- Jingchu University of Technology, Jingmen, China
Chromodomain-Helicase-DNA (CHD)-binding proteins have been characterized in various species as important transcription regulators by their chromatin remodeling activity. However, in plant the function of these proteins has hardly been analyzed before except that Arabidopsis PIKLE and rice CHR729 are identified to play critical roles in the regulation of series of genes involved in developmental or stress responding process. In this review we focus on how plant CHD proteins regulate gene expression and the role of these proteins in controlling plant development and stress response.
Introduction
Chromodomain Helicase DNA (CHD)-binding proteins, which are Snf2 family ATP-dependent chromatin remodeling factors, play important roles in the regulation of gene expression. Sequence analysis indicates that CHD proteins contain double chromodomains and Helicase-like region comprising SNF2_N and Helicase_C domains, which are critical for chromatin remodeling activity of the proteins. Based on the structure and function conservation CHD proteins could be divided into three subfamilies: Chd1 subfamily (also named CHD1 protein), Mi-2 subfamily (also named CHD3 protein), and CHD7 subfamily (Flaus et al., 2006). In addition to the common domains described above each subfamily members bear diverse extra domains (Figure 1). Double chromodomains of human CHD1 are associated with methylated histone H3 lysine 4 (H3K4me) which is a hallmark of active chromatin (Flanagan et al., 2005). Yeast Chd1 regulates transcription by interacting with SAGA complex which contains histone acetyltransferase and Drosophila CHD1 is required for histone H3.3 deposition demonstrating that the proteins in Chd1 subfamily are positively involved in transcription regulation (Pray-Grant et al., 2005; Konev et al., 2007). In Mi-2 subfamily, two PHD finger domains at N-terminal of human CHD4 bind to histone tails, while the double chromodomains of dMi-2 display DNA binding activity (Bouazoune et al., 2002; Mansfield et al., 2011). The effect of CHD3 protein on gene transcription seems to be complicated and will be discussed later. The proteins in CHD7 subfamily are homologs of Drosophila Trithorax-group protein, kismet, which are exclusively found in animals and also implicated in the regulation of gene expression in concert with other proteins (Bajpai et al., 2010).
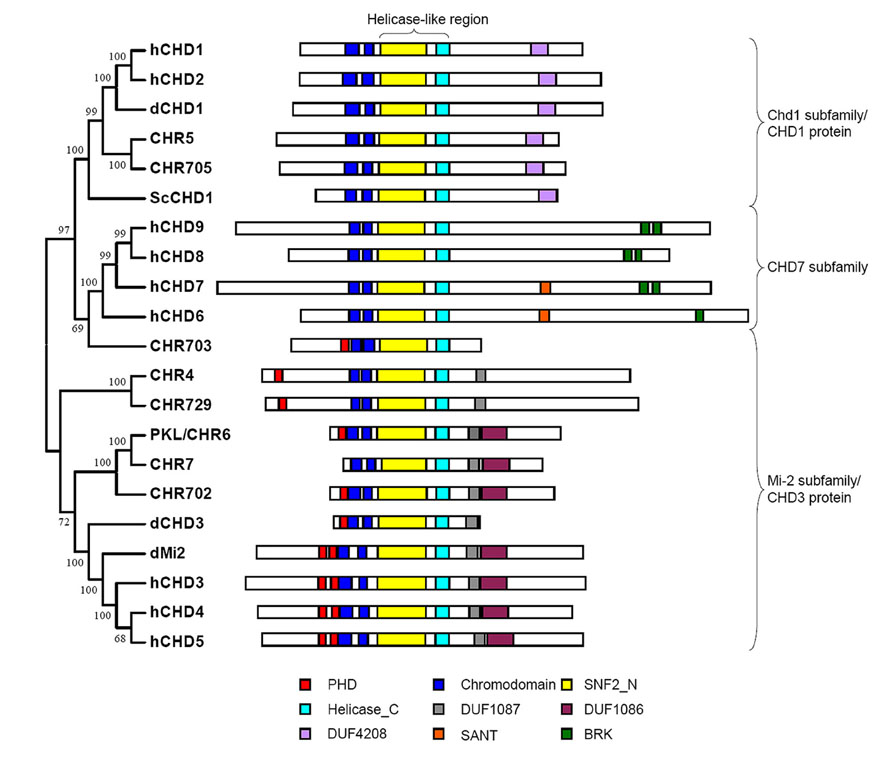
FIGURE 1. Structural domain of CHD proteins. Conserved domains of CHD proteins in yeast (Sc), fly (d), human (h), Arabidopsis (CHR4, CHR5, CHR6 and CHR7) and rice (CHR702, CHR703, CHR705 and CHR729) are represented by rectangle with different colors. The CHD proteins could be divided into three subfamilies. Each subfamily member has distinct extra domains besides double chromdomains and helicase-like region containing SNF2_N and Helicase_C domains.
