- Dipartimento di Bioscienze, Università degli Studi di Milano, Milan, Italy
In spermatophytes the ovules upon fertilization give rise to the seeds. It is essential to understand the mechanisms that control ovule number and development as they ultimately determine the final number of seeds and, thereby, the yield in crop plants. In Arabidopsis thaliana, ovules arise laterally from a meristematic tissue within the carpel referred to as placenta. For a correct determination of the number of ovules, a precise establishment of the positions where ovule primordia emerge is needed, and a tight definition of the boundaries between ovules is therefore also required. In the last decades, few factors have been identified to be involved in the determination of ovule number. Recently, plant hormones have also been revealed as fundamental players in the control of the initiation of ovule formation. In this review we summarize the current knowledge about both the molecular and hormonal mechanisms that control ovule formation in Arabidopsis thaliana.
Introduction
Fruits are a major evolutionary acquisition of flowering plants (Angiosperms). They likely evolved to protect the developing seeds and to ensure seed dispersal (Knapp, 2002). Fruits derive mostly from the fertilized mature gynoecium although, especially in fleshy fruits, additional floral components have frequently been recruited. The gynoecium (or pistil), the female reproductive organ, is composed of a single carpel or a number of carpels that are often fused. Carpels are essential for sexual plant reproduction because they house the ovules and upon fertilization the carpel develops into the fruit that protects, nourishes and ultimately disperses the seeds.
In Arabidopsis, the fundamental processes leading to the formation of a complete developed set of ovules can be summarized in a few main steps (Figure 1). First of all, the lateral margins of the carpels, containing a meristematic tissue named the medial ridge or carpel margin meristem (CMM), give rise to the placenta, the septum and transmitting track. The CMM formation is known to be controlled by the interaction of genetic and hormonal networks (reviewed by Reyes-Olalde et al., 2013). Once the placenta is formed, some mechanisms, still poorly understood, are needed for the definition of boundary regions that will separate the ovule primordia. The ovule primordia are initiated by periclinal divisions from the subepidermal tissue of the placenta. During the early growth phase of primordia formation a series of predominantly anticlinal divisions take place. Later on, the relatively homogenous mass of cells of the primordium will be organized in three different regions along the proximal-distal axis: the funiculus, the chalaza and the nucellus (Figure 1). Within the nucellus, megasporogenesis and megagametogenesis take place, and finally the mature haploid embryo sac is formed. From the chalaza region the two integuments, progenitors of the seed coat, develop, while the funiculus connects the ovule to the mother plant.
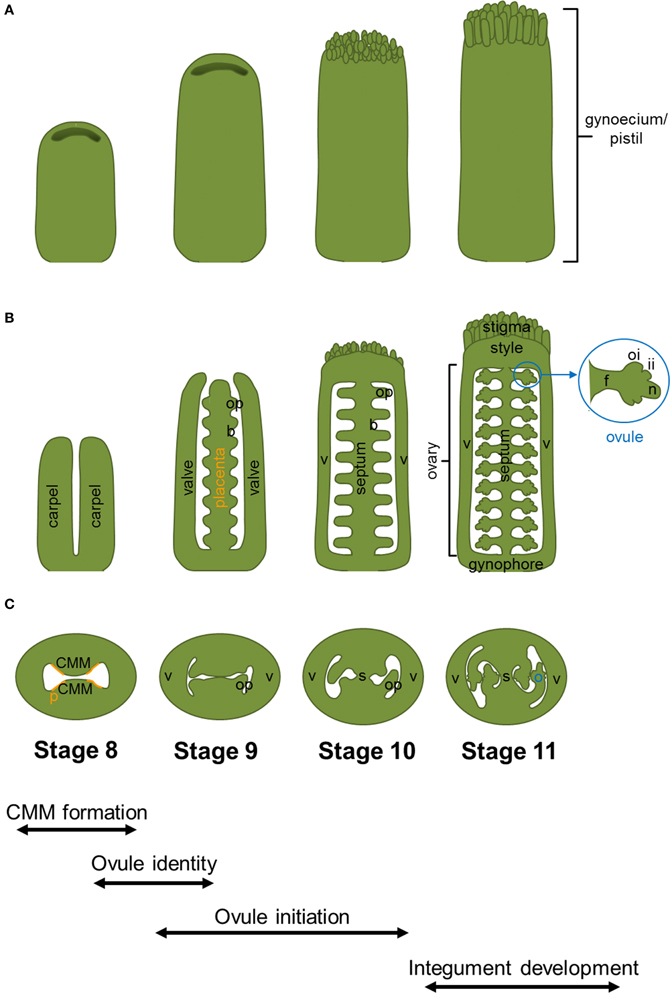
Figure 1. Arabidopsis thaliana gynoecium development. Cartoons displaying wild-type gynoecia (A), in longitudinal sections (B), and transversal sections (C) from stage 8 to 11. The different stages and processes of gynoecium and ovule development are indicated at the bottom of the figure. Abbreviations: b, boundary; CMM, carpel margin meristem; f, funiculus; ii, inner integument; n, nucellus; o, ovule; oi, outer integument; op, ovule primordium; p, placenta; s, septum; v, valve. The region of the CMM where placenta is formed is indicated with orange lines.
In the last decades, several studies have identified genes involved in ovule identity determination and development in different species such as Arabidopsis, Petunia and rice (Bowman et al., 1991; Angenent et al., 1995; Colombo et al., 1995; Angenent and Colombo, 1996; Pinyopich et al., 2003; Dreni et al., 2007). However, the players that determine the number of ovules are largely unknown, due to the difficulties that the studies tempting to genetically dissect ovule initiation and development have encountered. On one hand, many genes that control ovule development are also involved in initiation and growth of other floral organs, masking their effects on ovules. On the other hand, it is difficult to establish if a mutation in a gene causes a reduction in ovule number in mutants that already display an altered gynoecium phenotype.
Nonetheless, understanding the factors that control ovule initiation is of great importance from an agricultural and economical point of view, as the ovule number will determine the number of seeds that develop in a fruit, and thus the crop yield.
With this review on ovule initiation we aim to summarize the current knowledge about the factors and the hormonal pathways that have been identified to be involved in the determination of ovule numbers in Arabidopsis thaliana, and the cross-talk between these hormonal and regulatory pathways.
Carpel Margin Meristem Formation
Genetic Factors Controlling Carpel Margin Meristem Formation
As already mentioned, the establishment and maintenance of the meristematic tissues of the CMM is inherently correlated to the generation of ovule primordia. CMM development is known to be controlled at the transcriptional level and by hormones as reviewed by Reyes-Olalde et al. (2013). Several single and higher order mutant combinations with strongly reduced carpel marginal tissue development have been described in literature. One of the them is the aintegumenta (ant) mutant. ANT is a transcription factor that contains two AP2 domains that controls organ initiation and promotes cellular divisions during organ development (Klucher et al., 1996). Interestingly, the ant-9 mutant has medial ridges that are frequently unfused to each other with a consequent reduction in functional CMM tissue. It has also been reported that the ant mutant displays enhanced morphological defects when combined with a mutation in REVOLUTA (REV), a member of the class III Homeodomain-Leucine Zipper (HD-ZIP III) family. In the ant rev double mutant a partial disruption of CMM and placenta development causes the reduced development of ovule primordia (Nole-Wilson et al., 2010).
An unfused carpels phenotype due to the compromised fusion between the two medial ridges was also observed in the mutants for LEUNIG (LUG), a floral organ identity gene that encodes a glutamine-rich protein with seven WD repeats, typical of transcriptional co-repressors (Liu et al., 2000). Despite this failure in ridge fusion, ovules are formed from the placenta although in a markedly decreased number in both lug-1 (intermediate-strength allele) and lug-3 (strong allele) mutants (Table 1). The simultaneous loss of LUG and ANT functions enhanced the defects in flower development in respect to the single lug and ant mutants. While the double mutant lug-3 ant-9 did not form any ovules, septum or stigma, nearly 50% of the lug-1 ant-9 pistils could develop normal medial ridges, that gave rise to partially formed septal tissues, although ovules, stigma and style were never present (Liu et al., 2000) (Table 1).
