- Graduate School of Biosphere Sciences, Hiroshima University, Higashi-Hiroshima, Japan
CAPRICE (CPC) encodes a small protein with an R3 MYB motif and regulates root hair and trichome cell differentiation in Arabidopsis thaliana. Six additional CPC-like MYB proteins including TRIPTYCHON (TRY), ENHANCER OF TRY AND CPC1 (ETC1), ENHANCER OF TRY AND CPC2 (ETC2), ENHANCER OF TRY AND CPC3/CPC-LIKE MYB3 (ETC3/CPL3), TRICHOMELESS1 (TCL1), and TRICHOMELESS2/CPC-LIKE MYB4 (TCL2/CPL4) also have the ability to regulate root hair and/or trichome cell differentiation in Arabidopsis. In this review, we describe our latest findings on how CPC-like MYB transcription factors regulate root hair cell differentiation. Recently, we identified the tomato SlTRY gene as an ortholog of the Arabidopsis TRY gene. Transgenic Arabidopsis plants harboring SlTRY produced more root hairs, a phenotype similar to that of 35S::CPC transgenic plants. CPC is also known to be involved in anthocyanin biosynthesis. Anthocyanin accumulation was repressed in the SlTRY transgenic plants, suggesting that SlTRY can also influence anthocyanin biosynthesis. We concluded that tomato and Arabidopsis partially use similar transcription factors for root hair cell differentiation, and that a CPC-like R3 MYB may be a key common regulator of plant root-hair development.
Brief Background
Cell fate determination is a critical step in plant development. In growing roots, epidermal cells differentiate into two cell types, root-hair cells, and non-hair cells in a file-specific manner. In Arabidopsis roots, epidermal cells in eight symmetrically positioned files differentiate into root-hair cells, and the cells of the other files become non-hair cells. Morphological analysis has shown the positional relationship between cortical cells and epidermal cells. Epidermal cells in contact with the junction of two underlying cortical cells differentiate into root-hair cells, whereas the cells in contact with only one cortical cell differentiate into non-hair cells (Dolan et al., 1993, 1994; Galway et al., 1994; Berger et al., 1998). Several regulatory factors are involved in root-hair or non-hair cell differentiation. The glabra 2 (gl2) and werewolf (wer) mutants convert non-hair cells to root hair cells (Masucci et al., 1996; Lee and Schiefelbein, 1999). The GL2 gene encodes a homeodomain leucine-zipper protein, and the WER gene encodes an R2R3-type MYB transcription factor that activates GL2 expression preferentially in differentiating non-hair cells (Rerie et al., 1994; Di Cristina et al., 1996; Masucci et al., 1996; Lee and Schiefelbein, 1999). GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) encode basic helix-loop-helix (bHLH) transcription factors that affect non-hair cell differentiation in a redundant manner, as evidenced by the conversion of non-hair cells to root-hair cells in the gl3 egl3 double mutant (Bernhardt et al., 2003). Although, obvious increase in the number of root-hair cells was hardly observed in both gl3 and egl3 single mutants (Bernhardt et al., 2003). The TRANSPARENT TESTA GLABRA1 (TTG1) gene is also involved in non-hair cell fate determination, as shown by the conversion of non-hair cells to root-hair cells in the ttg1 mutant (Galway et al., 1994). The TTG1 gene encodes a WD40-repeat protein (Walker et al., 1999). GL3 and EGL3 interact with WER (Bernhardt et al., 2003) and with TTG1 (Payne et al., 2000; Esch et al., 2003; Zhang et al., 2003) in yeast cells. A protein complex including WER, GL3/EGL3, and TTG1 acts upstream of the GL2 gene in the root-hair regulatory pathway and promotes GL2 gene expression (Galway et al., 1994; Rerie et al., 1994; Wada et al., 1997; Hung et al., 1998; Lee and Schiefelbein, 1999; Bernhardt et al., 2003, 2005). The cells expressing GL2 differentiate into non-hair cells (Figure 1). In contrast, the root-hair cell differentiation is controlled by CAPRICE (CPC) as shown by a few root-hair phenotype of the cpc mutant (Wada et al., 1997). The CPC gene encodes R3-type MYB protein (Wada et al., 1997). The TTG1-GL3/EGL3-WER protein complex also up-regulates CPC gene expression in non-hair cells (Koshino-Kimura et al., 2005). The CPC protein moves from non-hair cells to neighboring cells and disturbs the formation of the TTG1-GL3/ETC3-WER transcriptional complex by competitively binding with WER (Wada et al., 2002; Koshino-Kimura et al., 2005; Kurata et al., 2005; Tominaga et al., 2007). The formation of the TTG1-GL3/EGL3-CPC protein complex represses expression of GL2, thereby inhibiting non-hair cell differentiation (Wada et al., 2002; Kurata et al., 2005; Figure 1).
