- Génétique, Reproduction et Développement, UMR CNRS 6293/U1103 INSERM/Clermont Université, Université Blaise Pascal, Aubiére cedex, France
Failure to repair DNA double strand breaks (DSB) can lead to chromosomal rearrangements and eventually to cancer or cell death. Radiation and environmental pollutants induce DSB and this is of particular relevance to plants due to their sessile life style. DSB also occur naturally in cells during DNA replication and programmed induction of DSB initiates the meiotic recombination essential for gametogenesis in most eukaryotes. The linear nature of most eukaryotic chromosomes means that each chromosome has two “broken” ends. Chromosome ends, or telomeres, are protected by nucleoprotein caps which avoid their recognition as DSB by the cellular DNA repair machinery. Deprotected telomeres are recognized as DSB and become substrates for recombination leading to chromosome fusions, the “bridge-breakage-fusion” cycle, genome rearrangements and cell death. The importance of repair of DSB and the severity of the consequences of their misrepair have led to the presence of multiple, robust mechanisms for their detection and repair. After a brief overview of DSB repair pathways to set the context, we present here an update of current understanding of the detection and signaling of DSB in the plant, Arabidopsis thaliana.
DSBs Repair Pathways in Arabidopsis Thaliana
Double strand breaks (DSB) repair pathways are classed as either homologous recombination (HR) or non-homologous end-joining (NHEJ), depending upon the dependence or not on DNA sequence homology between the recombining molecules. HR requires the presence of an intact homologous DNA template and is most active in S/G2 phase when the sister chromatid is present. The critical step during HR is the formation of RAD51 filament on the 3′ ended single-stranded DNA (ssDNA) produced by resection of the breaks. The nucleofilament formed by RAD51 on the broken DNA molecule catalyzes the invasion of a homologous DNA template sequence by the 3′ ended DNA strand(s), which are extended through DNA synthesis, and finally the joint recombination intermediate is resolved to complete the process (for review, Heyer and Liu, 2010). The major players in HR are very highly conserved and most have been identified and characterized in Arabidopsis thaliana (Mannuss et al., 2011).
The participation, or not, of the KU complex permits classification of NHEJ pathways into two categories: direct joining of breaks through the KU-dependent pathway and end-joining involving microhomologies by the KU-independent microhomology-mediated (MMEJ) and “alternative” or “back-up” end-joining (Alt-NHEJ or B-NHEJ; for review, Decottignies, 2013). In Arabidopsis the KU-dependent pathway has been the subject of a number of studies (Riha et al., 2002; Friesner and Britt, 2003; Gallego et al., 2003; Van Attikum et al., 2003). The distinction between KU-independent pathways is not clear because both imply the use of microhomology sequence to repair the break. In vertebrates, it is known that Alt-NHEJ is based on the action of proteins usually known for their role in single strand breaks repair XRCC1, PARP1 and LIG3 (Decottignies, 2013). In Arabidopsis, the conservation of this pathway has been confirmed through studies of XRCC1 (Charbonnel et al., 2010) and PARP1/PARP2 (Jia et al., 2013). Concerning the MMEJ pathway, the first actors identified were the MRX (MRN) and the Rad1/Rad10 (ERCC1/XPF) complexes in yeast (Ma et al., 2003). Similarly in Arabidopsis, MRE11 has been implicated in the use of microhomologies in telomere fusions (Heacock et al., 2004) and XPF has been shown to be involved in a third NHEJ pathway of DSB repair independent of the KU complex and XRCC1 (Charbonnel et al., 2011).
The viability of the single and multiple mutants for each of these pathways in Arabidopsis permitted study of the kinetics of DSB repair in planta, establishing a hierarchy of DSB repair pathways in Arabidopsis (Charbonnel et al., 2011). A surprising result of this study was the ability of quadruple ku80 xrcc1 xpf xrcc2 mutants (invalidated for all known HR and NHEJ pathways) to repair ionising radiation (IR)-induced DSB, but at a very reduced rate. Although this “repair” is accompanied by high levels of anaphase chromosome bridging, plants cells are thus able to repair DSB in the absence of all four major DSB repair pathways. This results points to another end-joining pathway that would be activated in case of extreme stress and could be one part of the explanation of the striking ability of plants to develop in presence of high levels of genome damage.
The choice of repair mechanisms is tightly regulated with respect to the cell cycle phase and the nature of the break (Chapman et al., 2012). DSB end resection has been shown to be an essential step for the choice of repair pathway, with recent reports showing the implication of 53BP1-RIF1 in blocking resection and thus stimulating NHEJ, and BRCA1-CtIP promoting DNA resection and HR in mammals (Chapman et al., 2013; Escribano-Díaz et al., 2013; Zimmermann et al., 2013). CtIP (Uanschou et al., 2007) and BRCA1 (Lafarge and Montane, 2003; Trapp et al., 2011) orthologs, but not 53BP1 or RIF1, have been described in Arabidopsis, but no detail of their roles in these processes have been reported.
ncRNA (non-coding RNA) are clearly involved in multiple aspects of DNA repair. miRNA (microRNA) transcription is induced after DNA damage and these small RNA are believed to be involved in the regulation of DNA damage repair proteins (reviewed by Chowdhury et al., 2013). Recent work shows links directly to DSB repair in Arabidopsis as well as in mammalian cells. Small RNA (diRNA) are produced directly at break sites and are required for correct repair, probably through chromatin modifications or through the recruitment of repair proteins to facilitate repair (Wei et al., 2012).
