Interaction of PLS and PIN and hormonal crosstalk in Arabidopsis root development
- The Integrative Cell Biology Laboratory, School of Biological and Biomedical Sciences, The Biophysical Sciences Institute, Durham University, Durham, UK
Plants are sessile organisms and therefore they must adapt their growth and architecture to a changing environment. Understanding how hormones and genes interact to coordinate plant growth in a changing environment is a major challenge in developmental biology. Although a localized auxin concentration maximum in the root tip is important for root development, auxin concentration cannot change independently of multiple interacting hormones and genes. In this review, we discuss the experimental evidence showing that the POLARIS peptide of Arabidopsis plays an important role in hormonal crosstalk and root growth, and review the crosstalk between auxin and other hormones for root growth with and without osmotic stress. Moreover, we discuss that experimental evidence showing that, in root development, hormones and the associated regulatory and target genes form a network, in which relevant genes regulate hormone activities and hormones regulate gene expression. We further discuss how it is increasingly evident that mathematical modeling is a valuable tool for studying hormonal crosstalk. Therefore, a combined experimental and modeling study on hormonal crosstalk is important for elucidating the complexity of root development.
Introduction
Hormone signaling systems coordinate plant growth and development through a range of complex interactions. The activities of hormones such as auxin, ethylene, cytokinin, abscisic acid, gibberellin, and brassinosteroids depend on cellular context and exhibit either synergistic or antagonistic interactions. Additionally, auxin is directionally transported through plant tissues, providing positional and vectorial information during development (Vanneste and Friml, 2009). Patterning in Arabidopsis root development is coordinated via a localized auxin concentration maximum in the root tip (Sabatini et al., 1999), requiring the regulated expression of specific genes. This auxin gradient has been hypothesized to be sink-driven (Friml et al., 2002) and computational modeling suggests that auxin efflux carrier activity may be sufficient to generate the gradient in the absence of auxin biosynthesis in the root (Grieneisen et al., 2007; Wabnik et al., 2010). However, other experimental studies show that local auxin biosynthesis modulates gradient-directed planar polarity in Arabidopsis, and a local source of auxin biosynthesis contributes to auxin gradient homeostasis (Ikeda et al., 2009). Thus genetic studies show that auxin biosynthesis (Ikeda et al., 2009; Normanly, 2010; Zhao, 2010), the AUX1/LAX influx carriers (Swarup et al., 2005, 2008; Jones et al., 2008; Krupinski and Jonsson, 2010), and the PIN auxin efflux carriers (Petrásek et al., 2006; Grieneisen et al., 2007; Krupinski and Jonsson, 2010; Mironova et al., 2010) all play important roles in the formation of auxin gradients.
In addition, experimental evidence shows that, in root development, hormones and the associated regulatory and target genes form a network, in which relevant genes regulate hormone activities and hormones regulate gene expression. For example, ethylene promotes auxin flux in the root, in a process dependent on the POLARIS (PLS) peptide (Ruzicka et al., 2007; Swarup et al., 2007; Liu et al., 2010a). Furthermore, PIN levels are positively regulated by ethylene and auxin in Arabidopsis roots (Ruzicka et al., 2007). Interestingly, cytokinin can negatively regulate PIN levels (Ruzicka et al., 2009), while repressing auxin biosynthesis and promoting ethylene responses (Nordstrom et al., 2004; Chandler, 2009; Liu et al., 2010a). Cytokinin also has the capacity to modulate auxin transport, by transcriptional regulation of the PIN genes (Ruzicka et al., 2009).
KEY CONCEPT 1. POLARIS (PLS) peptide
The POLARIS (PLS) peptide was identified in the plant species Arabidopsis thaliana by promoter trapping. The gene encodes a mRNA of ca. 600 bases, at the 3'-end of which is a 36 amino acid open reading frame (ORF). Translation of the ORF is required for biological activity. The transcriptional start of the gene overlaps with the 3'-UTR of an upstream gene. Expression of the PLS gene is strongest in root tips, but is also detectable in young leaves, and is induced by auxin. Mutation of POLARIS leads to several developmental defects, including a short root phenotype and reduced vascular complexity in leaves. Recent results show a role for PLS in repressing ethylene responses, and in promoting ethylene-mediated auxin biosynthesis.
Different aspects of hormonal crosstalk in root development have been reviewed recently (Bishopp et al., 2011; Depuydt and Hardtke, 2011; Ross et al., 2011; Garay-Arroyo et al., 2012; Hwang et al., 2012; Vanstraelen and Eva Benkova, 2012). These review articles have concentrated on the interactions of either a wide range of hormones or the specific interactions of a couple of hormones. For example, the review by Garay-Arroyo et al. (2012) covers auxin, ethylene, cytokinin, gibberellins, brassinosteroids and abscisic acid. The review by Hwang et al. (2012) mainly discusses the interaction between cytokinin and auxin in detail. The readers may consult those reviews for different aspects of hormonal crosstalk. Here our focused review concentrates on a combined experimental and modeling perspective of hormonal crosstalk in root development.
KEY CONCEPT 2. Hormonal crosstalk
Hormonal crosstalk refers to the phenomenon whereby the activities of hormones such as auxin, ethylene, cytokinin, abscisic acid, gibberellin, and brassinosteroids exhibit either synergistic or antagonistic interactions, depending on cellular context.