In plants, there is little functional information about CHD proteins. Most studies concentrate on Arabidopsis PKL, which is a CHD3 protein. In recent years two papers reported the function of CHR729, a rice CHD3 protein (Hu et al., 2012; Zhao et al., 2012). From these studies we found that CHD proteins act as important epigenetic regulators to control series of genes expression and participate in the processes of development and stress response.
Plant CHD Proteins
Phylogenetic analysis of Snf2 family proteins in Arabidopsis and rice identified two subfamilies of CHD proteins in plant: Chd1 subfamily and Mi-2 subfamily (Hu et al., 2013). Arabidopsis MORPHEUS’ MOLECULE1 (MOM1) and its homologs in rice evolved from CHD3 chromatin remodelers were not considered to be CHD protein as they do not have conserved Helicase_C domain in the helicase-like region (Hu et al., 2013). Only one protein was found in Chd1 subfamily in both Arabidopsis (CHR5) and rice (CHR705; Hu et al., 2013). The function of these proteins has not been analyzed yet. Sequence analysis of plant CHD1 proteins indicates that they have conserved tandem chromodomain but lack the aromatic residues responsible for recognition of methylated H3 tail (Hu et al., 2013). However, the possibility could not be excluded that plant CHD1 binds to methylated H3 via the other residues. There are three members of Mi-2 subfamily in Arabidopsis (PKL, CHR4, and CHR7) and rice (CHR702, CHR729, and CHR703). All of these proteins contain single PHD domain and two chromodomains except Arabidopsis CHR7 which lacks PHD domain at N-terminal part of the protein (Figure 1). Phylogenetic analysis using helicase-like region showed that CHR7 was the close homolog of PKL (Hu et al., 2013). Functional analysis suggests that they act redundantly to regulate gene expression and control plant development, giving rise to the question whether PHD domain is vital for the function of PKL (Aichinger et al., 2009). Although containing both PHD domain and chromodomain rice CHR703 seems to be less homologous to the other CHD proteins and the homolog of the protein in Arabidopsis does not exist demonstrating that it might be evolved after the divergence of monocot and dicot (Hu et al., 2013).
Regulatory Function of CHD3 Protein in Plant
It has long been found that dMi-2 is involved in the repression of hox genes. The NuRD (Nucleosome Remodeling Deacetylase) complex which contains CHD3 protein exhibits histone deacetylase activity further improving the repressive function of the protein (Kehle et al., 1998; Xue et al., 1998; Figure 2). However, later results show that dCHD3 which is homologous to dMi-2 but lacks one PHD domain is associated with actively transcribed sites as well as dMi-2 (Murawska et al., 2008; Figures 1 and 2). This indicates that CHD3 protein may also positively participate in the process of transcription of series of genes.