Signaling of DSBs
The first essential step of the repair process is the recognition and the signaling of the DNA break. This step is critical as it allows cell-cycle arrest, recruitment of DSB repair proteins, chromatin remodeling and eventually cell death or senescence (Goodarzi et al., 2010). In yeast as well as in mammals, the main factors involved in the sensing of the DSB are the MRX/N (Mre11, Rad50 and Xrs2/Nbs1) and the KU (Ku70/Ku80) complexes that compete for binding to unprocessed DSBs (Hiom, 2010). Together with DNA-PKcs, the human KU complex, forming the DNA-PK holoenzyme, functions as a DNA end-bridging factor leading to repair via NHEJ, essentially in G1 phase (Lieber, 2010). In G2 phase, the binding of KU is inhibited and the MRN complex initiates repair via HR (Heyer and Liu, 2010). In plants as well as in yeast, the DNA-PKcs enzyme is not conserved, hence the tethering of the DNA ends is presumably carried out by the MRN complex or by other proteins.
The signaling role is then assumed by specific kinases belonging to the PI3K-like protein kinase family (PIKK): Tel1/ATM and Mec1/ATR. The binding of the yeast MRX complex to the DSB promotes the recruitment of Tel1 leading to Tel1-dependent cell cycle checkpoint activation prior to DNA processing (Usui et al., 2001). Absence of Tel1 can be compensated for by Mec1 (Morrow et al., 1995), with the yeast tel1 mutant being checkpoint sufficient and not hyper-sensitive to DNA damaging agents (Mantiero et al., 2007). In vertebrates, ATM is activated by DNA double-strand breaks, while ATR is activated by ssDNA, formed notably in processing blocked replication forks (Cimprich and Cortez, 2008). Once bound to DNA, MRN recruits and activates ATM via interaction with Nbs1 (Lavin, 2007) and Mre11 nuclease activity leads to the formation of single strand oligonucleotides that further promote ATM activation (Jazayeri et al., 2008). Further maturation of the DNA extremities can also lead to ssDNA formation and ATR activation (Jazayeri et al., 2006). Mutation of ATM in humans leads to Ataxia-telangectasia (A-T), a genomic instability disorder characterized by neurodegeneration, immunodeficiency and sensibility to ionizing radiation. At the cellular level, the hallmarks of ATM deficiency are increased chromosomal breakage and premature senescence (Shiloh and Ziv, 2013). In the absence of ATM (in A-T cells), signaling of DNA breaks can be accomplished by ATR helped by EXO1, however, the absence of both kinases results in the absence of cell cycle arrest due to defects in signaling of breaks (Tomimatsu et al., 2009).
Mec1/ATR is considered to be the specific sensor of DNA replication fork stalling and DNA replication damage, and is more generally activated by a variety of lesions that have in common the generation of ssDNA. Irrespective of the origin of the ssDNA, ATR is recruited by its cofactor ATRIP, which indirectly recognizes ssDNA through interaction with the ssDNA-binding protein, RPA. The 9-1-1 checkpoint clamp has also been implicated in activation of the ATR/Mec1 kinase (Majka et al., 2006). Mec1 is an essential gene in yeast (Weinert et al., 1994) and even in the absence of exogenous genotoxic stress, Mec1 mutants accumulate gross spontaneous chromosomal rearrangements (GCRs; Myung and Kolodner, 2002). ATR deficiency is lethal in mammalian cells but hypomorphic atr mutations have been described in a few patients with the rare Seckel syndrome, characterized by microcephaly and growth retardation (O’Driscoll et al., 2003).
The presence of ATM and ATR is well conserved while, as for yeast, no DNA-PK ortholog has been identified in plants. IR-induced gamma-H2AX foci are mediated essentially by ATM and less so by ATR, with no foci observed in irradiated atm atr mutant cells (Friesner et al., 2005), confirming that AtATM and AtATR are the only DSB signaling PIKK kinases in plants. The presence in Arabidopsis of the protein AtATRIP, necessary for AtATR activation as seen in mammals, further reinforces the idea that DNA damage signaling in plants is conserved (Sweeney et al., 2009). The role of the MRN complex in DNA damage detection and activation of kinase mediated signaling is conserved in Arabidopsis (Amiard et al., 2010) and plant homologs of the genes encoding the 9-1-1 (Rad9/Rad1/Hus1) sensor complex have been identified and are required for resistance to the DNA damaging agents Bleomycin and Mitomycin C (MMC; Heitzeberg et al., 2004).
Arabidopsis atm mutants are phenotypically wild type, except for a partial sterility (Culligan and Britt, 2008). These plants are however hypersensitive to ionizing irradiation and methyl methane sulphonate (MMS), but not to UV irradiation. Arabidopsis atr mutants are viable, fertile, and like atm mutants, phenotypically wild-type in the absence of exogenous DNA damaging agents. atr mutants are hypersensitive to hydroxyurea and aphidicolin, due to a defective G2 checkpoint response to blocked replication forks (Culligan et al., 2004). ATR can however partially compensate for the ATM response, as the double atm atr mutant is completely sterile due to meiotic prophase genome fragmentation (Culligan and Britt, 2008).
