- 1Tianjin Key Laboratory of Protein Science and Department of Genetics and Cell Biology, College of Life Sciences, Nankai University, Tianjin, China
- 2National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China
The endoplasmic reticulum (ER) is a membrane-bounded organelle whose membrane comprises a network of tubules and sheets. The formation of these characteristic shapes and maintenance of their continuity through homotypic membrane fusion appears to be critical for the proper functioning of the ER. The atlastins (ATLs), a family of ER-localized dynamin-like GTPases, have been identified as fusogens of the ER membranes in metazoans. Mutations of the ATL proteins in mammalian cells cause morphological defects in the ER, and purified Drosophila ATL mediates membrane fusion in vitro. Plant cells do not possess ATL, but a family of similar GTPases, named root hair defective 3 (RHD3), are likely the functional orthologs of ATLs. In this review, we summarize recent advances in our understanding of how RHD3 proteins play a role in homotypic ER fusion. We also discuss the possible physiological significance of forming a tubular ER network in plant cells.
Introduction
The endoplasmic reticulum (ER) is the origin of the endomembrane system in eukaryotic cells. Secretory proteins and most of the integral membrane proteins are synthesized and folded by the ER, cellular membrane sources are generated on the ER, and calcium ions are stored in the lumen of the ER. Morphologically, the ER membranes can adopt a tubular shape or form flattened cisternal structures, called ER sheets (Shibata et al., 2006). Despite the ER representing one of the largest intracellular membrane surfaces, the ER membranes in each cell are continuous as one entity. Though the sheets may be stacked by helicoidal membrane motifs (Terasaki et al., 2013), tubules often extend from sheets and are connected via three-way junctions into a reticular network (Lee and Chen, 1988; Hu et al., 2008). In some areas of the ER, tubules and sheets are interspersed in fenestrated structures (West et al., 2011).
The formation and maintenance of a continuous membrane system requires constant fusion of identical membranes. A similar fusion process includes the merger of mitochondrial membranes, in which dynamin-like GTPases mitofusin/Fzo1 and OPA1/Mgm1 play important roles (Chan, 2006; Hoppins et al., 2007). However, how such homotypic fusion occurs is poorly understood. In contrast, the merger of heterotypic membranes, such as the fusion of viral and cellular membranes or transport vesicles with target membranes, has been studied intensively. In viral fusion, the membranes are pulled together by an intramolecular conformational change in a single protein (Harrison, 2008). In vesicular fusion, three t-SNARE proteins in one membrane and a v-SNARE partner in the other zipper up to form a four-helix bundle in the fused lipid bilayer (Jahn and Scheller, 2006; Martens and McMahon, 2008; Wickner and Schekman, 2008; Sudhof and Rothman, 2009).
In mammalian cells, defects in branch formation of the ER network, a sign of a lack of sufficient homotypic fusion, was recently linked to a class of membrane-bound, dynamin-like GTPases named atlastins (ATLs; Rismanchi et al., 2008; Hu et al., 2009). Lipid bilayer fusion can be achieved with purified Drosophila ATL (Orso et al., 2009; Bian et al., 2011). Following the discovery of ATLs, Sey1p in yeast cells was identified as a functional ortholog (Anwar et al., 2012). The deletion of Sey1p drastically delays ER fusion during mating, and the re-introduction of Sey1p restores the defects (Anwar et al., 2012). Similar to ATL, reconstituted Sey1p is capable of fusing vesicles in vitro (Anwar et al., 2012).
Plant cells do not possess ATL homologs; however, a GTPase called root hair defective 3 (RHD3) is related to Sey1p in regards to sequence (Brands and Ho, 2002) and has been suggested to mediate the fusion of ER membranes (Hu et al., 2009; Chen et al., 2011). Although the mechanisms of ER fusion may be conserved in plant cells, the plant ER exhibits several unique features: a prominent cortical ER (West et al., 2011); participation in plasmodesma formation, a specialized intercellular ER connection (Gupton et al., 2006); and the movement of Golgi bodies along ER tubules (Boevink et al., 1998; Faso et al., 2009; Sparkes et al., 2009). These characteristics imply that homotypic ER fusion in plant cells may play distinct roles.