Roles of the Polaris Peptide of Arabidopsis in Hormonal Crosstalk and Root Growth
The POLARIS (PLS) gene of Arabidopsis transcribes a short mRNA encoding a 36-amino acid peptide (Casson et al., 2002). Expression of the PLS gene of Arabidopsis is repressed by ethylene and induced by auxin (Casson et al., 2002; Chilley et al., 2006). It was also experimentally shown that pls mutant roots are short, with reduced cell elongation, and they are hyper-responsive to exogenous cytokinins. Moreover, pls mutant roots show increased expression of the cytokinin-inducible gene, ARR5/IBC6, compared with the wild type (Casson et al., 2002). On the other hand, in the pls mutant, auxin concentration is reduced (Figure 1), cytokinin concentration is enhanced and ethylene production remains approximately unchanged compared to wild-type (Casson et al., 2002; Chilley et al., 2006; Liu et al., 2010a). In the PLS overexpressing transgenic PLOSox, auxin concentration is increased, while ethylene production remains approximately unchanged. In the ethylene resistant pls etr1 double mutant, auxin concentration is approximately recovered to the same level as that in wild-type seedlings (Casson et al., 2002; Chilley et al., 2006; Liu et al., 2010a). In addition, immunolocalization studies reveal that both PIN1 (Figure 1) and PIN2 protein levels increase in the pls mutant, and decrease in PLSox (Liu et al., 2013). In the ethylene-insensitive etr1 mutant, PIN1 and PIN2 levels are lower than those in wild-type. In addition, the double mutant pls etr1 exhibits reduced PIN1 and PIN2 levels compared to pls and slightly lower PIN1 and PIN2 levels compared to wild-type (Liu et al., 2013). Therefore, experimental data have shown that the PLS gene plays important roles in the crosstalk between auxin, ethylene and cytokinin.
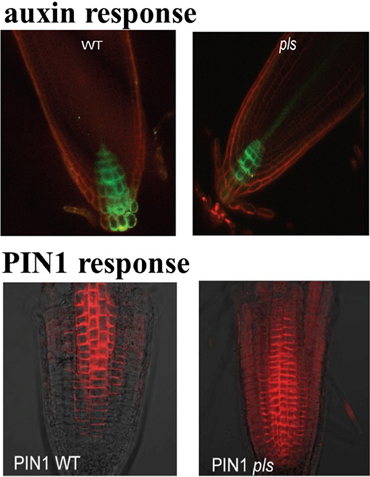
Figure 1. DR5::GFP expression in wild type and pls mutant, showing difference in auxin gradients (upper panel), and PIN1 immunolocalization in wildtype and pls mutant, showing differences in PIN protein levels (lower panel). This figure is adapted with permission from the Figure 4 of Liu et al. (2010a) and the Figure 1 of Liu et al. (2013).
By combining the experimental data relating to the PLS gene with a variety of other experimental data in the literature, we have revealed that PLS, PIN1/PIN2, and three hormones (auxin, ethylene and cytokinin) form an interacting network (Figure 2), in which expression of PLS and PIN1/PIN2 levels regulate auxin, ethylene and cytokinin responses, which in turn regulate expression of PLS and PIN1/PIN2 (Liu et al., 2013). In addition, changing the concentration of, or response to a given hormone may also change the concentrations of/responses to other hormones. Therefore, functions of hormones and the associated genes in root development must be analyzed as an integrative system, as exemplified in Figure 2.
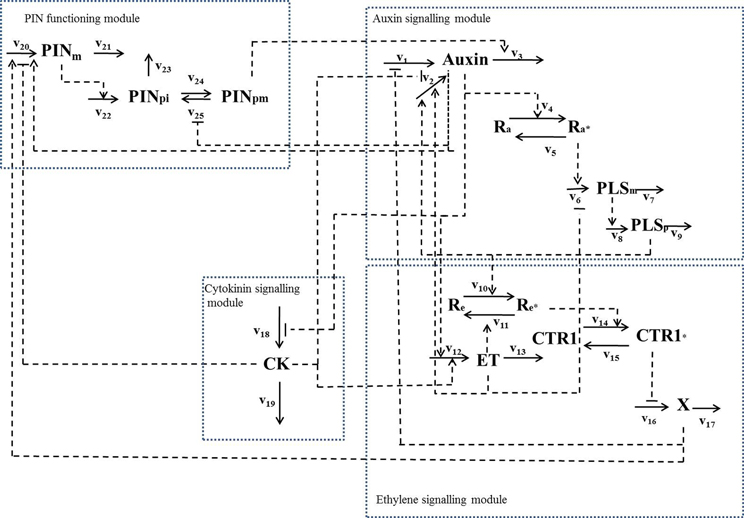
Figure 2. A hormonal crosstalk network of auxin, ethylene and cytokinin for root development, showing that change in one signaling component leads to change in other signaling components in the network (modified with permission from Liu et al., 2013). The reaction rates are: v1, total auxin influx from all neighboring; v2, auxin biosynthesis rate in the cell; v3, total auxin efflux from the cell; v4, rate for conversion of the inactive form of the auxin receptor, Ra, to its active form, Ra*; v5, rate for conversion of the active form of the auxin receptor, Ra*, to its inactive form, Ra; v6, transcription rate of the POLARIS (PLS)gene; v7, decay rate of PLS mRNA; v8, translation rate of the PLS protein; v9, decay rate of PLS protein; v10, rate for conversion of the inactive form of the ethylene receptor, Re, to its active form by PLS protein (PLSp), Re*; v11, rate for conversion of the active form of ethylene receptor, Re*, to its inactive form, Re; v12, ethylene biosynthesis rate; v13, rate for removal of ethylene; v14, rate for conversion of the inactive form of the CONSTITUTIVE TRIPLE RESPONSE 1 (CTR1) protein, CTR1, to its active form, CTR1*; v15, rate for conversion of the active form of CTR1 protein, CTR1*, to its inactive form, CTR1; v16, rate for activation of the ethylene signaling response; v17, rate for removal of the unknown ethylene signaling component, X; v18, rate for cytokinin biosynthesis; v19, rate for removal of cytokinin; v20, transcription rate of the PIN gene; v21, rare for the decay of PIN mRNA; v22, translation rate of PIN protein; v23, rate for decay of PIN protein in cytosol; v24, rate for transport of PIN protein from cytosol to plasma membrane; v25, rate for internalization of PIN protein. When exogenous hormones are applied: v26, rate for uptake of IAA when exogenous IAA is applied; v27, rate for uptake of ACC when exogenous ACC is applied; v28, rate for uptake of cytokinin when exogenous cytokinin is applied.