Neither ATR nor ATM signaling is thus essential during normal plant development – a surprising result given the conservation of the roles of these proteins in plants and the lethality of the corresponding mutants in mammals. A hint to a possible explanation for this could come from the ability of DSBs to be repaired in plants in the double rad50 atr mutant, which combines absence of ATM and ATR activities and absence of H2AX phosphorylation (see next section). Spontaneous DSBs appear in consequence of replication defects in these plants and result in high levels of anaphase bridging, showing that Arabidopsis can repair DSB in the absence of PIKK activation (Amiard et al., 2010).
Once activated, PIKK can activate many targets necessary to maintain genomic integrity (Culligan et al., 2006; Matsuoka et al., 2007). Phosphorylation of the histone variant H2A/H2AX around the break by PIKK is an early cellular response to the induction of DSBs and occurs over 50 kb in yeast to 2 Mb for H2AX in mammals. H2AX phosphorylation is easily detected using phospho-specific antisera and has emerged as a highly specific and sensitive molecular marker for monitoring DNA damage and its repair (Kinner et al., 2008). Although not required for the initial recruitment of signaling and repair factors, H2AX phosphorylation is essential for their accumulation at the breaks (Celeste et al., 2003; Fernandez-Capetillo et al., 2003; Fillingham et al., 2006). The importance of this is seen in the sensitivity to DSB damaging agents, impaired DSB repair and defects in G1 checkpoint activation of yeast mutants of the H2A gene (Downs et al., 2000; Redon et al., 2003; Hammet et al., 2007) and similar phenotypes of mammalian cells and mice deficient for H2AX (Celeste et al., 2002). Moreover H2AX deficient mice were radiation sensitive, growth retarded, immune deficient and males were infertile.
In contrast, Arabidopsis mutants for this histone develop normally and only a slight defect in DSB repair has been reported in RNAi knock-down lines (Lang et al., 2012). Moreover, the phosphorylation of this histone does not seem required for DSB repair in plants, as seen in the chromosome fusions observed in the rad50 atr double mutant (Amiard et al., 2010). This being so, how is DSB signaling mediated in the absence of H2AX phosphorylation in Arabidopsis? A possible answer comes from reports showing roles of modifications of other histones around DSB in mammals: ubiquitinylation of H2A by RNF8 is required for proper 53BP1 recruitment (Marteijn et al., 2009; Rossetto et al., 2010) and a role for histone lysine methylation in DSB repair is supported by the observation that H3K36me2 enhances DNA repair by NHEJ (Fnu et al., 2011). H3K36me2, once formed at DSB site, may create docking sites for other repair proteins, recruiting them for transcription and DNA repair. It will be of great interest to see whether such modifications also play important roles in repair of DSBs in plants.
Signaling of Deprotected Telomeres
Telomeres consist of an elaborate, higher-order assembly of specific DNA sequence and proteins that cooperatively provide protection against degradation and recombination of the ends of linear eukaryotic chromosomes. In vertebrates, telomere protection is provided mainly by Shelterin, a complex of six telomeric proteins (TRF1, TRF2, POT1, TIN2, TPP1 and RAP1) that prevents inappropriate recombination and fusion between telomeres, and also has complementary roles in telomere replication and length regulation (Palm and De Lange, 2008; Martínez and Blasco, 2011). TRF1 and TRF2 bind to the duplex region of the telomere and searches for TRF-like proteins in Arabidopsis have identified many proteins able to bind double-stranded telomeric DNA (Zellinger and Riha, 2007; Amiard et al., 2011b; Peška et al., 2011). None of these seems however to be essential for telomere protection, suggesting redundancy of double-stranded DNA binding telomeric proteins in plants. POT1 binds to the natural single-stranded (ss) extension of the G-rich strand of chromosome ends (G-overhang or 3′-overhang) and in both humans and Saccharomyces pombe, POT1 plays a key role in telomere end protection (Baumann and Cech, 2001). Arabidopsis has two POT1 orthologs, POT1a and POT1b, both of which associate with the telomerase ribonucleoprotein but do not bind telomeric ssDNA and are not essential for telomere capping (Surovtseva et al., 2007; Shakirov et al., 2009; Cifuentes-Rojas et al., 2011).
In S. cerevisiae there has been no shelterin-like complex identified to date and a somewhat simpler protection complex, consisting mainly of the CST complex (Cdc13, Stn1 and Ten1), is present (Garvik et al., 1995; Grandin et al., 2001; Shore and Bianchi, 2009). Yeast Cdc13, together with Stn1 and Ten1, plays a dual role in telomere end protection and regulation of telomere replication. Orthologs of the S. cerevisiae CST proteins have been found in humans and mouse, as well as in Arabidopsis (Miyake et al., 2009; Surovtseva et al., 2009). Recent studies in mammalian cells reveal that the CST complex seems to be implicated in facilitating telomere replication by rescuing replication after fork stalling (Stewart et al., 2012) and that this complex is involved in the regulation of the telomeric 3′ overhang by C-strand fill-in by Polymerase alpha (Wang et al., 2012). Plants appear to represent an evolutionary intermediate between S. cerevisiae, which has only CST as a capping complex, and vertebrates which use both shelterin and CST complex for telomere capping and correct telomeric replication (Giraud-Panis et al., 2010; Price et al., 2010).