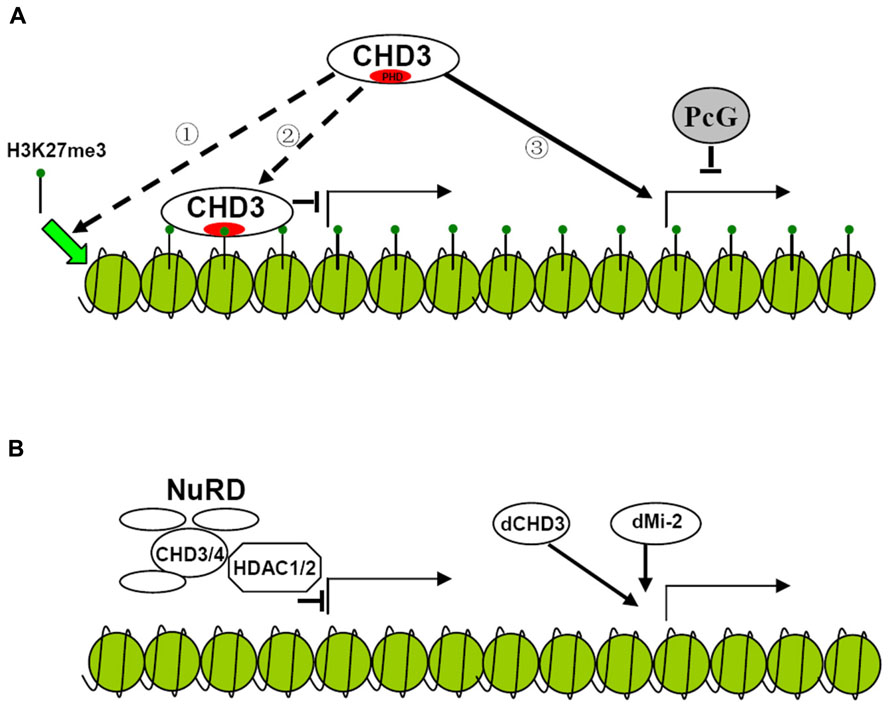
FIGURE 2. Possible models for the regulation of gene transcription by CHD3 protein in plant and animal. (A) Hypothesized model for the regulation of gene transcription by plant CHD3 proteins. Plant CHD3 proteins act as monomer to repress or activate gene expression. The repression involves the association with H3K27me3 by PHD domain. However, it is not known whether the binding of CHD3 protein to H3K27me3 is necessary for the deposition of the mark ① or serves as a recognition mechanism for the protein to repress target gene expression ②. Plant CHD3 proteins are also required for the expression of many PcG target genes ③. (B) Model for the regulation of gene transcription by animal CHD3 proteins. Animal CHD3 proteins repress gene expression by composing NuRD complex with Histone deacetylase and other subunits. In Drosophila dMi-2 and dCHD3 also localize to actively transcribed regions to participate in the process of transcription.
In plant, there are also controversial arguments on the function of PKL. It was initially characterized to repress embryonic identity related genes and later was found to negatively regulate several genes expression. The repression mediated by PKL was found to involve trimethylation of histone H3 lysine 27 (H3K27me3), a repressive epigenetic mark. PKL target genes were enriched for H3K27me3 and mutation of PKL leads to the loss of H3K27me3 at these loci (Zhang et al., 2008, 2012). Recent study indicates that unlike animal CHD3 protein, which is a component of multisubunit complex, PKL acts as a monomer suggesting that distinct mechanism may be adopted by plant CHD3 protein to repress gene expression (Ho et al., 2013; Figure 2). In rice, CHR729, which is the homolog of Arabidopsis CHR4, was found to bind to H3K27me3 via its PHD domain in vitro. Loss-of-function of CHR729 results in genome-wide decrease of H3K27me3 level (Hu et al., 2012). This indicates that plant CHD3 proteins may have conserved function to repress gene expression involving H3K27me3. However, it is not clear the association of CHD3 protein with H3K27me3 is necessary for the deposition of the mark or serves as a recognition mechanism for the protein to repress target genes expression (Figure 2).
