc.109.073239
Hagen, G., and Guilfoyle, T. (2002). Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol. Biol. 49, 373–385.
Haywood, V., Yu, T. S., Huang, N. C., and Lucas, W. J. (2005). Phloem long-distance trafficking of GIBBERELLIC ACID-INSENSITIVE RNA regulates leaf development. Plant J. 42, 49–68. doi: 10.1111/j.1365-313X.2005.02351.x
Jain, M., Kaur, N., Garg, R., Thakur, J. K., Tyagi, A. K., and Khurana, J. P. (2006). Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct. Integr. Genomics 6, 47–59.
Kanehira, A., Yamada, K., Iwaya, T., Tsuwamoto, R., Kasai, A., Nakazono, M., et al. (2010). Apple phloem cells contain some mRNAs transported over long distances. Tree Genet. Genomes 5, 635–642.
Kehr, J., and Buhtz, A. (2008). Long distance transport and movement of RNA through the phloem. J. Exp. Bot. 59, 85–92.
Kim, M., Canio, W., Kessler, S., and Sinha, N. (2001). Developmental changes due to long- distance movement of a homeobox fusion transcript in tomato. Science 293, 287–289. doi: 10.1126/science.1059805
Li, C., Gu, M., Shi, N., Zhang, H., Yang, X., Osman, T., et al. (2011). Mobile FT mRNA contributes to the systemic florigen signalling in floral induction. Sci. Rep. 1:73. (Epub Aug 24).
Lin, S. I., Chiang, S. F., Lin, W. Y., Chen, J. W., Tseng, C. Y., Wu, P. C., et al. (2008). Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol. 147, 732–746. doi: 10.1104/pp.108.116269
Lin, T., Sharma, P., Gonzalez, D. H., Viola, I. L., and Hannapel, D. J. (2013). The impact of the long-distance transport of a BEL1-like mRNA on development. Plant Physiol. 161, 760–772. doi: 10.1104/pp.112.209429
Liscum, E., and Reed, J. W. (2002). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49, 387–400.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, K. J., Huang, N. C., Liu, Y. S., Lu, C. A., and Yu, T. S. (2012). Long-distance movement of Arabidopsis FLOWERING LOCUS T RNA participates in systemic floral regulation. RNA Biol. 9, 653–662. doi: 10.4161/rna.19965
Mahajan, A., Bhogale, S., Kang, I. H., Hannapel, D. J., and Banerjee, A. K. (2012). The mRNA of a Knotted1-like transcription factor of potato is phloem mobile. Plant Mol. Biol. 79, 595–608.
Matthewman, C. A., Kawashima, C. G., Húska, D., Csorba, T., Dalmay, T., and Kopriva, S. (2012). miR395 is a general component of the sulfate assimilation regulatory network in Arabidopsis. FEBS Lett. 586, 3242–3248. doi: 10.1016/j.febslet.2012.06.044
Melnyk, C. W., Molnar, A., Bassett, A., and Baulcombe, D. C. (2011). Mobile 24 nt small RNAs direct transcriptional gene silencing in the root meristems of Arabidopsis thaliana. Curr. Biol. 21, 1678–1683.
Molnar, A., Melnyk, C. W., Bassett, A., Hardcastle, T. J., Dunn, R., and Baulcombe, D. C. (2010). Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328, 872–875. doi: 10.1126/science.1187959
Müller, J., Wang, Y., Franzen, R., Santi, L., Salamini, F., and Rohde, W. (2001). In vitro interactions between barley TALE homeodomain proteins suggest a role for protein-protein associations in the regulation of Knox gene function. Plant J. 27, 13–23.
Notaguchi, M., Wolf, S., and Lucas, W. J. (2012). Phloem-mobile Aux/IAA transcripts target to the root tip and modify root architecture. J. Integr. Plant Biol. 54, 760–772.