Deprotected telomeres are recognized by cells as DSB, and their “repair” results in chromosome fusions/rearrangements and genomic instability (De Lange, 2009). As for other DSB, deprotected telomeres are substrates for kinase activation and are characterized by the appearance of TIFs (telomere induced foci), DNA damage response factors that coincide with telomere signals. In mammals the absence of TRF2 or POT1 leads to the appearance of TIFs and this depends upon ATM and ATR, respectively (De Lange, 2009). In plants, we have shown that the appearance of TIFs in ctc1 or stn1 mutants are exclusively ATR-dependent and that in absence of the catalytic subunit of the telomerase (TERT), the short deprotected telomeres are recognized as DSBs through the activation of both ATM and ATR (Amiard et al., 2011a). Surprisingly, we have shown that in the Arabidopsis ctc1 atr mutant, which does not form TIFs, telomeres are still able to fuse. This result contrast clearly with the situation in vertebrates, where ATM and ATR are absolutely required for fusion of deprotected telomeres in absence of the TRF2 or POT1, respectively (Denchi and De Lange, 2007). Hence here again, plant repair pathways can still be activated in absence of the kinase activity.
Conclusion
This short review summarizes knowledge concerning DNA break signaling in Arabidopsis thaliana. A list of genes discussed here is presented in Table 1 and we refer interested readers to a recent compilation of Arabidopsis DNA repair/recombination genes (http://www.plb.ucdavis.edu/labs/britt/Plant_DNA_Repair_Genes.html). Given the crucial importance of the signaling step in DNA repair it is not surprising to find strong conservation of these mechanisms in higher eukaryotes. Nevertheless, evidence points to a particular ability of plants to repair even in absence of signaling and the presence of an unknown plant specific repair pathway(s) is now suspected. Plants possess a not fully understood ability to resist and develop in presence of DNA damaging agents and the implication of plant specific recombination events could provide part of the explanation for this. The increased spontaneous recombination rates seen in plants subjected to biotic or abiotic stresses (review by Waterworth et al., 2011) has been proposed to be a programmed response increasing the plasticity of plant genome leading to acceleration of plant evolution (Molinier et al., 2006; Boyko and Kovalchuk, 2011).
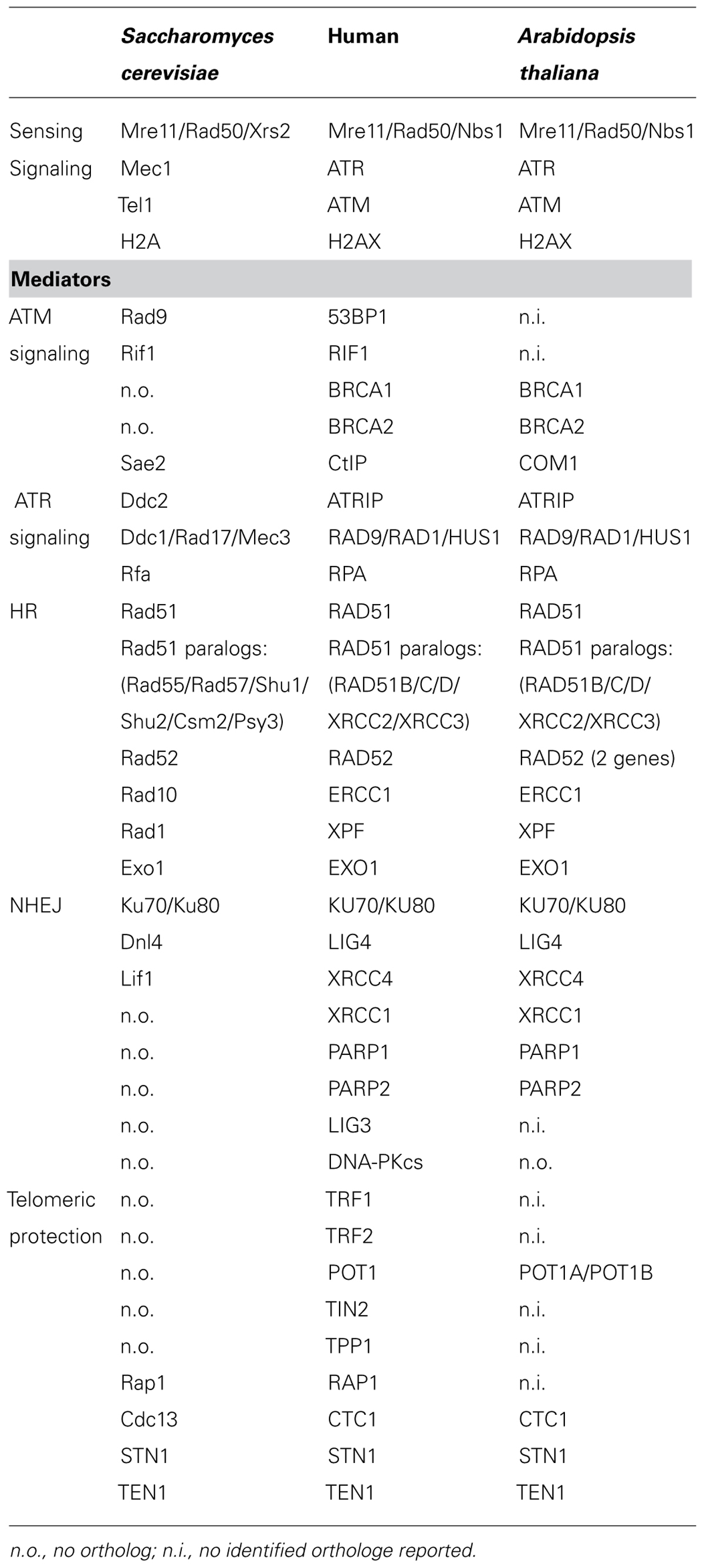
TABLE 1. Major factors involved in DNA double strand break signaling and repair and telomere protection in budding yeast, human and Arabidopsis thaliana.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from European Union research grant (LSHG-CT-2005-018785), the Centre National de la Recherche Scientifique, the Université Blaise Pascal, the Université d’Auvergne, and the Institut National de la Santé et la Recherche Medicale.