Homotypic ER Fusion in Mammalian and Yeast Cells
The first clue of homotypic fusion of the ER membranes in mammalian cells came from overexpression of mutant forms of ATL, a membrane-bound GTPase (Rismanchi et al., 2008). ATL mutations cause unbranched ER morphology, indicating a lack of fusion between ER tubules. ATL is anchored in the membrane by two closely spaced transmembrane (TM) segments, exposing both the N-terminal GTPase domain and the C-terminal tail (CT) to the cytosol (Figure 1A). ATL localizes mostly in the tubular region of the ER and interacts with the ER tubule resident proteins reticulons and DP1/Yop1p (Hu et al., 2009), two families of integral membranes that induce high curvature in the ER membranes to form tubules (Voeltz et al., 2006; Hu et al., 2009). ATL belongs to the dynamin superfamily of GTPases. A related family member, mitofusin (MFN), shares membrane topology and domain structures with ATL (Figure 1B) and is known to mediate fusion of the outer mitochondrial membranes (Hermann et al., 1998; Chen et al., 2003). Thus, ATL is likely responsible for fusion of the ER membranes. In fact, depletion of ATL causes unbranched ER in mammalian cells (Hu et al., 2009), and ER fragmentation in Drosophila (Orso et al., 2009). In addition, antibodies against ATL block ER network formation in vitro in Xenopus egg membrane extracts (Hu et al., 2009). Most convincingly, purified Drosophila ATL is able to mediate vesicle fusion in vitro when reconstituted into proteoliposomes (Orso et al., 2009).
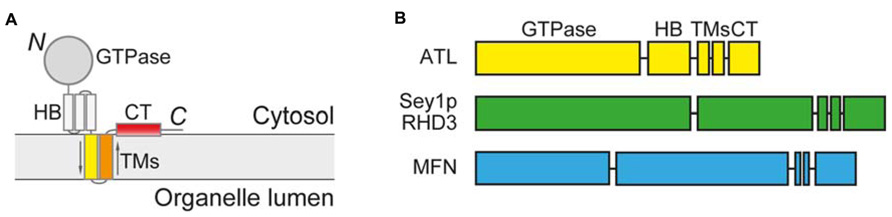
FIGURE 1. Membrane topology and domain structure of ATL-related GTPases. (A) Membrane topology of ATL-related GTPases. 3HB, three-helix bundle; TMs, transmembrane segments; CT, C-terminal tail. (B) Domain structure of ATL-related GTPases.
Atlastins is conserved in most metazoa, but it is not found in many organisms in which the tubular ER network is properly formed. Saccharomyces cerevisiae is among these organisms, but a similar GTPase named Sey1p was recently identified as an ER fusogen (Anwar et al., 2012). Sey1p is a synthetic enhancer for Yop1p (Brands and Ho, 2002), one of the ER tubule shaping proteins, and it not so surprisingly plays a role in tubular ER network formation. Cells lacking Sey1p exhibit minor morphological defects in the ER (Hu et al., 2009), but an ER SNARE Ufe1p is required when SEY1 is deleted and may represent an alternative ER fusogen in yeast (Anwar et al., 2012).
Assays for Homotypic ER Fusion
Several assays have been developed or adapted to verify candidates for homotypic ER fusion. The first assay visualizes the integrity of the ER in yeast cells (Hu et al., 2009). Deletion of Sey1p and either Rtn1p or Yop1p, membrane proteins that shape the ER tubules, causes drastic ER defects; in particular, the tubular ER network is mostly converted to sheets and the large areas of the cortex are void of ER, indicating a lack of ER fusion. When wild-type Sey1p is re-introduced, fusion activities in the ER resume and ER morphology, visualized by GFP-labeled ER-resident protein Sec63p, restored. The rescue of the ER defects in sey1Δ rtn1Δ or sey1Δ yop1Δ cells by certain proteins indirectly indicates its ability to mediate ER fusion.
