- Plant Breeding Institute, University of Kiel, Kiel, Germany
Rapeseed (Brassica napus L.) is grown in different geographical regions of the world. It is adapted to different environments by modification of flowering time and requirement for cold. A broad variation exists from very early-flowering spring-type to late-flowering winter cultivars which only flower after exposure to an extended cold period. B. napus is an allopolyploid species which resulted from the hybridization between B. rapa and B. oleracea. In Arabidopsis thaliana, the PEBP-domain genes FLOWERING LOCUS-T (FT) and TERMINAL FLOWER-1 (TFL1) are important integrators of different flowering pathways. Six FT and four TFL1 paralogs have been identified in B. napus. However, their role in flowering time control is unknown. We identified EMS mutants of the B. napus winter-type inbreed line Express 617. In total, 103 mutant alleles have been determined for BnC6FTb, BnC6FTa, and BnTFL1-2 paralogs. We chose three non-sense and 15 missense mutant lines (M3) which were grown in the greenhouse. Although only two out of 6 FT paralogs were mutated, 6 out of 8 BnC6FTb mutant lines flowered later as the control, whereas all five BnC6FTa mutant lines started flowering as the non-mutated parent. Mutations within the BnTFL1-2 paralog had no large effects on flowering time but on yield components. F1 hybrids between BnTFL1-2 mutants and non-mutated parents had increased seed number per pod and total seeds per plant suggesting that heterozygous mutations in a TFL1 paralog may impact heterosis in rapeseed. We demonstrate that single point-mutations in BnFT and BnTFL1 paralogs have effects on flowering time despite the redundancy of the rapeseed genome. Moreover, our results suggest pleiotropic effects of BnTFL1 paralogs beyond the regulation of flowering time.
Introduction
Rapeseed (Brassica napus L.) is a major oil crop accounting for more than 60 million tons of seed and more than 20 million tons of extracted oil worldwide (http://www.worldoil.com/). This crop is widely cultivated in several temperate regions of the world such as northern Europe, Canada, China and Australia. Adapting flowering time to regional environmental conditions has been a major target of rapeseed breeding. A broad variation exists from very early-flowering spring-type to late-flowering winter cultivars that only flower after exposure to an extended cold period, a process known as vernalization (Iniguez-Luy and Federico, 2011). In rapeseed, flowering time and yield are closely linked to each other. Several genomic regions have been reported to contain major QTL for both traits. In a doubled haploid (DH) population derived from a cross between the Chinese semi-winter line Ningyou7 and the European winter-type Tapidor, at least four flowering time QTL were suggested as indicator QTL for yield (Long et al., 2007; Shi et al., 2009). Genetic variation within the different rapeseed types is relatively small, implying a need for wide crosses between non-adapted ecotypes to introduce traits of interest into elite materials (Girke et al., 2012). However, the introgression of genes from non-adapted ecotypes into elite cultivars is difficult due non-adapted flowering time. Therefore, it is of great interest to measure the effects of different paralogs on flowering and other characters.
Brassica napus and its close relatives Brassica oleracea and Brassica rapa belong to the family Brassicaceae which also includes the model plant Arabidopsis thaliana. Rapeseed is an allotetraploid species originating from the spontaneous hybridization between B. rapa (AA) and B. oleracea (CC) less than 5000 years ago (Ziolkowski et al., 2006; Wang et al., 2011). These two progenitor species are ancient polyploids that underwent genome triplication between the Brassica-Arabidopsis split (~13 MYA) and their actual divergence event (~two MYA). Comparative mapping between the Arabidopsis and Brassica genomes revealed numerous homologous regions arranged in highly syntenic chromosome blocks. Many Arabidopsis genes are represented in the B. napus genome by at least three paralogs (Schranz et al., 2006). Due to their close phylogenetic relationship and the high economic importance of rapeseed, knowledge transfer from the model species Arabidopsis to the complex Brassica genomes constitutes a worthwhile challenge for genomics research.
