- 1Department of Energy—Plant Research Laboratory, Michigan State University, East Lansing, MI, USA
- 2Department of Biochemistry and Molecular Biology, Michigan State University, East Lansing, MI, USA
Light perception by photoreceptors impacts plastid transcription, development, and differentiation. This photoreceptor-dependent activity suggests a mechanism for photoregulation of gene expression in the nucleus and plastid that serves to coordinate expression of critical genes of these two organelles. This coordinate expression is required for proper stoichiometric accumulation of components needed for assembly of plastids, photosynthetic light-harvesting complexes and components such as phytochromes. Chloroplast-targeted sigma factors, which function together with the plastid-encoded RNA polymerase to regulate expression of plastid-encoded genes, and nuclear-encoded plastid development factors, such as GLK1 and GLK2, are targets of phytochrome regulation. Such phytochrome-dependent functions are hypothesized to allow light-dependent regulation, and feasibly tuning, of plastid components and function in response to changes in the external environment, which directly affects photosynthesis and the potential for light-induced damage. When the size and protein composition of the light-harvesting complexes are not tuned to the external environment, imbalances in electron transport can impact the cellular redox state and cause cellular damage. We show that phytochromes specifically regulate the expression of multiple factors that function to modulate plastid transcription and, thus, provide a paradigm for coordinate expression of the nuclear and plastid genomes in response to changes in external light conditions. As phytochromes respond to changes in the prevalent wavelengths of light and light intensity, we propose that specific phytochrome-dependent molecular mechanisms are used during light-dependent signaling between the nucleus and chloroplast during photomorphogenesis to coordinate chloroplast development with plant developmental stage and the external environment.
Introduction
The involvement of regulatory factors or transcriptional regulators in organismal responses to environmental signals is well known. However, specific information about the distinct factors involved is limited in many cases. For example, light quality and quantity are known to stimulate proplastid or dark-prevalent etioplast to light-dependent chloroplast transitions and the expression of many nuclear-encoded, chloroplast-targeted genes (Pogson and Albrecht, 2011). The general importance for a role of some photoreceptors in this process is known. Early studies demonstrated a role for phytochrome-mediated detection of distinct wavelengths in the light-dependent regulation of chloroplast development (Wellburn and Wellburn, 1973). Additional studies demonstrated that both blue light-responsive cryptochrome and red/far-red light-responsive phytochrome photoreceptors perceive light and stimulate plastid development (Thum et al., 2001), and phytochromes are involved in regulating chloroplast gene transcription in mature leaves (Chun et al., 2001). Consistent with these observations, phytochrome-deficient mutants, including phyB (Reed et al., 1994) and chromophore-deficient hy1 and hy2 mutants (Chory et al., 1989), exhibit defects in chloroplast development and/or differentiation. However, insights into the identity and functions of specific photoreceptor-dependent effectors that impact chloroplast development and function are limited.
Functional plastid development depends upon tight regulation of the expression of nuclear-encoded and plastid genome-encoded photosynthetic genes in the proper stoichiometry, together with the synthesis of photosynthetic pigments (Pogson and Albrecht, 2011). A limited number of factors known to function downstream of the photoreceptors in this process have been identified, including two transcription factors linked to phytochrome function, i.e., PIF1 and PIF3, that have been shown to function as regulators of light-dependent chloroplast development (Monte et al., 2004; Moon et al., 2008; Stephenson et al., 2009). Other transcription factors previously shown to impact the transcription of photosynthesis genes, and thereby chloroplast development, include nuclear Golden2-like (GLK) factors, i.e., GLK1 and GLK2 (Waters et al., 2008), and two plastid-targeted sigma factor (SIG) proteins, i.e., SIG2 and SIG6 (Kanamaru et al., 2001; Ishizaki et al., 2005). GLKs regulate the expression of nuclear-encoded photosynthetic genes (Waters et al., 2008, 2009). SIGs serve as promoter specificity factors that regulate the activity of the plastid-encoded RNA polymerase or PEP (Kanamaru et al., 1999; Hanaoka et al., 2003), which serves to drive expression of genes encoding photosynthetic proteins (Jarvis and López-Juez, 2013).
The expression of GLK (Fitter et al., 2002) and SIG (Isono et al., 1997; Tsunoyama et al., 2002; Privat et al., 2003; Ishizaki et al., 2005) genes is light-dependent. Notably, GLK1 and GLK2 exhibit distinctions, i.e., GLK1 is primarily regulated by light, whereas GLK2 appears to be regulated both by light and diurnal cues (Fitter et al., 2002). Furthermore, GLK genes have been shown to have a role in acclimation to light intensity (Waters et al., 2009). The only prior reported connection of these genes to a specific photoreceptor was the regulation of SIG5 primarily by cryptochromes (Onda et al., 2008) and our recent connection of phytochromes to the regulation of SIG2 expression (Oh and Montgomery, 2013). The photoreceptor-dependent regulation of such factors is presumed to be critical for “adjusting” or matching chloroplast development and composition to the external environment and to integrating this process with the organismal energy budget.