Crosstalk Between Auxin and Other Hormones for Root Growth
Figure 2 describes the crosstalk network for three major hormones: auxin, cytokinin and ethylene. This network can be further expanded by including other crosstalk components. At early developmental stages, the balance between auxin and cytokinin signaling is crucial: for example, in the Arabidopsis root meristem, auxin promotes cell division (Dello Ioio et al., 2008) and cytokinin promotes cell differentiation (Perilli et al., 2010). Moubayidin et al. (2010) found that higher gibberellin (GA) levels in the young meristem repress ARR1 expression, which is accompanied by a drop in IAA3/SHY2 transcription. The ARR1 cytokinin-responsive transcription factor activates the gene SHY2, which negatively regulates the PIN genes encoding auxin transport facilitators. Thus, GA forms a circuit regulating the balance between auxin and cytokinin signaling.
Brassinosteroids (BRs) also interact with auxin. For example, both BRs and auxin pathways synergistically regulate the expression of several auxin-responsive genes (Mouchel et al., 2006). BRs also regulate root development and this activity is concentration-dependent (Mussig et al., 2003). Thus, a complex relationship between auxin and BRs is formed (Hardtke, 2007).
Abscisic acid (ABA) is an isoprenoid hormone that is involved in the regulation of seed development and dormancy, as well as plant responses to various environmental stresses (Finkelstein and Rock, 2002). It has been experimentally shown that the gene VIVIPAROUS1 (VP1) in maize and its Arabidopsis ortholog ABI3, which encodes a transcription factor involved in ABA signaling, is auxin-inducible (Suzuki et al., 2001; Brady et al., 2003). Therefore, ABA interacts with the crosstalk network via the action of auxin (Figure 2).
The interaction between auxin, cytokinin and ethylene in Figure 2 may also be extended to include other signals that regulate root development. For example, glucose signals regulate Arabidopsis seedling root directional growth by interacting with auxin, cytokinin and ethylene (Kushwah et al., 2011). A specific hexokinase in Arabidopsis (HXK1) has a predominant role in glucose signaling. It has been experimentally shown that catalytically inactive HXK1 restores auxin sensitivity for callus and root induction, indicating that the action of the HKX-dependent pathway is closely associated with the action of the auxin signaling pathway (Moore et al., 2003). Therefore, glucose signals can be integrated into the hormonal crosstalk network via interplay with auxin signaling.
KEY CONCEPT 3. Hormonal crosstalk network
A hormonal crosstalk network is a type of biological network that describes gene expression, signal transduction and metabolic conversion complexities associated with hormonal crosstalk activity in plant development. A hormonal crosstalk network is therefore a mixed-type network that integrates transcriptomic, proteomic and metabolic networks.
Hormonal Crosstalk Under Osmotic Stress
Plants remodel their root architecture to deal with osmotic stress, inhibiting lateral root initiation and altering root growth rates (van der Weele et al., 2000; Deak and Malamy, 2005). At low to moderate levels of osmotic stress root growth is increased and at higher levels it is inhibited (van der Weele et al., 2000; Xu et al., 2013).
Hormone crosstalk integrates stress responses with developmental control and as with most abiotic stresses, osmotic stress is characterized by an increase in abscisic acid levels. Increases in abscisic acid are concentrated in the root cap and are less significant than in aerial tissues, but are essential to normal growth under osmotic stress (Christmann et al., 2005; Deak and Malamy, 2005; Xu et al., 2013).
The increase in root growth rate under moderate osmotic stress occurs in an ABA-dependent manner (van der Weele et al., 2000; Xu et al., 2013). Osmotically induced ABA increases basipetal auxin transport through elevated PIN2 and AUX1 levels, increasing H+-ATPase activity and root elongation (Xu et al., 2013). High levels of applied ABA also induce expression of ARF2, a negative regulator of auxin responses and arf2 mutants display altered auxin transport and shorter roots under ABA application (Wang et al., 2011).
Cytokinin is also thought to play a role in osmotic stress responses. Mature plants alter expression of cytokinin biosynthesis and metabolism genes, decreasing cytokinin levels under dehydration stress (Nishiyama et al., 2011). The AHK2 and AHK3 cytokinin receptor kinases negatively regulate ABA and osmotic stress responsive gene expression, and cytokinin-deficient mutants have increased survival rates under stress (Tran et al., 2007; Werner et al., 2010; Nishiyama et al., 2011). Intriguingly, cytokinin-deficient mutants display lower levels of ABA, implying that a positive feedback on ABA may exist (Nishiyama et al., 2011).