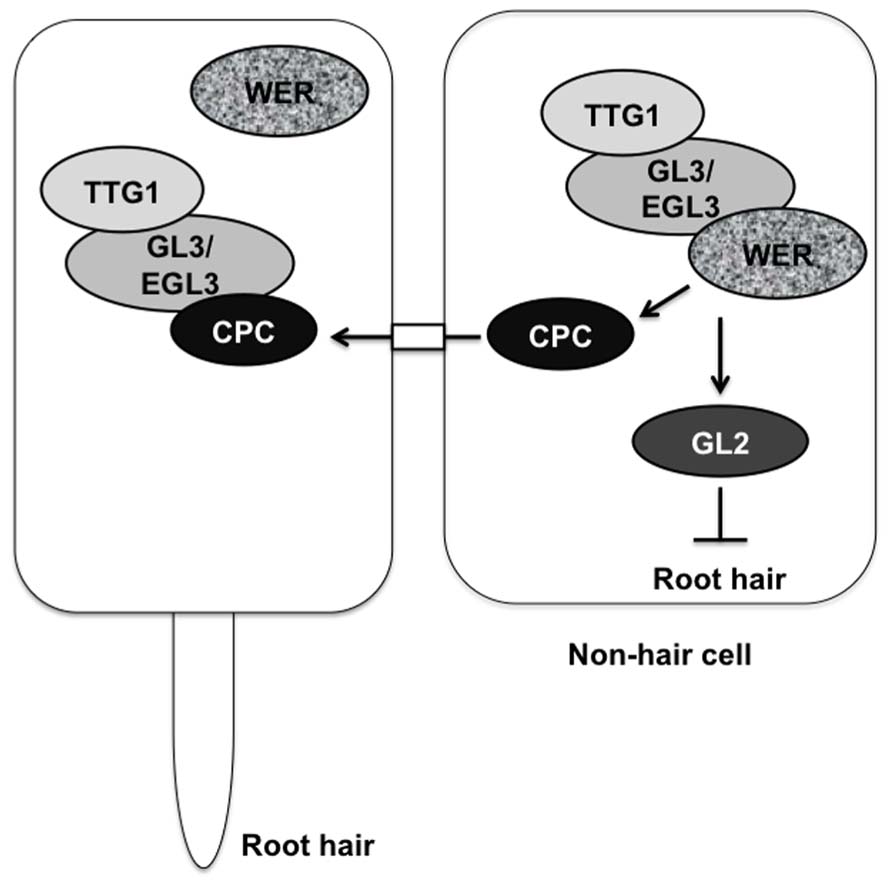
FIGURE 1. Regulation of root-hair cell and non-hair cell fate determination by transcription factors. The TTG1-GL3/EGL3-WER complex promotes GL2 and CPC expression. The cell expressing GL2 differentiates into a non-hair cell. The CPC protein moves from the non-hair cell to a neighboring cell and competes with WER for binding to GL3/EGL3. The TTG1-GL3/EGL3-CPC complex cannot promote GL2 expression, resulting in the differentiation of a root-hair cell.
