- 1Biological Resources and Post-harvest Division, Japan International Research Center for Agricultural Sciences, Tsukuba, Japan
- 2Laboratory of Plant Molecular Physiology, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo, Japan
- 3Gene Discovery Research Group, RIKEN Center for Sustainable Resource Science, Yokohama, Japan
Drought negatively impacts plant growth and the productivity of crops around the world. Understanding the molecular mechanisms in the drought response is important for improvement of drought tolerance using molecular techniques. In plants, abscisic acid (ABA) is accumulated under osmotic stress conditions caused by drought, and has a key role in stress responses and tolerance. Comprehensive molecular analyses have shown that ABA regulates the expression of many genes under osmotic stress conditions, and the ABA-responsive element (ABRE) is the major cis-element for ABA-responsive gene expression. Transcription factors (TFs) are master regulators of gene expression. ABRE-binding protein and ABRE-binding factor TFs control gene expression in an ABA-dependent manner. SNF1-related protein kinases 2, group A 2C-type protein phosphatases, and ABA receptors were shown to control the ABA signaling pathway. ABA-independent signaling pathways such as dehydration-responsive element-binding protein TFs and NAC TFs are also involved in stress responses including drought, heat, and cold. Recent studies have suggested that there are interactions between the major ABA signaling pathway and other signaling factors in stress responses. The important roles of these TFs in crosstalk among abiotic stress responses will be discussed. Control of ABA or stress signaling factor expression can improve tolerance to environmental stresses. Recent studies using crops have shown that stress-specific overexpression of TFs improves drought tolerance and grain yield compared with controls in the field.
Introduction
The world population is expected to reach nine billion by 2050. Considering this population increase, crop yields need to be improved by 40% in areas where drought is likely to occur by 2025 (Pennisi, 2008). In addition, frequent occurrences of drought and abnormal weather events have lately been observed all over the world. Drought negatively impacts plant growth and crop production (Bray et al., 2000). Almost every year, some region of the earth is hit by drought, damaging crops, and disrupting agricultural production. Severe drought affected the central and south of the US Corn Belt during 2012 (Edmeades, 2013). Drought also causes great damage to the production of other crops such as rice, wheat, and soybean. The southern states of Brazil, which account for 40% of the soybean production by the second leading producer worldwide, lost more than 20% of their production because of drought during the 2003/2004 and 2004/2005 seasons (Polizel et al., 2011). The development of stress-tolerant crops will be significantly advantageous in areas where such stresses occur frequently. Recently, some progress has been made toward identification of stress-related genes potentially capable of increasing the tolerance of plants to abiotic stress. Understanding the molecular mechanisms in the drought response is important to improve drought tolerance using molecular techniques. ABA accumulates under osmotic stress caused by drought, but also by other water limiting conditions, and plays an important role in stress responses and tolerance in plants (reviewed in Finkelstein et al., 2002; Yamaguchi-Shinozaki and Shinozaki, 2006; Nakashima et al., 2009b; Figure 1). Molecular studies have revealed that ABA-independent gene expression is also important in stress tolerance in plants (Figure 1). In this review, we summarize some of the most important TFs in drought responses and discuss their regulatory networks and crosstalk in abiotic stress responses. By applying current knowledge of stress-regulated TFs and their target genes, improvement of drought stress tolerance is in progress in various crops using transgenic technology.
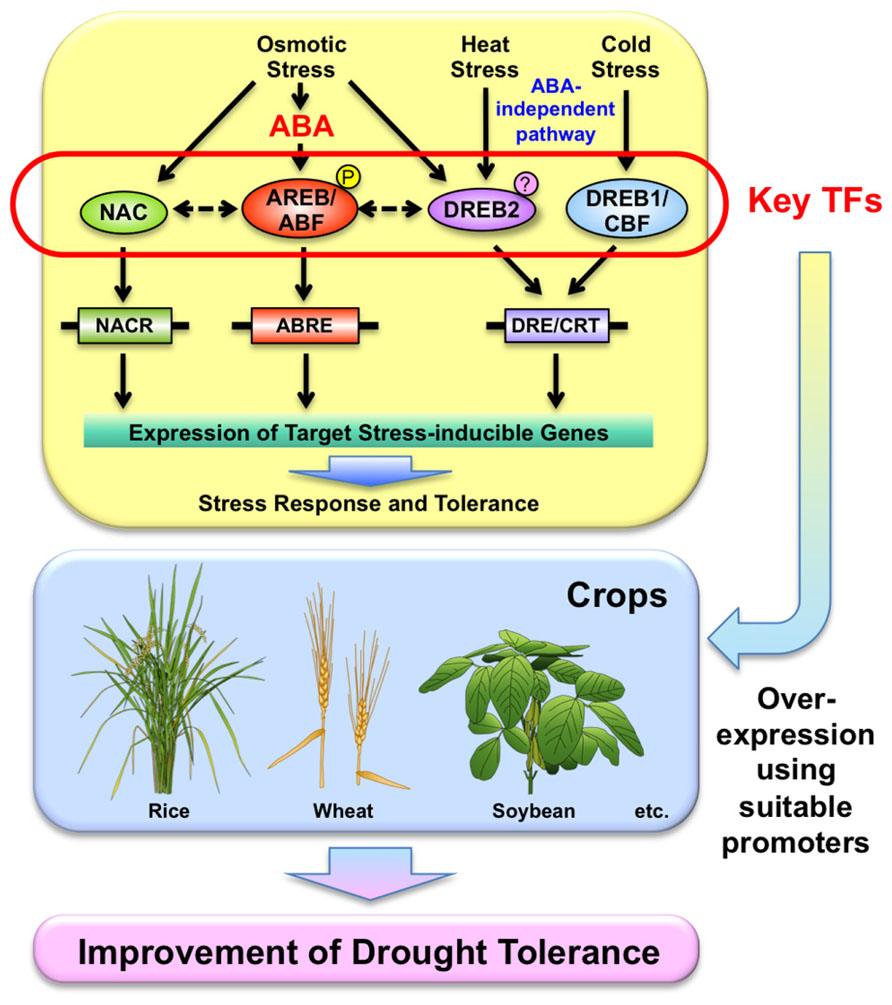
FIGURE 1. Utilization of transcription factors (TFs) involved in stress-responsive pathways in stress responses for the improvement of drought tolerance of crops. Usage of suitable promoters might be necessary to control their expression.