The second assay monitors ER fusion during the mating of yeast cells (Anwar et al., 2012). Similar assays have been used to study nuclear fusion or mitochondrial fusion. Haploid yeast cells expressing cytosolic GFP remated with cells expressing a red fluorescent protein (RFP)-containing ER marker (ss-RFP-HDEL). When cell fusion occurs between two types of cells, the cytosolic GFP of one cell rapidly diffuses to the other cell, marking the starting point for ER fusion. The efficiency of ER fusion is monitored by the equilibration of the RFP signal between two cells. Cells lacking Sey1p exhibit a significant delay in ER fusion, but plasmid-driven expression of Sey1p would restore such a defect. To test whether certain molecules mediate ER fusion in vivo, they are expressed in haploid cells with either cytosolic or ER marker, both of which lack Sey1p. ER fusion is then measured and compared to that of untransformed sey1Δ cells.
Finally, in vitro fusion assays have been adapted from studies of SNARE-mediated fusion (Weber et al., 1998; Scott et al., 2003). For lipid mixing tests, full-length fusion candidates are purified in detergent and reconstituted into proteoliposomes upon detergent removal. A group of vesicles incorporates lipids labeled with two fluorophores (NBD and rhodamine) at quenching concentrations. When these vesicles fuse with vesicles containing unlabeled lipids, the labeled lipids are diluted and subsequently dequenched. The increase in fluorescence correlates with the level of lipid mixing resulting from fusion. To further distinguish hemi-fusion and full fusion, two fluorescent dyes are incorporated as a FRET pair into reconstituted vesicles and the FRET signal measured as an indicator of the content mixing resulting from full fusion of the two bilayers (Zucchi and Zick, 2011).
Combining these three assays, ATL and Sey1p proteins were tested and confirmed to mediate fusion of the ER membranes (Table 1). Based on the results for Drosophila ATL, the fusion reaction can lead to efficient content mixing with nearly no lysis of the membranes (Liu et al., 2012). Using the same criteria, RHD3 has recently joined the list of ER fusogens as a plant ortholog of ATL and Sey1p (Zhang et al., 2013).
RHD3 Family Proteins as Plant ER Fusogens
The components involved in shaping ER tubules are conserved among eukaryotes. Shortly after reticulons and DP1/Yop1p were found in mammals and yeast cells (Voeltz et al., 2006), the plant orthologs were analyzed and confirmed to have the same role (Nziengui et al., 2007; Sparkes et al., 2010). Similarly, when ATL and Sey1p were shown to mediate ER fusion (Hu et al., 2009), a related protein family, RHD3, became very plausible candidates for ER fusogens in plant cells.
RHD3 was initially discovered by a genetic screen of root hair development (Schiefelbein and Somerville, 1990). Mutations in RHD3 proteins cause short and wavy root hairs and a dwarf phenotype (Schiefelbein and Somerville, 1990; Wang et al., 1997). A role for RHD3 in ER morphogenesis was indicated, even before the characterization of ATL and Sey1p, when rhd3-1 plants (A575V) were found to contain “cable-like” ER (Zheng et al., 2004), a defect reminiscent of ATL mutations or depletion in mammalian cells. Subsequently, several other RHD3 point mutants or null mutants were found to result in the same ER defects (Zheng et al., 2004; Chen et al., 2011; Stefano et al., 2012; Zhang et al., 2013), supporting the notion that RHD3 plays a role in connecting ER tubules. In addition to RHD3, two RHD3-like proteins were found in Arabidopsis (Hu et al., 2003). RL1 is expressed only in pollen, whereas RL2 is expressed ubiquitously, but both are present at very low levels. Individual deletions of the RL proteins show no detectable defects in plant development. However, over-expression of RL2 rescues the rhd3-1 mutant (Chen et al., 2011), suggesting a redundant role among these proteins.