In contrast, the other group showed that PKL was not directly involved in the repression of the genes. Derepression of these genes and loss of H3K27me3 result from down-regulation of polycomb (PcG) genes which are activated by PKL and CHR7 synergistically (Aichinger et al., 2009). They also found that PKL was directly required for activation of several other PcG target genes demonstrating the positive role of the protein in gene transcription in plant (Aichinger et al., 2009; Figure 2). Since many transcription factors were down-regulated in chr729 plants it would be interesting to test if these genes are direct targets of CHR729 to clarify if the protein also participate in the process of transcription (Hu et al., 2012). As Arabidopsis CHR7 which lacks PHD domain acts redundantly with PKL to activate gene expression it is plausible that PHD domain is not necessary for the activation. It is not known whether CHR7 and PKL have redundant function to repress gene expression and whether mutation of PHD domain alone would affect repressive function of the protein. Further experiments are required to confirm whether PHD domain is necessary for CHD3 protein to repress gene expression.
Plant CHD3 Proteins Control Multiple Developmental Processes and Stress Response
In Arabidopsis, PKL was found to play important roles in plant development as reviewed recently (Gentry and Hennig, 2014). It was initially reported to be involved in suppressing embryonic traits. The primary root meristem of pkl showed characteristics of embryonic tissue (Ogas et al., 1997). The genes specifically expressed in embryo such as LEAFY COTYLEDON 1 (LEC1), LEAFY COTYLEDON 2 (LEC2), FUSCA3 (FUS3) and PHERES1 were derepressed in the mutants (Ogas et al., 1997, 1999; Dean Rider et al., 2003; Li et al., 2005). Then it was proposed that GYMNOS, which is the same gene as PKL, temporally regulated target genes expression which was also regulated by CRABS CLAW spatially to promote polarity establishment of carpel (Eshed et al., 1999). Later studies revealed that PKL may play roles in several hormones responsive genes repression. For example, repression of auxin responsive transcription activators Auxin Response Factor 7 (ARF7) and ARF9 by SOLITARY-ROOT (SLR)/IAA14 requires PKL, also named SSL2 in the study (Fukaki et al., 2006); Cytokinin-hypersensitive 2 (CKH2) which is also PKL encoded product act together with CKH1/AtTAF12b to regulate cytokinin responsive genes and play negative roles in cytokinin responses (Kubo and Kakimoto, 2000; Furuta et al., 2011); A subset of gibberellin-dependent responses is mediated by PKL during shoot development (Henderson et al., 2004). It was also reported that PKL control meristematic activity discrepantly in leaf and root. It acts to restrict meristematic activity in leaf but was required for maintaining root meristematic activity (Ori et al., 2000; Hay et al., 2002; Aichinger et al., 2011). Recent paper showed that transcription repressor HY5 could recruit PKL to the promoter of cell elongation genes to repress H3K27me3 at the target loci which facilitates gene transcription, demonstrating involvement of the protein in cell elongation of hypocotyl responding to light (Jing et al., 2013). However, the other CHD3 proteins in Arabidopsis have hardly been characterized to date. As mentioned above, Arabidopsis CHR7 has only been reported to suppress embryonic identity and maintain root cell identity redundantly with PKL (Aichinger et al., 2009). Although detailed analysis was not performed it has been shown that down-regulation of CHR4 by RNAi (RNA interference) technique affects plant growth, demonstrating that this gene is also important in controlling plant development (Shaked et al., 2006).
In rice, the homolog of PKL, CHR702 seems not to be critical in developmental processes. CHR702 T-DNA insertion mutants or RNAi plants of the gene does not show any visible phenotype (Hu et al., 2012). However, mutation or down-regulation of CHR729 affects many aspects of plant development. chr729 plants showed short and narrow leaves, reduced stem elongation, thinner culm, and short and narrow seeds, suggesting that it might be involved in controlling organ size (Hu et al., 2012). The other study also showed that CHR729 was required for chloroplast development in adaxial leaves (Zhao et al., 2012). Function of the other CHD3 genes in rice have not been studied before. Expression analysis indicates that three genes are regulated during endosperm development especially CHR703, which is specifically expressed in early endosperm and down-regulated with the mature of endosperm (Hu et al., 2013). This implies that rice CHD3 proteins might be related to endosperm development.