Osmotic stress can also promote ethylene biosynthesis, which can have an antagonistic role with ABA on root growth (Ichimura et al., 2000; Sharp, 2002; Liu and Zhang, 2004; Joo et al., 2008; Cheng et al., 2009). ABA inhibits ethylene biosynthesis by positively regulating ERF11 via HY5 to repress expression of ACS5, which catalyses the rate limiting enzyme in ethylene biosynthesis (Vogel et al., 1998; Li et al., 2011). Ethylene can also limit ABA biosynthesis, with ethylene-insensitive mutants hyperaccumulating ABA, associated with increased expression of ABA biosynthetic genes such as NCED3 (Ghassemian et al., 2000; Chiwocha et al., 2005; Wang et al., 2007; Cheng et al., 2009).
Genetically or pharmacologically impairing ethylene signaling makes plants insensitive to root growth inhibition by ABA, but plants impaired in ethylene signaling have shorter roots under severe osmotic stress (Beaudoin et al., 2000; Ghassemian et al., 2000; Wang et al., 2007, 2008). Where limiting ethylene perception makes plants less sensitive to ABA-induced root shortening, pharmacologically limiting ethylene biosynthesis makes plants more sensitive (Ghassemian et al., 2000). This implies that both hormones are required for correct root growth under osmotic stress but the antagonistic regulation of each other's biosynthesis is insufficient to explain inhibition of root growth responses alone.
Mutant analysis has revealed little interaction between the two signal transduction pathways (Cheng et al., 2009). However EIN2, an essential component of ethylene signaling, shows reduced expression under osmotic stress but not ABA treatment, indicating ethylene signaling may be mediated by stress independently of ABA (Wang et al., 2007).
Osmotic stress clearly affects hormone levels in a series of complex interactions which cannot explain root growth adequately in isolation. Hormones and the associated regulatory and target genes in plant root form a network in which relevant genes regulate hormone activities and hormones regulate gene expression. An important question for understanding these complex interactions is: what are the mechanisms that regulate the fluxes of plant hormones and levels of the proteins encoded by the regulatory and target genes? To address this question, it is increasingly evident that mathematical modeling is becoming a valuable tool to tackle the complexity of hormonal crosstalk.
Mathematical Modeling as a Valuable Tool for Studying Hormonal Crosstalk
A hormonal crosstalk network is a type of network consisting of gene expression, signal transduction and metabolic conversions. For example, in Figure 2, hormonal signals are transduced, auxin, ethylene, and cytokinin are synthesized and decayed through metabolic processes, and expression of PIN and PLS genes is realized. Therefore, a hormonal crosstalk network is a mixed-type network that integrates all these components. There are different mathematical tools available for analysing plant biological networks at different levels (Liu et al., 2010b). In particular, kinetic modeling (Rohwer, 2012) is useful to analyse quantitatively hormonal crosstalk networks.
KEY CONCEPT 4. Kinetic modeling
Kinetic modeling is a modeling method that uses differential equations to analyse how each component (concentration, reaction rate) in a hormonal crosstalk network changes in space and time. In kinetic modeling, the rate of a reaction is described by the concentrations of all chemicals involved in the reaction, and the mass balance of all chemicals is described using differential equations.
The hormonal crosstalk between auxin, ethylene and cytokinin via the action of the PLS gene was analyzed using kinetic modeling (Liu et al., 2010a). On the basis of the model structure of the hormonal crosstalk network, relationships between auxin biosynthesis pathway(s) and PLS-regulated hormonal crosstalk can be analyzed. Although the molecular basis for auxin biosynthesis is poorly characterized, mathematical modeling reveals that the regulation of auxin biosynthesis by PLS peptide (PLSp) must be realized through its compounded effects with ethylene and cytokinin: PLSp cannot regulate auxin biosynthesis independently of the regulation of ethylene and cytokinin. This demonstrates that different structures of a hormonal crosstalk network may have different auxin concentration responses, revealing that a combined modeling and experimental analysis is a powerful tool for dissecting the causal relationship for the interactions between genes and hormonal crosstalk (Liu et al., 2010a). In addition, the hormonal crosstalk network constructed by iteratively combining experimental data and mathematical modeling (Liu et al., 2010a) can further integrate novel experimental data (Liu et al., 2013). Thus, through the cycle of modeling predictions and novel experimental measurements, the hormonal crosstalk network can be used to analyse the functions of hormone signals in root development and predict new experiments.
The role of the crosstalk between auxin and cytokinin signaling in specifying the root architecture of Arabidopsis has also been modeled using kinetic modeling (Muraro et al., 2011, 2013). It is found that tissue-specific oscillations in gene expression can be understood based on the joint activity of auxin and cytokinin (Muraro et al., 2013). These results reveal that hormonal crosstalk can be a mechanism explaining time-dependent dynamics such as oscillations, although other mechanisms may also generate oscillations in gene expression (Bujdoso and Davis, 2013; Rue and Garcia-Ojalvo, 2013).
The crosstalk between auxin and BRs signaling has also been analyzed using a Boolean logic approach (Sankar et al., 2011). An advantage of this approach is that it does not require kinetic parameters, which are usually not available (Liu et al., 2010b). However, the underlying assumptions of Boolean logic and their relevance to biological reality should be carefully assessed when applied to the modeling of hormonal crosstalk networks.
Perspectives
This review has focused on recent progress in (a) experimental measurements relating to hormonal interactions in the Arabidopsis root; (b) construction of hormonal crosstalk networks based on experimental data; and (c) combination of experimental and mathematical modeling for elucidating and predicting the roles of hormonal crosstalk in root development.