ANT also interacts synergistically with SEUSS (SEU), a transcriptional coregulator functionally similar to LEU, in the control of organ size of the flower. While the seu-3 single mutant shows on average ovule numbers not significantly different from wild-type Col-0, the double mutant seu-3 ant-1 results in a complete loss of ovule initiation, caused by severe defects in early gynoecia development. In the weaker allelic combination seu-3 ant-3, employing the ant-3 hypomorphic allele, placenta formation is not compromised but defects such as ovule initiation and gametogenesis are present at later stages (Table 1) (Azhakanandam et al., 2008).
Other two players in CMM development are CUP-SHAPED COTYLEDON1 (CUC1) and CUC2, two transcription factors that belong to the NAC transcription factor family. The cuc1 and cuc2 single mutants display almost no phenotype, while the cuc1 cuc2 double mutant completely lacks the shoot apical meristem (SAM) and the cotyledons are fused along their margin forming a cup-shaped structure. These seedlings die a few days after germination (Aida et al., 1997). Studying gynoecium development in the cuc1 cuc2 double mutant was only possible using plants obtained by in vitro regeneration. They presented defects in the formation of the septum and in ovule development (Ishida et al., 2000). A gene that has been described to play a role with CUC1 and CUC2 in promoting the formation of carpel marginal structures and thus facilitating septum and ovule development is SPATULA (SPT), which encodes a basic helix–loop–helix (bHLH) transcription factor. Mutations in SPT cause a split carpel phenotype in the apical part of the gynoecium. Moreover, spt plants have slightly fewer ovules than the wild type, from which only a small fraction develop into seeds (Nahar et al., 2012). When combined with cuc1 and cuc2 single mutants, the average number of ovules decreases. Thus, while the spt single mutant shows an average of 48 ovules per carpel, spt cuc1 and spt cuc2 present 36 and 32 respectively (Table 1), indicating that CUC1, CUC2, and SPT are together required for ovule development. Another mutant that displays an unfused gynoecium at the apex is crabs claw (crc) (Alvarez and Smyth, 1999). CRC encodes a transcription factor of the YABBY family and the characterization of different mutant alleles showed that, besides the failure of the fusion of the stylar region, crc mutants present a gradation of phenotypes with wider and shorter gynoecia that contain fewer ovules compared to the wild type (Alvarez and Smyth, 1999; Bowman and Smyth, 1999).
Thereby, the phenotype of ovule reduction that we frequently observe in the mutants defective in medial ridge fusion and thus in CMM formation could be due, at least in part, to their role in regulating cell proliferation in the medial ridges, from which septum and ovules originate.
CMM Formation and the Auxin Gradient
Auxin is a key hormone for plant development, and it is also fundamental for gynoecium and thereby CMM and ovule development. In the last two decades several studies have demonstrated that local auxin biosynthesis and polar transport are responsible for the correct apical–basal patterning of the gynoecium. The auxin gradient hypothesis supports that high levels of auxin in the gynoecium apical regions control stigma and style formation; medium levels direct ovary formation whereas low levels of the hormone are responsible of gynophore development at the gynoecium base (Nemhauser et al., 2000). Indeed, all mutants in which the auxin synthetic pathway or transport are compromised have a similar severe gynoecium phenotype forming a pistil-like structure with reduction/absence of the valves, expansion of the gynophore and stylar regions and serious vasculature defects (reviewed in Balanzá et al., 2006; Larsson et al., 2013). This phenotype was characterized for the first time in the flowers of pin-formed1-1 (pin1-1), a strong mutant allele of the auxin efflux carrier PIN1 (Okada et al., 1991) and in the pinoid mutant, a knock-out line for a serine/threonine kinase that regulates PINs polarity (Bennett et al., 1995). Other examples of mutants with similar pistil-like structure phenotypes are the yucca1 yucca4 (yuc1 yuc4) and weak ethylene insensitive8 tryptophan aminotransferase related2 (wei8 tar2) double mutants, in which local auxin production is impaired (Cheng et al., 2006; Stepanova et al., 2008). Predictably, in most of these auxin-related mutants the severe defects in gynoecium formation lead to a pistil with a reduction or complete absence of ovules and the consequent complete sterility.
Nemhauser et al. (2000) confirmed the importance of polar auxin transport (PAT) in gynoecium development through an experiment in which they used 1-napthylphthalamic acid (NPA), an inhibitor of the auxin transport. They showed that NPA application caused significant loss of ovules. The authors also highlighted that ovules seemed more sensitive to disruption in PAT, with respect to the other tissues of the gynoecium. Indeed, treated carpels were largely devoid of ovules but were still able to produce valves. In 2010 Nole-Wilson and collaborators proposed the connection between ANT and the hormone auxin on the base of the observation that the ant mutant is more sensitive than the wild type to alteration in PAT. Moreover, the expression of a subset of auxin-related genes was altered in the ant single and ant rev double mutant gynoecia, indicating that the morphological defects of the ant rev double mutants, at least in part, are due to an alteration in auxin homeostasis in these plants.
Auxin signaling is primarily regulated by the AUXIN RESPONSE FACTOR (ARF) gene family products, together with the AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) proteins. The phenotype of ARF5/MONOPTEROS (MP) strong mutant alleles results in an embryo lethal phenotype, while mp partial loss of function mutants have normal embryo development whereas that their reproductive development is compromised (Hardtke and Berleth, 1998). In the pistil of the mpS319 weak allele the CMM does not develop, and placenta and ovules are completely missing (Cole et al., 2009; Galbiati et al., 2013). Interestingly, MP has been demonstrated to directly activate the ANT, CUC1 and CUC2 transcription factors encoding genes (Galbiati et al., 2013) Their role as major players in ovule primordia initiation and ovule number determination will be discussed in the following sections.
Ovule Identity Establishment
The ovule cell fate is controlled by the ovule identity genes SHATTERPROOF1 (SHP1), SHP2, and SEEDSTICK (STK), that belong to the MADS-box gene family of transcription factors. While in the single and double mutant combinations of these genes there is no detectable ovule phenotype, in the stk shp1 shp2 triple mutant the ovule integuments are converted into leaf/carpel-like structures (Pinyopich et al., 2003; Brambilla et al., 2007). Moreover, ectopic expression of these MADS box genes results in ovule formation on sepals (Favaro et al., 2003; Pinyopich et al., 2003; Battaglia et al., 2006). STK, SHP1, and SHP2 have overlapping expression patterns in the placenta and ovule primordia also with AGAMOUS (AG) (Rounsley et al., 1995; Savidge et al., 1995; Theißen et al., 1996; Pinyopich et al., 2003), one of the first identified MADS-box factors that determines stamen and carpel identity (Yanofsky et al., 1990). It has been shown that also AG plays a role in ovule development by experiments in which the apetala2 (ap2) single mutant was compared with the ap2 ag double mutant. Thus, in the ap2 single mutant petals were mostly absent, while sepals were converted into carpel structures bearing ectopic ovules, some of which were transformed into carpelloid structures. Interestingly, the sepals (or first-whorl organs) of the ap2 ag double mutant still presented carpel identity, and the number of ovules converted into carpel structures was significantly higher, indicating that AG activity also contributes to ovule identity establishment (Bowman et al., 1991; Pinyopich et al., 2003). Interestingly, Skinner et al. (2004) suggested that when the functions of stk, shp1, and shp2 were lost in a triple mutant, fewer ovules initiated and ovule development is severely disrupted.
Ovule Primordia Initiation
The Establishment of the Boundaries
When new organ primordia are originated in the plant, two different regions, the boundaries and the zone of primordia outgrowth, need to be defined. The organ boundary is defined as the region between the meristem and the developing organ, or, as in the case of ovules, as the region between two adjacent ovule primordia. As Aida and Tasaka nicely reviewed in 2006, the “boundary cells” need to have peculiar characteristics respect to the surrounding cells, usually displaying reduced cell division and expansion. Another important aspect is the arrangement of the plasmodesmata that regulates the movement of transcription factors between cells. For example, the boundaries in the inflorescence meristem seem to restrict the passage of proteins into flower primordia (Wu, 2003).