Direct evidence that plant CHD3 proteins are involved in stress response has not been present yet. However, in Arabidopsis PKL was found to be necessary for the repression of ABSCISIC ACID–INSENSITIVE3 (ABI3) and ABI5 in response to ABA implicating the protein may play a role in osmoic stress (Perruc et al., 2007). PKL and CHR4 are also involved in DNA damage response as mutants or RNAi plants of the genes showed sensitive or resistant to γ-irradiation or UV-C (Shaked et al., 2006). In rice, microarray data revealed that mutation of CHR729 resulted in up-regulation of many stress-responsive genes suggesting the possible involvement of the protein in stress response (Hu et al., 2012).
Conclusion and Perspectives
Studies on Arabidopsis PKL and rice CHR729 have revealed that the proteins contribute to epigenetic regulation of gene expression which involves polycomb mediated H3K27me3 albeit the mechanism remains to be explored. PKL was found to control expression of several developmental genes. Despite direct targets are not clear mutants of CHR729 showed multiple developmental defects suggesting that both genes have effects on plant development. In addition, Arabidopsis CHD3 proteins are also involved in stress response. In light of the idea that H3K27me3 is an epigenetic mark associated with regulation of developmental and stress responsive genes it would be intriguing to disclose how CHD3 proteins and H3K27me3 are related in the matter of repressing gene expression (Zhang et al., 2007; Li et al., 2013). Plant CHD1 protein has never been analyzed before although only one protein was identified in rice and Arabidopsis respectively. Since distinct molecular function of CHD1 protein has been found in human, fly and yeast it is necessary to analyzed whether the protein in plant functions in the same way or adopt a novel mechanism to activate gene expression. Genetic analysis is also required to show whether plant CHD1 protein play important roles in development or stress response as CHD3 protein.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
Our work is supported by the National Natural Science Foundation of China (31301300).
References
Aichinger, E., Villar, C. B., Di Mambro, R., Sabatini, S., and Kohler, C. (2011). The CHD3 chromatin remodeler PICKLE and polycomb group proteins antagonistically regulate meristem activity in the Arabidopsis root. Plant Cell 23, 1047–1060. doi: 10.1105/tpc.111.083352
Aichinger, E., Villar, C. B., Farrona, S., Reyes, J. C., Hennig, L., and Kohler, C. (2009). CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis. PLoS Genet. 5:e1000605. doi: 10.1371/journal.pgen.1000605
Bajpai, R., Chen, D. A., Rada-Iglesias, A., Zhang, J., Xiong, Y., Helms, J., et al. (2010). CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature 463, 958–962. doi: 10.1038/nature08733
Bouazoune, K., Mitterweger, A., Langst, G., Imhof, A., Akhtar, A., Becker, P. B., et al. (2002). The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization. EMBO J. 21, 2430–2440. doi: 10.1093/emboj/21.10.2430
Dean Rider, S. Jr., Henderson, J. T., Jerome, R. E., Edenberg, H. J., Romero-Severson, J., and Ogas, J. (2003). Coordinate repression of regulators of embryonic identity by PICKLE during germination in Arabidopsis. Plant J. 35, 33–43. doi: 10.1046/j.1365-313X.2003.01783.x