One of the important features for hormonal crosstalk in root development (Figure 2) is that change in one signaling component leads to change in other signaling components. Therefore, elucidating the regulation of one signaling component requires the development of novel modeling methodology (Liu et al., 2014).
An additional important aspect of hormonal signaling is its spatiotemporal dynamics. Understanding the roles of hormones in root development needs an analysis of the dynamics of hormonal crosstalk networks in spatial settings within a root. Here we propose a methodology that combines experimental data and mathematical modeling to study the spatiotemporal dynamics of hormonal signaling in root development (Figure 3). The study of hormonal crosstalk of auxin, ethylene and cytokinin in the non-spatial setting of a single cell has demonstrated that the methodology described in Figure 3 is a powerful tool (Liu et al., 2010a, 2013).
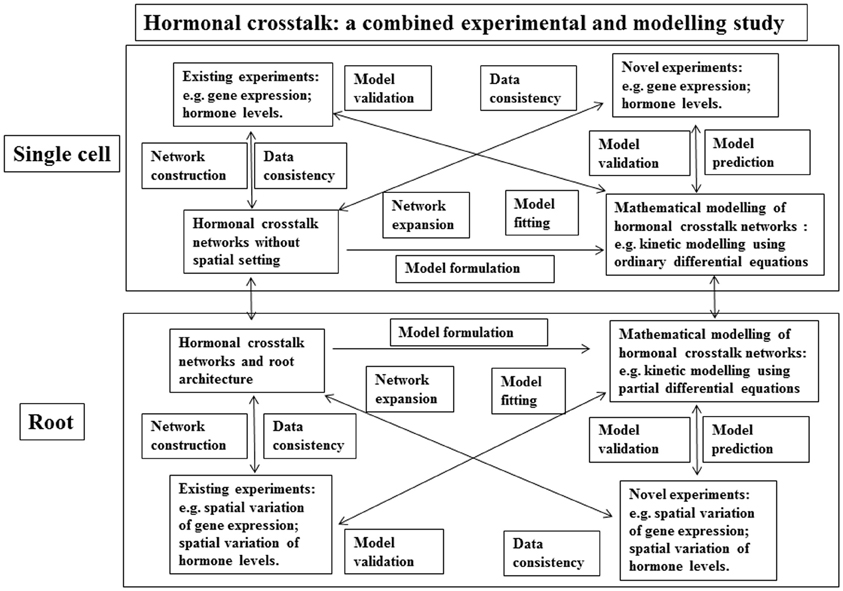
Figure 3. A schematic description of a methodology for a combined experimental and modeling study on hormonal crosstalk in root development. Upper panel: Hormonal crosstalk networks are constructed using existing experimental data. The networks are used to check the consistency of existing experimental data and they are also used to formulate mathematical models such as models described using ordinary differential equations. The mathematical models are validated using the existing experimental data. Then the mathematical models are used to predict novel experiments. The data acquired using the novel experiments are used to expand the hormonal crosstalk networks and to validate the mathematical models again. The expanded hormonal crosstalk networks are used to check the consistency of the novel experiments, and they are also used to develop novel mathematical models. Lower panel: the same as the upper panel, but root architecture is included.
For the spatiotemporal modeling of hormonal crosstalk in the root, the important spatial aspects include (a) mechanisms of PIN polarity and auxin transport (van Berkel et al., 2013); (b) the relationship between auxin distribution and experimental observation of PIN polarity (Grieneisen et al., 2007); and (c) integration of multi-scale root systems (Hill et al., 2013). In addition to what we have discussed in the review, these aspects should be integrated when the root is modeled as an integrative system in space and time.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors gratefully acknowledge Research Councils UK and the Biotechnology and Biological Sciences Research Council for funding in support of this study.