In Arabidopsis, four pathways controlling flowering time have been extensively studied (Amasino, 2010). All these pathways converge at the CO/FT regulon (Andres and Coupland, 2012). Under long day (LD) conditions, the CONSTANS (CO) protein accumulates in leaves and induces expression of the floral integrator gene FLOWERING LOCUS T (FT) in the phloem companion cells (Moon et al., 2003; Turck et al., 2008). FT is the long-sought “florigen” and it is reported to be a strong mobile signal triggering activation of floral identity genes in the Arabidopsis shoot apical meristem (Andres and Coupland, 2012). The FT protein is transported via the sieve tubes to the shoot apex, where it forms a heterodimer with the FD (FLOWERING LOCUS D) protein (Abe et al., 2005; Wigge et al., 2005). Interestingly, a very closely related gene, TERMINAL FLOWER 1 (TFL1) plays an FT-antagonistic role by competing for FD, leading to a repression of floral transition (Valverde, 2011; Andres and Coupland, 2012). In Arabidopsis, FT-overexpressing plants and TFL1 non-sense mutants show the same early-flowering phenotype and produce terminal flowers in the shoot apex. TFL1 represses transcription of genes which are activated by FT (Hanano and Goto, 2011). In Arabidopsis, these two highly similar polypeptides belong to a family of six members characterized by the phosphatidylethanolamine-binding domain (PEBP) (Kardailsky et al., 1999). Substitutions of crucial amino acids from the FT and TFL1 exon II, as well as the exchange of the exon IV led to contrasting protein functions for both polypeptides (Hanzawa et al., 2005; Ahn et al., 2006).
Apart from their major role to control flowering time, FT and TFL1 orthologs have been shown to alter a variety of phenotypic characters. In tomato, the SINGLE FLOWER TRUSS (FT ortholog) and SELF PRUNING (TFL1 ortholog) genes impact fruit yield heterosis. F1 hybrids generated by crosses between loss of function SFT mutants and tomato wild type (WT) plants of the non-mutagenized line M82 have shown strong increment in fruit production (Molinero-Rosales et al., 2004; Krieger et al., 2010). FT/TFL1 gene orthologs have been characterized in diploid crops such as rice (Kojima et al., 2002), pea (Hecht et al., 2011), barley (Faure et al., 2007), poplar (Böhlenius et al., 2006), and sugar beet (Pin et al., 2010) and in two polyploids, wheat (Yan et al., 2006) and potato (Navarro et al., 2011). The characterization of the FT/TFL1 gene orthologs in polyploid plants is a special challenge because duplicated genes can build new regulation networks leading to sub- or neo-functionalization (Pin and Nilsson, 2012).
Rapeseed has six BnFT paralogs (BnA2FT, BnC2FT, BnC6FTa, BnC6FTb, BnA7FTa, and BnC7FTb) sharing high sequence similarity (92–99%) in their four exons (Wang et al., 2009). It has been shown that the BnC2FT copy is silenced in B. napus and B. oleracea due to the insertion of a miniature inverted-repeat transposable element (MITE) in its promoter region, whereas the remaining five copies are detectable in B. napus, B. rapa and B. oleracea (Wang et al., 2012a). There are at least four TFL1 paralogs in the B. napus genome (Mimida et al., 1999). Among them, the BnTFL1-2 paralog shares high homology with the B. rapa ortholog on chromosome A10, whereas BnTFL1-1, BnTFL1-3 are highly similar to their B. oleracea counterparts. The BnC6FTa and BnC6FTb paralogs were co-located to a major flowering time QTL detected in nine winter-cropped environments which could support their function as flowering time regulators in B. napus (Qiu et al., 2006; Shi et al., 2009). Until now, B. napus FT/TFL1 homologs have not been functionally characterized.
This study had two major aims. First, we aimed to uncover the role in flowering time control of different FT and TFL1 paralogs in B. napus by analyzing EMS-treated offspring with missense and splice-site mutations within selected paralogs. We demonstrate that single mutations can change the onset of flowering in B. napus despite the redundancy of its allopolyploid genome. Moreover, we postulated that BnTFL1 mutations also affect seed yield components in rapeseed. We found increased seed yield in F1 plants carrying a mutated BnTFL1 allele on the Express 617 background. Our data suggest that EMS-generated alleles may constitute a new resource to broaden the genetic basis of rapeseed breeding.
Materials and Methods
Plant Materials and Greenhouse Experiments
Seedlings of M3 lines and Express 617 (controls) were grown in the greenhouse at constant temperature (22°C) under long days (LD, 16 h light/8 h dark) for 4 weeks. Express 617 is an inbreed line (F11) originated from the rapeseed winter-type cultivar Express (Harloff et al., 2012). Subsequently, plants were vernalized for 8 weeks at 4°C under LD conditions in a cold chamber. Of each M3 line, 30 plants were grown. After vernalization, plants were returned to the initial greenhouse conditions and transplanted to 11 × 11 cm pots. M3 plants and Express 617 controls were arranged in randomized blocks. Plant positions on the greenhouse were indexed and linked to randomly generated numbers using the Microsoft Excel software. Selected M3 lines were crossed with the male sterile (MS) line MSL007 (NPZ, Hohenlieth, Germany) using homozygous M3 plants as a pollinators. F1 plants and Express 617 controls were grown in the greenhouse under the conditions mentioned above. F2 populations were produced by crossing M3 homozygous plants from a selected BnC6FTb mutation (BnC6FTbG2154A) and non-mutagenized Express 617 plants. In each greenhouse experiment, the following phenotypic characters were measured according to the BBCH scale (http://www.jki.bund.de/en/startseite/veroeffentlichungen/bbch-codes.html): first non-cotyledonal leaves (NCL, BBCH10), rosette plant (BBCH30), visible floral buds (BBCH50), first open flower (BBCH60), and end of flowering (BBCH69). Plants that did not grow beyond NCL (BBCH 10) were excluded from the experiment. Plant height, number of branches, initial flowers, filled pods, seed number and seed weight were recorded for each plant separately.