Eshed, Y., Baum, S. F., and Bowman, J. L. (1999). Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99, 199–209. doi: 10.1016/S0092-8674(00)81651-7
Flanagan, J. F., Mi, L. Z., Chruszcz, M., Cymborowski, M., Clines, K. L., Kim, Y., et al. (2005). Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature 438, 1181–1185. doi: 10.1038/nature04290
Flaus, A., Martin, D. M., Barton, G. J., and Owen-Hughes, T. (2006). Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 34, 2887–2905. doi: 10.1093/nar/gkl295
Fukaki, H., Taniguchi, N., and Tasaka, M. (2006). PICKLE is required for SOLITARY-ROOT/IAA14-mediated repression of ARF7 and ARF19 activity during Arabidopsis lateral root initiation. Plant J. 48, 380–389. doi: 10.1111/j.1365-313X.2006.02882.x
Furuta, K., Kubo, M., Sano, K., Demura, T., Fukuda, H., Liu, Y. G., et al. (2011). The CKH2/PKL chromatin remodeling factor negatively regulates cytokinin responses in Arabidopsis calli. Plant Cell Physiol. 52, 618–628. doi: 10.1093/pcp/pcr022
Gentry, M., and Hennig, L. (2014). Remodelling chromatin to shape development of plants. Exp. Cell Res. 321, 40–46. doi: 10.1016/j.yexcr.2013.11.010
Hay, A., Kaur, H., Phillips, A., Hedden, P., Hake, S., and Tsiantis, M. (2002). The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr. Biol. 12, 1557–1565. doi: 10.1016/S0960-9822(02)01125-9
Henderson, J. T., Li, H. C., Rider, S. D., Mordhorst, A. P., Romero-Severson, J., Cheng, J. C., et al. (2004). PICKLE acts throughout the plant to repress expression of embryonic traits and may play a role in gibberellin-dependent responses. Plant Physiol. 134, 995–1005. doi: 10.1104/pp.103.030148
Ho, K. K., Zhang, H., Golden, B. L., and Ogas, J. (2013). PICKLE is a CHD subfamily II ATP-dependent chromatin remodeling factor. Biochim. Biophys. Acta 1829, 199–210. doi: 10.1016/j.bbagrm.2012.10.011
Hu, Y., Liu, D., Zhong, X., Zhang, C., Zhang, Q., and Zhou, D. X. (2012). CHD3 protein recognizes and regulates methylated histone H3 lysines 4 and 27 over a subset of targets in the rice genome. Proc. Natl. Acad. Sci. U.S.A. 109, 5773–5778. doi: 10.1073/pnas.1203148109
Hu, Y., Zhu, N., Wang, X., Yi, Q., Zhu, D., Lai, Y., et al. (2013). Analysis of rice Snf2 family proteins and their potential roles in epigenetic regulation. Plant Physiol. Biochem. 70, 33–42. doi: 10.1016/j.plaphy.2013.05.001
Jing, Y., Zhang, D., Wang, X., Tang, W., Wang, W., Huai, J., et al. (2013). Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell 25, 242–256. doi: 10.1105/tpc.112.105742
Kehle, J., Beuchle, D., Treuheit, S., Christen, B., Kennison, J. A., Bienz, M., et al. (1998). dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science 282, 1897–1900. doi: 10.1126/science.282.5395.1897
Konev, A. Y., Tribus, M., Park, S. Y., Podhraski, V., Lim, C. Y., Emelyanov, A. V., et al. (2007). CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science 317, 1087–1090. doi: 10.1126/science.1145339
Kubo, M., and Kakimoto, T. (2000). The Cytokinin-hypersensitive genes of Arabidopsis negatively regulate the cytokinin-signaling pathway for cell division and chloroplast development. Plant J. 23, 385–394. doi: 10.1046/j.1365-313x.2000.00796.x
Li, H. C., Chuang, K., Henderson, J. T., Rider, S. D. Jr., Bai, Y., Zhang, H., et al. (2005). PICKLE acts during germination to repress expression of embryonic traits. Plant J. 44, 1010–1022. doi: 10.1111/j.1365-313X.2005.02602.x