Similar to ATL and Sey1p, RHD3 localizes mainly to the tubular ER network; colocalizes with HVA22 (Chen et al., 2011), a plant ortholog of DP1/Yop1p; and its homolog RL2 interacts with plant reticulons (Lee et al., 2013). However, RHD3 and Sey1p are not thought to be interchangeable (Chen et al., 2011), i.e. Sey1p cannot rescue the rhd3 mutant, and RHD3 cannot replace Sey1p in yeast. To test the possibility that RHD3 and Sey1p act differently, the yeast complementation assay was recently revisited. Either RHD3 or the RL proteins was expressed under the control of the endogenous SEY1 promoter in sey1Δ yop1Δ cells, and the results indicated that RHD3 family members are capable of restoring ER defects in sey1Δ yop1Δ cells (Zhang et al., 2013). Though Sey1p might not be functional in the setting of plant cells, these findings suggest that RHD3 and Sey1p act similarly in yeast cells and in vitro as purified proteins. Using the same assays that are applicable to ATL and Sey1p, the RHD3 proteins fuse the ER in cells and lipid membranes in vitro, confirming that they are ER fusogens in plant cells(Zhang et al., 2013).
Some fusion events have been observed between peripheral ER tubules in plant cells lacking RHD3 (Stefano et al., 2012). As neither RL is dispensable on the rhd3-null background (Stefano et al., 2012), these fusion activities are likely carried out by RL proteins, even though their levels are very low compared to RHD3. The strong ER-branching defect in rhd3 mutants suggests that RHD3 is the major force to connect ER membranes, but whether another ER fusogen exists in plant cells remains to be tested.
Mechanisms for Homotypic ER Fusion in Plant Cells
Given the domain structure and functional similarity among ATL, Sey1p, and RHD3, these GTPases likely utilize conserved mechanisms to mediate fusion. How RHD3 performs the fusion reaction in plant cells is not clear, but significant progress has been made in understanding ATL-mediated homotypic fusion. Crystal structures of the N-terminal cytosolic domain of human ATL1 have been determined (Bian et al., 2011; Byrnes and Sondermann, 2011), revealing a GTPase domain and three-helix bundle (3HB) connected by a linker region. The GTPase domains face each other to form a nucleotide-dependent dimer in all structures, but the 3HBs are positioned differently. In the structure obtained when only GDP is added, the 3HBs following the GTPase domains point in opposite directions; in another structure in which GDP and phosphate are present, the 3HBs are parallel to one another and crossover to dock against the GTPase domain of the partner molecule; and in the recent structures in which GMPPNP, a non-hydrolysable analog of GTP, or GDP and aluminum fluoride were used, the 3HBs come even closer in the crossed over conformation (Byrnes et al., 2013). Taken together, the evidence indicates that GTP binding induces interactions between ATL molecules across the apposing membranes, and subsequent GTP hydrolysis causes conformational changes through steps that are not entirely clear to force the 3HBs of engaging ATL molecules come very close, pulling the two membranes together.
In addition to the GTP-dependent mechanism, the CT of ATL forms an amphipathic helix that binds and perturbs the membrane bilayer, facilitating the fusion process, and the TM segments are required for efficient fusion, probably by mediating nucleotide-independent oligomerization of ATL molecules (Liu et al., 2012).
The homotypic interactions of RHD3 protein have been confirmed (Chen et al., 2011), but the mechanisms of RHD3-mediated membrane fusion remain to be tested. Notably, the region between the GTPase domain of RHD3 and the TM segments is much longer than that of the 3HB in ATL. Based on secondary structure prediction, this region likely forms a helical bundle, but whether it binds to the GTPase domain, or even in a similar manner as ATL, is largely unknown.
Perspective
Homotypic ER fusion appears to be a conserved process among eukaryotic cells. In plants, the process is mediated primarily by the RHD3 family of proteins. Like the ATL family of proteins, members of the RHD3 family are present ubiquitously. However, the prominent defects caused by mutations in RHD3 occur in cells with long protrusions, namely the root hairs, which is reminiscent of ATL1 with cortical spinal motor neurons. Though complete deletion of ATLs in mammals is yet to be achieved, the loss of RHD3 and either of the RL proteins results in lethality (Zhang et al., 2013). These results suggest that, at least in plants, more cell types require the presence of the RHD3 family than is previously thought.