The CPC Family Promotes Root-Hair Cell Differentiation
CAPRICE encodes a small protein with an R3 MYB motif and strongly promotes root-hair cell differentiation in Arabidopsis (Wada et al., 1997). In addition, we presented a model in which CPC was proposed to have evolved from WER (Tominaga et al., 2007). Chimeric constructs made from the R3 MYB regions of CPC and WER and used in reciprocal complementation tests revealed that the CPC R3 could not functionally substitute for WER R3 in the differentiation of non-hair cells (Tominaga et al., 2007). In contrast, WER R3 can substitute for CPC R3 (Tominaga et al., 2007). Our results suggest that CPC evolved from WER after truncation of the activation domain and loss of DNA binding ability (Tominaga et al., 2007). Arabidopsis has six additional CPC-like MYB sequences in its genome, including TRY, ETC1, ETC2, ETC3/CPL3, TCL1, and TCL2/CPL4 (Hulskamp et al., 1994; Schellmann et al., 2002; Esch et al., 2004; Kirik et al., 2004a,b; Simon et al., 2007; Wang et al., 2007, 2008, 2010; Tominaga et al., 2008; Wester et al., 2009; Gan et al., 2011; Tominaga-Wada and Nukumizu, 2012). These seven CPC-like MYB transcription factors act as positive regulators of root-hair cell differentiation and as negative regulators of trichome differentiation in a partially redundant manner (Tominaga-Wada et al., 2011; Tominaga-Wada and Nukumizu, 2012). The try mutant forms trichome clusters on leaves indicating that TRY functions in trichome differentiation (Hulskamp et al., 1994; Schellmann et al., 2002). ETC1 and ETC2 have redundant and enhancer functions with CPC and TRY in root-hair and trichome differentiation (Esch et al., 2004; Kirik et al., 2004a,b). Therefore, these genes were named ENHANCER OF TRY AND CPC (ETC; Esch et al., 2004; Kirik et al., 2004a,b). TCL1 and TCL2 negatively regulate trichome formation on the inflorescence stems and pedicels (Wang et al., 2007; Gan et al., 2011). These findings suggest functional divergence among CPC family genes.
Recent Findings on the Functions of the CPC Family
We have identified the CPL4 gene between At2g30430 and ETC2 (At2g30420) independently of Gan et al. (Gan et al., 2011; Tominaga-Wada and Nukumizu, 2012). Between CPL4 and ETC2, there were several chimeric transcripts generated through alternative splicing (Tominaga-Wada and Nukumizu, 2012). Our study proposed that inter-genic alterative splicing also characterizes the CPC-like MYB gene family (Tominaga-Wada and Nukumizu, 2012).
A lateral inhibition mechanism mediated by cell-to-cell movement of CPC was thought to cause cell fate specification (Lee and Schiefelbein, 2002; Kwak and Schiefelbein, 2007, 2008). However, it is unclear how CPC, which is preferentially expressed in non-hair cells, specifically acts in the root-hair cells rather than in non-hair cells. Recently, nuclear trapping of CPC in the root-hair cells by EGL3 was suggested to be involved in root-hair cell differentiation (Kang et al., 2013). CPC protein accumulates predominantly in the nuclei of root-hair cells in the early meristematic region, and this localization requires specific expression of EGL3 in the root-hair cells (Kang et al., 2013). These results suggest that cell-to-cell movement of CPC occurs within the meristem of root epidermal cells and that EGL3 traps the CPC protein in the root-hair cells (Kang et al., 2013). CPC and TRY were reported to recruit AtMYC1 into the nucleus, suggesting mutual control of the intracellular localization of patterning proteins (Pesch et al., 2013). AtMYC1, a homologue of GL3 and EGL3, encodes a bHLH transcription factor predominantly localized in the cytoplasm (Urao et al., 1996; Pesch et al., 2013). AtMYC1 regulates the distribution of GL1 protein between the nucleus and the cytoplasm. On the other hand, AtMYC1 is recruited into the nucleus by TRY and CPC, subsequent to significant accumulation of TRY and CPC in the nucleus (Pesch et al., 2013). These results and genetic analyses imply that AtMYC1 represses the activity of TRY and CPC (Pesch et al., 2013).
Tissue-specific transcript profiling also indicated that there were some redundancies between CPC and TRY at the transcriptional level (Simon et al., 2013). We have extended the characterization of CPC-like MYB genes to include the identification of inter-genic alterative splicing and precise expression patterns using tissue-specific transcript profiling (Tominaga-Wada and Nukumizu, 2012; Simon et al., 2013). Recent findings have also revealed that in addition to the formation of the transcription complex, each type of transcription factor can regulate the inter- and intra-cellular localization of the other types to regulate root hair and trichome formation (Kang et al., 2013; Pesch et al., 2013).