Mutation Screening
A total of 3488 M2 plants of the Express 617 EMS-population were screened by TILLING as described by Harloff et al. (2012). Gene specific primers were designed for BnC6FTa (FJ848915.1), BnC6FTb (FJ848917.1), and BnTFL1-2 (AB017526.1) (Supplementary Table S1). For primer design and comparative analysis, B. rapa and B. oleracea genome sequences were downloaded from (http://brassicadb.org/brad/downloadOverview.php) and (http://ocri-genomics.org/bolbase/), respectively. Plant genomic DNA arrayed in two dimensional 8-fold pools was amplified by direct or nested PCR. Forward and reverse primers were 5′-end labeled with 700 nm (DY-681) or 800 nm (DY-781) IRD fluorescence dyes, respectively (Biomers, Ulm, Germany, www.biomers.net). PCR amplifications with labeled oligos were done using the following profile: 95°C 5 min; 35 cycles of 95°C 30 s, 60°C 45 s, 72°C 90 s, 72°C 10 min. Heteroduplex-specific restriction endonuclease CEL1 was extracted from celery and stored at −80°C as reported by Frerichmann et al. (2013). Labeled fragments were separated by a LI-COR 4300 DNA analyzer (LI-COR Biosciences) for 3:15 to 4:15 h at 1,500 V, 40 mA and 40 W. Gel images were analyzed using the software GelBuddy (http://www.proweb.org/gelbuddy/). After positive pools had been identified, single plant DNA was amplified with unlabeled oligos and sequenced for SNPs confirmation. Sequences were analyzed with the CLC-bio Main Workbench sequence alignment tool (CLC bio, Aarhus, Denmark).
DNA Isolation and Genotyping
Total DNA was extracted from young leaves using a CTAB protocol (Morjane et al., 1994). Total DNA was treated with RNAse I (Fermentas, www.fermentas.de), and DNA concentration was determined by spectrometry (NanoDrop, www.nanodrop.com). DNA quality was checked by 1% agarose gel electrophoresis. For genotyping mutant lines, genomic DNA from single plants was amplified by PCR using unlabeled primers. PCR was done essentially as described in the previous paragraph. Five micro litter of each PCR product were loaded on 1% (w/v) agarose gels. Upon band size confirmation, the remaining 25 μl of PCR product were sequenced via Sanger capillary sequencing. The sequences were analyzed with the CLC-Bio software (CLC bio, Aarhus, Denmark) using the sequence assembly viewer tool.
Tissues Sampling and RT-qPCR
Young leaves of M3 plants and Express 617 controls were sampled at four developmental stages, as described above. Genomic DNA sequences from the different flowering time genes analyzed were retrieved from the non-redundant NCBI nucleotide database (http://www.ncbi.nlm.nih.gov/). Individual sequences were loaded to the CLC-bio main workbench version 6.0 (http://www.clcbio.com), and groups of paralogs were aligned with the help of the internal alignment routine. Two main strategies were applied for expression analysis: (i) primers were designed in conserved regions within groups of paralogs for detecting joint gene expression, and (ii) copy-specific primers were designed for the members of selected paralog genes (Supplementary Table S1). Total RNA was extracted using the RNeasy kit (QIAGEN, www.qiagen.com) according to the manufacturer's protocol. The RNA concentration was determined by spectrometry (Nano Drop; Thermo Scientific, Wilmington, USA) and quality was checked by agarose gel electrophoresis. Total RNA was treated with DNAse I (Fermentas Inc., Maryland, USA). First-strand cDNA was synthesized using Oligo(dT)18 primers and the M-MuLV Reverse Transcriptase (Fermentas).