AREB/ABF TFs for ABA-Dependent Gene Expression
The promoter regions of ABA-responsive genes contain a conserved cis-element, named the ABRE (PyACGTGG/TC), which controls gene expression (Figure 1). Studies have revealed that expression of ABA-responsive genes requires more than one ABRE or a combination of an ABRE and a CE for a functional promoter (reviewed in Fujita et al., 2011, 2013; Nakashima and Yamaguchi-Shinozaki, 2013). Comprehensive and molecular analyses showed that ABA regulates the expression of many genes under osmotic stress conditions, and that the ABRE is the major cis-element for ABA-responsive gene expression (Maruyama et al., 2012). AREB/ABFs are bZIP TFs that regulate ABA-dependent gene expression, acting as major TFs under abiotic stress conditions in Arabidopsis (reviewed in Fujita et al., 2011, 2013; Figure 1). Among the nine members of the AREB/ABF TF family identified in Arabidopsis, AREB1/ABF2 has been reported to control ABA signaling and environmental stress responses during the vegetative growth stage. The AREB/ABF TFs are induced by abiotic stress and their transcriptional activities are controlled by ABA-dependent phosphorylation. ABA is required for full activation of AREB1 (Fujita et al., 2005; Yoshida et al., 2010) and its activity is regulated by the ABA-dependent phosphorylation of multiple sites within conserved domains (Furihata et al., 2006). Transgenic Arabidopsis plants overexpressing deleted and active forms of AREB1 showed enhanced drought tolerance and ABA hypersensitivity (Fujita et al., 2005). Overexpression of AREB1 also improved drought tolerance in rice and soybean (Oh et al., 2005; Barbosa et al., 2013). Progress in understanding ABA perception and signal transduction has been made recently (reviewed in Cutler et al., 2010; Raghavendra et al., 2010; Umezawa et al., 2010; Weiner et al., 2010; Nakashima and Yamaguchi-Shinozaki, 2013). It was revealed that SnRK2, group A PP2Cs, and RCAR/PYR/PYL ABA receptors control the ABA signaling pathway including AREB/ABFs in land plants (reviewed in Umezawa et al., 2010; Miyakawa et al., 2013; Nakashima and Yamaguchi-Shinozaki, 2013). The phosphorylation of AREB/ABFs by SnRK2s is critical in the ABA-dependent signaling network (Fujita et al., 2009; Nakashima et al., 2009a; Umezawa et al., 2013). Recent studies have indicated that group A PP2Cs evolved early in land plants as key regulators of intrinsic desiccation tolerance, such as in the moss Physcomitrella patens (Komatsu et al., 2013). Perception and signaling factors such as PYL4 can also be used to improve stress tolerance (Pizzio et al., 2013).
DREB1/CBF TFs for Cold-Responsive Gene Expression to Improve Drought Tolerance
Analysis of the promoter regions of genes showing ABA-independent expression in stress responses and tolerance has shown a cis-element with the sequence A/GCCGAC, designated the DRE/CRT (Figure 1). Two groups of AP2/ERF TFs were identified as DREB; DREB1/CBF and DREB2 in Arabidopsis (Liu et al., 1998). DREB1/CBF TFs specifically interact with the DRE/CRT and control the expression of a large number of stress-responsive genes in Arabidopsis. Improvements in tolerance to drought, salinity and freezing stresses have been reported in transgenic Arabidopsis overexpressing DREB1/CBF TFs, although their constitutive expression causes growth defects (Liu et al., 1998; Kasuga et al., 1999). However, overexpression of DREB1 under the control of the Arabidopsis stress-responsive RD29A promoter improved stress tolerance in Arabidopsis without growth defects (Kasuga et al., 1999). Cold-inducible DREB1/CBF genes have also been isolated from a number of plant species, such as maize, oilseed rape, rye (Secale cereale), rice, tomato, and wheat (Triticum aestivum; reviewed in Mizoi et al., 2012). Interestingly, the major QTLs for tolerance to frost in Arabidopsis, diploid wheat (T. monococcum) and barley map to DREB1/CBF genes, and the expression levels of DREB1/CBF genes are correlated with frost tolerance (Vágújfalvi et al., 2003; Alonso-Blanco et al., 2005; Francia et al., 2007; Knox et al., 2008). Thus, the function of the DREB1/CBF regulon in the regulation of cold stress responses is widely conserved in angiosperms. Overexpression of DREB/CBF TFs has been reported to enhance drought tolerance in transgenic crops including chrysanthemum (Hong et al., 2006), peanut (Bhatnagar-Mathur et al., 2007, Bhatnagar-Mathur et al., 2013), potato (Behnam et al., 2007; Iwaki et al., 2013), rice (Oh et al., 2005; Ito et al., 2006; Datta et al., 2012), soybean (Polizel et al., 2011; de Paiva Rolla et al., 2013), tobacco (Kasuga et al., 2004), tomato (Hsieh et al., 2002a,b), and wheat (Pellegrineschi et al., 2004; Saint Pierre et al., 2012). For example, rice DREB1/CBF-type TFs involved in cold-responsive gene expression also conferred improved tolerance to drought in transgenic rice (Ito et al., 2006). The rice DREB1/CBF-type genes, OsDREB1A and OsDREB1B, are induced by cold stress. Transgenic Arabidopsis and rice plants overexpressing rice OsDREB1 or Arabidopsis DREB1 genes showed improved tolerance to drought, high-salt and cold stresses but defective growth under normal growth conditions. Elevated contents of osmoprotectants including free proline and soluble sugars were detected in the transgenic rice. These results indicate that the DREB1/CBF regulon is conserved in rice, and that DREB1/CBF-type genes may be useful for improvement of tolerance to different environmental stresses in various kinds of transgenic monocot plants as well as dicot plants.