Author Biography

References
Beaudoin, N., Serizet, C., Gosti, F., and Giraudat, J. (2000). Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 12, 1103–1115. doi: 10.2307/3871258
Bishopp, A., Benkova, E., and Helariutta, Y. (2011). Sending mixed messages: auxin-cytokinin crosstalk in roots. Curr. Opin. Plant Biol. 14, 10–16. doi: 10.1016/j.pbi.2010.08.014
Brady, S. M., Sarkar, S. F., Bonetta, D., and McCourt, P. (2003). The ABSCISIC ACID INSENSITIVE3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in Arabidopsis. Plant J. 34, 67–75. doi: 10.1046/j.1365-313X.2003.01707.x
Bujdoso, N., and Davis, S. J. (2013). Mathematical modelling of an oscillating gene circuit to unravel the circadian clock network of Arabidopsis thaliana. Front. Plant Sci. 4:3. doi: 10.3389/fpls.2013.00003
Casson, S. A., Chilley, P. M., Topping, J. F., Evans, I. M., Souter, M. A., and Lindsey, K. (2002). The POLARIS gene of Arabidopsis encodes a predicted peptide required for correct root growth and leaf vascular patterning. Plant Cell 14, 1705–1721. doi: 10.1105/tpc.002618
Chandler, J. W. (2009). Auxin as compère in plant hormone crosstalk. Planta 231, 1–12. doi: 10.1007/s00425-009-1036-x
Cheng, W. H., Chiang, M. H., Hwang, S. G., and Lin, P. C. (2009). Antagonism between abscisic acid and ethylene in Arabidopsis acts in parallel with the reciprocal regulation of their metabolism and signaling pathways. Plant Mol. Biol. 71, 61–80. doi: 10.1007/s11103-009-9509-7
Chilley, P. M., Casson, S. A., Tarkowski, P., Hawkins, N., Wang, K. L., Hussey, P. J., et al. (2006). The POLARIS peptide of Arabidopsis regulates auxin transport and root growth via effects on ethylene signaling. Plant Cell 18, 3058–3072. doi: 10.1105/tpc.106.040790
Chiwocha, S. D. S., Cutler, A. J., Abrams, S. R., Ambrose, S. J., Yang, J., Ross, A. R. et al. (2005). The etr1-2 mutation in Arabidopsis thaliana affects the abscisic acid, auxin, cytokinin and gibberellin metabolic pathways during maintenance of seed dormancy, moist-chilling and germination. Plant J. 42, 35–48. doi: 10.1111/j.1365-313X.2005.02359.x
Christmann, A., Hoffmann, T., Teplova, I., Grill, E., and Muller, A. (2005). Generation of active pools of abscisic acid revealed by in vivo Imaging of water-stressed Arabidopsis. Plant Physiol. 137, 209–219. doi: 10.1104/pp.104.053082
Deak, K. I., and Malamy, J., (2005). Osmotic regulation of root system architecture. Plant J. 43, 17–28. doi: 10.1111/j.1365-313X.2005.02425.x
Dello Ioio, R., Nakamura, K., Moubayidin, L., Perilli, S., Taniguchi, M., Morita, M. T., et al. (2008). A genetic framework for the control of cell division and differentiation in the root meristem. Science 322, 1380–1384. doi: 10.1126/science.1164147
Depuydt, S., and Hardtke, C. S. (2011). Hormone signalling crosstalk in plant growth regulation. Curr. Biol. 21, R365–R373. doi: 10.1016/j.cub.2011.03.013
Finkelstein, R. R., and Rock, C. D. (2002). Abscisic Acid biosynthesis and response. Arabidopsis Book 1, e0058. doi: 10.1199/tab.0058
Friml, J., Benkova, E., Blilou, I., Wisniewska, J., Hamann, T., Ljung, K., et al. (2002). AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108, 661–673. doi: 10.1016/S0092-8674(02)00656-6
Garay-Arroyo, A., De La Paz Sanchez, M., Garcıa-Ponce, B., Azpeitia, E., and Alvarez-Buylla, E. R. (2012). Hormone symphony during root growth and development. Dev. Dyn. 241, 1867–1885. doi: 10.1002/dvdy.23878
Ghassemian, M., Nambara, E., Cutler, S., Kawaide, H., Kamiya, Y., and McCourt, P. (2000). Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 12, 1117–1126. doi: 10.2307/3871259
Grieneisen, V. A., Xu, J., Marée, A. F. M., Hogeweg, P., and Scheres, B. (2007). Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449, 1008–1013. doi: 10.1038/nature06215
Hardtke, C. S. (2007). Transcriptional auxin-brassinosteroid crosstalk: who's talking? Bioessays 29, 1115–1123. doi: 10.1002/bies.20653
Hill, K., Porco, S., Lobet, G., Zappala, S., Mooney, S., Draye, X., and Bennett, M. J. (2013). Root systems biology: integrative modeling across scales, from gene regulatory networks to the rhizosphere. Plant Physiol. 163, 1487–1503. doi: 10.1104/pp.113.227215
Hwang, I., Sheen, J., and Muller, B. (2012). Cytokinin signaling networks. Annu. Rev. Plant Biol. 63, 353–380. doi: 10.1146/annurev-arplant-042811-105503
Ichimura, K., Mizoguchi, T., Yoshida, R., Yuasa, T., and Shinozaki, K. (2000). Various abiotic stresses vapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J. 24, 655–665. doi: 10.1046/j.1365-313x.2000.00913.x
Ikeda, Y., Men, S., Fischer, U., Stepanova, A. N., Alonso, J. M., Ljung, K., et al. (2009). Local auxin biosynthesis modulates gradient directed planar polarity in Arabidopsis. Nat. Cell Biol. 11, 731–738. doi: 10.1038/ncb1879
Jones, A. R., Kramer, E. M., Knox, K., Swarup, R., Bennett, M. J., Lazarus, C. M., et al. (2008). Auxin transport through non-hair cells sustains root-hair development. Nat. Cell Biol. 11, 78–84. doi: 10.1038/ncb1815