Quantitative real-time RT-PCR (RT-qPCR) was performed with SYBR qPCR Super mix w/ROX (Invitrogen Corporation, Carlsbad, USA) using a CFX96 Real-Time System (Bio-Rad Laboratories GmbH, München, Germany). Reactions were performed in a total volume of 15 μ l containing 100 nM of each primer and 2 μ l of diluted cDNA templates, and amplified using the following cycling conditions: 95°C for 3 min, 40 cycles of 95°C for 10 s, 60°C for 30 s, and 72°C for 30 s, followed by 95°C for 10 min. A melting curve was generated using a temperature range from 65°C to 95°C with increments of 0.5°C every 5 s. For each sample at least three technical replications were performed. For data analysis, the mean Ct value of the target gene was normalized against the average Ct value of two housekeeping genes (BnGADPH-3 and BnB-Tub). Calculation of relative expression values was carried out following Pfaffl (2001) after extracting main Ct values via CFX manager software (Bio-Rad Laboratories GmbH, München, Germany). In each analysis, the relative expression value for the reference sample has been set to 1. Normalized expression was averaged over two biological replicates and three technical repetitions in each case. Standard curves for the target and housekeeping genes are based on dilution series of purified cloned fragments for each gene.
Sequence Diversity Analysis
For analyzing sequence diversity within the BnC6FTb and BnTFL1-2 genes, genomic DNA from one-hundred accessions of the B. napus ASSYST panel was amplified with paralog-specific primers and sequenced via Sanger method. We selected 117 lines from the B. napus ASSYST diversity set (Bus et al., 2011) including winter, semi-winter and spring types which had been phenotyped in several environments worldwide (Supplementary Table S2). Lyophilized leaf samples harvested from young plants were used for DNA isolation with the NucleoSpin Plant II DNA isolation kit (Macherei & Nagel, Germany), following the manufacturer's instructions. PCR amplifications were carried out with paralog-specific primers as follows: 95°C 5 min; 35 cycles of 95°C 30 s, 60°C 45 s, 72°C 90 s, 72°C 10 min. Sequences resulting from single band amplicons were assembled and aligned using the CLC-bio main workbench software (CLC bio, Aarhus, Denmark) and the resulting FASTA alignment was loaded into the software TASSEL (http://www.maizegenetics.net) for identification of polymorphic SNPs.
Results
Paralog-Specific Expression of Five BnFT Genes
We carried out a RT-qPCR experiment to measure the paralog specific expression of six BnFT paralogs in leaves of the winter-type inbred line Express 617 during the transition to reproductive stages. Samples were taken from greenhouse-grown plants at three different stages of development (BBCH30 before and after vernalization and BBCH50). Relative expression values for each paralog were calculated after Ct normalization using BnGAPDH as a reference gene. Leaf samples at BBCH30 before vernalization (preV) were used as reference samples for relative expression calculations. At BBCH30 before vernalization (BBCH30-preV), four BnFT paralogs (BnC6FTa/b and BnC6FT7a/b) were weakly expressed (Figure 1), whereas two transcripts were not expressed (BnC2FT and BnA2FT). Moreover, BnA2FT was only highly expressed at BBCH60 after floral transition (data not shown), whereas BnC2FT showed no expression at all. After vernalization (BBCH30-postV), BnC6FTa/b and BnA7FTa/b expression was higher in rosette plants, but differences between paralogs were obvious. BnC6FTb showed the largest relative expression level (~9-fold). At BBCH50 (visible floral buds), the BnC6FTb and BnA7FTb paralogs showed the largest relative expression levels (~13-fold) (Figure 1). In leaves at BBCH 60 (first flower open), all paralogs with exception of BnC2FT showed very high relative expression levels (>2000-fold), where BnC6FTa showed the highest relative expression (data not shown).
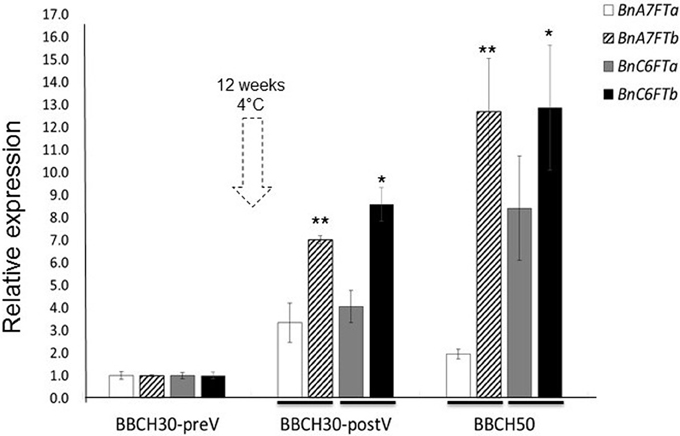
Figure 1. Relative expression of five BnFT paralogs in Express 617 plants at three developmental stages before and after vernalization (dotted arrow). Plants at BBCH30 were analyzed before vernalization (preV) and after vernalization (postV). The time point BBCH30-preV was taken as reference for calculation of relative expression in all target genes. Two biological replicates and three technical repetitions were analyzed for each time point. Error bars indicate the standard error of the mean of the relative expression values. Expression levels of target BnFT genes were normalized against BnGAPDH total expression. Ct values of the paralogs BnA2FT and BnC2FT were below the detection threshold at BBCH30 and BBCH50. Significant differences (P < 0.05) are depicted by asterisks. Differences in relative expression between the BnC6FTa/BnC6FTb (*) and BnA7FTa/BnA7FTb paralogs (**) at each time point were tested via t-test. Lines at the base of the bars indicate the comparison pairs. All samples were taken between zeitgeber 11 h and 12 h in each developmental stage. Express 617 plants reached BBCH30-preV ~30 days after sowing. BBCH30-postV was registered ~90 days after sowing. BBCH50 was registered ~107 days after sowing.