The boundary-specific regulatory genes play a critical role in orchestrating several morphogenetic and patterning events and their spatial coordination. When this coordination is missing, fusion between organs is the most frequent observed phenotype (Aida et al., 1997). The CUC gene family was the first discovered to have a fundamental role in organ boundary establishment. In fact, in the cuc1cuc2 double mutant embryo the cotyledons do not separate (Aida et al., 1997).
The transcripts of CUC1 and CUC2 were detected by in situ hybridization in the anlagen placenta and in ovules at stage 1-II and later on, starting from stage 2-I, restricted to the boundary between two ovules (Ishida et al., 2000; Galbiati et al., 2013). As we already mentioned, the study of the gynoecium phenotype of the cuc1 cuc2 double mutant was only possible on plants regenerated in vitro. They showed defects in the formation of the septum and in ovule development; most of the gynoecia having less than 10 ovules (Table 1). However, the cuc1 cuc2 double mutant plants never gave seeds (Ishida et al., 2000). A further demonstration that CUC1 and CUC2 are directly linked to the determination of ovule number in a direct way came from the work of Galbiati et al. (2013). In order to study the ovule phenotype in absence of both CUC1 and CUC2, CUC1 was silenced in a cuc2-1 mutant background using a CUC1 specific RNAi construct under the control of the ovule-specific SEEDSTICK promoter (pSTK:CUC1_RNAi) which is already active in the placenta before ovule primordia arise. The analysis of cuc2-1 pSTK:CUC1_RNAi plants revealed a reduction in ovule number of 20% (Table 1). Furthermore, ant-4 cuc2-1 pSTK::CUC1_RNAi plants were generated in order to analyze the possible additive role of ANT to CUC function in the regulation of ovule primordia formation. The ant-4 cuc2-1 pSTK::CUC1_RNAi plants displayed a further dramatic reduction in the number of developing ovules (a mean of seven ovule primordia per pistil), while the single mutant ant-4 and the plants cuc2-1 pSTK::CUC1_RNAi showed 20 and 30 ovules per pistil, respectively (Table 1). Despite the reduction in ovule number in the different mutant backgrounds, the size of the pistils was not reduced. Therefore, the ovules were more distantly spaced compared to those in wild-type pistils (Galbiati et al., 2013). These studies of the characterization of the ant single and cuc1 cuc2 double mutants, as well as ant-4 cuc2-1 pSTK::CUC1_RNAi plants prove that ANT, CUC1 and CUC2 are key players in the control of the number of ovule primordia that develop from the placenta and that they act additively (Elliott et al., 1996; Ishida et al., 2000; Galbiati et al., 2013). All the information about these factors taken together indicates that they work in different ways: while ANT promotes ovule primordia growth, the CUCs play a role in the establishment of the ovule primordia boundaries (Figure 2).
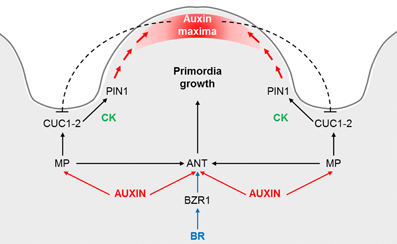
Figure 2. Proposed model for the control of the ovule primordia initiation. Auxin triggers ANT and MP expression, which in its turn is required for ANT, CUC1 and CUC2 expression during the early stages of placenta development and ovule primordia formation. ANT expression is also regulated by brassinosteroids (BR), specifically being directly regulated by BZR1. ANT controls cell proliferation in the placenta and ovules, whereas CUC1 and CUC2 establish the boundaries and control PIN1 expression, which is required for primordia formation. Cytokinin (CK) may act downstream of CUC proteins in promoting PIN1 expression. Once the primordia have formed, auxin accumulates at the edge of the developing ovule. An inhibitory loop of auxins on CUC1 and CUC2, as it is postulated for the leaf serration, could be happening at the ovule boundaries. Adapted from Galbiati et al. (2013).
CUC3, another putative NAC-domain transcription factor member of the CUC family, is expressed in an extensive range of boundaries in adult plants. Besides, the function of CUC3 is partially redundant with that of its homologous CUC1 and CUC2 in the establishment of the cotyledon boundary (Vroemen et al., 2003). Several studies revealed that CUC expression is controlled and restricted to the boundaries in several ways. For instance, in the SAM CUC1 and CUC2 but not CUC3 are regulated by miR164, which restricts the expression of CUC1 and CUC2 mRNAs to the boundary domain (Laufs et al., 2004). In the carpel, in a similar way to the pattern already described for CUC1 and CUC2, CUC3 expression marks the boundaries between ovule primordia. Therefore, it would be interesting to study also the contribution of CUC3 in the regulation of defining ovule boundaries.
In 2008, Xu and Shen showed that three different transcription factors, ASSYMETRIC LEAVES 1 (AS1), AS2, and JAGGED (JAG), support normal sepal and petal growth by restricting the expression domain of the boundary-specifying genes CUC1 and CUC2. AS1 and AS2 were already suggested to have roles in boundary control, given that they positive regulate, within the shoot apex, the members of the LATERAL ORGAN BOUNDARIES (LOB) gene family, a plant-specific family of transcription factors that are expressed in the boundaries (Byrne et al., 2002). LOB, the gene that names the family, is expressed at the base of all lateral organs. Interestingly, plants overexpressing LOB produced abnormal flowers with reduced floral organs and they were sterile even when fertilized with wild-type pollen (Shuai et al., 2002). Lee et al. in 2009 identified two new MYB transcription factors involved in lateral organ separation: LATERAL ORGAN FUSION 1 (LOF1) and LOF2. The single mutant lof1 exhibits a novel fusion between the axillary stem and the cauline leaf. Additional fusions resulted when lof1 was combined with lof2, cuc2 or cuc3, indicating the existence of overlapping roles for LOF1, CUC2, and CUC3 to control organ separation during reproductive development.
Despite the identification of a number of boundary-specific transcription factors, boundary formation and maintenance is still a poorly understood process, and only CUC1 and CUC2 have been demonstrated to have a role in ovule boundary establishment. The factors that have been described to regulate or interact with the CUCs in a different developmental context could also have a role during ovule initiation, and some of them, like AS1 and AS2 are already known to be expressed in the gynoecium and ovules (Xu and Shen, 2008).
Aintegumenta, a Master Regulator of Primordia Formation
In Arabidopsis many genes have been described to play roles in the different phases of ovule development, although most of them do not determine directly the number of ovules (Schneitz et al., 1997; reviewed in Shi and Yang, 2011). However, the ANT transcription factor has been described to have a clear role in ovule primordia formation. In situ hybridization experiments showed that within the carpel it is expressed in the placenta and in the integuments of the developing ovules. In ant plants ovules do not develop integuments and megasporogenesis is blocked at the tetrad stage leading to complete female-sterility (Elliott et al., 1996). ANT is not only required for ovule development but it is also involved in ovule primordia formation. Indeed, in the ant-9 mutant the number of ovules per carpel is reduced by more than half in respect to the wild-type (Table 1). Given that the ant gynoecia have the same length as those of wild type, the ovules that do arise in ant are more distantly spaced than in wild-type plants (Liu et al., 2000).