References
Amiard, S., Charbonnel, C., Allain, E., Depeiges, A., White, C. I., and Gallego, M. E. (2010). Distinct roles of the ATR kinase and the Mre11-Rad50-Nbs1 complex in the maintenance of chromosomal stability in Arabidopsis. Plant Cell 22, 3020–3033. doi: 10.1105/tpc.110.078527
Amiard, S., Depeiges, A., Allain, E., White, C. I., and Gallego, M. E. (2011a). Arabidopsis ATM and ATR kinases prevent propagation of genome damage caused by telomere dysfunction. Plant Cell 23, 4254–4265. doi: 10.1105/tpc.111.092387
Amiard, S., White, C., and Gallego, M. E. (2011b). Recombination proteins and telomere stability in plants. Curr. Protein Pept. Sci 12, 84–92. doi: 10.2174/138920311795684931
Baumann, P., and Cech, T. R. (2001). Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292, 1171–1175. doi: 10.1126/science.1060036
Boyko, A., and Kovalchuk, I. (2011). Genome instability and epigenetic modification – heritable responses to environmental stress? Curr. Opin. Plant Biol. 14, 260–266. doi: 10.1016/j.pbi.2011.03.003
Celeste, A., Fernandez-Capetillo, O., Kruhlak, M. J., Pilch, D. R., Staudt, D. W., Lee, A., et al. (2003). Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5, 675–679. doi: 10.1038/ncb1004
Celeste, A., Petersen, S., Romanienko, P. J., Fernandez-Capetillo, O., Chen, H. T., Sedelnikova, O. A., et al. (2002). Genomic instability in mice lacking histone H2AX. Science 296, 922–927. doi: 10.1126/science.1069398
Chapman, J. R., Barral, P., Vannier, J.-B., Borel, V., Steger, M., Tomas-Loba, A., et al. (2013). RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol. Cell 49, 858–871. doi: 10.1016/j.molcel.2013.01.002
Chapman, J. R., Taylor, M. R. G., and Boulton, S. J. (2012). Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 47, 497–510. doi: 10.1016/j.molcel.2012.07.029
Charbonnel, C., Allain, E., Gallego, M. E., and White, C. I. (2011). Kinetic analysis of DNA double-strand break repair pathways in Arabidopsis. DNA Repair 10, 611–619. doi: 10.1016/j.dnarep.2011.04.002
Charbonnel, C., Gallego, M. E., and White, C. I. (2010). Xrcc1-dependent and Ku-dependent DNA double-strand break repair kinetics in Arabidopsis plants. Plant J. 64, 280–290. doi: 10.1111/j.1365-313X.2010.04331.x
Chowdhury, D., Choi, Y. E., and Brault, M. E. (2013). Charity begins at home: non-coding RNA functions in DNA repair. Nat. Rev. Mol. Cell Biol. 14, 181–189. doi: 10.1038/nrm3523
Cifuentes-Rojas, C., Kannan, K., Tseng, L., and Shippen, D. E. (2011). Two RNA subunits and POT1a are components of Arabidopsis telomerase. Proc. Natl. Acad. Sci. U.S.A. 108, 73–78. doi: 10.1073/pnas.1013021107
Cimprich, K. A., and Cortez, D. (2008). ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9, 616–627. doi: 10.1038/nrm2450
Culligan, K., Tissier, A., and Britt, A. (2004). ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 16, 1091–1104. doi: 10.1105/tpc.018903
Culligan, K. M., and Britt, A. B. (2008). Both ATM and ATR promote the efficient and accurate processing of programmed meiotic double-strand breaks. Plant J. 55, 629–638. doi: 10.1111/j.1365-313X.2008.03530.x
Culligan, K. M., Robertson, C. E., Foreman, J., Doerner, P., and Britt, A. B. (2006). ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 48, 947–961. doi: 10.1111/j.1365-313X.2006.02931.x
Decottignies, A. (2013). Alternative end-joining mechanisms: a historical perspective. Front. Genet. 4:48–54. doi: 10.3389/fgene.2013.00048
De Lange, T. (2009). How telomeres solve the end-protection problem. Science 326, 948–952. doi: 10.1126/science.1170633
Denchi, E. L., and De Lange, T. (2007). Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448, 1068–1071. doi: 10.1038/nature06065
Downs, J. A., Lowndes, N. F., and Jackson, S. P. (2000). A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408, 1001–1004. doi: 10.1038/35050000
Escribano-Díaz, C., Orthwein, A., Fradet-Turcotte, A., Xing, M., Young, J. T. F., Tkáè, J., et al. (2013). A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell 49, 872–883. doi: 10.1016/j.molcel.2013.01.001