Joo, S., Liu, Y., Lueth, A., and Zhang, S. (2008). MAPK phosphorylation-induced stabilization of ACS6 protein is mediated by the non-catalytic C-terminal domain, which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway. Plant J. 54, 129–140. doi: 10.1111/j.1365-313X.2008.03404.x
Krupinski, P., and Jonsson, H. (2010). Modeling auxin-regulated development. Cold Spring Harb. Perspect. Biol. 2, a001560. doi: 10.1101/cshperspect.a001560
Kushwah, S., Jones, A. M., and Laxmi, A. (2011). Cytokinin interplay with ethylene, auxin, and glucose signaling controls Arabidopsis seedling root directional growth. Plant Physiol. 156, 1851–1866. doi: 10.1104/pp.111.175794
Li, Z., Zhang, L., Yu, Y., Quan, R., Zhang, Z., Zhang, H., et al. (2011). The ethylene response factor AtERF11 that is transcriptionally modulated by the bZIP transcription factor HY5 is a crucial repressor for ethylene biosynthesis in Arabidopsis. Plant J. 68, 88–99. doi: 10.1111/j.1365-313X.2011.04670.x
Liu, J. L., Grieson, C. S., Webb, A. A. R., and Hussey, P. J. (2010b). Modelling dynamic plant cells. Curr. Opin. Plant Biol. 13, 744–749. doi: 10.1016/j.pbi.2010.10.002
Liu, J. L., Lindsey, K., and Hussey, P. J. (2014). Elucidating the regulation of complex signalling systems in plant cells. Biochem. Soc. Trans. 42, 219–223. doi: 10.1042/BST20130090
Liu, J. L., Mehdi, S., Topping, J., Friml, J., and Lindsey, K. (2013). Interaction of PLS and PIN and hormonal crosstalk in Arabidopsis root development. Front. Plant Sci. 4:75. doi: 10.3389/fpls.2013.00075
Liu, J. L., Mehdi, S., Topping, J., Tarkowski, P., and Lindsey, K. (2010a). Modelling and experimental analysis of hormonal crosstalk in Arabidopsis. Mol. Syst. Biol. 6, 373. doi: 10.1038/msb.2010.26
Liu, Y. D., and Zhang, S. Q. (2004). Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16, 3386–3399. doi: 10.1105/tpc.104.026609
Mironova, V. V., Omelyanchuk, N. A., Yosiphon, G., Fadeev, S. I., Kolchanov, N. A., Mjolsness, E., et al. (2010). A plausible mechanism for auxin patterning along the developing root. BMC Syst. Biol. 4:98. doi: 10.1186/1752-0509-4-98
Moore, B., Zhou, L., Rolland, F., Hall, Q., Cheng, W. H., Liu, Y. X., et al. (2003). Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300, 332–336. doi: 10.1126/science.1080585
Moubayidin, L., Perilli, S., Dello Ioio, R., Di Mambro, R., Costantino, P., and Sabatini, S. (2010). The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Curr. Biol. 20, 1138–1143. doi: 10.1016/j.cub.2010.05.035
Mouchel, C. F., Osmont, K. S., and Hardtke, C. S. (2006). BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature 443, 458–461. doi: 10.1038/nature05130
Muraro, D., Byrne, H., King, J., and Bennett, M. (2013). The role of auxin and cytokinin signalling in specifying the root architecture of Arabidopsis thaliana. J. Theor. Biol. 317, 71–86. doi: 10.1016/j.jtbi.2012.08.032
Muraro, D., Byrne, H., King, J., Vob, U., Kieber, J., and Bennett, M. (2011). The influence of cytokinin–auxin cross-regulation on cell-fate determination in Arabidopsis thaliana root development. J. Theor. Biol. 283, 152–167. doi: 10.1016/j.jtbi.2011.05.011
Mussig, C., Shin, G. H., and Altmann, T. (2003). Brassinosteroids promote root growth in Arabidopsis. Plant Physiol. 133, 1261–1271. doi: 10.1104/pp.103.028662
Nishiyama, R., Watanabe, Y., Fujita, Y., Dung Tien, L., Kojima, M., Werner, T., et al. (2011). Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 23, 2169–2183. doi: 10.1105/tpc.111.087395
Nordstrom, A., Tarkowski, P., Tarkowska, D., Norbaek, R., Åstot, C., Dolezal, K., et al. (2004). Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: a factor of potential importance for auxin–cytokinin-regulated development. Proc. Natl. Acad. Sci. U.S.A. 101, 8039–8044. doi: 10.1073/pnas.0402504101
Normanly, J. (2010). Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harb. Perspect. Biol. 2, a001594. doi: 10.1101/cshperspect.a001594
Perilli, S., Moubayidin, L., and Sabatini, S. (2010). The molecular basis ofcytokinin function. Curr. Opin. Plant Biol. 13, 21–26. doi: 10.1016/j.pbi.2009.09.018
Petrásek, J., Petrášek, J., Mravec, J., Bouchard, R., Blakeslee, J. J., Abas, M., et al., (2006). PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312, 914–918. doi: 10.1126/science.1123542
Rohwer, J. M. (2012). Kinetic modelling of plant metabolic pathways. J. Exp. Bot. 63, 2275–2292. doi: 10.1093/jxb/ers080
Ross, J. J., Weston, D. E., Davidson, S. E., and Reid, J. B. (2011). Plant hormone interactions: how complex are they? Physiol. Plant. 141, 299–309. doi: 10.1111/j.1399-3054.2011.01444.x
Rue, P., and Garcia-Ojalvo, J. (2013). Modeling gene expression in time and space. Annu. Rev. Biophys. 42, 605–27. doi: 10.1146/annurev-biophys-083012-130335
Ruzicka, K., Ljung, K., Vanneste, S., Podhorska, R., Beeckman, T., Friml, J., et al. (2007). Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19, 2197–2212. doi: 10.1105/tpc.107.052126
Ruzicka, K., Simásková, M., Duclercq, J., Petrásek, J., Zazímalová, E., Simon, S., et al. (2009). Cytokinin regulates root meristem activity via modulation of the polar auxin transport. Proc. Natl. Acad. Sci. U.S.A. 106, 4284–4289. doi: 10.1073/pnas.0900060106