EMS Mutations in BnFT and BnTFL1 Paralogs
We screened our EMS population to measure the flowering time effect of mutations within the BnFT paralogs BnC6FTa (FJ848915.1) and BnC6FTb (FJ848917.1). In Arabidopsis, apart from FT, other PEBP proteins such as TERMINAL FLOWER-1 (TFL1) regulate flowering by competing with FT for its binding targets (Mimida et al., 2001). Therefore, we developed primers for the BnTFL1-2 (ABO17526) gene assigned to B. rapa chromosome A10. In total, 3488 M2 plants were screened by TILLING for EMS-induced mutations in BnC6FTa/b and BnTFL1-2.
We generated paralog-specific PCR amplicons covering between 50% (BnC6FTa) and 100% (BnC6FTb and BnTFL1-2) of the open reading frames. The BnC6FTa fragment covered exon I and intron I. The two BnC6FTb fragments covered exon I / intron I and exons III / IV (Figure 2). The BnTFL1-2 fragment covered all four exons. We identified 55, 14, and 34 single nucleotide mutations in the BnC6FTb, BnC6FTa, and BnTFL1-2 genes, respectively. Forty-three mutations are located in introns, 19 are silent mutations, and three are located within the UTRs (Table 1). Mutation rates ranged between 1/72 kb and 1/24 kb per 1000 plants. The names of the mutant alleles contain the nucleotide substitution and nucleotide position (Table 2).
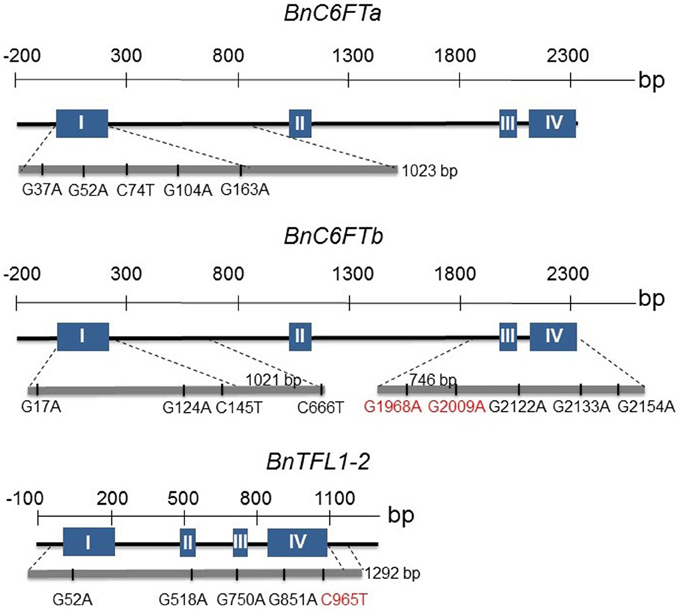
Figure 2. Graphical presentation of 18 point mutations in two FT (BnC6FTa, BnC6FTb) and one TFL1 (BnTFL1-2) paralog of the B. napus winter-type cultivar Express 617. The exon (blue boxes) - intron (black lines) structure of each gene is shown. Locations of SNPs are depicted by the gray bar below. Numbers refer to the position of the mutations from the START codon. Only STOP/splice-mutations (red) and selected missense mutations are shown.