Plastid biogenesis and development are coordinated with the external environment to optimize plastid-dependent processes such as photosynthesis (Pogson and Albrecht, 2011). Coordinating plastid development with external light impacts the utilization of light for the production of chemical energy during photosynthesis and/or the limitation of photodamage/photoprotection in plants (Pogson and Albrecht, 2011). A failure to coordinate the composition and size of the light-harvesting complexes with the environment can result in the generation of damaging reactive oxygen species. Maintaining the proper stoichiometry of nuclear proteins, plastid proteins, chlorophylls, and carotenoids is critical for assembly of functional photosynthetic and photoprotective complexes in plastids. Thus, factors which coordinate expression of nuclear- and plastid-encoded components of the light-harvesting complexes must have central roles in organismal light responses and photoregulation of chloroplast development. Yet little insight has been gained into the molecular nature of factors that serve as central components of the light-dependent mechanism(s) utilized to coordinate transcription of nuclear- and chloroplast-encoded genes.
Based on recent results which demonstrated that phytochromes regulate the accumulation of photosynthetic proteins encoded by genes from both the nuclear genome and the chloroplast genome (Oh and Montgomery, 2011) and the expression of SIG2 that encodes a chloroplast transcriptional regulator (Oh and Montgomery, 2013), we investigated the role of phytochromes in regulating other factors that affect plastid development and/or transcription. We sought to elucidate the phytochrome-dependent photoregulation of anterograde signaling between the nucleus and plastid. We determined that phytochromes regulate the expression of another chloroplast transcriptional regulator, SIG6, and a suite of other regulatory and developmental factors that impact plastid transcription and development. These results provide insights into the molecular basis of the central role of phytochromes in coordinating gene expression in the nucleus and chloroplasts during photomorphogenesis. Furthermore, these findings support a hypothesis that phytochromes serve as central integrators of information about the external light environment over time and space to allow plants to finely coordinate plastid function to optimize light capture for photosynthesis, while simultaneously minimizing the potential for light-associated damage and phototoxicity.
Materials and Methods
Experimental Organism and Growth Conditions
Transgenic BVR lines, i.e., 35S::pBVR3 (Montgomery et al., 1999) and CAB3::pBVR2 (Warnasooriya and Montgomery, 2009), and T-DNA insertion mutants, i.e., phyA (Mayfield et al., 2007; Ruckle et al., 2007), phyB (Mayfield et al., 2007; Ruckle et al., 2007), and double mutant phyAphyB (Oh and Montgomery, 2013), were previously described. Sterilized seeds were planted and seedlings grown on MS medium containing 1% (w/v) sucrose and 0.7% (w/v) Phytoblend agar (Caisson Labs, UT) at 22°C for 7 days under the indicated light condition as previously described (Oh and Montgomery, 2013). Light sources utilized for far-red (FR; λ max ~735 nm), red (R; λmax ~670 nm), and white (W) light were described previously (Warnasooriya and Montgomery, 2009). Fluence rates of R, and W were measured using a LI-250A Light Meter (LI-COR) connected to a LI-COR quantum sensor and for FR using a StellarNet EPP2000 spectroradiometer (Apogee Instruments).
Total RNA Extraction and qRT-PCR Analysis
RNA samples were extracted from 7-days-old whole seedlings grown under continuous darkness (Dc), continuous FR (FRc) illumination (5 μmol m−2s−1), continuous R (Rc) illumination (50 μmol m−2s−1), or white (W; 100 μmol m−2s−1) using the RNeasy® Plant Minikit (Qiagen, CA) as previously described (Oh and Montgomery, 2013). Quantitative RT-PCR (qRT-PCR) was performed essentially as described previously (Oh and Montgomery, 2013). Briefly, cDNA was synthesized using total RNA (100 ng) and random primers using a Reverse Transcription System (Promega, WI) by following the manufacturer's instructions. The cDNA was then mixed with Fast SYBR® Green Master Mix (Applied Biosystems) and qPCR performed in three technical and three biological replicates using an ABI 7500 Fast Real-Time PCR System (Applied Biosystems). UBC21 was used as a normalization standard in all qRT-PCR experiments. The primers used for qRT-PCR are indicated in Table 1.
Results
Phytochromes Impact the Expression of Multiple Nuclear-Encoded Genes Encoding Chloroplast-Targeted Sigma Factors
We previously demonstrated that the expression of chloroplast-targeted transcriptional regulator SIG2 is regulated by phytochromes (Oh and Montgomery, 2013). SIG2 is one of six Arabidopsis sigma factors targeted to the chloroplast, whose activity regulates the plastid-encoded RNA polymerase, or PEP (Kanamaru et al., 1999; Allison, 2000). These factors generally serve as promoter-specificity factors (Hanaoka et al., 2003). The level of SIG6 mRNA also is phytochrome dependent, as its accumulation was significantly lower in a transgenic Arabidopsis line depleted of mesophyll-localized phytochromes through CAB3-promoter-driven expression of a gene encoding a phytochrome chromophore-degrading enzyme biliverdin reductase (BVR) (Figure 1A). As observed for SIG2 mRNA accumulation (Oh and Montgomery, 2013), we confirmed the downregulation of SIG6 expression by qRT-PCR in lines depleted of mesophyll-localized phytochromes relative to WT (Figure 1B). We also demonstrate roles for phyA and phyB in the photoregulation of SIG6 expression by assessing accumulation of SIG6 mRNA in phyA, phyB, and phyAphyB mutants. SIG6 was downregulated by ~2-fold in either single phy mutants or by 3-fold in the phyAphyB double mutant relative to WT (Figure 1C).