A CPC-Like MYB in Tomato
Recently, we identified the tomato SlTRY gene as an ortholog of an Arabidopsis CPC-like MYB gene (Tominaga-Wada et al., 2013b). The CPC::SlTRY construct in cpc-2 transgenic plants increased the number of root-hairs compared with that of the cpc-2 mutant plants (Figure 2; Tominaga-Wada et al., 2013b). These results suggest that tomato and Arabidopsis use common transcription factors for root-hair differentiation. In addition to root-hair cell differentiation, the Arabidopsis CPC gene is known to regulate anthocyanin biosynthesis (Zhu et al., 2009). Anthocyanin accumulation was repressed in the CPC::SlTRY transgenic plants as was observed in the 35S::CPC transgenic plants, suggesting that SlTRY also influences anthocyanin pigment synthesis (Tominaga-Wada et al., 2013a). Tomato and Arabidopsis partially use similar transcription factors for root hair cell differentiation, and a CPC-like R3 MYB may be a key common regulator of plant root-hair development. Further analysis of CPC-like gene function in tomato is on-going.
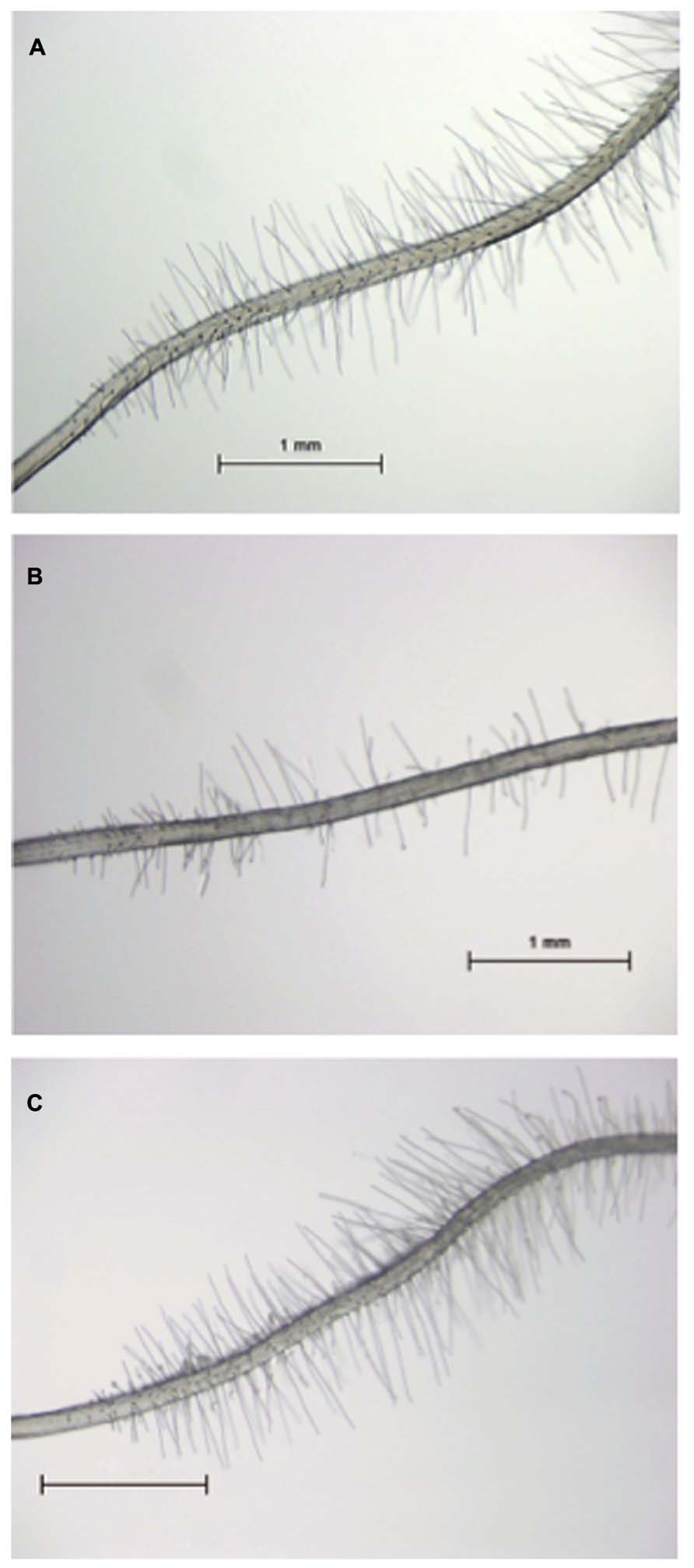
FIGURE 2. Root-hair phenotype of CPC::SlTRY transgenic plants. (A) Root-hair phenotype of a 5-day-old Arabidopsis wild type (Col-0) seedling. (B) Root-hair phenotype of a 5-day-old Arabidopsis cpc-2 mutant seedling. (C) Root-hair phenotype of a 5-day-old Arabidopsis cpc-2 mutant seedling transformed with CPC::SlTRY. Scale bars: 1 mm.