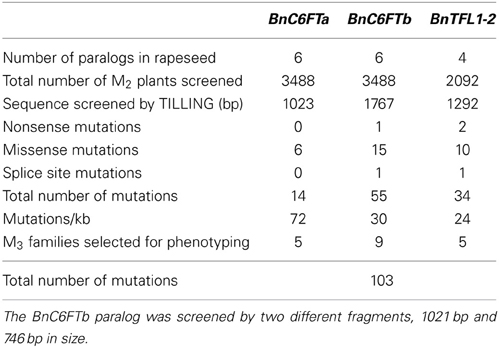
Table 1. EMS mutations in three flowering time genes detected by TILLING of the winter-type inbred line Express 617.
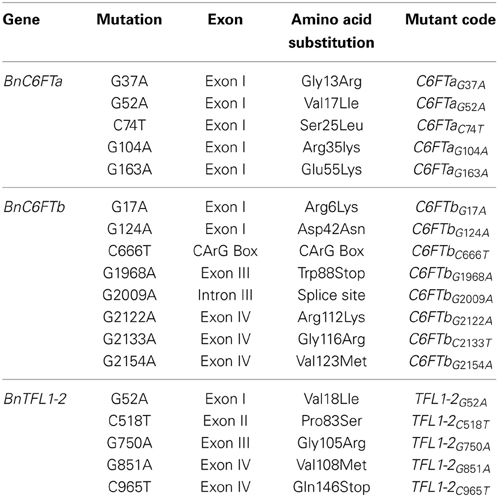
Table 2. Nucleotide position and amino acid changes due to EMS mutations in 18 missense/non-sense mutations in three B. napus flowering time regulators.
We identified one non-sense mutation in exon III of the BnC6FTb gene (BnC6FTbG1968A) leading to a truncated protein by substitution of a tryptophan by a stop codon (position 88). Another mutation (BnC6FTbG2009A) resulted in a splice-site deletion leading to a truncated protein by interrupting the junction between exons III and IV. The BnTFL1-2C965T mutation in Exon III results in the substitution of a glutamine by a stop codon (position 146). Moreover, we detected numerous missense mutations in BnC6FTa (15), BnC6FTb (16), and BnTFL1-2 (10).
We decided to focus on splice site- and missense-mutations which are most likely to affect the protein function. All observed missense mutations were compared to the SIFT database (http://sift.jcvi.org) in order to evaluate the impact of the amino acid substitutions on the protein function (data not shown). According to this analysis, we selected 18 M3 lines for growth experiments in the greenhouse (5 BnC6FTa, 8 BnC6FTb, and 5 BnTFL1-2 mutations) (Supplementary Table S3) (Figure 2).
Phenotypic Characterization of BnFT and BnTFL1-2 Mutants
First, we confirmed the genotype of each selected M2 plant by Sanger sequencing. Then, M3 lines were grown in the greenhouse together with non-mutagenized Express 617 plants as a control. The phenological development of BnC6FTa and BnC6FTb lines was clearly different. All five BnC6FTa mutants flowered as the control, whereas six out of eight BnC6FTb mutants flowered later (Figure 3). The C6FTbG1968A mutants (stop mutation) showed a flowering delay of ca. 18 days, while C6FTbG2009A splice-site mutants flowered 29 days later as the control. Interestingly, 40 and 54% of the C6FTbG1968A and C6FTbG2009A M3 mutants, respectively, did not bolt at all (Figure 4). BnC6FTb missense mutants started flowering 7 days (BnC6FTbC2122A) up to 26 days (BnC6FTbG17A) later as the control. Apart from flowering time, reduced fertility was also apparent, mostly in BnC6FTb M3 plants (Supplementary Figure 1).
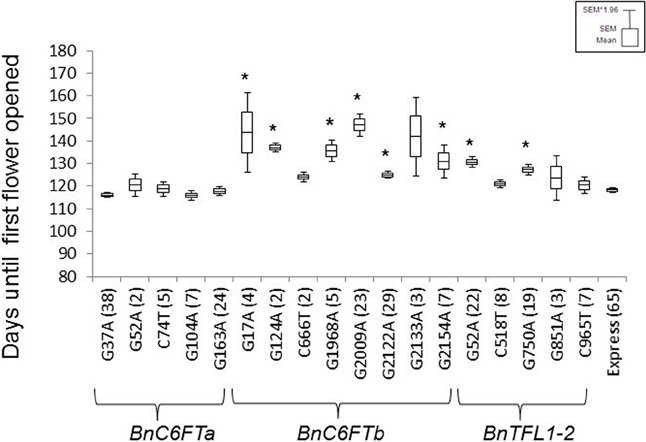
Figure 3. Flowering time point of 18 BnC6FT- and BnTFL1-2 mutants grown in the greenhouse at constant temperature (22°C), and LD (16 h light) after vernalization (4°C, 16 h light, 8 weeks). Days to flowering (BBCH 60) was measured in M3 plants homozygous for the EMS allele. The non-mutagenized donor line Express 617 was used as a control. The number of plants analyzed is written in brackets. Differences in flowering time between homozygous mutants and control plants were tested via t-test. Significant differences (P < 0.05) are depicted by asterisks.