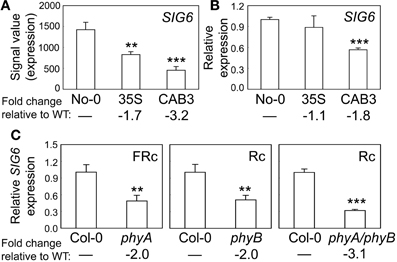
Figure 1. Phytochrome-dependent regulation of SIG6 expression. (A) Expression levels (signal value) of SIG6 in No-0 wild-type (WT), 35S::pBVR3 (35S), and CAB3::pBVR2 (CAB3) under continuous far-red (FRc) are shown (±SD, n = 3). Signal value indicates signal intensity on the ATH1 array as calculated by Affymetrix Microarray Suite (MAS). (B) Validation of microarray data for SIG6 using quantitative RT-PCR (qRT-PCR) analysis. Relative SIG6 expression level compared to UBC21 is shown (±SD, n = 3). (C) qRT-PCR analysis of SIG6 expression in Col-0 WT, phyA (SALK_014575), phyB (SALK_022035), or phyA/phyB double mutant seedlings under FRc or continuous red (Rc). Relative SIG6 expression level compared to UBC21 is shown (±SD, n = 3). Statistics: Unpaired, two-tailed Student's t-test comparing mutants or transgenic lines to WT, **p < 0.01, ***p < 0.005.
The expression of SIG6 in regards to developmental stage and tissue specificity is very similar to that of SIG2 (Oh and Montgomery, 2013), i.e., highest expression for SIG6 was observed in cotyledons, young leaf tissue, and adult rosette leaves (Figure 2A). Light regulation of SIG6 expression was also similar to SIG2 (Figure 2B; Oh and Montgomery, 2013). For 7-days-old seedlings, the highest accumulation of SIG6 mRNA occurred under Rc and W illumination (Figure 2C), again very similar to what was reported for SIG2 (Oh and Montgomery, 2013).
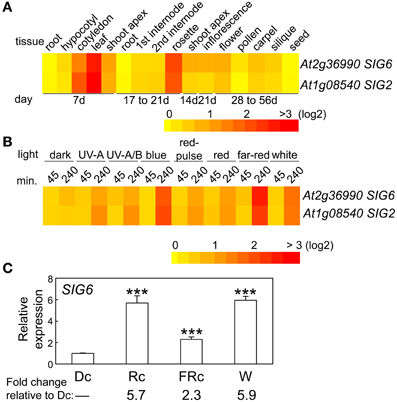
Figure 2. Expression of SIG6 in different tissues and light conditions. (A) Heat map showing the expression of SIG6 in different tissues indicating mean-normalized values of Col-0 WT from AtGenExpress expression library (www.weigelworld.org) and BAR Heatmapper Plus (bar.utoronto.ca). (B) Heat map showing the expression of SIG6 in different light conditions for aerial parts (hypocotyl and cotyledons) of 4-days-old Col-0 WT seedlings grown on MS medium treated with different light for either 45 or 240 min. (C) qRT-PCR analysis of SIG6 expression in Col-0 WT under dark (Dc), Rc, FRc, or white (W). Relative SIG6 expression level compared to UBC21 is shown (±SD, n = 3). Statistics: Unpaired, two-tailed Student's t-test comparing different growth conditions ***p < 0.005.
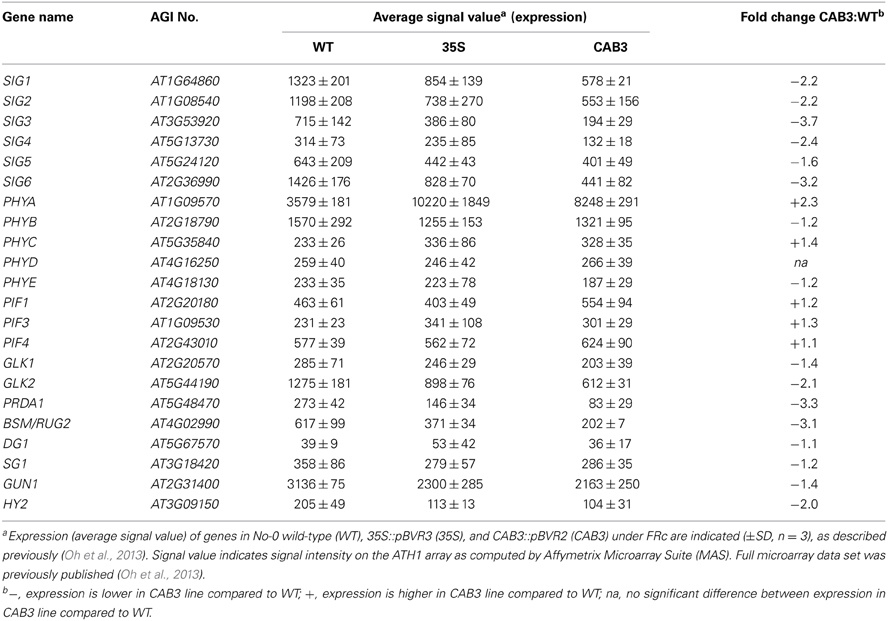
Although SIG2 and SIG6 have been shown previously to have the most significant effect on chlorophyll accumulation and plastid development among SIG family members (Kanamaru et al., 2001; Ishizaki et al., 2005), we assessed the impact of functional phytochromes on the expression of the remaining SIG family members. The expression of additional SIG genes was also impacted by mesophyll phytochrome depletion (Table 2). SIG1, SIG3, and SIG4 were downregulated by 2.2-, 3.7-, and 2.4-fold, respectively, in the CAB3::pBVR2 line relative to WT (Table 2). SIG5 was less impacted than other SIG genes for the FRc conditions under which we grew plants, i.e., it was downregulated by only 1.6-fold (Table 2). However, expression of the SIG5 gene has been previously reported to be mostly regulated by blue light (Tsunoyama et al., 2002, 2004; Onda et al., 2008). The blue light effects on the expression of SIG5-dependent chloroplast genes were characterized as regulated by both cryptochromes and phytochrome A (Thum et al., 2001; Onda et al., 2008).