In addition to ANT, another essential gene for the regulation of ovule primordia outgrowth and for the control of integument formation is HUELLENLOS (HLL), a gene that encodes a mitochondrial ribosomal protein. Thus, plants presenting mutations in HLL display a phenotype similar to ant at the level of ovule integuments (Schneitz et al., 1997, 1998). Moreover, hll-1 and hll-3 mutant alleles display a reduction of about 10% in the number of ovules, although the authors also describe that hll plants display smaller gynoecia, which could contribute to the development of fewer ovules (Table 1). The phenotype of the double mutant hll ant was more severe at the level of primordia outgrowth however, nothing was described regarding ovule number (Schneitz et al., 1998; Skinner et al., 2001). A similar phenotype to hll was observed in the short integuments 2 (sin2) mutants. Apart of an arrest in cell division in both ovule integuments, sin2 plants presented shorter pistils bearing less ovules than the wild type (Table 1). Moreover, the authors describe an abnormal distribution of the ovules along the placenta, being the distance between ovules bigger than in wild-type plants (Broadhvest et al., 2000). Thus, in this particular case the shorter carpel might not be the only cause of reduced ovule numbers. The double mutant sin2 ant-5 was not different from ant-5 single mutant, indicating that ANT is epistatic to SIN2 with respect to ovule development. On the contrary, sin2 hll-1 double mutant had a stronger effect on ovule development than sin2 or hll-1 single mutants (Broadhvest et al., 2000). All these experiments taken together indicate that although ANT plays a master role, SIN2 and HLL also contribute to ovule primordia formation.
The Role of Hormones in Ovule Primordia Formation
Auxin is Required for Ovule Primordia Formation
As we previously underlined, the boundary region and the primordia formation zone are highly interconnected. It has been demonstrated that a fundamental role of the “boundary transcription factors” is to organize PAT, mediated by PIN proteins, in order to create a zone of auxin maximum where organ founder cells will be selected. Auxin maxima are fundamental for the formation of primordia, and auxin action has been well described for lateral roots (LR) and flower primordia (reviewed in Benková et al., 2003, 2009; Yamaguchi et al., 2013). The directionality of auxin flux depends principally on the polar localization of the PIN proteins. In Arabidopsis there are eight PIN proteins (PIN1-8), from which only PIN1 and PIN3 are expressed in the pistil and ovules (Benková et al., 2003; Ceccato et al., 2013). PIN1 protein is localized at the membrane of placenta cells and later on, in the developing ovules, it is restricted to the lateral-apical membranes of nucellus cells. PIN3 is also present in few cells at the tip of the developing nucellus shortly after ovule primordia emergence but, contrary to PIN1, it is not expressed in the placenta cells (Ceccato et al., 2013). PIN-dependent efflux mediates primordium development by supplying auxin to the tip creating an auxin maxima; indeed in plants expressing the GFP reporter gene downstream the auxin-responsive DR5 promoter (pDR5::GFP), the GFP signal is detected at the tip of all ovule primordia (Benková et al., 2003). The weak pin1-5 mutant allele is able to develop some flowers in which the pistils have slightly reduced valves but normal styles and stigmas (Sohlberg et al., 2006). The pistils of the pin1-5 weak allele have an average of 9 ovules per carpel (Table 1) (Bencivenga et al., 2012). In addition, Galbiati et al. (2013) demonstrated that the reduced number of ovules in cuc2-1 pSTK:CUC1_RNAi was caused by a down-regulation of PIN1 and an incorrect PIN1 protein localization. CUC1 and CUC2 promote PIN1 expression and localization to correctly form the auxin maximum where primordium will form (Figure 2). In the same way, a CUC2-dependent regulatory pathway controlling PIN1-mediated auxin efflux has been described to explain leaf serrations (Bilsborough et al., 2011). Moreover, in the newly formed primordia of the SAM the auxin maxima, in a negative feed-back loop, repress CUC2 expression and restricts it to the boundaries (Vernoux et al., 2000; Heisler et al., 2005; and reviewed in Aida and Tasaka, 2006; Rast and Simon, 2008). A similar inhibitory loop could control CUC expression at the ovule boundaries (Figure 2). The phenotype of cuc2-1 pSTK:CUC1_RNAi was completely recovered by cytokinin (CK) application, since CK has been demonstrated to increase PIN1 expression in the ovules (Bencivenga et al., 2012). These experiments evidence a convergence of two different plant hormones in the regulation of ovule primordia formation. In the next paragraph we will delve deeper into the role of CK in the formation and determination of ovule number.
Cytokinin Positively Regulates Ovule Number
CK is an essential hormone for plant growth and development as it has a central role in the regulation of cell division and differentiation. In the last 10 years, several studies have clearly proven that CK has also a significant role during ovule development. As it will be explained in this paragraph, it has been demonstrated that in plants that are defective in the production or perception of this hormone, correct ovule formation is compromised and/or the number of ovule is drastically reduced. CK signaling, which has been recently summarized in a detailed review article (Hwang et al., 2012), is mediated by a two-component signaling pathway: histidine protein kinases (AHKs) work as CK receptors, while histidine phosphotransfer proteins (AHPs) transmit the signal from AHKs to nuclear response regulators (ARRs), which are able to regulate transcription. In Arabidopsis the CK signal is perceived by three histidine kinases: ARABIDOPSIS HISTIDINE KINASE4 (AHK4, also known as CYTOKININ RESPONSE1, CRE1/WOODEN LEG, WOL), AHK2 and AHK3. These three genes are all expressed in inflorescences, carpels and developing ovules (Higuchi et al., 2004; Nishimura et al., 2004). More precisely, AHK2 and AHK3 are expressed during all stages of ovule development, starting from early primordia stages to ovule maturity, whereas CRE1 expression remains restricted to the chalazal region and later to the integuments of ovules during all the developmental stages (Bencivenga et al., 2012). The single and double mutants of AHKs do not present any phenotype at the level of the ovules (Higuchi et al., 2004). However, mutants lacking all three receptors exhibit no perception of CK and present a strong slowdown of shoot and root growth. The resulting miniature plants also show delayed flower induction and impaired fertility (Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006). Thus, the triple mutant cre1-12 ahk2-2 ahk3-3 do not produce seeds (Higuchi et al., 2004) because the gametophyte arrests at stage FG1-FG2 (Bencivenga et al., 2012). Moreover, a severe reduction in the ovule number, an average of 5 ovule per pistil, was noticed in these triple mutant plants (Table 1) (Bencivenga et al., 2012). A similar sterile phenotype was also observed for another allelic combination: the ahk4-1 ahk2-1 ahk3-1 triple mutant (Nishimura et al., 2004). Differently, Riefler et al. (2006) obtained a weaker triple mutant cre1-2 ahk2-5 ahk3-7 that self-fertilized and formed few seeds, suggesting that infertility of the histidine kinase triple mutants is a phenotype associated with specific mutant alleles.
Attention has also been given to the importance of CK catabolism. In Arabidopsis the irreversible degradation of CK is catalyzed by the oxidase/dehydrogenase (CKX). The CKX gene family of Arabidopsis consists of seven members (CKX1 to CKX7), and by promoter:GUS fusion constructs it was shown that CKX1, CKX5, and CKX6 (At3g63440, previously called AtCKX7) are expressed in flower tissues, being CKX6 the only one reported to be expressed in the carpel and ovules, in particular in the funiculus (Werner et al., 2003). Werner and colleagues engineered transgenic Arabidopsis plants that individually overexpressed six different CKXs in order to enhance CK degradation. As expected, these plants manifested phenotypes linked to CK deficiency, like delayed vegetative growth and leaf expansion, diminished activity and size of the SAM but increased overall root system. The reproductive development of CK-deficient plants was also altered. In 35S::CKX1 and 35S::CKX3 plants, flowering was strongly delayed and furthermore the fertility of flowers was heavily reduced, partially due to the lack of pollen. 35S::CKX1 and 35S::CKX3 siliques were not filled completely and they formed approximately 8–20 viable seeds, whereas the wild-type siliques harbored up to 60 seeds. Although the number of ovules formed in these plants was not reported in this work, the expression patterns together with the phenotypes in the flowers and fruits indicate once more that CK play a role during reproduction. Moreover, the authors suggest a role for ANT in the observed reduced cell division in the leaves of ckx plants. Considering the documented role of ANT in ovule primordia initiation already introduced in this article, it will be very interesting to analyze also its role in the reproductive tissues of these plants.