Figure 4. Phenotypes of 18 B. napus BnC6FTa/b and BnTFL1-2 EMS M3 lines. Photos were taken as the non-mutagenized Express 617 plants started flowering (top left). Plants were grown in the greenhouse at constant temperature (22°C), and LD (16 h light) after vernalization (4°C, 16 h light, 8 weeks).
To evaluate the effect of background mutations on flowering time, we produced an F2 population by crossing BnC6FTbG2154A M3 plants with non-mutagenized Express 617. BnC6FTbG2154A M3 missense mutants gave higher hybrid seed yield as the stop mutants and they flowered 15 days later as the control. BnC6FTbG2154A M3 plants showed a reduced number of initial flowers in comparison to other M3 mutants, however most flowers were fertile. A total of 26 F2 plants encompassing all three genotypic classes were grown in the greenhouse together with Express 617. In agreement with M3 observations, homozygous F2 mutants (ft ft) flowered 13 days later than F2 siblings homozygous for the wild-type allele (FT FT) which did not show any significant differences in flowering time as compared to non-mutagenized Express 617 (Supplementary Figure 2).
The stop mutation BnTFL1-2C965T did not lead to a major delay in flowering time. In contrast, the missense mutants BnTFL1-2G52A and BnTFL1-2G750A flowered ~10 days later than the control (Figure 3). Since, the stop mutation is close to the end of the BnTFL1-2 gene, a functional protein may still arise after translation. Furthermore, BnTFL1G750A mutants exhibited modifications in plant architecture which gave us a reason to select them for crossing experiments.BnTFL1G750A mutants developed normally during the early growth phase until reaching BBCH50 (visible floral buds). The internode elongation phase was much longer as compared to Express 617, as a consequence, mutant plants were not able to stand by themselves after BBCH50. In this M3 line, the flower development limited the continuous growth of the floral meristem, whereas side branches continued flowering.
The EMS Mutations in BnC6FTb and BnTFL1-2 Paralogs are Located in Highly Monomorphic Regions of Exon III and Exon IV
To investigate the genetic structure of those BnFT and BnTFL1 paralogs with paramount impact on flowering time, we analyzed the sequence diversity of BnC6FTb and BnTFL1-2 in B. napus by sequencing their complete exons III and IV in 117 B. napus inbreed lines from different geographic origins and growth types. Sequences selected for analysis in each gene after quality trimming are deposited in Supplementary Table S2. While BnC6FTb exon III turned out to be highly conserved, exon IV exhibited larger sequence diversity. Within 41 bp exon III of the BnC6FTb gene, only a single polymorphism was found at position 2004 which corresponds to an allele frequency of 1%. The EMS mutation G1968A resides within a sequence domain which is monomorphic among all accessions investigated. For BnC6FTb (exon IV), six polymorphic regions were found with minor allele frequencies of <5.0%. The EMS-generated alleles (positions G2122, G2133, and G2154) are residing in monomorphic sequences (Supplementary Figure 3).
In contrast to BnFT genes, a higher variability in BnTFL1-2 exon III than in exon IV was found. In exon III the SNP showing the largest variation was a C insertion at position 731. The EMS mutation G750A which is also located within this exon, was located in a fully monomorphic domain. For BnTFL1-2 exon IV, only a T/C polymorphism was found at position 1030. In conclusion, there is a high degree of sequence conservation within the analyzed sequences. Our EMS treatment created novel sequence variations within these highly conserved regions (Supplementary Figure 4). FASTA-formatted sequences for each gene are deposited in Supplementary File 1. All sequences have been submitted to NCBI (www.ncbi.nlm.nih.gov) (accession numbers KJ533546 - KJ533625 and KJ533626 - KJ533728).
A BnC6FTb-Splice-Site Mutation Impacts the Expression of other Flowering Time Genes in Leaves
We reasoned that a loss of function of the BnC6FTb paralog directly impacts the transcriptional activity of other major flowering time regulators downstream of BnFT. To test this hypothesis, we selected the BnC6FTbG2009A mutant because, first the G2009A SNP causes a splice-site mutation that leads to a truncated protein, and second, C6FTbG2009A M3 plants are characterized by a marked flowering delay of about 29 days compared to the Express 617 control (Figure 3).