DREB2 TFs for Osmotic- and Heat-Responsive Gene Expression To Improve Drought Tolerance
The DREB2 gene encoding a DRE/CRT-binding protein is induced by osmotic stress (Liu et al., 1998; Figure 1). However, transgenic plants overexpressing DREB2A did not show any changes in phenotype. Domain analysis of DREB2A using Arabidopsis protoplasts showed that deletion of the central region makes DREB2A constitutively active (DREB2Aca), indicating that this region contains a negative regulatory domain (NRD; Sakuma et al., 2006a). Overexpression of DREB2Aca induced growth defects, up-regulation of stress-inducible genes, and enhanced drought tolerance (Sakuma et al., 2006a). Stress-inducible overexpression of DREB2ca improved drought tolerance in Arabidopsis and soybean without growth defects (Sakuma et al., 2006a; Engels et al., 2013). The NRD region of DREB2A is required for regulation of DREB2A protein stability. As mentioned above, overexpression of DREB1A improves freezing and dehydration stress tolerance in transgenic plants. By contrast, overexpression of DREB2Aca improves dehydration stress tolerance but only slightly improves freezing stress tolerance in transgenic plants. Integrated analysis of transcripts and metabolites was conducted to see the difference in the downstream gene products of DREB1A and DREB2A in Arabidopsis (Maruyama et al., 2009). Microarray analysis indicated that the downstream gene products of DREB1A and those of DREB2A have similar putative functions, but the expression of genes for carbohydrate metabolism in DREB1A and DREB2A transgenic plants is very different. Under dehydration and cold conditions, expression of genes for starch-degradation, sucrose metabolism and sugar alcohol synthesis changes dynamically. As a result, many kinds of mono-, di-, and trisaccharides, and sugar alcohols accumulate in plants. Overexpression of DREB1A caused similar changes in these metabolic processes, and these changes might improve dehydration and freezing stress tolerance in transgenic plants. By contrast, overexpression of DREB2Aca did not increase the level of these metabolites in transgenic plants. In addition, degradation of DREB2A is mediated by DRIPs, which are C3HC4 RING domain-containing proteins. DRIPs bind to DREB2A and function as E3 ubiquitin ligases mediating ubiquitination of DREB2A (Qin et al., 2008). Overexpression of DREB2Aca also induced expression of genes related to heat shock stress and improved thermotolerance in transgenic plants (Sakuma et al., 2006b). These results indicate that DREB2s function in both dehydration and heat shock stress responses. DREB2-type proteins have been isolated from a number of other plant species such as barley, rice, sunflower, maize, and wheat (Mizoi et al., 2012). GmDREB2A;2 is a DREB2A ortholog in soybean (Mizoi et al., 2013), but there are differences between DREB2A and GmDREB2A;2 in the NRD sequence. The effects on gene expression in transgenic plants overexpressing GmDREB2A;2 are different from those in transgenic plants overexpressing DREB2A. This suggests that specialization in DREB2 regulons has occurred, although their basic functions are conserved between Arabidopsis and soybean. Recently, GWAS of ZmDREB2 and natural variations in the drought tolerance of maize (Zea mays) indicated that natural variation in the promoter region of ZmDREB2.7 contributes to drought tolerance in maize (Liu et al., 2013). The favorable ZmDREB2.7 allele may be a good resource for improving drought tolerance in maize. Recent studies suggest that DREB2 has important functions in drought tolerance, and that it can be used for improvement of drought tolerance in crops.
NAC TFs for Drought-Responsive Gene Expression to Improve Drought Tolerance
NAM, ATAF, and CUC TF proteins are plant-specific TFs. More than 100 NAC genes have been identified in Arabidopsis and rice (reviewed in Nakashima et al., 2012). Phylogenetic analyses indicate that six groups were established in an ancient moss. NAC TFs have a variety of important functions in development and stress responses. The genes in the SNAC group have important roles in the control of environmental stress tolerance (reviewed in Nakashima et al., 2012; Figure 1), and can bind to the NACR (NAC recognition sequence; CACG core). Stress-responsive Arabidopsis SNAC genes such as RD26 and ATAF1, and rice SNAC genes such as SNAC1, OsNAC6/SNAC2, and OsNAC5 can improve drought and/or high-salt stress tolerance when overexpressed (Tran et al., 2004; Hu et al., 2006; Nakashima et al., 2007; Takasaki et al., 2010; reviewed in Nakashima et al., 2012). Stress-responsive overexpression of NACs utilizing rice stress-responsive LIP9, OsNAC6, or OsHox24 promoters is effective in inducing stress tolerance without the inhibitory effects of NAC on plant growth (Nakashima et al., 2007, 2012, 2014; Takasaki et al., 2010). Recent studies have suggested that the root-specific promoter RCc3 is useful for the overexpression of SNACs such as SNAC1 and OsNAC10 to enhance the abiotic stress tolerance of rice in field conditions (Jeong et al., 2010, 2013; Redillas et al., 2012). These results indicate that SNACs have important roles in the control of abiotic stress responses and tolerance and that it is possible to improve stress tolerance by overexpressing SNACs using suitable promoters in the field. The many kinds of drought-responsive or tissue/organ-specific promoters reported for roots and stomata might be effective tools to control the expression of drought-responsive factors that cause growth defects at the right time and right position (Nakashima et al., 2007, 2014; Rai et al., 2009; Wu et al., 2009; Xiao et al., 2009; Yi et al., 2010; Ganguly et al., 2011; Yang and Xiong, 2011; Bang et al., 2013; Rusconi et al., 2013).