Fernandez-Capetillo, O., Celeste, A., and Nussenzweig, A. (2003). Focusing on foci: H2AX and the recruitment of DNA-damage response factors. Cell Cycle 2, 426–427. doi: 10.4161/cc.2.5.509
Fillingham, J., Keogh, M.-C., and Krogan, N. J. (2006). GammaH2AX and its role in DNA double-strand break repair. Biochem. Cell Biol. 84, 568–577. doi: 10.1139/o06-072
Fnu, S., Williamson, E. A., De Haro, L. P., Brenneman, M., Wray, J., Shaheen, M., et al. (2011). Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc. Natl. Acad. Sci. U.S.A. 108, 540–545. doi: 10.1073/pnas.1013571108
Friesner, J., and Britt, A. B. (2003). Ku80- and DNA ligase IV-deficient plants are sensitive to ionizing radiation and defective in T-DNA integration. Plant J. 34, 427–440. doi: 10.1046/j.1365-313X.2003.01738.x
Friesner, J. D., Liu, B., Culligan, K., and Britt, A. B. (2005). Ionizing radiation-dependent gamma-H2AX focus formation requires ataxia telangiectasia mutated and ataxia telangiectasia mutated and Rad3-related. Mol. Biol. Cell 16, 2566–2576. doi: 10.1091/mbc.E04-10-0890
Gallego, M. E., Bleuyard, J. Y., Daoudal-Cotterell, S., Jallut, N., and White, C. I. (2003). Ku80 plays a role in non-homologous recombination but is not required for T-DNA integration in Arabidopsis. Plant J. 35, 557–565. doi: 10.1046/j.1365-313X.2003.01827.x
Garvik, B., Carson, M., and Hartwell, L. (1995). Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 15, 6128–6138.
Giraud-Panis, M.-J., Teixeira, M. T., Géli, V., and Gilson, E. (2010). CST meets shelterin to keep telomeres in check. Mol. Cell 39, 665–676. doi: 10.1016/j.molcel.2010.08.024
Goodarzi, A. A., Jeggo, P., and Löbrich, M. (2010). The influence of heterochromatin on DNA double strand break repair: getting the strong, silent type to relax. DNA Repair 9, 1273–1282. doi: 10.1016/j.dnarep.2010.09.013
Grandin, N., Damon, C., and Charbonneau, M. (2001). Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 20, 1173–1183. doi: 10.1093/emboj/20.5.1173
Hammet, A., Magill, C., Heierhorst, J., and Jackson, S. P. (2007). Rad9 BRCT domain interaction with phosphorylated H2AX regulates the G1 checkpoint in budding yeast. EMBO Rep. 8, 851–857. doi: 10.1038/sj.embor.7401036
Heacock, M., Spangler, E., Riha, K., Puizina, J., and Shippen, D. E. (2004). Molecular analysis of telomere fusions in Arabidopsis: multiple pathways for chromosome end-joining. EMBO J. 23, 2304–2313. doi: 10.1038/sj.emboj.7600236
Heitzeberg, F., Chen, I.-P., Hartung, F., Orel, N., Angelis, K. J., and Puchta, H. (2004). The Rad17 homologue of Arabidopsis is involved in the regulation of DNA damage repair and homologous recombination. Plant J. 38, 954–968. doi: 10.1111/j.1365-313X.2004.02097.x
Heyer, W.-D., and Liu, J. (2010). Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 44, 113–139. doi: 10.1146/annurev-genet-051710-150955
Hiom, K. (2010). Coping with DNA double strand breaks. DNA Repair 9, 1256–1263. doi: 10.1016/j.dnarep.2010.09.018
Jazayeri, A., Balestrini, A., Garner, E., Haber, J. E., and Costanzo, V. (2008). Mre11-Rad50-Nbs1-dependent processing of DNA breaks generates oligonucleotides that stimulate ATM activity. EMBO J. 27, 1953–1962. doi: 10.1038/emboj.2008.128
Jazayeri, A., Falck, J., Lukas, C., Bartek, J., Smith, G. C. M., Lukas, J., et al. (2006). ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 8, 37–45. doi: 10.1038/ncb1337
Jia, Q., Den Dulk-Ras, A., Shen, H., Hooykaas, P. J., and De Pater, S. (2013). Poly(ADP-ribose)polymerases are involved in microhomology mediated back-up non-homologous end joining in Arabidopsis thaliana. Plant Mol. Biol. 82, 339–351. doi: 10.1007/s11103-013-0065-9
Kinner, A., Wu, W., Staudt, C., and Iliakis, G. (2008). Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 36, 5678–5694. doi: 10.1093/nar/gkn550
Lafarge, S., and Montane, M. H. (2003). Characterization of Arabidopsis thaliana ortholog of the human breast cancer susceptibility gene 1: AtBRCA1, strongly induced by gamma rays. Nucleic Acids Res. 31, 1148–1155. doi: 10.1093/nar/gkg202
Lang, J., Smetana, O., Sanchez-Calderon, L., Lincker, F., Genestier, J., Schmit, A.-C., et al. (2012). Plant γH2AX foci are required for proper DNA DSB repair responses and colocalize with E2F factors. New Phytol. 194, 353–363. doi: 10.1111/j.1469-8137.2012.04062.x