With an opposite experimental approach, the simultaneous mutations of two CKX genes, it was demonstrated that plants with an increased level of CK had an enhanced activity of the reproductive meristem (Bartrina et al., 2011). Indeed, the ckx3-1 ckx5-1 double mutant produced more flowers due to a larger inflorescence meristem with more cells than the wild type. Moreover, flowers were bigger and so were the gynoecia. Besides, double mutant gynoecia contained twice as many ovules as wild-type ones, indicating an increased activity of their placental tissue. The ckx3-1 and ckx5-1 single mutants already developed more ovules than the wild-type, and the flower size and the number of ovules was reflected into the length of the fruits (siliques of ckx3 ckx5 were 20 mm long compared with the 17 mm of the wild type) and the seed number (110 seeds in the ckx3 ckx5 mutant siliques compared with an average of 65 seeds in wild-type siliques, Table 1) The authors suggested that CKX3 and CKX5 may regulate the activity of meristematic cells in the placenta thus affecting organogenic capacity and ovule primordia formation.
A conclusive evidence about the relationship between the levels of CK and the initiation of ovule formation was obtained from experiments in which inflorescences were treated with synthetic CK (6-Benzylaminopurine, BAP). The treatment resulted in the formation of new primordia, 20 ± 3 primordia in average in each pistil, positioned between the ovules already formed before the CK application (Bencivenga et al., 2012). An equivalent CK treatment was also able to increase the ovule number in pSTK::CUC1_RNAi cuc2 plants already described in this review, by acting on the expression and localization of the auxin efflux carrier PIN1 (Galbiati et al., 2013). These results point out the importance of the cross-talk between CK and auxin during ovule primordia formation. However, the hormonal cross-talk is not limited to auxin and CK since very recently it has been demonstrated that also brassinosteroids (BR) play a crucial role in ovule and seed formation by regulating the expression of genes that control ovule development (Huang et al., 2012), as will be explained in the next paragraph.
The Role of Brassinosteroids
BRs are hormones known to control general plant development. More specifically, they have been described as involved in the control of the initiation and formation of reproductive organs (Szekeres et al., 1996; Kim et al., 2005). Huang et al. (2012) found that the BR-deficient and -insensitive mutants have smaller and less seeds, while BR-enhanced mutants have more seeds. The analysis of the number of ovules and seeds and the morphological analysis of the siliques of det-2 (a BR-deficient mutant involved in BR biosynthesis), bri1-5 (the mutant for the BR receptor), heterozygous plants for bin2-1 (a gain of function mutant deficient in BR signaling) and bzr1-1D (a BR signal-enhanced mutant) leaded to the conclusion that BR signaling positively regulates ovule number (Table 1) (Huang et al., 2012). Specifically, it was found that the transcription factor BRZ1 plays an important role in ovule and seed number determination, depending on its state of phosphorylation/dephosphorylation (more dephosphorylation implying more activity and more ovules and seeds).
By treating plants with BR it was shown that BR influences ovule development through regulating the transcription of genes such as HLL and ANT, which are redundant in the control of ovule primordia growth as already introduced in this review (Schneitz et al., 1998), and AP2, that affects floral organ (including ovule) pattern formation (Modrusan et al., 1994). HLL and ANT are clearly induced by BR, while AP2 is slightly repressed by BR. These genes appeared to be targets of BRZ1, and its state of phosphorylation/dephosphorylation influences the expression of these genes. Further analysis indicated that AP2 and ANT are direct targets of BRZ1, while HLL is regulated by an indirect way. The analysis of ovule number of bzr1-1D and ap2-5 single mutants and bzr1-1D ap2-5 double mutant (Table 1), together with other molecular proofs, indicate that BZR1 and AP2 play antagonistic effects in ovule number determination, being BZR1 (and HLL and ANT) promoters and AP2 inhibitor of ovule primordia formation (Huang et al., 2012).
A model for ovule primordia formation that integrates the molecular and hormonal networks has been proposed by Galbiati et al. (2013): MP is required for ANT, CUC1 and CUC2 expression during the early stages of placenta development and ovule primordia formation, being ANT expressed in the ovule primordia, whereas CUC1 and CUC2 in the ovule boundaries. CUC1 and CUC2 may be involved in the increase of CKs required for proper PIN1 expression needed for primordia formation. Once the primordia have formed, auxin accumulates at the edge of the developing ovule. This model can be easily extended with the recently discovered role of the plant hormones BR, which positively regulate the number of ovule primordia, in part by the direct regulation of ANT by BZR1 (Figure 2).
Other Mechanisms Controlling Ovule Number: The Epigenetic Regulation
Interestingly, in different Arabidopsis ecotypes (diploid accessions) a variation in ovule numbers can be observed. Alonso-Blanco et al. (1999) found that the Lansberg erecta ecotype presents 20% more ovules than the Cape Verde Islands (Cvi) one (Table 1). Recently a considerable genetic variation in ovule number was described in selfed F1 triploids of different A. thaliana genotypes (Duszynska et al., 2013). Triploids were obtained by crossing a tetraploid Ler-0 line (used as a male or female parent) with different diploid accessions. Interestingly, it was observed an effect of the parental genome excess (2m:1p vs. 1m:2p) in the determination of the total ovule number in genetically identical F1 hybrid offsprings. These were the first parent-of-origin effects on ovule number in reciprocal triploids of plants. The authors postulate that such effects may represent epigenetic effects, because changes in DNA sequence cannot explain mitotically and/or meiotically heritable changes in gene function but they might be due to changes in DNA methylation, for example (Duszynska et al., 2013). Indeed, in Arabidopsis the ASH1 class of proteins, that can methylate lysine residues on histone tails, maintains an active transcriptional state during development. One of its members, ASH1 HOMOLOG 2 (ASHH2), has been described as a controller of reproductive development via H3K36 trimethylation. Plants homozygous for ashh2 null alleles presented an 80% reduction in ovule numbers when compared to wild-type plants (Table 1) (Grini et al., 2009). These data altogether indicate that epigenetics may also play a role in the control of ovule number, and they open up a new interesting field of research.
Concluding Remarks and Further Perspectives
In the past years, several genes such as AG, STK, SHP1, and SHP2 have been identified as ovule identity genes, and ANT, HLL, SIN2, INNER NO OUTER (INO), and SUPERMAN (SUP) as regulators of ovule outgrowth (Elliott et al., 1996; Schneitz et al., 1997, 1998; Broadhvest et al., 2000; Pinyopich et al., 2003). Nevertheless, most of their targets, which might be the genes that determine the correct development of the ovule, remain to be uncovered. Another quite unknown process is the regulation of the ovule primordia initiation. As explained in this review, only a few regulators, such as the transcription factors ANT, CUC1, CUC2, AP2 and the mitochondrial ribosomal protein HLL have been identified (Elliott et al., 1996; Schneitz et al., 1998; Galbiati et al., 2013). The majority of them are transcription factors, and the transcriptional cascades triggered by them, that will determine the regulation of the morphogenetic parameters such as cell division and expansion, or expression patterns of identity genes of particular organs, are also largely unknown. Therefore, one of the next challenges would be the identification of downstream targets of these transcription factors by genetic or molecular biological approaches, including suppressor/enhancer mutant screenings or RNA-sequencing transcriptome analyses. It is worth to highlight that these regulators are not exclusively transcription factors, but also mitochondrial proteins or chromatin remodeling factors, indicating that a correct ovule initiation depends on a complex genetic and molecular network.