Sabatini, S., Sabatini, S., Beis, D., Wolkenfelt, H., Murfett, J., Guilfoyle, T., et al. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99, 463–472. doi: 10.1016/S0092-8674(00)81535-4
Sankar, M., Osmont, K. S., Rolcik, J., Gujas, B., Tarkowska, D., Strnad, M., et al. (2011). A qualitative continuous model of cellular auxin and brassinosteroid signalling and their crosstalk. Bioinformatics 27, 1404–1412. doi: 10.1093/bioinformatics/btr158
Sharp, R. E. (2002). Interaction with ethylene: changing views on the role of abscisic acid in root and shoot growth responses to water stress. Plant Cell Environ. 25, 211–222. doi: 10.1046/j.1365-3040.2002.00798.x
Suzuki, M., Kao, C. Y., Cocciolone, S., and McCarty, D. R. (2001). Maize VP1 complements Arabidopsis abi3 and confers a novel ABA/auxin interaction in roots. Plant J. 28, 409–418. doi: 10.1046/j.1365-313X.2001.01165.x
Swarup, K., Benková, E., Swarup, R., Casimiro, I., Péret, B., Yang, Y., et al. (2008). The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 10, 946–954. doi: 10.1038/ncb1754
Swarup, R., Kramer, E. M., Perry, P., Knox, K., Leyser, H. M., Haseloff, J., et al. (2005). Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat. Cell Biol. 7, 1057–1065. doi: 10.1038/ncb1316
Swarup, R., Perry, P., Hagenbeek, D., Van Der Straeten, D., Beemster, G. T., Sandberg, G., et al. (2007). Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19, 2186–2196. doi: 10.1105/tpc.107.052100
Tran, L. S. P., Urao, T., Qin, F., Maruyama, K., Kakimoto, T., Shinozaki, K., et al. (2007). Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 20623–20628. doi: 10.1073/pnas.0706547105
van Berkel, K., de Boer, R. J., Scheres, B., and ten Tusscher, K. (2013). Polar auxin transport: models and mechanisms. Development 140, 2253–2268. doi: 10.1242/dev.079111
van der Weele, C. M., Spollen, W. G., Sharp, R. E., and Baskin, T. I. (2000). Growth of Arabidopsis thaliana seedlings under water deficit studied by control of water potential in nutrient-agar media. J. Exp. Bot. 51, 1555–1562. doi: 10.1093/jexbot/51.350.1555
Vanneste, S., and Friml, J. (2009) Auxin: A trigger for change in plant development. Cell 136, 1005–1016. doi: 10.1016/j.cell.2009.03.001
Vanstraelen, M., and Eva Benkova, E. (2012). Hormonal interactions in the regulation of plant development. Annu. Rev. Cell Dev. Biol. 28, 22.1–22.25. doi: 10.1146/annurev-cellbio-101011-155741
Vogel, J. P., Woeste, K. E., Theologis, A., and Kieber, J. J. (1998). Recessive and dominant mutations in the ethylene biosynthetic gene ACS5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proc. Natl. Acad. Sci. U.S.A. 95, 4766–4771. doi: 10.1073/pnas.95.8.4766
Wabnik, K., Kleine-Vehn, J., Balla, J., Sauer, M., Naramoto, S., Reinöhl, V., et al. (2010). Emergence of tissue polarization from synergy of intracellular and extracellular auxin signaling. Mol. Syst. Biol. 6, 447. doi: 10.1038/msb.2010.103
Wang, L., Hua, D. P., He, J. N., Duan, Y., Chen, Z. Z., Hong, X. H., et al. (2011). Auxin response factor2 (ARF2) and its regulated homeodomain gene HB33 mediate abscisic acid response in Arabidopsis. PLoS Genet. 7:e1002172. doi: 10.1371/journal.pgen.1002172
Wang, Y., Liu, C., Li, K., Sun, F., Hu, H., Li, X., et al. (2007). Arabidopsis EIN2 modulates stress response through abscisic acid response pathway. Plant Mol. Biol. 64, 633–644. doi: 10.1007/s11103-007-9182-7
Wang, Y. N., Wang, T., Li, K. X., and Li, X. (2008). Genetic analysis of involvement of ETR1 in plant response to salt and osmotic stress. Plant Growth Regul. 54, 261–269. doi: 10.1007/s10725-007-9249-0
Werner, T., Nehnevajova, E., Kollmer, I., Novak, O., Strnad, M., Kramer, U., et al. (2010). Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in arabidopsis and tobacco. Plant Cell 22, 3905–3920. doi: 10.1105/tpc.109.072694
Xu, W., Jia, L., Shi, W., Liang, J., Zhou, F., Li, Q., et al. (2013). Abscisic acid accumulation modulates auxin transport in the root tip to enhance proton secretion for maintaining root growth under moderate water stress. New Phytol. 197, 139–150. doi: 10.1111/nph.12004
Keywords: root development, POLARIS peptide, hormonal crosstalk, osmotic stress, kinetic modeling
Citation: Liu J, Rowe J and Lindsey K (2014) Hormonal crosstalk for root development: a combined experimental and modeling perspective. Front. Plant Sci. 5:116. doi: 10.3389/fpls.2014.00116
Received: 06 February 2014; Paper pending published: 27 February 2014;
Accepted: 11 March 2014; Published online: 27 March 2014.
Edited by:
Wagner L. Araújo, Universidade Federal de Viçosa, BrazilReviewed by:
Jedrzej Jakub Szymanski, Max Planck Institute of Molecular Plant Physiology, GermanyDimas Mendes Ribeiro, Universidade Federal de Viçosa, Brazil
Copyright © 2014 Liu, Rowe and Lindsey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: keith.lindsey@durham.ac.uk
† Joint corresponding authors
 Junli Liu
Junli Liu James Rowe
James Rowe Keith Lindsey
Keith Lindsey