Interactions Between Multiple TFs in Drought Responses
Evidence for interaction between the AREB/ABFs and DREB/CBFs has been reported. The DRE/CRT motif in the promoters of drought-responsive genes is a binding region for an ABA-independent DREB/CBF TF and functions as a CE for ABRE in ABA-dependent gene expression (Narusaka et al., 2003). Lee et al. (2010) showed that the DREB1A/CBF3, DREB2A, and DREB2C proteins interact physically with AREB/ABF proteins. These data suggest crosstalk between elements of the ABA-dependent and -independent response pathways. Moreover, interactions in the signaling pathways have also been indicated. Kim et al. (2011) reported that an ABRE promoter sequence, AREB/ABF TFs, and SnRK2s are involved in expression of the DREB2A gene under osmotic stress conditions, suggesting complex interaction between the AREB and DREB regulons at the gene expression level as well as the protein level.
Interaction between the AREB/ABFs and NACs has also been indicated at the gene expression level. Jensen et al. (2013) reported that Arabidopsis SNAC TF ATAF1 directly regulates the ABA biosynthetic gene NCED3 in Arabidopsis, suggesting that SNAC TFs may regulate ABA-dependent gene expression of ABRE regulons. On the other hand, the promoters of SNAC genes contain ABRE sequences (Nakashima et al., 2012). Recently, Xu et al. (2013) reported that Arabidopsis ANAC096 cooperates with AREB/ABF factors (ABF2/AREB1 and ABF4/AREB2) in dehydration and osmotic stress responses. These results indicate complex interaction between the AREB/ABF and NAC regulons.
Finally, interaction between DREB/CBFs and other kinds of AP2/ERFs at the gene expression level has also been suggested. Cheng et al. (2013) reported that the Arabidopsis ERF1 regulates gene expression by binding to two kinds of cis-elements, the GCC box and DRE/CRT, in response to different stress signals. ERF1 is an upstream TF in both ethylene and jasmonate signaling and is involved in resistance to pathogens. Their results suggested that ERF1 bound to the GCC box but not the DRE/CRT in response to biotic stress, and to the DRE/CRT under abiotic stress. These results suggest that ERF1 may integrate ethylene, jasmonate, and ABA signaling and play an important role in biotic and abiotic stress responses.
Conclusion
Molecular analysis has suggested that drought-responsive TFs such as DREB1/CBF, DREB2, AREB/ABF, and NAC TFs function in drought responses and tolerance (Figure 1). These TFs also function in crosstalk in abiotic stress responses, such as drought, cold, and heat. As mentioned above, these factors can be used to improve drought tolerance in a variety of crops. Our group has utilized these key TFs for the improvement of drought tolerance in crops including rice, wheat, and soybean in collaboration with international and domestic institutes (Pellegrineschi et al., 2004; Hong et al., 2006; Behnam et al., 2007; Bhatnagar-Mathur et al., 2007; Polizel et al., 2011; Datta et al., 2012; Ishizaki et al., 2012; Saint Pierre et al., 2012; Barbosa et al., 2013; Bhatnagar-Mathur et al., 2013; de Paiva Rolla et al., 2013; Engels et al., 2013; Iwaki et al., 2013). Some results using crops including rice and peanut have shown that stress-specific overexpression of DREB1A improves drought tolerance and grain yield compared with controls in the field (Datta et al., 2012; Bhatnagar-Mathur et al., 2013). These results suggest that overexpression of key TFs under the control of suitable promoters can improve stress tolerance, although the regulatory network in the plant response is complex in water limiting environments (Figure 1). Since TFs function in balanced crosstalk in abiotic stress responses, overexpression of a certain TF may affect other signaling pathways. Thus, we should examine the molecular effects of overexpressing TFs in addition to conducting stress tolerance assays. In addition, the effects of a transgene may depend on the genetic background of the species or cultivar used for transformation. Furthermore, since the degree of drought varies in actual fields (strength, timing, and period of stress, complex stresses such as drought with heat stress etc.), the effect of a transgene may differ depending on environmental conditions. Continuous field experiments might be necessary to see the effects of transgene-encoded TFs in the field using a variety of genotypes and environments. Recently, QTL analyses have revealed novel genes involved in drought resistance. DEEPER ROOTING 1 (DRO1), a QTL controlling root growth angle in rice, was cloned and characterized (Uga et al., 2013). This study revealed that changes in root system architecture can improve drought avoidance. Other drought resistant QTLs have also been reported in rice. Multiple QTLs were reported in the rice mega-variety IR64 that enhance the yield under drought conditions (Swamy et al., 2013). Combinations/pyramiding of transgenic plants and QTL drought resistant varieties by marker-assist selection (MAS) may promote drought tolerance.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Masami Toyoshima for skillful editorial assistance. Research in our laboratories was supported by the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry (BRAIN); the Ministry of Agriculture, Forestry and Fisheries (MAFF); the Science and Technology Research Partnership for Sustainable Development (SATREPS) of the Japan Science and Technology Agency (JST)/Japan International Cooperation Agency (JICA); Grants-in-Aid for Scientific Research by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) and the Japan Society for the Promotion of Science (JSPS).