One of the difficulties of the genetic dissection of ovule initiation and development is that many mutations that affect ovule initiation have already pleiotropic effects on earlier stages of the development of the reproductive tissues, causing floral aberrations that may mask their effects on ovules. Thus, many genes that control ovule development are also involved in primordium initiation and growth of other floral organs (Elliott et al., 1996; Schneitz et al., 1998; Alvarez and Smyth, 1999). Moreover, it is difficult to establish if a mutation in a gene causes a reduction in ovule number if this mutant already has an altered gynoecium phenotype (Alvarez and Smyth, 1999; Western and Haughn, 1999; Broadhvest et al., 2000; Liu et al., 2000; Pinyopich et al., 2003; Nole-Wilson et al., 2010; Nahar et al., 2012). The reanalysis of these carpel mutants, measuring the space between ovules, or expressing the ovule number as the ovule number per millimeter of gynoecium, as some authors already presented (Huang et al., 2012), could contribute to resolve this uncertainty. The use of specialized vectors, for instance containing placenta-specific promoters to obtain milder vegetative and/or floral effects of these mutations would help to uncover the role of specific factors in ovule development. Besides, a reverse-genetic strategy using RNA interference or insertional mutants can be used to identify new regulators of ovule numbers determination.
Ovule boundary establishment is still a poorly understood process, and only CUC1 and CUC2 have been demonstrated to play a role (Galbiati et al., 2013). The contribution to the determination of ovule boundaries of the genes that have been described to regulate or interact with the CUCs in other organ boundaries would be worth to be analyzed, by means of the study of their patterns of expression and how these are accurately determined. The identification and characterization of single and multiple mutants, as has been done for the CUC genes (Aida et al., 1997; Galbiati et al., 2013) is also key to study their roles. Moreover, the analysis of their incidence at the cellular level will help to define the effects on cell behavior (i.e., division or expansion) that these factors could have. It has also been widely demonstrated that hormones play a role in the regulation of ovule primordia initiation, being auxin, CK and more recently also BR identified as the important hormonal players in this process. The crosstalk between these hormones, as Bencivenga et al. (2012) and Galbiati et al. (2013) present in their works, is starting to be revealed (Figure 2) and it will be very interesting to investigate in the future how auxin, CK and BR interact. Moreover, it will be important to explore if hormone and gene expression levels are responsible for the variation in ovule numbers described for the different ecotypes (Alonso-Blanco et al., 1999), and to identify QTLs linked to this trait.
Based on the experimental data exposed in this review, a similarity between ovule initiation and the initiation of other lateral organs in the plant can be proposed. The strongest pieces of evidence are the triggering role of auxins and the conservation in the genes that establish the boundaries and promote new organ growth. Although further studies will be needed in order to identify the common and specific players of the different lateral organ initiation processes, conserved modules can be already suggested. In the case of flower primordia initiation, similarly to what happens during ovule primordia formation, the coordinate action of MP and ANT is required. In particular, at the reproductive shoot apex, auxin-activated MP directly induces ANT, other two key regulators of floral growth, LEAFY (LFY) and AINTEGUMENTA LIKE-6 (AIL6), and probably a forth unknown factor, which together lead the flower primordium initiation (Yamaguchi et al., 2013). Also the factors determining the new organ boundary seem conserved between ovule and flower primordia initiation: the coordinated spatial and temporal action of auxin, PIN transporters and CUC proteins is required (Heisler et al., 2005; Galbiati et al., 2013). If we instead compare the initiation of ovule primordia with the initiation of LR we also find many common players, despite the clear fact that ovule primordia arise from the naked placenta while LR have to pass through several cell layers to emerge. Thus, we find an auxin maxima that precedes organ formation (Benková et al., 2003). Also other hormones, such as BR and CK play a role in both ovule and LR initiation, although CK play opposite roles (it activates ovule primordia formation while inhibits LR initiation) (Werner et al., 2003; Higuchi et al., 2004; Bartrina et al., 2011; Huang et al., 2012; Bianco et al., 2013; Chang et al., 2013). Besides, the participation of IAA/AUX-ARF modules exists in both processes, and MP seems to be a regulator of the two of them (De Smet et al., 2010; Galbiati et al., 2013). Other members of the ARF family, as well as the NAC and the MADS-box transcription factors could be conserved in both processes, as some introductory works seem to indicate (Pinyopich et al., 2003; Moreno-Risueno et al., 2010; reviewed in Benková and Bielach, 2010). Finally, downstream the auxin signaling cascades, the activation of cell cycle genes will take place in order to promote organ growth, as it is starting to be revealed in the case of LR (Rast and Simon, 2008). Thereby, the analysis of the expression of cell cycle genes during ovule primordia formation would be very revealing. Apart of the hormonal and molecular pathways controlling LR formation, the influence of the environmental factors on this process is of extreme importance for the plant. How environment influences ovule primordia formation would be for sure a very challenging topic of research.
With this work we wanted to point out the little specific information available about the factors that control ovule primordia initiation, due to the difficulties to identify mutants presenting defects only in this particular step of ovule formation. Here we propose different experimental approaches to overcome the severity of some mutant phenotypes as well as to investigate these processes from a new point of view. The contribution and conservation of chromatin remodeling changes to the regulation of ovule number is starting to be elucidated and opens an extremely interesting field of research. Moreover, the most recent progresses in the fields of ovule, flower and root development strongly suggest common hormonal and molecular signals in all these organ initiation processes; such as a crosstalk between auxin and CK and probably also BR and the factors that establish organ boundaries and those that promote new organ outgrowth.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize for references not cited because of space limitations. The authors are grateful to Prof. M.M. Kater for critical reading of the manuscript. Mara Cucinotta was funded by a Ph.D fellowship from the Università degli Studi di Milano, Irma Roig-Villanova was funded by MIUR-Fondo per gli Investimenti della Ricerca di Base-Futuro in Ricerca. The work of Lucia Colombo is supported by MIUR-PRIN 2012.
References
Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841–857. doi: 10.1105/tpc.9.6.841
Aida, M., and Tasaka, M. (2006). Morphogenesis and patterning at the organ boundaries in the higher plant shoot apex. Plant Mol. Biol. 60, 915–928. doi: 10.1007/s11103-005-2760-7
Alonso-Blanco, C., Blankestijn-de Vries, H., Hanhart, C. J., and Koornneef, M. (1999). Natural allelic variation at seed size loci in relation to other life history traits of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 96, 4710–4717. doi: 10.1073/pnas.96.8.4710
Alvarez, J., and Smyth, D. R. (1999). CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126, 2377–2386.
Angenent, G. C., and Colombo, L. (1996). Molecular control of ovule development. Trends Plant Sci. 1, 228–232. doi: 10.1016/1360-1385(96)86900-7
Angenent, G. C., Franken, J., Busscher, M., Van Dijken, A., Van Went, J. L., Dons, H. J., et al. (1995) A novel class of MADS box genes is involved in ovule development in petunia. Plant Cell 7, 1569–1582.
Azhakanandam, S., Nole-Wilson, S., Bao, F., and Franks, R. G. (2008). SEUSS and AINTEGUMENTA mediate patterning and ovule initiation during gynoecium medial domain development. Plant Physiol. 146, 1165–1181. doi: 10.1104/pp.107.114751
Balanzá, V., Navarrete, M., Trigueros, M., and Ferrándiz, C. (2006). Patterning the female side of Arabidopsis: the importance of hormones. J. Exp. Bot. 57, 3457–3469. doi: 10.1093/jxb/erl188
Bartrina, I., Otto, E., Strnad, M., Werner, T., and Schmülling, T. (2011). Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 23, 69–80. doi: 10.1105/tpc.110.079079
Battaglia, R., Brambilla, V., Colombo, L., Stuitje, A. R., and Kater, M. M. (2006). Functional analysis of MADS-box genes controlling ovule development in Arabidopsis using the ethanol-inducible alc gene-expression system. Mech. Dev. 123, 267–276. doi: 10.1016/j.mod.2006.01.002
Bencivenga, S., Simonini, S., Benková, E., and Colombo, L. (2012). The transcription factors BEL1 and SPL are required for cytokinin and auxin signaling during ovule development in Arabidopsis. Plant Cell 24, 2886–2897. doi: 10.1105/tpc.112.100164
Benková, E., and Bielach, A. (2010). Lateral root organogenesis - from cell to organ. Curr. Opin. Plant Biol. 13, 677–683. doi: 10.1016/j.pbi.2010.09.006
Benková, E., Ivanchenko, M. G., Friml, J., Shishkova, S., and Dubrovsky, J. G. (2009). A morphogenetic trigger: is there an emerging concept in plant developmental biology? Trends Plant Sci. 14, 189–193. doi: 10.1016/j.tplants.2009.01.006
Benková, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertová, D., Jürgens, G., et al. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602. doi: 10.1016/S0092-8674(03)00924-3
Bennett, S. R. M., Alvarez, J., Bossinger, G., and Smyth, D. R. (1995). Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J. 8, 505–520. doi: 10.1046/j.1365-313X.1995.8040505.x
Bianco, M. D., Giustini, L., and Sabatini, S. (2013). Spatiotemporal changes in the role of cytokinin during root development. New Phytol. 199, 324–338. doi: 10.1111/nph.12338
Bilsborough, G. D., Runions, A., Barkoulas, M., Jenkins, H. W., Hasson, A., and Galinha, C. (2011). Model for the regulation of Arabidopsis thaliana leaf margin development. Proc. Natl. Acad. Sci. U.S.A. 108, 3424–3429. doi: 10.1073/pnas.1015162108
Bowman, J. L., Drews, G. N., and Meyerowitz, E. M. (1991). Expression of the Arabidopsis floral homeotic gene AGAMOUS is restricted to specific cell types late in flower development. Plant Cell 3, 749–758.