Phytochromes Impact the Expression of Nuclear GLK Genes, which Regulate Chloroplast Development
A number of genes previously demonstrated to be involved in chloroplast development also were identified as misregulated in our microarray analysis assessing lines depleted of phytochromes (Table 2). There were minimal effects on the expression of PIF1 and PIF3 under FRc. However, other genes previously shown to impact plastid development were downregulated when mesophyll phytochromes were depleted. Specifically, we identified misregulation of the expression of two nuclear transcription factors, i.e., Golden2-like 1 (GLK1) and Golden2-like 2 (GLK2) (Table 2), which have been previously shown to impact expression of nuclear photosynthetic genes linked to chloroplast development (Waters et al., 2008, 2009). In agreement with our observed link to phytochrome activity, prior analysis of publicly available microarray data indicated that expression of GLK1 and GLK2 is promoted by red and blue light (Waters et al., 2008). Expression of GLK2 was significantly downregulated in lines depleted of mesophyll-localized phytochromes (Figure 3A; Oh and Montgomery, 2013). A greater impact on GLK2 expression than GLK1 was confirmed by qRT-PCR analyses (Figure 3B). Notably, both GLK1 and GLK2 expression was significantly downregulated in a phyAphyB double mutant under Rc light (Figure 3C), definitively indicating that expression of both GLK genes is impacted by phytochromes in vivo.
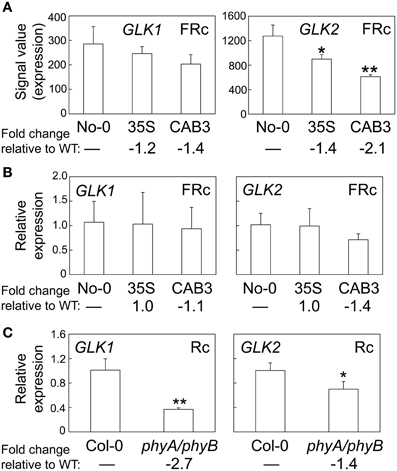
Figure 3. Phytochrome-dependent regulation of GLK1 and GLK2 expression. (A) Expression levels (signal value) of GLK1 or GLK2 in No-0 WT, 35S, and CAB3 under FRc are shown (±SD, n = 3). (B) qRT-PCR validation of microarray data for GLK1 or GLK2 under FRc. Relative GLK1 or GLK2 expression level compared to UBC21 is shown (±SD, n = 3). (C) qRT-PCR quantification of GLK1 or GLK2 expression in Col-0 WT or phyA/phyB double mutant seedlings under Rc. Relative GLK1 or GLK2 expression level compared to UBC21 is shown (±SD, n = 3). Statistics: Unpaired, two-tailed Student's t-test comparing mutants or transgenic lines to WT, *p < 0.05, **p < 0.01.
The two GLK genes are predicted to have a similar distribution of mRNA accumulation in different tissues and at different developmental stages (Figure 4A). Both genes are highly expressed in cotyledons, young leaf tissues, adult rosette leaves, and internodes (Figure 4A). However, expression of GLK2 appears to be more significantly impacted by light than GLK1 (Figure 4B), at least in the 4-days-old seedlings used for the experiments included in the heat map. We analyzed expression of the two GLK genes in 7-days-old seedlings to assess the impact of light on their expression for seedlings of the same age as those used in the microarray analysis in which we noted a phytochrome-dependent regulation of GLK expression. In these analyses, we observed a significant impact of Rc and W light on GLK1 expression, whereas FRc light had no significant effect on GLK1 expression (Figure 4C). By comparison, GLK2 was significantly upregulated by Rc, FRc, and W light relative to darkness, although the impact of Rc and W were much more robust than FRc (Figure 4D).
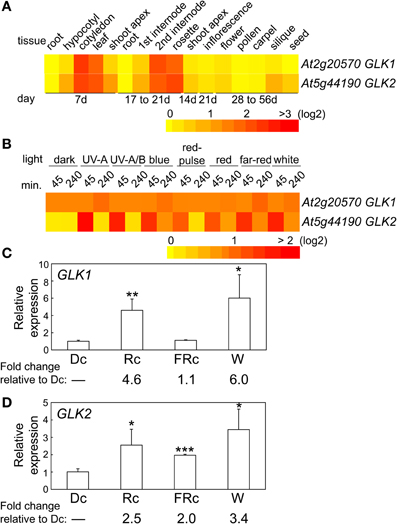
Figure 4. Expression of GLK1 and GLK2 in different tissues and light conditions. (A) Heat map showing the expression of GLK1 or GLK2 in different tissues indicating mean-normalized values of Col-0 WT from AtGenExpress expression library (www.weigelworld.org) and BAR Heatmapper Plus (bar.utoronto.ca). (B) Heat map showing the expression of GLK1 or GLK2 in different light conditions for aerial parts (hypocotyl and cotyledons) of 4-days-old Col-0 WT seedlings grown on MS medium treated with different light for either 45 or 240 min. qRT-PCR analysis of (C) GLK1 or (D) GLK2 expression in Col-0 WT under dark (Dc), Rc, FRc, or white (W). Relative GLK1 or GLK2 expression level compared to UBC21 is shown (±SD, n = 3). Statistics: Unpaired, two-tailed Student's t-test comparing different growth conditions *p < 0.05, **p < 0.01, ***p < 0.005.