Future Perspectives
The cell-to-cell movement of CPC from non-hair cells to root-hair cells is important for root-hair cell specification; however, the precise mechanism of CPC movement is unknown. How CPC is targeted, transported through plasmodesmata, and trapped in the nucleus of the root-hair cells to define cell fate is an intriguing problem.
Transcriptome analyses provide detailed characterizations of transcription factors involved in root epidermal cell differentiation. Further molecular characterization of individual genes and mutant phenotypes is necessary to fully assess the precise mechanism for root epidermal cell differentiation, including an analysis of redundancies in the epidermal cell regulatory pathway.
TRY and GL3 homologous genes were identified from the tomato genome and named SlTRY and SlGL3, respectively (Tominaga-Wada et al., 2013b). SlTRY showed a similar function to TRY, including inhibition of trichome formation and enhancement of root-hair differentiation. On the other hand, SlGL3 did not show any obvious effect on trichome or non-hair cell differentiation (Tominaga-Wada et al., 2013b). There may be other GL3 ortholog(s) in the unannotated tomato genomes, or tomato uses other pathways to regulate epidermal cell differentiation. Further studies to determine the functions of R3-MYB and bHLH in epidermal cell differentiation in tomato are required.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
JSPS KAKENHI Grant numbers 24658032, 23570057, and 25114513 financially supported this work.
References
Berger, F., Haseloff, J., Schiefelbein, J., and Dolan, L. (1998). Positional information in root epidermis is defined during embryogenesis and acts in domains with strict boundaries. Curr. Biol. 8, 421–430.
Bernhardt, C., Lee, M. M., Gonzalez, A., Zhang, F., Lloyd, A., and Schiefelbein, J. (2003). The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130, 6431–6439. doi: 10.1242/dev.00880
Bernhardt, C., Zhao, M., Gonzalez, A., Lloyd, A., and Schiefelbein, J. (2005). The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132, 291–298. doi: 10.1242/dev.01565
Di Cristina, M., Sessa, G., Dolan, L., Linstead, P., Baima, S., Ruberti, I., et al. (1996). The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J. 10, 393–402. doi: 10.1046/j.1365-313X.1996.10030393.x
Dolan, L., Duckett, C. M., Grierson, C., Linstead, P., Schneider, K., Lawson, E., et al. (1994). Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development 120, 2465–2474.
Dolan, L., Janmaat, K., Willemsen, V., Linstead, P., Poethig, S., Roberts, K., et al. (1993). Cellular organisation of the Arabidopsis thaliana root. Development 119, 71–84.
Esch, J. J., Chen, M. A., Hillestad, M., and Marks, M. D. (2004). Comparison of TRY and the closely related At1g01380 gene in controlling Arabidopsis trichome patterning. Plant J. 40, 860–869.
Esch, J. J., Chen, M., Sanders, M., Hillestad, M., Ndkium, S., Idelkope, B., et al. (2003). A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis. Development 130, 5885–5894. doi: 10.1242/dev.00812
Galway, M. E., Masucci, J. D., Lloyd, A. M., Walbot, V., Davis, R. W., and Schiefelbein, J. W. (1994). The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 166, 740–754.