Lavin, M. F. (2007). ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene 26, 7749–7758. doi: 10.1038/sj.onc.1210880
Lieber, M. R. (2010). The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211. doi: 10.1146/annurev.biochem.052308.093131
Ma, J.-L., Kim, E. M., Haber, J. E., and Lee, S. E. (2003). Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 23, 8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003
Majka, J., Niedziela-Majka, A., and Burgers, P. M. J. (2006). The checkpoint clamp activates Mec1 kKinase during initiation of the DNA damage checkpoint. Mol. Cell 24, 891–901. doi: 10.1016/j.molcel.2006.11.027
Mannuss, A., Trapp, O., and Puchta, H. (2011). Gene regulation in response to DNA damage. Biochim. Biophys. Acta 1819, 154–165. doi: 10.1016/j.bbagrm.2011.08.003
Mantiero, D., Clerici, M., Lucchini, G., and Longhese, M. P. (2007). Dual role for Saccharomyces cerevisiae Tel1 in the checkpoint response to double-strand breaks. EMBO Rep. 8, 380–387. doi: 10.1038/sj.embor.7400911
Marteijn, J. A., Bekker-Jensen, S., Mailand, N., Lans, H., Schwertman, P., Gourdin, A. M., et al. (2009). Nucleotide excision repair-induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. J. Cell Biol. 186, 835–847. doi: 10.1083/jcb.200902150
Martínez, P., and Blasco, M. A. (2011). Telomeric and extra-telomeric roles for telomerase and the telomere-binding proteins. Nat. Rev. Cancer 11, 161–176. doi: 10.1038/nrc3025
Matsuoka, S., Ballif, B. A., Smogorzewska, A., Mcdonald, E. R., Hurov, K. E., Luo, J., et al. (2007). ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316, 1160–1166. doi: 10.1126/science.1140321
Miyake, Y., Nakamura, M., Nabetani, A., Shimamura, S., Tamura, M., Yonehara, S., et al. (2009). RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol. Cell 36, 193–206. doi: 10.1016/j.molcel.2009.08.009
Molinier, J., Ries, G., Zipfel, C., and Hohn, B. (2006). Transgeneration memory of stress in plants. Nature 442, 1046–1049. doi: 10.1038/nature05022
Morrow, D. M., Tagle, D. A., Shiloh, Y., Collins, F. S., and Hieter, P. (1995). TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell 82, 831–840. doi: 10.1016/0092-8674(95)90480-8
Myung, K., and Kolodner, R. D. (2002). Suppression of genome instability by redundant S-phase checkpoint pathways in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 99, 4500–4507. doi: 10.1073/pnas.062702199
O’Driscoll, M., Ruiz-Perez, V. L., Woods, C. G., Jeggo, P. A., and Goodship, J. A. (2003). A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat. Genet. 33, 497–501. doi: 10.1038/ng1129
Palm, W., and De Lange, T. (2008). How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42, 301–334. doi: 10.1146/annurev.genet.41.110306.130350
Peška, V., Procházková Schrumpfová, P., and Fajkus, J. (2011). Using the telobox to search for plant telomere binding proteins. Curr. Protein Pept. Sci. 12, 75–83. doi: 10.2174/138920311795684968
Price, C. M., Boltz, K. A., Chaiken, M. F., Stewart, J. A., Beilstein, M. A., and Shippen, D. E. (2010). Evolution of CST function in telomere maintenance. Cell Cycle 9, 3157–3165. doi: 10.4161/cc.9.16.12547
Redon, C., Pilch, D. R., Rogakou, E. P., Orr, A. H., Lowndes, N. F., and Bonner, W. M. (2003). Yeast histone 2A serine 129 is essential for the efficient repair of checkpoint-blind DNA damage. EMBO Rep. 4, 678–684. doi: 10.1038/sj.embor.embor871
Riha, K., Matthew Watson, J., Parkey, J., and Shippen, D. E. (2002). Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO J. 21, 2819–2826. doi: 10.1093/emboj/21.11.2819
Rossetto, D., Truman, A. W., Kron, S. J., and Cote, J. (2010). Epigenetic modifications in double-strand break DNA damage signaling and repair. Clin. Cancer Res. 16, 4543–4552. doi: 10.1158/1078-0432.CCR-10-0513
Shakirov, E. V., Song, X., Joseph, J. A., and Shippen, D. E. (2009). POT1 proteins in green algae and land plants: DNA-binding properties and evidence of co-evolution with telomeric DNA. Nucleic Acids Res. 37, 7455–7467. doi: 10.1093/nar/gkp785