Bowman, J. L., and Smyth, D. R. (1999). CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126, 2387–2396.
Brambilla, V., Battaglia, R., Colombo, M., Masiero, S., Bencivenga, S., Kater, M. M., et al. (2007). Genetic and molecular interactions between BELL1 and MADS box factors support ovule development in Arabidopsis. Plant Cell 19, 2544–2556. doi: 10.1105/tpc.107.051797
Broadhvest, J., Baker, S. C., and Gasser, C. S. (2000). SHORT INTEGUMENTS 2 promotes growth during Arabidopsis reproductive development. Genetics 155, 899–907.
Byrne, M. E., Simorowski, J., and Martienssen, R. A. (2002). ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129, 1957–1965.
Ceccato, L., Masiero, S., Sinha Roy, D., Bencivenga, S., Roig-Villanova, I., Ditengou, F. A., et al. (2013). Maternal control of PIN1 is required for female gametophyte development in arabidopsis. PLoS ONE 8:e66148. doi: 10.1371/journal.pone.0066148
Chang, L., Ramireddy, E., and Schmülling, T. (2013). Lateral root formation and growth of Arabidopsis is redundantly regulated by cytokinin metabolism and signalling genes. J. Exp. Bot. 64, 5021–5032. doi: 10.1093/jxb/ert291
Cheng, Y., Dai, X., and Zhao, Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20, 1790–1799. doi: 10.1101/gad.1415106
Cole, M., Chandler, J., Weijers, D., Jacobs, B., Comelli, P., and Werr, W. (2009). DORNROSCHEN is a direct target of the auxin response factor MONOPTEROS in the Arabidopsis embryo. Development 136, 1643–1651. doi: 10.1242/dev.032177
Colombo, L., Franken, J., Koetje, E., Van Went, J., Dons, H. J., Angenent, G. C., et al. (1995) The petunia MADS box gene FBP11 determines ovule identity. Plant Cell 7, 1859–1868.
De Smet, I., Lau, S., Voss, U., Vanneste, S., Benjamins, R., Rademacher, E. H., et al. (2010). Bimodular auxin response controls organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 107, 2705–2710. doi: 10.1073/pnas.0915001107
Dreni, L., Jacchia, S., Fornara, F., Fornari, M., Ouwerkerk, P. B., An, G., et al. (2007) The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J. 52, 690–699. doi: 10.1111/j.1365-313X.2007.03272.x
Duszynska, D., McKeown, P. C., Juenger, T. E., Pietraszewska-bogiel, A., Geelen, D., and Spillane, C. (2013). Gamete fertility and ovule number variation in selfed reciprocal F1 hybrid triploid plants are heritable and display epigenetic parent-of-origin effects. New Phytol. 198, 71–81. doi: 10.1111/nph.12147
Elliott, R. C., Betzner, A. S., Huttner, E., Oakes, M. P., Tucker, W. Q. J., Gerentes, D., et al. (1996). AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8, 155–168.
Favaro, R., Pinyopich, A., Battaglia, R., Kooiker, M., Borghi, L., Ditta, G., et al. (2003). MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 15, 2603–2611. doi: 10.1105/tpc.015123
Galbiati, F., Sinha Roy, D., Simonini, S., Cucinotta, M., Ceccato, L., Cuesta, C., et al. (2013). An integrative model of the control of ovule primordia formation. Plant J. 12, 1–10. doi: 10.1111/tpj.12309
Grini, P. E., Thorstensen, T., Alm, V., Vizcay-Barrena, G., Windju, S. S., Jørstad, T. S., et al. (2009). The ASH1 HOMOLOG 2 (ASHH2) histone H3 methyltransferase is required for ovule and anther development in Arabidopsis. PLoS ONE 4:e7817. doi: 10.1371/journal.pone.0007817
Hardtke, C. S., and Berleth, T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17, 1405–1411. doi: 10.1093/emboj/17.5.1405
Heisler, M. G., Ohno, C., Das, P., Sieber, P., Reddy, G. V., Long, J. A., et al. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15, 1899–1911. doi: 10.1016/j.cub.2005.09.052
Higuchi, M., Pischke, M. S., Mähönen, A. P., Miyawaki, K., Hashimoto, Y., Seki, M., et al. (2004). In planta functions of the Arabidopsis cytokinin receptor family. Proc. Natl. Acad. Sci. U.S.A. 101, 8821–8826. doi: 10.1073/pnas.0402887101
Huang, H.-Y., Jiang, W.-B., Hu, Y.-W., Wu, P., Zhu, J.-Y., Liang, W.-Q., et al. (2012). BR signal influences arabidopsis ovule and seed number through regulating related genes expression by BZR1. Mol. Plant 6, 456–469. doi: 10.1093/mp/sss070
Hwang, I., Sheen, J., and Müller, B. (2012). Cytokinin signaling networks. Annu. Rev. Plant Biol. 63, 353–380. doi: 10.1146/annurev-arplant-042811-105503
Ishida, T., Aida, M., Takada, S., and Tasaka, M. (2000). Involvement of CUP-SHAPED COTYLEDON genes in gynoecium and ovule development in Arabidopsis thaliana. Plant Cell Physiol. 41, 60–67. doi: 10.1093/pcp/41.1.60
Kim, T. W., Hwang, J. Y., Kim, Y. S., Joo, S. H., Chang, S. C., Lee, J. S., et al. (2005). Arabidopsis CYP85A2, a cytochrome P450, mediates the Baeyer-Villiger oxidation of castasterone to brassinolide in brassinosteroid biosynthesis. Plant Cell 17, v2397–v2412. doi: 10.1105/tpc.105.033738
Klucher, K. M., Chow, H., Reiser, L., and Fischer, R. L. (1996). The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8, 137–153.
Knapp, S. (2002). Tobacco to tomatoes: a phylogenetic perspective on fruit diversity in the Solanaceae. J. Exp. Bot. 53, 2001–2022. doi: 10.1093/jxb/erf068
Larsson, E., Franks, R. G., and Sundberg, E. (2013). Auxin and the Arabidopsis thaliana gynoecium. J. Exp. Bot. 64, 2619–2627. doi: 10.1093/jxb/ert099
Laufs, P., Peaucelle, A., Morin, H., and Traas, J. (2004). MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131, 4311–4322. doi: 10.1242/dev.01320
Lee, D.-K., Geisler, M., and Springer, P. S. (2009). LATERAL ORGAN FUSION1 and LATERAL ORGAN FUSION2 function in lateral organ separation and axillary meristem formation in Arabidopsis. Development 136, 2423–2432. doi: 10.1242/dev.031971
Liu, Z., Franks, R. G., and Klink, V. P. (2000). Regulation of gynoecium marginal tissue formation by LEUNIG and AINTEGUMENTA. Plant Cell 12, 1879–1892. doi: 10.1105/tpc.12.10.1879
Modrusan, Z., Reiser, L., Feldmann, K. A., Fischer, R. L., and Haughn, G. W. (1994). Homeotic transformation of ovules into carpel-like structures in Arabidopsis. Plant Cell 6, 333–349.