One important question that remains to be addressed is the role of the RHD3 family in vivo. For example, there may be a link between these GTPases and the ER-plasma membrane (PM) contact sites. The non-branched or “cable-like” ER is often clustered in the cell body rather than present in the cortex. It is reasonable to speculate that RHD3 mutations perturb the general functions of the ER-PM contact sites, such as calcium signaling or lipid sensing and transfer. The formation of plasmodesmata also relies on coordination of the PM and cortical ER, and ER tubules from neighboring cells need to be fused in the nanopores. Will plasmodesmata be properly generated in RHD3 mutants? If not, what happens to the nanopores? Finally, Golgi body distribution and movement along the tubular ER network has been shown to be affected in rhd3-1 (Chen et al., 2011) or gom8 (Stefano et al., 2012), an EMS mutant of RHD3 (P701S). Does this defect directly lead to impaired plant development? The hope is that answering these questions will have an impact on our understanding of the correlation between the morphology of organelles and their functions.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Anwar, K., Klemm, R. W., Condon, A., Severin, K. N., Zhang, M., Ghirlando, R., et al. (2012). The dynamin-like GTPase Sey1p mediates homotypic ER fusion in S. cerevisiae. J. Cell Biol. 197, 209–217. doi: 10.1083/jcb.201111115
Bian, X., Klemm, R. W., Liu, T. Y., Zhang, M., Sun, S., Sui, X., et al. (2011). Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc. Natl. Acad. Sci. U.S.A. 108, 3976–3981. doi: 10.1073/pnas.1101643108
Boevink, P., Oparka, K., Santa Cruz, S., Martin, B., Betteridge, A., and Hawes, C. (1998). Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J. 15, 441–447. doi: 10.1046/j.1365-313X.1998.00208.x
Brands, A., and Ho, T. H. (2002). Function of a plant stress-induced gene, HVA22. Synthetic enhancement screen with its yeast homolog reveals its role in vesicular traffic. Plant Physiol. 130, 1121–1131. doi: 10.1104/pp.007716
Byrnes, L. J., Singh, A., Szeto, K., Benvin, N. M., O’Donnell, J. P., Zipfel, W. R., et al. (2013). Structural basis for conformational switching and GTP loading of the large G protein atlastin. EMBO J. 32, 369–384. doi: 10.1038/emboj.2012.353
Byrnes, L. J., and Sondermann, H. (2011). Structural basis for the nucleotide-dependent dimerization of the large G protein atlastin-1/SPG3A. Proc. Natl. Acad. Sci. U.S.A. 108, 2216–2221. doi: 10.1073/pnas.1012792108
Chan, D. C. (2006). Mitochondrial fusion and fission in mammals. Annu. Rev. Cell Dev. Biol. 22, 79–99. doi: 10.1146/annurev.cellbio.22.010305.104638
Chen, H., Detmer, S. A., Ewald, A. J., Griffin, E. E., Fraser, S. E., and Chan, D. C. (2003). Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 160, 189–200. doi: 10.1083/jcb.200211046
Chen, J., Stefano, G., Brandizzi, F., and Zheng, H. (2011). Arabidopsis RHD3 mediates the generation of the tubular ER network and is required for Golgi distribution and motility in plant cells. J. Cell Sci. 124, 2241–2252. doi: 10.1242/jcs.084624
Faso, C., Boulaflous, A., and Brandizzi, F. (2009). The plant Golgi apparatus: last 10 years of answered and open questions. FEBS Lett. 583, 3752–3757. doi: 10.1016/j.febslet.2009.09.046