Gan, L., Xia, K., Chen, J. G., and Wang, S. (2011). Functional Characterization of TRICHOMELESS2, a new single-repeat R3 MYB transcription factor in the regulation of trichome patterning in Arabidopsis. BMC Plant Biol. 11:176. doi: 10.1186/1471-2229-11-176
Hulskamp, M., Misra, S., and Jurgens, G. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell 76, 555–566. doi: 10.1016/0092-8674(94)90118-X
Hung, C. Y., Lin, Y., Zhang, M., Pollock, S., Marks, M. D., and Schiefelbein, J. (1998). A common position-dependent mechanism controls cell-type patterning and GLABRA2 regulation in the root and hypocotyl epidermis of Arabidopsis. Plant Physiol. 117, 73–84. doi: 10.1104/pp.117.1.73
Kang, Y. H., Song, S. K., Schiefelbein, J., and Lee, M. M. (2013). Nuclear trapping controls the position-dependent localization of CAPRICE in the root epidermis of Arabidopsis. Plant Physiol. 163, 193–204. doi: 10.1104/pp.113.221028
Kirik, V., Simon, M., Huelskamp, M., and Schiefelbein, J. (2004a). The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev. Biol. 268, 506–513.
Kirik, V., Simon, M., Wester, K., Schiefelbein, J., and Hulskamp, M. (2004b). ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol. Biol. 55, 389–398. doi: 10.1007/s11103-004-0893-8
Koshino-Kimura, Y., Wada, T., Tachibana, T., Tsugeki, R., Ishiguro, S., and Okada, K. (2005). Regulation of CAPRICE transcription by MYB proteins for root epidermis differentiation in Arabidopsis. Plant Cell Physiol. 46, 817–826.
Kurata, T., Ishida, T., Kawabata-Awai, C., Noguchi, M., Hattori, S., Sano, R., et al. (2005). Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 132, 5387–5398.
Kwak, S. H., and Schiefelbein, J. (2007). The role of the SCRAMBLED receptor-like kinase in patterning the Arabidopsis root epidermis. Dev. Biol. 302, 118–131. doi: 10.1016/j.ydbio.2006.09.009
Kwak, S. H., and Schiefelbein, J. (2008). Cellular pattern formation by SCRAMBLED, a leucine-rich repeat receptor-like kinase in Arabidopsis. Plant Signal. Behav. 3, 110–112.
Lee, M. M., and Schiefelbein, J. (1999). WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99, 473–483. doi: 10.1016/S0092-8674(00)81536-6
Lee, M. M., and Schiefelbein, J. (2002). Cell pattern in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell 14, 611–618. doi: 10.1105/tpc.010434
Masucci, J. D., Rerie, W. G., Foreman, D. R., Zhang, M., Galway, M. E., Marks, M. D., et al. (1996). The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122, 1253–1260.
Payne, C. T., Zhang, F., and Lloyd, A. M. (2000). GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156, 1349–1362.
Pesch, M., Schultheiss, I., Digiuni, S., Uhrig, J. F., and Hulskamp, M. (2013). Mutual control of intracellular localisation of the patterning proteins AtMYC1, GL1 and TRY/CPC in Arabidopsis. Development 140, 3456–3467. doi: 10.1242/dev.094698
Rerie, W. G., Feldmann, K. A., and Marks, M. D. (1994). The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 8, 1388–1399.
Schellmann, S., Schnittger, A., Kirik, V., Wada, T., Okada, K., Beermann, A., et al. (2002). TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 21, 5036–5046. doi: 10.1093/emboj/cdf524
Simon, M., Bruex, A., Kainkaryam, R. M., Zheng, X., Huang, L., Woolf, P. J., et al. (2013). Tissue-specific profiling reveals transcriptome alterations in Arabidopsis mutants lacking morphological phenotypes. Plant Cell 25, 3175–3185. doi: 10.1105/tpc.113.115121
Simon, M., Lee, M. M., Lin, Y., Gish, L., and Schiefelbein, J. (2007). Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev. Biol. 311, 566–578. doi: 10.1016/j.ydbio.2007.09.001
Tominaga, R., Iwata, M., Okada, K., and Wada, T. (2007). Functional analysis of the epidermal-specific MYB genes CAPRICE and WEREWOLF in Arabidopsis. Plant Cell 19, 2264–2277. doi: 10.1105/tpc.106.045732