Moreno-Risueno, M. A., Norman, J. M. V., Moreno, A., Zhang, J., Ahnert, S. E., and Benfey, P. N. (2010). Oscillating gene expression Arabidopsis root branching. Science 329, 1306–1311. doi: 10.1126/science.1191937
Nahar, M. A.-U., Ishida, T., Smyth, D. R., Tasaka, M., and Aida, M. (2012). Interactions of CUP-SHAPED COTYLEDON and SPATULA genes control carpel margin development in Arabidopsis thaliana. Plant Cell Physiol. 53, 1134–1143. doi: 10.1093/pcp/pcs057
Nemhauser, J. L., Feldman, L. J., and Zambryski, P. C. (2000). Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 127, 3877–3888.
Nishimura, C., Ohashi, Y., Sato, S., Kato, T., Tabata, S., and Ueguchi, C. (2004). Histidine kinase Homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16, 1365–1377. doi: 10.1105/tpc.021477
Nole-Wilson, S., Azhakanandam, S., and Franks, R. G. (2010). Polar auxin transport together with AINTEGUMENTA and REVOLUTA coordinate early Arabidopsis gynoecium development. Dev. Biol. 346, 181–195. doi: 10.1016/j.ydbio.2010.07.016
Okada, K., Ueda, J., Komaki, M. K., and Bell, C. J. (1991). Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 3, 677–684.
Pinyopich, A., Ditta, G. S., Savidge, B., Liljegren, S. J., Baumann, E., Wisman, E., et al. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424, 85–88. doi: 10.1038/nature01741
Rast, M. I., and Simon, R. (2008). The meristem-to-organ boundary: more than an extremity of anything. Curr. Opin. Genet. Dev. 18, 287–294. doi: 10.1016/j.gde.2008.05.005
Reyes-Olalde, J. I., Zuñiga-Mayo, V. M., Chávez Montes, R. A., Marsch-Martínez, N., and de Folter, S. (2013). Inside the gynoecium: at the carpel margin. Trends Plant Sci. 18, 644–655. doi: 10.1016/j.tplants.2013.08.002
Riefler, M., Novak, O., Strnad, M., and Schmu, T. (2006). Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18, 40–54. doi: 10.1105/tpc.105.037796
Rounsley, S. D., Ditta, G. S., and Yanofsky, M. F. (1995). Diverse roles for MADS box genes in Arabidopsis development. Plant Cell 7, 1259–1269.
Savidge, B., Rounsley, S. D., and Yanofsky, M. F. (1995). Temporal relationship between the transcription of two Arabidopsis MADS box genes and the floral organ identity genes. Plant Cell 7, 721–733.
Schneitz, K., Baker, S. C., Gasser, C. S., and Redweik, A. (1998). Pattern formation and growth during floral organogenesis: HUELLENLOS and AINTEGUMENTA are required for the formation of the proximal region of the ovule primordium in Arabidopsis thaliana. Development 125, 2555–2563.
Schneitz, K., Hülskamp, M., Kopczak, S. D., and Pruitt, R. E. (1997). Dissection of sexual organ ontogenesis: a genetic analysis of ovule development in Arabidopsis thaliana. Development 124, 1367–1376.
Shi, D. Q., and Yang, W. C. (2011) Ovule development in Arabidopsis: progress and challenge. Curr. Opin. Plant Biol. 14, 74–80. doi: 10.1016/j.pbi.2010.09.001
Shuai, B., Reynaga-pen, C. G., and Springer, P. S. (2002). The LATERAL ORGAN BOUNDARIES gene defines a novel, plant-specific gene family. Plant Physiol. 129, 747–761. doi: 10.1104/pp.010926
Skinner, D. J., Baker, S. C., Meister, R. J., Broadhvest, J., Schneitz, K., and Gasser, C. S. (2001). The Arabidopsis HUELLENLOS gene, which is essential for normal ovule development, encodes a mitochondrial ribosomal protein. Plant Cell 13, 2719–2730. doi: 10.1105/tpc.010323
Skinner, D. J., Hill, T. A., and Gasser, C. S. (2004). Regulation of ovule development. Plant Cell 16, 32–45. doi: 10.1105/tpc.015933
Sohlberg, J. J., Myrenås, M., Kuusk, S., Lagercrantz, U., Kowalczyk, M., Sandberg, G., et al. (2006). STY1 regulates auxin homeostasis and affects apical-basal patterning of the Arabidopsis gynoecium. Plant J. 47, 112–123. doi: 10.1111/j.1365-313X.2006.02775.x
Stepanova, A. N., Robertson-Hoyt, J., Yun, J., Benavente, L. M., Xie, D.-Y., Dolezal, K., et al. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133, 177–191. doi: 10.1016/j.cell.2008.01.047
Szekeres, M., Németh, K., Koncz-Kálmán, Z., Mathur, J., Kauschmann, A., Altmann, T., et al. (1996). Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85, 171–182. doi: 10.1016/S0092-8674(00)81094-6
Theißen, G., Kim, J. T., and Saedler, H. (1996). Molecular classification and phylogeny of the MADS-Box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J. Mol. Evol. 43, 484–516. doi: 10.1007/BF02337521
Vernoux, T., Kronenberger, J., Grandjean, O., Laufs, P., and Traas, J. (2000). PIN-FORMED 1 regulates cell fate at the periphery of the shoot apical meristem. Development 127, 5157–5165.
Vroemen, C. W., Mordhorst, A. P., Albrecht, C., Kwaaitaal, M. A. C. J., and Vries, S. C. D. (2003). The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15, 1563–1577. doi: 10.1105/tpc.012203
Werner, T., Motyka, V., Laucou, V., Smets, R., Van Onckelen, H., and Schmülling, T. (2003). Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15, 2532–2550. doi: 10.1105/tpc.014928
Western, T. L., and Haughn, G. W. (1999). BELL1 and AGAMOUS genes promote ovule identity in Arabidopsis thaliana. Plant J. 18, 329–336. doi: 10.1046/j.1365-313X.1999.00448.x
Wu, X. (2003). Modes of intercellular transcription factor movement in the Arabidopsis apex. Development 130, 3735–3745. doi: 10.1242/dev.00577
Xu, L., and Shen, W.-H. (2008). Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr. Biol. 18, 1966–1971. doi: 10.1016/j.cub.2008.11.019
Yamaguchi, A., Wu, M.-F., Yang, L., Wu, G., Poethig, R. S., and Wagner, D. (2013). A molecular framework for auxin-mediated initiation of flower primordial. Dev. Cell 17, 268–278. doi: 10.1016/j.devcel.2009.06.007
Keywords: Arabidopsis, ovule primordia, ovule number, development, transcription factors, hormones
Citation: Cucinotta M, Colombo L and Roig-Villanova I (2014) Ovule development, a new model for lateral organ formation. Front. Plant Sci. 5:117. doi: 10.3389/fpls.2014.00117
Received: 24 January 2014; Paper pending published: 21 February 2014;
Accepted: 11 March 2014; Published online: 27 March 2014.
Edited by:
Robert G. Franks, North Carolina State University, USAReviewed by:
Richard Macknight, University of Otago, New ZealandFrancois Parcy, Centre National de la Recherche Scientifique, France
Copyright © 2014 Cucinotta, Colombo and Roig-Villanova. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Irma Roig-Villanova, Dipartimento di Bioscienze, Università degli Studi di Milano, Via Celoria 26, 20133, Milano, Italy e-mail: irma.roig@unimi.it
 Mara Cucinotta
Mara Cucinotta Lucia Colombo
Lucia Colombo Irma Roig-Villanova
Irma Roig-Villanova