Gupton, S. L., Collings, D. A., and Allen, N. S. (2006). Endoplasmic reticulum targeted GFP reveals ER organization in tobacco NT-1 cells during cell division. Plant Physiol. Biochem. 44, 95–105. doi: 10.1016/j.plaphy.2006.03.003
Harrison, S. C. (2008). Viral membrane fusion. Nat. Struct. Mol. Biol. 15, 690–698. doi: 10.1038/nsmb.1456
Hermann, G. J., Thatcher, J. W., Mills, J. P., Hales, K. G., Fuller, M. T., Nunnari, J., et al. (1998). Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 143, 359–373. doi: 10.1083/jcb.143.2.359
Hoppins, S., Lackner, L., and Nunnari, J. (2007). The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 76, 751–780. doi: 10.1146/annurev.biochem.76.071905.090048
Hu, J., Shibata, Y., Voss, C., Shemesh, T., Li, Z., Coughlin, M., et al. (2008). Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science 319, 1247–1250. doi: 10.1126/science.1153634
Hu, J., Shibata, Y., Zhu, P. P., Voss, C., Rismanchi, N., Prinz, W. A., et al. (2009). A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell 138, 549–561. doi: 10.1016/j.cell.2009.05.025
Hu, Y., Zhong, R., Morrison, W. H. III, and Ye, Z. H. (2003). The Arabidopsis RHD3 gene is required for cell wall biosynthesis and actin organization. Planta 217, 912–921. doi: 10.1007/s00425-003-1067–1067
Jahn, R., and Scheller, R. H. (2006). SNAREs – engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7, 631–643. doi: 10.1038/nrm2002
Lee, C., and Chen, L. B. (1988). Dynamic behavior of endoplasmic reticulum in living cells. Cell 54, 37–46. doi: 10.1016/0092-8674(88)90177-8
Lee, H., Sparkes, I., Gattolin, S., Dzimitrowicz, N., Roberts, L. M., Hawes, C., et al. (2013). An Arabidopsis reticulon and the atlastin homologue RHD3-like2 act together in shaping the tubular endoplasmic reticulum. New Phytol. 197, 481–489. doi: 10.1111/nph.12038
Liu, T. Y., Bian, X., Sun, S., Hu, X., Klemm, R. W., Prinz, W. A., et al. (2012). Lipid interaction of the C terminus and association of the transmembrane segments facilitate atlastin-mediated homotypic endoplasmic reticulum fusion. Proc. Natl. Acad. Sci. U.S.A. 109, E2146–E2154. doi: 10.1073/pnas.1208385109
Martens, S., and McMahon, H. T. (2008). Mechanisms of membrane fusion: disparate players and common principles. Nat. Rev. Mol. Cell Biol. 9, 543–556. doi: 10.1038/nrm2417
Nziengui, H., Bouhidel, K., Pillon, D., Der, C., Marty, F., and Schoefs, B. (2007). Reticulon-like proteins in Arabidopsis thaliana: structural organization and ER localization. FEBS Lett. 581, 3356–3362. doi: 10.1016/j.febslet.2007.06.032
Orso, G., Pendin, D., Liu, S., Tosetto, J., Moss, T. J., Faust, J. E., et al. (2009). Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature 460, 978–983. doi: 10.1038/nature08280
Rismanchi, N., Soderblom, C., Stadler, J., Zhu, P. P., and Blackstone, C. (2008). Atlastin GTPases are required for Golgi apparatus and ER morphogenesis. Hum. Mol. Genet. 17, 1591–1604. doi: 10.1093/hmg/ddn046
Schiefelbein, J. W., and Somerville, C. (1990). Genetic control of root hair development in Arabidopsis thaliana. Plant Cell 2, 235–243. doi: 10.1105/tpc.2.3.235
Scott, B. L., Van Komen, J. S., Liu, S., Weber, T., Melia, T. J., and McNew, J. A. (2003). Liposome fusion assay to monitor intracellular membrane fusion machines. Methods Enzymol. 372, 274–300. doi: 10.1016/S0076-6879(03)72016–72013
Shibata, Y., Voeltz, G. K., and Rapoport, T. A. (2006). Rough sheets and smooth tubules. Cell 126, 435–439. doi: 10.1016/j.cell.2006.07.019