Phytochromes Impact the Expression of Additional Chloroplast Development Genes
Additional plastid-targeted factors that have been associated with light-dependent regulation of plastid development were impacted by mesophyll-localized phytochrome function. These genes include PEP-Related Development Arrested 1 (PRDA1) and BELAYA SMERT/RUGOSA2 (BSM/RUG2). PRDA1 is a nuclear genome-encoded gene which encodes a plastid-targeted protein that impacts plastid transcription (Qiao et al., 2013). Co-expression analyses and protein interaction studies indicated that PRDA1 functions in PEP-related chloroplast transcriptional regulation to affect plastid development (Qiao et al., 2013). Notably, a prda1 mutant exhibits a similar change in plastid gene expression to other PEP mutants, including sig6 (Qiao et al., 2013). The accumulation of mRNA for PRDA1 was downregulated by 3.3-fold in lines depleted of mesophyll phytochromes under FRc conditions (Table 2), suggesting that this chloroplast developmental regulator is also a target of phytochrome regulation.
BSM/RUG2 is a nuclear gene that encodes a plastid-targeted protein of the mitochondrial transcription termination factor (mTERF) family, which impacts plastid gene expression (Babiychuk et al., 2011; Quesada et al., 2011). Expression of BSM/RUG2 is positively regulated by phytochromes, as the gene is downregulated by 3.1-fold in a mesophyll phytochrome-depleted line (Table 2). The only light condition previously reported to impact BSM/RUG2 expression was high light (Quesada et al., 2011). Thus, the regulation of expression of this gene by phytochrome may be correlated with the ability of this photoreceptor family to respond to light intensity, in addition to light quality (Fankhauser, 2001).
It should be noted that the impact of phytochrome appears to be specific to some chloroplast development genes, while apparently having no influence on others. Two genes encoding a pentratricopeptide repeat (PPR) protein and a tetratricopeptide protein (TPR) that have been shown to impact chloroplast development show distinct patterns in our array data for lines depleted of mesophyll phytochromes. The expression of PPR-encoding gene DELAYED GREENING1 (DG1) has been shown to be light-dependent and to impact PEP-dependent transcription of chloroplast genes (Chi et al., 2008). TPR-encoding SLOW GREEN1 (SG1) also impacts expression of chloroplast- and nuclear-encoded genes (Hu et al., 2014). However, the expression of DG1 and SG1 are not significantly impacted in the CAB3::pBVR2 line relative to WT (Table 2). These results suggest that phytochromes selectively regulate the expression of particular genes which impact chloroplast development and differentiation, while having no direct impact on others. It should be noted, however, that DG1 has been shown to interact with SIG6, and thus DG1 function may be indirectly impacted by phytochromes through the phytochrome-dependent regulation of SIG6 expression (Chi et al., 2010).
Discussion
We have identified a number of nuclear factors and nuclear-encoded, plastid-targeted factors that impact chloroplast development and whose expression is regulated in a light-dependent manner by phytochromes (Figures 1–4; Oh and Montgomery, 2013). The expression of factors centrally involved in the early stages of light-dependent chloroplast development, including SIG2, SIG6, GLK1, and GLK2, is most highly upregulated under red and white light (Figures 1–4; Oh and Montgomery, 2013). Notably, red light was defined very early as the condition under which chloroplast development is most significantly stimulated (Wellburn and Wellburn, 1973). Chlorophyll-deficient phenotypes (Kanamaru et al., 2001; Ishizaki et al., 2005; Waters et al., 2008, 2009; Babiychuk et al., 2011; Quesada et al., 2011; Oh and Montgomery, 2013; Oh et al., 2013; Qiao et al., 2013; Woodson et al., 2013) or high light-sensitive phenotypes (Warnasooriya and Montgomery, 2009; Waters et al., 2009; Quesada et al., 2011) have been noted for lines with attenuated or complete knockdown of the expression of phytochrome-regulated genes described here. These findings support solid hypotheses about the mechanistic bases of interactions between photomorphogenesis and chloroplast development at the molecular level. We hypothesize that phytochromes regulate a number of factors that serve as a central part of the mechanism to coordinate transcription of nuclear genes and chloroplast genes to ensure the correct stoichiometric production of components of the photosynthetic light harvesting complexes and the production of components, such as phytochromes themselves, which depend upon products from these two organelles (Figure 5).