Omid, A., Keilin, T., Glass, A., Leshkowitz, D., and Wolf, S. (2007). Characterization of phloem-sap transcription profile in melon plants. J. Expt. Bot. 58, 3645–3656.
Pattison, R. J., and Catala, C. (2012). Evaluating auxin distribution in tomato (Solanum lycopersicum) through an analysis of the PIN and AUX/LAX gene families. Plant J. 70, 585–598. doi: 10.1111/j.1365-313X.2011.04895.x
Péret, B., Larrieu, A., and Bennett, M. J. (2009). Lateral root emergence: a difficult birth. J. Exp. Bot. 60, 3637–3643.
Rogg, L. E., and Bartel, B. (2001). Auxin signaling: derepression through regulated proteolysis. Develop. Cell 1, 595–604.
Rogg, L. E., Lasswell, J., and Bartel, B. (2001). A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13, 465–480.
Rosin, F. M., Hart, J. K., Van Onckelen, H., and Hannapel, D. J. (2003). Suppression of a vegetative MADS box gene of potato activates axillary meristem development. Plant Physiol. 131, 1613–1622. doi: 10.1104/pp.102.012500
Tiwari, S. B., Hagen, G., and Guilfoyle, T. J. (2004). Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16, 533–543. doi: 10.1105/tpc.017384
Tiwari, S. B., Wang, X. J., Hagen, G., and Guilfoyle, T. J. (2001). AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13, 2809–2822.
Turck, F., Fornara, F., and Coupland, G. (2008). Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Ann. Rev. Plant Biol. 59, 573–594. doi: 10.1146/annurev.arplant.59.032607.092755
Uehara, T., Okushima, Y., Mimura, T., Tasaka, M., and Fukaki, H. (2008). Domain II mutations in CRANE/IAA18 suppress lateral root formation and affect shoot development in Arabidopsis thaliana. Plant Cell Physiol. 49, 1025–1038. doi: 10.1093/pcp/pcn079
Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T. J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963–1971.
Vanneste, S., and Friml, J. (2009). Auxin: a trigger for change in plant development. Cell 136, 1005–1016. doi: 10.1016/j.cell.2009.03.001
Wang, Y., Deng, D., Bian, Y., Lv, Y., and Xie, Q. (2010). Genome-wide analysis of primary auxin-responsive Aux/IAA gene family in maize (Zea mays L.). Mol. Biol. Rep. 37, 3991–4001.
Wu, J., Peng, Z., Liu, S., He, Y., Cheng, L., Kong, F., et al. (2012). Genome-wide analysis of Aux/IAA gene family in Solanaceae species using tomato as a model. Mol. Genet. Genomics 287, 295–311.
Xu, H., Zhang, W., Li, M., Harada, T., Han, Z., and Li, T. (2010). Gibberellic acid insensitive mRNA transport in both directions between stock and scion in Malus. Tree Genet. Genomes 6, 1013–1019.
Yang, H. W., and Yu, T. S. (2010). Arabidopsis floral regulators FVE and AGL24 are phloem-mobile RNAs. Bot. Stud. 51, 17–26.
Keywords: auxin, hormones, mobile RNAs, non-cell-autonomous, potato, StBEL5
Citation: Hannapel DJ, Sharma P and Lin T (2013) Phloem-mobile messenger RNAs and root development. Front. Plant Sci. 4:257. doi: 10.3389/fpls.2013.00257
Received: 12 March 2013; Accepted: 25 June 2013;
Published online: 17 July 2013.
Edited by:
Sylvie Dinant, Institut National de la Recherche Agronomique, FranceReviewed by:
Jurriaan Ton, University of Sheffield, UKSylvie Dinant, Institut National de la Recherche Agronomique, France
Copyright © 2013 Hannapel, Sharma and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: David J. Hannapel, Plant Biology Major, Iowa State University, 253 Horticulture Hall, Ames, IA 50011-1100, USA e-mail: djh@iastate.edu