Shiloh, Y., and Ziv, Y. (2013). The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 14, 197–210. doi: 10.1038/nrm3546
Shore, D., and Bianchi, A. (2009). Telomere length regulation: coupling DNA end processing to feedback regulation of telomerase. 28, 2309–2322. doi: 10.1038/emboj.2009.195
Stewart, J. A., Wang, F., Chaiken, M. F., Kasbek, C., Chastain, P. D., Wright, W. E., et al. (2012). Human CST promotes telomere duplex replication and general replication restart after fork stalling. EMBO J. 31, 3537–3549. doi: 10.1038/emboj.2012.215
Surovtseva, Y. V., Churikov, D., Boltz, K. A., Song, X., Lamb, J. C., Warrington, R., et al. (2009). Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol. Cell 36, 207–218. doi: 10.1016/j.molcel.2009.09.017
Surovtseva, Y. V., Shakirov, E. V., Vespa, L., Osbun, N., Song, X., and Shippen, D. E. (2007). Arabidopsis POT1 associates with the telomerase RNP and is required for telomere maintenance. EMBO J. 26, 3653–3661. doi: 10.1038/sj.emboj.7601792
Sweeney, P. R., Britt, A. B., and Culligan, K. M. (2009). The Arabidopsis ATRIP ortholog is required for a programmed response to replication inhibitors. Plant J. 60, 518–526. doi: 10.1111/j.1365-313X.2009.03975.x
Tomimatsu, N., Mukherjee, B., and Burma, S. (2009). Distinct roles of ATR and DNA-PKcs in triggering DNA damage responses in ATM-deficient cells. EMBO Rep. 10, 629–635. doi: 10.1038/embor.2009.60
Trapp, O., Seeliger, K., and Puchta, H. (2011). Homologs of breast cancer genes in plants. Front. Plant Sci. 2:19. doi: 10.3389/fpls.2011.00019
Uanschou, C., Siwiec, T., Pedrosa-Harand, A., Kerzendorfer, C., Sanchez-Moran, E., Novatchkova, M., et al. (2007). A novel plant gene essential for meiosis is related to the human CtIP and the yeast COM1/SAE2 gene. EMBO J. 26, 5061–5070. doi: 10.1038/sj.emboj.7601913
Usui, T., Ogawa, H., and Petrini, J. H. (2001). A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell 7, 1255–1266. doi: 10.1016/S1097-2765(01)00270-2
Van Attikum, H., Bundock, P., Overmeer, R. M., Lee, L.-Y., Gelvin, S. B., and Hooykaas, P. J. J. (2003). The Arabidopsis AtLIG4 gene is required for the repair of DNA damage, but not for the integration of Agrobacterium T-DNA. Nucleic Acids Res. 31, 4247–4255. doi: 10.1093/nar/gkg458
Wang, F., Stewart, J. A., Kasbek, C., Zhao, Y., Wright, W. E., and Price, C. M. (2012). Human CST has independent functions during telomere duplex replication and C-strand fill-in. Cell Rep. 2, 1096–1103. doi: 10.1016/j.celrep.2012.10.007
Waterworth, W. M., Drury, G. E., Bray, C. M., and West, C. E. (2011). Repairing breaks in the plant genome: the importance of keeping it together. New Phytol. 192, 805–822. doi: 10.1111/j.1469-8137.2011.03926.x
Wei, W., Ba, Z., Gao, M., Wu, Y., Ma, Y., Amiard, S., et al. (2012). A role for small RNAs in DNA double-strand break repair. Cell 149, 101–112. doi: 10.1016/j.cell.2012.03.002
Weinert, T. A., Kiser, G. L., and Hartwell, L. H. (1994). Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 8, 652–665. doi: 10.1101/gad.8.6.652
Zellinger, B., and Riha, K. (2007). Composition of plant telomeres. Biochim. Biophys. Acta 1769, 399–409. doi: 10.1016/j.bbaexp.2007.02.001
Keywords: signaling, sensing, double strand breaks, telomere, DNA repair
Citation: Amiard S, Gallego ME and White CI (2013) Signaling of double strand breaks and deprotected telomeres in Arabidopsis. Front. Plant Sci. 4:405. doi: 10.3389/fpls.2013.00405
Received: 23 August 2013; Paper pending published: 09 September 2013;
Accepted: 24 September 2013; Published online: 16 October 2013.
Edited by:
Alma Balestrazzi, University of Pavia, ItalyReviewed by:
Avraham Levy, Weizmann Institute of Science, IsraelWanda Waaterworth, University of Leeds, UK
Copyright © 2013 Amiard, Gallego and White. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Charles I. White, Génétique, Reproduction et Développement, UMR CNRS 6293/U1103 INSERM/Clermont Université, Université Blaise Pascal, BP80026, 63171 Aubiére cedex, France e-mail: chwhite@univ-bpclermont.fr
 Simon Amiard
Simon Amiard Maria E. Gallego
Maria E. Gallego Charles I. White*
Charles I. White*