Abbreviations
ABA, abscisic acid; ABF, ABRE-binding factor; ABRE, ABA-responsive element; AP2, APETALA 2; AREB, ABRE-binding protein; bZIP, basic leucine zipper; CBF, CRT binding factor; CE, coupling element; DRE, dehydration-responsive element; DREB, DRE-binding protein; DRIP, DREB2A-interacting protein; CRT, C-repeat; ERF, ethylene-responsive element binding factor; GWAS, genome-wide association study; NAC, NAM, ATAF, and CUC; PP2C, 2C-type protein phosphatase; PYL, PYR1-like; PYR, pyrabactin resistance; QTL, quantitative trait locus; RCAR, regulatory component of ABA receptor; SNAC, stress-responsive NAC; SnRK2, SNF1-related protein kinase 2; TF, transcription factor.
References
Alonso-Blanco, C., Gomez-Mena, C., Llorente, F., Koornneef, M., Salinas, J., and Martinez-Zapater, J. M. (2005). Genetic and molecular analyses of natural variation indicate CBF2 as a candidate gene for underlying a freezing tolerance quantitative trait locus in Arabidopsis. Plant Physiol. 139, 1304–1312. doi: 10.1104/pp.105.068510
Bang, S. W., Park, S. H., Jeong, J. S., Kim, Y. S., Jung, H., Ha, S. H., et al. (2013). Characterization of the stress-inducible OsNCED3 promoter in different transgenic rice organs and over three homozygous generations. Planta 237, 211–224. doi: 10.1007/s00425-012-1764-1
Barbosa, E. G. G., Leite, J. P., Marin, S. R. R., Marinho, J. P., Fátima Corrêa Carvalho, J., Fuganti-Pagliarini, R., et al. (2013). Overexpression of the ABA-dependent AREB1 transcription factor from Arabidopsis thaliana improves soybean tolerance to water deficit. Plant Mol. Biol. Rep. 31, 719–730. doi: 10.1007/s11105-012-0541-4
Behnam, B., Kikuchi, A., Celebi-Toprak, F., Kasuga, M., Yamaguchi-Shinozaki, K., and Watanabe, K. N. (2007). Arabidopsis rd29A::DREB1A enhances freezing tolerance in transgenic potato. Plant Cell Rep. 26, 1275–1282. doi: 10.1007/s00299-007-0360-5
Bhatnagar-Mathur, P., Devi, M. J., Reddy, D. S., Lavanya, M., Vadez, V., Serraj, R., et al. (2007). Stress-inducible expression of At DREB1A in transgenic peanut (Arachis hypogaea L.) increases transpiration efficiency under water-limiting conditions. Plant Cell Rep. 26, 2071–2082. doi: 10.1007/s00299-007-0406-8
Bhatnagar-Mathur, P., Rao, J. S., Vadez, V., Dumbala, S. R., Rathore, A., Yamaguchi-Shinozaki, K., et al. (2013). Transgenic peanut overexpressing the DREB1A transcription factor has higher yields under drought stress. Mol. Breed. 33, 327–340. doi: 10.1007/s11032-013-9952-7
Bray, E. A., Bailey-Serres, J., and Weretilnyk, E. (2000). “Responses to abiotic stresses,” in Biochemistry and Molecular Biology of Plants, eds B. B. Buchanan, W. Gruissem, and R. L. Jones (Rockville: American Society of Plant Physiologists), 1158–1203.
Cheng, M. C., Liao, P. M., Kuo, W. W., and Lin, T. P. (2013). The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 162, 1566–1582. doi: 10.1104/pp.113.221911
Cutler, S. R., Rodriguez, P. L., Finkelstein, R. R., and Abrams, S. R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. doi: 10.1146/annurev-arplant-042809-112122
Datta, K., Baisakh, N., Ganguly, M., Krishnan, S., Yamaguchi Shinozaki, K., and Datta, S. K. (2012). Overexpression of Arabidopsis and rice stress genes’ inducible transcription factor confers drought and salinity tolerance to rice. Plant Biotechnol. J. 10, 579–586. doi: 10.1111/j.1467-7652.2012.00688.x
de Paiva Rolla, A. A., de Fatima Correa Carvalho, J., Fuganti-Pagliarini, R., Engels, C., Do Rio, A., Marin, S. R., et al. (2013). Phenotyping soybean plants transformed with rd29A:AtDREB1A for drought tolerance in the greenhouse and field. Transgenic Res. 23, 75–87. doi: 10.1007/s11248-013-9723-6
Edmeades, G. O. (2013). Progress in Achieving and Delivering Drought Tolerance in Maize – An Update. Ithaca, NY: ISAAA
Engels, C., Fuganti-Pagliarini, R., Marin, S. R. R., Marcelino-Guimarães, F. C., Oliveira, M. C. N., Kanamori, N., et al. (2013). Introduction of the rd29A:AtDREB2A CA gene into soybean (Glycine max L. Merril) and its molecular characterization in the leaves and roots during dehydration. Genet. Mol. Biol. 36, 556–565. doi: 10.1590/S1415-47572013000400015
Finkelstein, R. R., Gampala, S. S., and Rock, C. D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14(Suppl.), S15–S45. doi: 10.1105/tpc.010441
Francia, E., Barabaschi, D., Tondelli, A., Laido, G., Rizza, F., Stanca, A. M., et al. (2007). Fine mapping of a HvCBF gene cluster at the frost resistance locus Fr-H2 in barley. Theor. Appl. Genet. 115, 1083–1091. doi: 10.1007/s00122-007-0634-x
Fujita, Y., Fujita, M., Satoh, R., Maruyama, K., Parvez, M. M., Seki, M., et al. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17, 3470–3488. doi: 10.1105/tpc.105.035659