Li, T., Chen, X., Zhong, X., Zhao, Y., Liu, X., Zhou, S., et al. (2013). Jumonji C domain protein JMJ705-mediated removal of histone H3 lysine 27 trimethylation is involved in defense-related gene activation in rice. Plant Cell 25, 4725–4736. doi: 10.1105/tpc.113.118802
Mansfield, R. E., Musselman, C. A., Kwan, A. H., Oliver, S. S., Garske, A. L., Davrazou, F., et al. (2011). Plant homeodomain (PHD) fingers of CHD4 are histone H3-binding modules with preference for unmodified H3K4 and methylated H3K9. J. Biol. Chem. 286, 11779–11791. doi: 10.1074/jbc.M110.208207
Murawska, M., Kunert, N., Van Vugt, J., Langst, G., Kremmer, E., Logie, C., et al. (2008). dCHD3, a novel ATP-dependent chromatin remodeler associated with sites of active transcription. Mol. Cell. Biol. 28, 2745–2757. doi: 10.1128/MCB.01839-07
Ogas, J., Cheng, J. C., Sung, Z. R., and Somerville, C. (1997). Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277, 91–94. doi: 10.1126/science.277.5322.91
Ogas, J., Kaufmann, S., Henderson, J., and Somerville, C. (1999). PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 96, 13839–13844. doi: 10.1073/pnas.96.24.13839
Ori, N., Eshed, Y., Chuck, G., Bowman, J. L., and Hake, S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127, 5523–5532.
Perruc, E., Kinoshita, N., and Lopez-Molina, L. (2007). The role of chromatin-remodeling factor PKL in balancing osmotic stress responses during Arabidopsis seed germination. Plant J. 52, 927–936. doi: 10.1111/j.1365-313X.2007.03288.x
Pray-Grant, M. G., Daniel, J. A., Schieltz, D., Yates, J. R. III., and Grant, P. A. (2005). Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433, 434–438. doi: 10.1038/nature03242
Shaked, H., Avivi-Ragolsky, N., and Levy, A. A. (2006). Involvement of the Arabidopsis SWI2/SNF2 chromatin remodeling gene family in DNA damage response and recombination. Genetics 173, 985–994. doi: 10.1534/genetics.105.051664
Xue, Y., Wong, J., Moreno, G. T., Young, M. K., Cote, J., and Wang, W. (1998). NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2, 851–861. doi: 10.1016/S1097-2765(00)80299-3
Zhang, H., Bishop, B., Ringenberg, W., Muir, W. M., and Ogas, J. (2012). The CHD3 remodeler PICKLE associates with genes enriched for trimethylation of histone H3 lysine 27. Plant Physiol. 159, 418–432. doi: 10.1104/pp.112.194878
Zhang, H., Rider, S. D. Jr., Henderson, J. T., Fountain, M., Chuang, K., Kandachar, V., et al. (2008). The CHD3 remodeler PICKLE promotes trimethylation of histone H3 lysine 27. J. Biol. Chem. 283, 22637–22648. doi: 10.1074/jbc.M802129200
Zhang, X., Clarenz, O., Cokus, S., Bernatavichute, Y. V., Pellegrini, M., Goodrich, J., et al. (2007). Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 5:e129. doi: 10.1371/journal.pbio.0050129
Keywords: plant, CHD, chromatin, regulation, development, stress
Citation: Hu Y, Lai Y and Zhu D (2014) Transcription regulation by CHD proteins to control plant development. Front. Plant Sci. 5:223. doi: 10.3389/fpls.2014.00223
Received: 25 March 2014; Accepted: 05 May 2014;
Published online: 28 May 2014.
Edited by:
Keqiang Wu, National Taiwan University, TaiwanReviewed by:
Manoj K. Sharma, Jawaharlal Nehru University, IndiaYuhai Cui, Agriculture and Agri-Food Canada, Canada
Copyright © 2014 Hu, Lai and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongfeng Hu, Jingchu University of Technology, Xiangshan Road 33, Jingmen, China e-mail: feng2602@sina.com
 Yongfeng Hu
Yongfeng Hu Yan Lai
Yan Lai