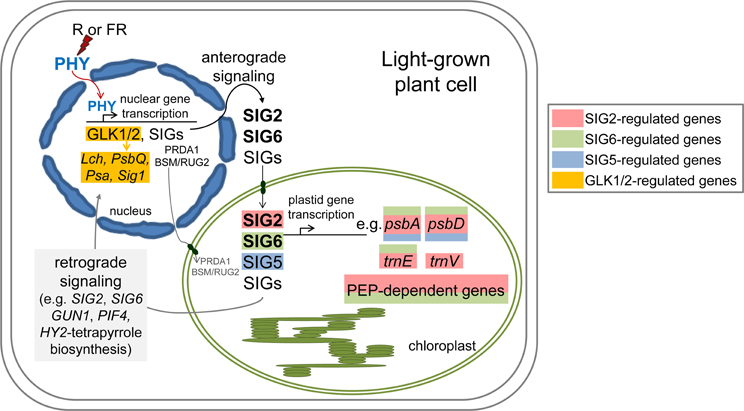
Figure 5. Model of light- and phytochrome-dependent regulation of anterograde signaling between the nucleus and plastid during chloroplast development and photomorphogenesis. PEP, Plastid-encoded plastid RNA polymerase; PHY, phytochrome; R, red; FR, far-red.
It was previously suggested that such light- and photoreceptor-dependent regulation of GLK expression may serve as a mechanism for adjusting photosynthesis in cells with environmental inputs (Waters et al., 2008). Notably, the phenotype of a double glk1glk2 mutant is distinct from BVR or sig phenotypes, as the small and large subunits of Rubisco accumulate to normal levels in a glk1glk2 mutant (Waters et al., 2009), but are disrupted in lines depleted of mesophyll phytochromes (Oh and Montgomery, 2011) and sig2 mutants (Kanamaru et al., 2001). Also, there is a defective etioplast phenotype for the double glk1glk2 mutant (Waters et al., 2009) and for the pif1pif3 double mutant (Stephenson et al., 2009), but no apparent dark-dependent phenotype for sig2 (Kanamaru et al., 2001) or sig6 mutants (Ishizaki et al., 2005). Interestingly, a role for SIG2 in coordinating expression of photosynthetic light-harvesting genes between nuclear and chloroplast genomes also has recently been observed in the red alga Cyanidioschyzon merolae (Fujii et al., 2013), extending a role of this protein in light-dependent anterograde signaling between the nucleus and chloroplasts beyond higher plants. SIG2 expression in C. merolae is also light-induced (Fujii et al., 2013), suggesting that a similar photoreceptor-dependent mechanism for regulating SIG accumulation (Figure 5), and thus coordination of light-harvesting complex assembly, may be employed in C. merolae. It has been suggested that photoregulation of the expression of SIG factors may also contribute to the observed light-dependent activation of some PEP-associated, plant-specific factors, including plastid transcriptionally active chromosome (pTAC) components, that impact plastid transcription (Yagi et al., 2012). Such regulatory mechanisms are hypothesized to allow modulation of chloroplast development in response to adverse environmental conditions (Yagi and Shiina, 2014).
Although not reported for all of the genes described to function in phytochrome-dependent anterograde signaling between the nucleus and plastids, SIG2 and SIG6 also have been reported to function in retrograde signaling, in which the functional state of plastids can feed back to impact expression of nuclear-encoded photosynthetic genes (Woodson et al., 2013; Figure 5). Thus, SIG2 and SIG6 have central roles in the establishment of plastid function during photomorphogenesis and in surveillance of plastid function during growth. Whether other proteins controlled by phytochromes during anterograde signaling have similar dual roles in retrograde signaling is not yet well understood.
Phytochrome-dependent regulation of anterograde signaling between the nucleus and plastids appears to be important for both development and differentiation of chloroplasts, as well as fine tuning of the organelles in response to fluctuations in environmental parameters such as light intensity. These results provide significant new information about the molecular mechanisms used for light-dependent anterograde signaling between the nucleus and chloroplast during photomorphogenesis. Additional studies to identify and characterize the distinct photoreceptors and photoreceptor-dependent effectors that impact the full complement of factors that function in the light-dependent coordination of gene expression in the nucleus and chloroplast will be required for a more complete understanding of this vital process.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the US Department of Energy, Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science [grant no. DE–FG02–91ER20021 to Beronda L. Montgomery].
References
Allison, L. A. (2000). The role of sigma factors in plastid transcription. Biochimie 82, 537–548. doi: 10.1016/S0300-9084(00)00611-8
Babiychuk, E., Vandepoele, K., Wissing, J., Garcia-Diaz, M., De Rycke, R., Akbari, H., et al. (2011). Plastid gene expression and plant development require a plastidic protein of the mitochondrial transcription termination factor family. Proc. Natl. Acad. Sci. U.S.A. 108, 6674–6679. doi: 10.1073/pnas.1103442108
Chi, W., Ma, J., Zhang, D., Guo, J., Chen, F., Lu, C., et al. (2008). The pentratricopeptide repeat protein DELAYED GREENING1 is involved in the of early chloroplast development and chloroplast gene expression in Arabidopsis. Plant Physiol. 147, 573–584. doi: 10.1104/pp.108.116194
Chi, W., Mao, J., Li, Q., Ji, D., Zou, M., Lu, C., et al. (2010). Interaction of the pentatricopeptide-repeat protein DELAYED GREENING 1 with sigma factor SIG6 in the regulation of chloroplast gene expression in Arabidopsis cotyledons. Plant J. 64, 14–25. doi: 10.1111/j.1365-313X.2010.04304.x
Chory, J., Peto, C. A., Ashbaugh, M., Saganich, R., Pratt, L., and Ausubel, F. (1989). Different roles for phytochrome in etiolated and green plants deduced from characterization of Arabidopsis thaliana mutants. Plant Cell 1, 867–880.