Fujita, Y., Fujita, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2011). ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124, 509–525. doi: 10.1007/s10265-011-0412-3
Fujita, Y., Nakashima, K., Yoshida, T., Katagiri, T., Kidokoro, S., Kanamori, N., et al. (2009). Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 50, 2123–2132. doi: 10.1093/pcp/pcp147
Fujita, Y., Yoshida, T., and Yamaguchi-Shinozaki, K. (2013). Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant. 147, 15–27. doi: 10.1111/j.1399-3054.2012.01635.x
Furihata, T., Maruyama, K., Fujita, Y., Umezawa, T., Yoshida, R., Shinozaki, K., et al. (2006). Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. U.S.A. 103, 1988–1993. doi: 10.1073/pnas.0505667103
Ganguly, M., Roychoudhury, A., Sarkar, S. N., Sengupta, D. N., Datta, S. K., and Datta, K. (2011). Inducibility of three salinity/abscisic acid-regulated promoters in transgenic rice with gusA reporter gene. Plant Cell Rep. 30, 1617–1625. doi: 10.1007/s00299-011-1072-4
Hong, B., Tong, Z., Ma, N., Li, J., Kasuga, M., Yamaguchi-Shinozaki, K., et al. (2006). Heterologous expression of the AtDREB1A gene in chrysanthemum increases drought and salt stress tolerance. Sci. China C Life Sci. 49, 436–445. doi: 10.1007/s11427-006-2014-1
Hsieh, T. H., Lee, J. T., Charng, Y. Y., and Chan, M. T. (2002a). Tomato plants ectopically expressing Arabidopsis CBF1 show enhanced resistance to water deficit stress. Plant Physiol. 130, 618–626. doi: 10.1104/pp.006783
Hsieh, T. H., Lee, J. T., Yang, P. T., Chiu, L. H., Charng, Y. Y., Wang, Y. C., et al. (2002b). Heterology expression of the Arabidopsis C-repeat/dehydration response element binding factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiol. 129, 1086–1094. doi: 10.1104/pp.003442
Hu, H., Dai, M., Yao, J., Xiao, B., Li, X., Zhang, Q., et al. (2006). Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. U.S.A. 103, 12987–12992. doi: 10.1073/pnas.0604882103
Ishizaki, T., Maruyama, K., Obara, M., Fukutani, A., Yamaguchi-Shinozaki, K., Ito, Y., et al. (2012). Expression of Arabidopsis DREB1C improves survival, growth, and yield of upland New Rice for Africa (NERICA) under drought. Mol. Breed. 31, 255–264. doi: 10.1007/s11032-012-9785-9
Ito, Y., Katsura, K., Maruyama, K., Taji, T., Kobayashi, M., Seki, M., et al. (2006). Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 47, 141–153. doi: 10.1093/pcp/pci230
Iwaki, T., Guo, L., Ryals, J. A., Yasuda, S., Shimazaki, T., Kikuchi, A., et al. (2013). Metabolic profiling of transgenic potato tubers expressing Arabidopsis dehydration response element-binding protein 1A (DREB1A). J. Agric. Food Chem. 61, 893–900. doi: 10.1021/jf304071n
Jensen, M. K., Lindemose, S., de Masi, F., Reimer, J. J., Nielsen, M., Perera, V., et al. (2013). ATAF1 transcription factor directly regulates abscisic acid biosynthetic gene NCED3 in Arabidopsis thaliana. FEBS Open Bio 3, 321–327. doi: 10.1016/j.fob.2013.07.006
Jeong, J. S., Kim, Y. S., Baek, K. H., Jung, H., Ha, S. H., Do Choi, Y., et al. (2010). Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 153, 185–197. doi: 10.1104/pp.110.154773
Jeong, J. S., Kim, Y. S., Redillas, M. C., Jang, G., Jung, H., Bang, S. W., et al. (2013). OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol. J. 11, 101–114. doi: 10.1111/pbi.12011
Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17, 287–291. doi: 10.1038/7036
Kasuga, M., Miura, S., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2004). A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol. 45, 346–350. doi: 10.1093/pcp/pch037
Kim, J. S., Mizoi, J., Yoshida, T., Fujita, Y., Nakajima, J., Ohori, T., et al. (2011). An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant Cell Physiol. 52, 2136–2146. doi: 10.1093/pcp/pcr143
Knox, A. K., Li, C., Vagujfalvi, A., Galiba, G., Stockinger, E. J., and Dubcovsky, J. (2008). Identification of candidate CBF genes for the frost tolerance locus Fr-Am2 in Triticum monococcum. Plant Mol. Biol. 67, 257–270. doi: 10.1007/s11103-008-9316-6
Komatsu, K., Suzuki, N., Kuwamura, M., Nishikawa, Y., Nakatani, M., Ohtawa, H., et al. (2013). Group A PP2Cs evolved in land plants as key regulators of intrinsic desiccation tolerance. Nat. Commun. 4, 2219. doi: 10.1038/ncomms3219
Lee, S. J., Kang, J. Y., Park, H. J., Kim, M. D., Bae, M. S., Choi, H. I., et al. (2010). DREB2C interacts with ABF2, a bZIP protein regulating abscisic acid-responsive gene expression, and its overexpression affects abscisic acid sensitivity. Plant Physiol. 153, 716–727. doi: 10.1104/pp.110.154617
Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., et al. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406. doi: 10.1105/tpc.10.8.1391