Sparkes, I. A., Ketelaar, T., De Ruijter, N. C., and Hawes, C. (2009). Grab a Golgi: laser trapping of Golgi bodies reveals in vivo interactions with the endoplasmic reticulum. Traffic 10, 567–571. doi: 10.1111/j.1600-0854.2009.00891.x
Sparkes, I., Tolley, N., Aller, I., Svozil, J., Osterrieder, A., Botchway, S., et al. (2010). Five Arabidopsis reticulon isoforms share endoplasmic reticulum location, topology, and membrane-shaping properties. Plant Cell 22, 1333–1343. doi: 10.1105/tpc.110.074385
Stefano, G., Renna, L., Moss, T., McNew, J. A., and Brandizzi, F. (2012). In Arabidopsis, the spatial and dynamic organization of the endoplasmic reticulum and Golgi apparatus is influenced by the integrity of the C-terminal domain of RHD3, a non-essential GTPase. Plant J. 69, 957–966. doi: 10.1111/j.1365-313X.2011.04846.x
Sudhof, T. C., and Rothman, J. E. (2009). Membrane fusion: grappling with SNARE and SM proteins. Science 323, 474–477. doi: 10.1126/science.1161748
Terasaki, M., Shemesh, T., Kasthuri, N., Klemm, R. W., Schalek, R., Hayworth, K. J., et al. (2013). Stacked endoplasmic reticulum sheets are connected by helicoidal membrane motifs. Cell 154, 285–296. doi: 10.1016/j.cell.2013.06.031
Voeltz, G. K., Prinz, W. A., Shibata, Y., Rist, J. M., and Rapoport, T. A. (2006). A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124, 573–586. doi: 10.1016/j.cell.2005.11.047
Wang, H., Lockwood, S. K., Hoeltzel, M. F., and Schiefelbein, J. W. (1997). The root hair defective 3 gene encodes an evolutionarily conserved protein with GTP-binding motifs and is required for regulated cell enlargement in Arabidopsis. Genes Dev. 11, 799–811. doi: 10.1101/gad.11.6.799
Weber, T., Zemelman, B. V., McNew, J. A., Westermann, B., Gmachl, M., Parlati, F., et al. (1998). SNAREpins: minimal machinery for membrane fusion. Cell 92, 759–772. doi: 10.1016/S0092-8674(00)81404-X
West, M., Zurek, N., Hoenger, A., and Voeltz, G. K. (2011). A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J. Cell Biol. 193, 333–346. doi: 10.1083/jcb.201011039
Wickner, W., and Schekman, R. (2008). Membrane fusion. Nat. Struct. Mol. Biol. 15, 658–664. doi: 10.1038/nsmb.1451
Zhang, M., Wu, F., Shi, J., Zhu, Y., Zhu, Z., Gong, Q., et al. (2013). Root hair defective family of dynamin-like GTPases mediates homotypic endoplasmic reticulum fusion and is essential for Arabidopsis development. Plant Physiol. 163, 713–720. doi: 10.1104/pp.113.224501
Zheng, H., Kunst, L., Hawes, C., and Moore, I. (2004). A GFP-based assay reveals a role for RHD3 in transport between the endoplasmic reticulum and Golgi apparatus. Plant J. 37, 398–414. doi: 10.1046/j.1365-313X.2003.01969.x
Keywords: endoplasmic reticulum, membrane proteins, membrane fusion, GTPase, plant development
Citation: Zhang M and Hu J (2013) Homotypic fusion of endoplasmic reticulum membranes in plant cells. Front. Plant Sci. 4:514. doi: 10.3389/fpls.2013.00514
Received: 14 November 2013; Paper pending published: 25 November 2013;
Accepted: 01 December 2013; Published online: 18 December 2013.
Edited by:
Jens Tilsner, University of St Andrews, UKCopyright © 2013 Zhang and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junjie Hu, Department of Genetics and Cell Biology, College of Life Sciences, Nankai University, 94 Weijin Road, New Life Science Building A408, Tianjin 300071, China e-mail: huj@nankai.edu.cn
 Miao Zhang
Miao Zhang Junjie Hu
Junjie Hu