Chun, L., Kawakami, A., and Christopher, D. A. (2001). Phytochrome A mediates blue light and UV-A-dependent chloroplast gene transcription in green leaves. Plant Physiol. 125, 1957–1966. doi: 10.1104/pp.125.4.1957
Fankhauser, C. (2001). The phytochromes, a family of red/far-red absorbing photoreceptors. J. Biol. Chem. 276, 11453–11456. doi: 10.1074/jbc.R100006200
Fitter, D. W., Martin, D. J., Copley, M. J., Scotland, R. W., and Langdale, J. A. (2002). GLK gene pairs regulate chloroplast development in diverse plant species. Plant J. 31, 713–727. doi: 10.1046/j.1365-313X.2002.01390.x
Fujii, G., Imamura, S., Hanaoka, M., and Tanaka, K. (2013). Nuclear-encoded chloroplast RNA polymerase sigma factor SIG2 activates chloroplast-encoded phycobilisome genes in a red alga, Cyanidioschyzon merolae. FEBS Lett. 587, 3354–3359. doi: 10.1016/j.febslet.2013.08.031
Hanaoka, M., Kanamaru, K., Takahashi, H., and Tanaka, K. (2003). Molecular genetic analysis of chloroplast gene promoters dependent on SIG2, a nucleus-encoded sigma factor for the plastid-encoded RNA polymerase, in Arabidopsis thaliana. Nucleic Acids Res. 31, 7090–7098. doi: 10.1093/nar/gkg935
Hu, Z., Xu, F., Guan, L., Qian, P., Liu, Y., Zhang, H., et al. (2014). The tetratricopeptide repeat-containing protein slow green1 is required for chloroplast development in Arabidopsis. J. Exp. Bot. 65, 1111–1123. doi: 10.1093/jxb/ert463
Ishizaki, Y., Tsunoyama, Y., Hatano, K., Ando, K., Kato, K., Shinmyo, A., et al. (2005). A nuclear-encoded sigma factor, Arabidopsis SIG6, recognizes sigma-70 type chloroplast promoters and regulates early chloroplast development in cotyledons. Plant J. 42, 133–144. doi: 10.1111/j.1365-313X.2005.02362.x
Isono, K., Shimizu, M., Yoshimoto, K., Niwa, Y., Satoh, K., Yokota, A., et al. (1997). Leaf-specifically expressed genes for polypeptides destined for chloroplasts with domains of sigma70 factors of bacterial RNA polymerases in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 94, 14948–14953. doi: 10.1073/pnas.94.26.14948
Jarvis, P., and López-Juez, E. (2013). Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14, 787–802. doi: 10.1038/nrm3702
Kanamaru, K., Fujiwara, M., Seki, M., Katagiri, T., Nakamura, M., Mochizuki, N., et al. (1999). Plastidic RNA polymerase sigma factors in Arabidopsis. Plant Cell Physiol. 40, 832–842. doi: 10.1093/oxfordjournals.pcp.a029612
Kanamaru, K., Nagashima, A., Fujiwara, M., Shimada, H., Shirano, Y., Nakabayashi, K., et al. (2001). An Arabidopsis sigma factor (SIG2)-dependent expression of plastid-encoded tRNAs in chloroplasts. Plant Cell Physiol. 42, 1034–1043. doi: 10.1093/pcp/pce155
Mayfield, J. D., Folta, K. M., Paul, A.-L., and Ferl, R. J. (2007). The 14-3-3 proteins m and u influence transition to flowering and early phytochrome response. Plant Physiol. 145, 1692–1702. doi: 10.1104/pp.107.108654
Monte, E., Tepperman, J. M., Al-Sady, B., Kaczorowski, K. A., Alonso, J. M., Ecker, J. R., et al. (2004). The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc. Natl. Acad. Sci. U.S.A. 101, 16091–16098. doi: 10.1073/pnas.0407107101
Montgomery, B. L., Yeh, K. C., Crepeau, M. W., and Lagarias, J. C. (1999). Modification of distinct aspects of photomorphogenesis via targeted expression of mammalian biliverdin reductase in transgenic Arabidopsis plants. Plant Physiol. 121, 629–639. doi: 10.1104/pp.121.2.629
Moon, J., Zhu, L., Shen, H., and Huq, E. (2008). PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 105, 9433–9438. doi: 10.1073/pnas.0803611105
Oh, S., and Montgomery, B. L. (2011). Identification of proteins associated with spatial-specific phytochrome-mediated light signaling in Arabidopsis thaliana by liquid chromatography-tandem mass spectrometry. Gravit. Space Biol. 25, 22–32.