Liu, S., Wang, X., Wang, H., Xin, H., Yang, X., Yan, J., et al. (2013). Genome-wide analysis of ZmDREB genes and their association with natural variation in drought tolerance at seedling stage of Zea mays L. PLoS Genet. 9:e1003790. doi: 10.1371/journal.pgen.1003790
Maruyama, K., Takeda, M., Kidokoro, S., Yamada, K., Sakuma, Y., Urano, K., et al. (2009). Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol. 150, 1972–1980. doi: 10.1104/pp.109.135327
Maruyama, K., Todaka, D., Mizoi, J., Yoshida, T., Kidokoro, S., Matsukura, S., et al. (2012). Identification of cis-acting promoter elements in cold- and dehydration-induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res. 19, 37–49. doi: 10.1093/dnares/dsr040
Miyakawa, T., Fujita, Y., Yamaguchi-Shinozaki, K., and Tanokura, M. (2013). Structure and function of abscisic acid receptors. Trends Plant Sci. 18, 259–266. doi: 10.1016/j.tplants.2012.11.002
Mizoi, J., Ohori, T., Moriwaki, T., Kidokoro, S., Todaka, D., Maruyama, K., et al. (2013). GmDREB2A;2, a canonical DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN2-type transcription factor in soybean, is posttranslationally regulated and mediates dehydration-responsive element-dependent gene expression. Plant Physiol. 161, 346–361. doi: 10.1104/pp.112.204875
Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 86–96. doi: 10.1016/j.bbagrm.2011.08.004
Nakashima, K., Fujita, Y., Kanamori, N., Katagiri, T., Umezawa, T., Kidokoro, S., et al. (2009a). Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50, 1345–1363. doi: 10.1093/pcp/pcp083
Nakashima, K., Ito, Y., and Yamaguchi-Shinozaki, K. (2009b). Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 149, 88–95. doi: 10.1104/pp.108.129791
Nakashima, K., Jan, A., Todaka, D., Maruyama, K., Goto, S., Shinozaki, K., et al. (2014). Comparative functional analysis of six drought-responsive promoters in transgenic rice. Planta 239, 47–60. doi: 10.1007/s00425-013-1960-7
Nakashima, K., Takasaki, H., Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 97–103. doi: 10.1016/j.bbagrm.2011.10.005
Nakashima, K., Tran, L. S., Van Nguyen, D., Fujita, M., Maruyama, K., Todaka, D., et al. (2007). Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 51, 617–630. doi: 10.1111/j.1365-313X.2007.03168.x
Nakashima, K., and Yamaguchi-Shinozaki, K. (2013). ABA signaling in stress-response and seed development. Plant Cell Rep. 32, 959–970. doi: 10.1007/s00299-013-1418-1
Narusaka, Y., Nakashima, K., Shinwari, Z. K., Sakuma, Y., Furihata, T., Abe, H., et al. (2003). Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 34, 137–148. doi: 10.1046/j.1365-313X.2003.01708.x
Oh, S. J., Song, S. I., Kim, Y. S., Jang, H. J., Kim, S. Y., Kim, M., et al. (2005). Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol. 138, 341–351. doi: 10.1104/pp.104.059147
Pellegrineschi, A., Reynolds, M., Pacheco, M., Brito, R. M., Almeraya, R., Yamaguchi-Shinozaki, K., et al. (2004). Stress-induced expression in wheat of the Arabidopsis thaliana DREB1A gene delays water stress symptoms under greenhouse conditions. Genome 47, 493–500. doi: 10.1139/g03-140
Pennisi, E. (2008). Plant genetics. The blue revolution, drop by drop, gene by gene. Science 320, 171–173. doi: 10.1126/science.320.5873.171
Pizzio, G. A., Rodriguez, L., Antoni, R., Gonzalez-Guzman, M., Yunta, C., Merilo, E., et al. (2013). The PYL4 A194T mutant uncovers a key role of PYR1-LIKE4/PROTEIN PHOSPHATASE 2CA interaction for abscisic acid signaling and plant drought resistance. Plant Physiol. 163, 441–455. doi: 10.1104/pp.113.224162
Polizel, A. M., Medri, M. E., Nakashima, K., Yamanaka, N., Farias, J. R., De Oliveira, M. C., et al. (2011). Molecular, anatomical and physiological properties of a genetically modified soybean line transformed with rd29A:AtDREB1A for the improvement of drought tolerance. Genet. Mol. Res. 10, 3641–3656. doi: 10.4238/2011
Qin, F., Sakuma, Y., Tran, L. S., Maruyama, K., Kidokoro, S., Fujita, Y., et al. (2008). Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20, 1693–1707. doi: 10.1105/tpc.107.057380
Raghavendra, A. S., Gonugunta, V. K., Christmann, A., and Grill, E. (2010). ABA perception and signalling. Trends Plant Sci. 15, 395–401. doi: 10.1016/j.tplants.2010.04.006
Rai, M., He, C., and Wu, R. (2009). Comparative functional analysis of three abiotic stress-inducible promoters in transgenic rice. Transgenic Res. 18, 787–799. doi: 10.1007/s11248-009-9263-2
Redillas, M. C., Jeong, J. S., Kim, Y. S., Jung, H., Bang, S. W., Choi, Y. D., et al. (2012). The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol. J. 10, 792–805. doi: 10.1111/j.1467-7652.2012.00697.x
Rusconi, F., Simeoni, F., Francia, P., Cominelli, E., Conti, L., Riboni, M., et al. (2013). The Arabidopsis thaliana MYB60 promoter provides a tool for the spatio-temporal control of gene expression in stomatal guard cells. J. Exp. Bot. 64, 3361–3371. doi: 10.1093/jxb/ert180