Tominaga, R., Iwata, M., Sano, R., Inoue, K., Okada, K., and Wada, T. (2008). Arabidopsis CAPRICE-LIKE MYB 3 (CPL3) controls endoreduplication and flowering development in addition to trichome and root hair formation. Development 135, 1335–1345. doi: 10.1242/dev.017947
Tominaga-Wada, R., and Nukumizu, Y. (2012). Expression analysis of an R3-type MYB transcription factor CPC-LIKE MYB4 (TRICHOMELESS2) and CPL4-related transcripts in Arabidopsis. Int. J. Mol. Sci. 13, 3478–3491. doi: 10.3390/ijms13033478
Tominaga-Wada, R., Ishida, T., and Wada, T. (2011). New insights into the mechanism of development of Arabidopsis root hairs and trichomes. Int. Rev. Cell Mol. Biol. 286, 67–106. doi: 10.1016/B978-0-12-385859-7.00002-1
Tominaga-Wada, R., Nukumizu, Y., and Wada, T. (2013a). Tomato (Solanum lycopersicum) homologs of TRIPTYCHON (SlTRY) and GLABRA3 (SlGL3) are involved in anthocyanin accumulation. Plant Signal. Behav. 8:e24575. doi: 10.4161/psb.24575
Tominaga-Wada, R., Nukumizu, Y., Sato, S., and Wada, T. (2013b). Control of plant trichome and root-hair development by a tomato (Solanum lycopersicum) R3 MYB transcription factor. PLoS ONE 8:e54019. doi: 10.1371/journal.pone.0054019
Urao, T., Yamaguchi-Shinozaki, K., Mitsukawa, N., Shibata, D., and Shinozaki, K. (1996). Molecular cloning and characterization of a gene that encodes a MYC-related protein in Arabidopsis. Plant Mol. Biol. 32, 571–576. doi: 10.1007/BF00019112
Wada, T., Kurata, T., Tominaga, R., Koshino-Kimura, Y., Tachibana, T., Goto, K., et al. (2002). Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development 129, 5409–5419.
Wada, T., Tachibana, T., Shimura, Y., and Okada, K. (1997). Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277, 1113–1116. doi: 10.1126/science.277.5329.1113
Walker, A. R., Davison, P. A., Bolognesi-Winfield, A. C., James, C. M., Srinivasan, N., Blundell, T. L., et al. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11, 1337–1350. doi: 10.1105/tpc.11.7.1337
Wang, S., Barron, C., Schiefelbein, J., and Chen, J. G. (2010). Distinct relationships between GLABRA2 and single-repeat R3 MYB transcription factors in the regulation of trichome and root hair patterning in Arabidopsis. New Phytol. 185, 387–400. doi: 10.1111/j.1469-8137.2009.03067.x
Wang, S., Hubbard, L., Chang, Y., Guo, J., Schiefelbein, J., and Chen, J. G. (2008). Comprehensive analysis of single-repeat R3 MYB proteins in epidermal cell patterning and their transcriptional regulation in Arabidopsis. BMC Plant Biol. 8:81. doi: 10.1186/1471-2229-8-81
Wang, S., Kwak, S. H., Zeng, Q., Ellis, B. E., Chen, X. Y., Schiefelbein, J., et al. (2007). TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development 134, 3873–3882. doi: 10.1242/dev.009597
Wester, K., Digiuni, S., Geier, F., Timmer, J., Fleck, C., and Hulskamp, M. (2009). Functional diversity of R3 single-repeat genes in trichome development. Development 136, 1487–1496. doi: 10.1242/dev.021733
Zhang, F., Gonzalez, A., Zhao, M., Payne, C. T., and Lloyd, A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130, 4859–4869. doi: 10.1242/dev.00681
Keywords: Arabidopsis, MYB, root-hair, tomato, transcription factors
Citation: Tominaga-Wada R and Wada T (2014) Regulation of root hair cell differentiation by R3 MYB transcription factors in tomato and Arabidopsis. Front. Plant Sci. 5:91. doi: 10.3389/fpls.2014.00091
Received: 27 November 2013; Accepted: 24 February 2014;
Published online: 13 March 2014.
Edited by:
John Schiefelbein, University of Michigan, USAReviewed by:
John Schiefelbein, University of Michigan, USAJin-Gui Chen, Oak Ridge National Laboratory, USA
Viktor Kirik, Illinois State University, USA
Copyright © 2014 Tominaga-Wada and Wada. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rumi Tominaga-Wada, Graduate School of Biosphere Sciences, Hiroshima University, 1-4-4 Kagamiyama, Higashi-Hiroshima 739-8528, Japan e-mail: rtomi@hiroshima-u.ac.jp
 Rumi Tominaga-Wada
Rumi Tominaga-Wada Takuji Wada
Takuji Wada