Oh, S., and Montgomery, B. L. (2013). Phytochrome-induced SIG2 expression contributes to photoregulation of phytochrome signaling and photomorphogenesis in Arabidopsis thaliana. J. Exp. Bot. 64, 5457–5472. doi: 10.1093/jxb/ert308
Oh, S., Warnasooriya, S. N., and Montgomery, B. L. (2013). Downstream effectors of light- and phytochrome-dependent regulation of hypocotyl elongation in Arabidopsis thaliana. Plant Mol. Biol. 81, 627–640. doi: 10.1007/s11103-013-0029-0
Onda, Y., Yagi, Y., Saito, Y., Takenaka, N., and Toyoshima, Y. (2008). Light induction of Arabidopsis SIG1 and SIG5 transcripts in mature leaves: differential roles of cryptochrome 1 and cryptochrome 2 and dual function of SIG5 in the recognition of plastid promoters. Plant J. 55, 968–978. doi: 10.1111/j.1365-313X.2008.03567.x
Pogson, B. J., and Albrecht, V. (2011). Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiol. 155, 1545–1551. doi: 10.1104/pp.110.170365
Privat, I., Hakimi, M. A., Buhot, L., Favory, J. J., and Mache-Lerbs, S. (2003). Characterization of Arabidopsis plastid sigma-like transcription factors SIG1, SIG2 and SIG3. Plant Mol. Biol. 51, 385–399. doi: 10.1023/A:1022095017355
Qiao, J., Li, J., Chu, W., and Luo, M. (2013). PRDA1, a novel chloroplast-nucleoid protein, is required for early chloroplast development and is involved in the regulation of plastid gene expression in Arabidopsis. Plant Cell Physiol. 54, 2071–2084. doi: 10.1093/pcp/pct148
Quesada, V., Sarmiento-Mañús, R., González-Bayón, R., Hricová, A., Pérez-Marcos, R., Graciá-Martínez, E., et al. (2011). Arabidopsis RUGOSA2 encodes an mTERF family member required for mitochondrion, chloroplast and leaf development. Plant J. 68, 738–753. doi: 10.1111/j.1365-313X.2011.04726.x
Reed, J. W., Nagatani, A., Elich, T. D., Fagan, M., and Chory, J. (1994). Phytochrome A and Phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 104, 1139–1149.
Ruckle, M. E., Demarco, S. M., and Larkin, R. M. (2007). Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 19, 3944–3960. doi: 10.1105/tpc.107.054312
Stephenson, P. G., Fankhauser, C., and Terry, M. J. (2009). PIF3 is a repressor of chloroplast development. Proc. Natl. Acad. Sci. U.S.A. 106, 7654–7659. doi: 10.1073/pnas.0811684106
Thum, K. E., Kim, M., Christopher, D. A., and Mullet, J. E. (2001). Cryptochrome 1, cryptochrome 2, and phytochrome A co-activate the chloroplast psbD blue light–responsive promoter. Plant Cell 13, 2747–2760. doi: 10.1105/tpc.010345
Tsunoyama, Y., Ishizaki, Y., Morikawa, K., Kobori, M., Nakahira, Y., Takeba, G., et al. (2004). Blue light-induced transcription of plastid-encoded psbD gene is mediated by a nuclear-encoded transcription initiation factor, AtSig5. Proc. Natl. Acad. Sci. U.S.A. 101, 3304–3309. doi: 10.1073/pnas.0308362101
Tsunoyama, Y., Morikawa, K., Shiina, T., and Toyoshima, Y. (2002). Blue light specific and differential expression of a plastid sigma factor, Sig5 in Arabidopsis thaliana. FEBS Lett. 516, 225–228. doi: 10.1016/S0014-5793(02)02538-3
Warnasooriya, S. N., and Montgomery, B. L. (2009). Detection of spatial-specific phytochrome responses using targeted expression of biliverdin reductase in Arabidopsis. Plant Physiol. 149, 424–433. doi: 10.1104/pp.108.127050
Waters, M. T., Moylan, E. C., and Langdale, J. A. (2008). GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant J. 56, 432–444. doi: 10.1111/j.1365-313X.2008.03616.x
Waters, M. T., Wang, P., Korkaric, M., Capper, R. G., Saunders, N. J., and Langdale, J. A. (2009). GLK transcription factors coordinated expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21, 1109–1128. doi: 10.1105/tpc.108.065250
Wellburn, F. A., and Wellburn, A. (1973). Response of etiplasts in situ and in isolated suspensions to pre-illumination with various combinations of red, far-red and blue light. New Phytol. 72, 55–60. doi: 10.1111/j.1469-8137.1973.tb02010.x
Woodson, J. D., Perez-Ruiz, J. M., Schmitz, R. J., Ecker, J. R., and Chory, J. (2013). Sigma factor-mediated plastid retrograde signals control nuclear gene expression. Plant J. 73, 1–13. doi: 10.1111/tpj.12011
Yagi, Y., Ishizaki, Y., Nakahira, Y., Tozawa, Y., and Shiina, T. (2012). Eukaryotic-type plastid nucleoid protein pTAC3 is essential for transcription by the bacterial-type plastid RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 109, 7541–7546. doi: 10.1073/pnas.1119403109
Keywords: anterograde signaling, light signaling, nuclear gene expression, plastid gene expression, phytochrome, sigma factor
Citation: Oh S and Montgomery BL (2014) Phytochrome-dependent coordinate control of distinct aspects of nuclear and plastid gene expression during anterograde signaling and photomorphogenesis. Front. Plant Sci. 5:171. doi: 10.3389/fpls.2014.00171
Received: 07 March 2014; Accepted: 10 April 2014;
Published online: 30 April 2014.
Edited by:
Zuhua He, Chinese Academy of Sciences, ChinaReviewed by:
Zhong-Nan Yang, Shanghai Normal University, ChinaHongtao Liu, Chinese Academy of Science, China
Copyright © 2014 Oh and Montgomery. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beronda L. Montgomery, MSU-DOE Plant Research Laboratory, Plant Biology Laboratories, Michigan State University, 612 Wilson Road, Rm. 106, East Lansing, MI 48824-1312, USA e-mail: montg133@msu.edu
 Sookyung Oh1
Sookyung Oh1 Beronda L. Montgomery
Beronda L. Montgomery