Saint Pierre, C., Crossa, J. L., Bonnett, D., Yamaguchi-Shinozaki, K., and Reynolds, M. P. (2012). Phenotyping transgenic wheat for drought resistance. J. Exp. Bot. 63, 1799–1808. doi: 10.1093/jxb/err385
Sakuma, Y., Maruyama, K., Osakabe, Y., Qin, F., Seki, M., Shinozaki, K., et al. (2006a). Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18, 1292–1309. doi: 10.1105/tpc.105.035881
Sakuma, Y., Maruyama, K., Qin, F., Osakabe, Y., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2006b). Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. U.S.A. 103, 18822–18827. doi: 10.1073/pnas.0605639103
Swamy, B. P. M., Ahmed, H. U., Henry, A., Mauleon, R., Dixit, S., Vikram, P., et al. (2013). Genetic, physiological, and gene expression analyses reveal that multiple QTL enhance yield of rice mega-variety IR64 under drought. PLoS ONE 8:e62795. doi: 10.1371/journal.pone.0062795
Takasaki, H., Maruyama, K., Kidokoro, S., Ito, Y., Fujita, Y., Shinozaki, K., et al. (2010). The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol. Genet. Genomics 284, 173–183. doi: 10.1007/s00438-010-0557-0
Tran, L. S., Nakashima, K., Sakuma, Y., Simpson, S. D., Fujita, Y., Maruyama, K., et al. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498. doi: 10.1105/tpc.104.022699
Uga, Y., Sugimoto, K., Ogawa, S., Rane, J., Ishitani, M., Hara, N., et al. (2013). Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45, 1097–1102. doi: 10.1038/ng.2725
Umezawa, T., Nakashima, K., Miyakawa, T., Kuromori, T., Tanokura, M., Shinozaki, K., et al. (2010). Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol. 51, 1821–1839. doi: 10.1093/pcp/pcq156
Umezawa, T., Sugiyama, N., Takahashi, F., Anderson, J. C., Ishihama, Y., Peck, S. C., et al. (2013). Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 6:rs8. doi: 10.1126/scisignal.2003509
Vágújfalvi, A., Galiba, G., Cattivelli, L., and Dubcovsky, J. (2003). The cold-regulated transcriptional activator Cbf3 is linked to the frost-tolerance locus Fr-A2 on wheat chromosome 5A. Mol. Genet. Genomics 269, 60–67. doi: 10.1007/s00438-003-0806-6
Weiner, J. J., Peterson, F. C., Volkman, B. F., and Cutler, S. R. (2010). Structural and functional insights into core ABA signaling. Curr. Opin. Plant Biol. 13, 495–502. doi: 10.1016/j.pbi.2010.09.007
Wu, X., Shiroto, Y., Kishitani, S., Ito, Y., and Toriyama, K. (2009). Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 28, 21–30. doi: 10.1007/s00299-008-0614-x
Xiao, B. Z., Chen, X., Xiang, C. B., Tang, N., Zhang, Q. F., and Xiong, L. Z. (2009). Evaluation of seven function-known candidate genes for their effects on improving drought resistance of transgenic rice under field conditions. Mol. Plant 2, 73–83. doi: 10.1093/mp/ssn068
Xu, Z. Y., Kim, S. Y., Hyeon Do, Y., Kim, D. H., Dong, T., Park, Y., et al. (2013). The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 25, 4708–4724. doi: 10.1105/tpc.113.119099
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803. doi: 10.1146/annurev.arplant.57.032905.105444
Yang, M., and Xiong, L. (2011). Isolation and characterization of a drought-inducible promoter Oshox24P in rice. J. Huazhong Agric. Univ. 30, 525–531.
Yi, N., Kim, Y. S., Jeong, M. H., Oh, S. J., Jeong, J. S., Park, S. H., et al. (2010). Functional analysis of six drought-inducible promoters in transgenic rice plants throughout all stages of plant growth. Planta 232, 743–754. doi: 10.1007/s00425-010-1212-z
Yoshida, T., Fujita, Y., Sayama, H., Kidokoro, S., Maruyama, K., Mizoi, J., et al. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61, 672–685. doi: 10.1111/j.1365-313X.2009.04092.x
Keywords: ABA, transcription factor, signal transduction, abiotic stress, drought
Citation: Nakashima K, Yamaguchi-Shinozaki K and Shinozaki K (2014) The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 5:170. doi: 10.3389/fpls.2014.00170
Received: 04 February 2014; Accepted: 10 April 2014;
Published online: 16 May 2014.
Edited by:
Mukesh Jain, National Institute of Plant Genome Research, IndiaReviewed by:
Alejandra A. Covarrubias, Universidad Nacional Autónoma de México, MexicoKemal Kazan, Commonwealth Scientific and Industrial Research Organization, Australia
Eiji Nambara, University of Toronto, Canada
Copyright © 2014 Nakashima, Yamaguchi-Shinozaki and Shinozaki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kazuo Shinozaki, Gene Discovery Research Group, RIKEN Center for Sustainable Resource Science, 1-7-22 Suehiro, Tsurumi, Yokohama, Kanagawa 230-0045, Japan e-mail: kazuo.shinozaki@riken.jp
 Kazuo Nakashima
Kazuo Nakashima Kazuko Yamaguchi-Shinozaki
Kazuko Yamaguchi-Shinozaki Kazuo Shinozaki
Kazuo Shinozaki