We chose BnAP1 and BnSOC1 as putative downstream targets of BnFT genes based on our knowledge from Arabidopsis (Yoo et al., 2005; Kaufmann et al., 2010). We measured their expression in leaves. BnC6FTbG2009A M3 plants were grown in the greenhouse under constant temperature and LD conditions. For expression analysis, young leaves of three different plants were taken at stages BBCH30 (pre and post-vernalization), BBCH50, and BBCH60. Arabidopsis AP1 and SOC1 sequences were BLASTed against B. oleracea and B. rapa. High homology hits were aligned and primers were designed from highly conserved regions. Subsequently, joint expression of all paralogs was measured by RT-qPCR. Gene expression levels of BnAP1 and BnSOC1 were normalized using BnGAPDH and BnB-Tub genes.
We detected altered transcriptional activities of BnAP1 in BnC6FTbG2009A mutants when compared to Express 617 control plants. Control Express 617 plants at BBCH30 (preV) were used as reference sample for relative expression calculations. At rosette stages (BBCH30), BnAP1 expression in the M3 mutantwas higher than in control plants, while at BBCH50 and BBCH60 relative expression levels were 10–40% lower (Figure 5). Before vernalization (BBCH30-preV), BnSOC1 expression in mutants was 2.5-fold higher than in Express 617. After vernalization, we detected a reduction of BnSOC1 in BnC6FTbG2009A mutants compared to the control. When the first flower opened, the difference in expression between BnC6FTbG2009A mutants and Express 617 was at its maximum. The altered expression in leaves indicates that a single BnC6FTb mutation may affect other major flowering time regulators. We expect that both genes are expressed in the shoot apical meristem as well.
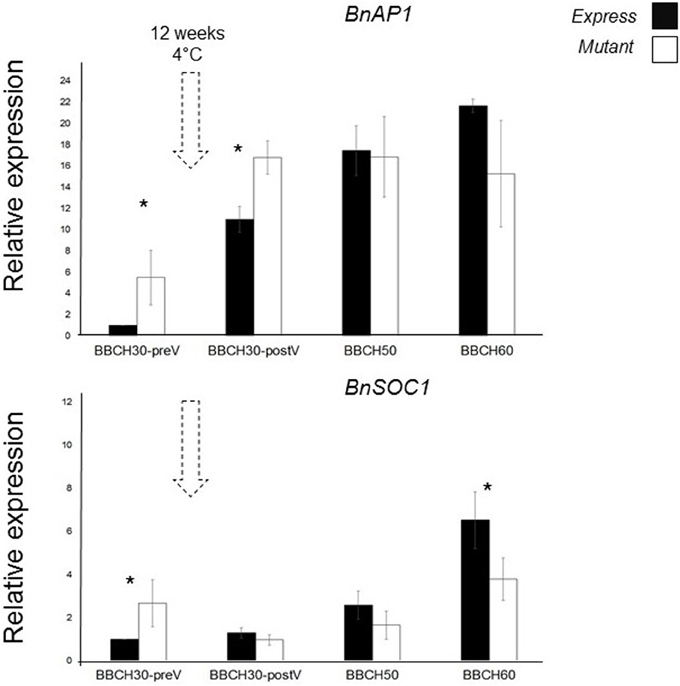
Figure 5. Joined expression analysis of two BnFT downstream targets in the BnC6FTbG2009 mutant (open boxes) and Express 617 (filled boxes). Four developmental stages were analyzed before and after vernalization (dotted arrows). Plants at BBCH30 were analyzed before vernalization (preV) and after vernalization (postV). Two biological replicates (M3 plants) and three technical repetitions were analyzed for each time point. Error bars: standard error of the mean for biological replicates. Ct of target genes were normalized against the BnGAPDH and BnB-Tub total expression. The time point BBCH30-preV in Express 617 control plants was taken as reference sample for calculation of relative expression. Differences in relative expression were pairwise tested (control Vs M3 line) via t-test. Significant differences (P < 0.05) are depicted by asterisks. All samples were taken between zeitgeber 11 h and 12 h.
Performance of F1 Hybrids Using the BnTFL1-2 Mutants as Parents
In tomato, mutations in FT and TFL1 orthologs accounted for fruit yield heterosis in F1 hybrids (Krieger et al., 2010). We made an initial experiment to address the question whether B. napus orthologs might have a similar function. For producing F1 hybrids, we selected BnC6FTbG2009A and BnTFL1-2G750A homozygous M3 mutants as pollinators due to their late flowering phenotype in conjunction with an altered inflorescence (lower number of fertile flowers, Supplementary Figure 1). We crossed homozygous M3 plants with the male-sterile (MS) line MSL007. The MSL007 line (MSL-Express) is an isogenic line of Express that carries the male sterility Lembke (MSL) genic male sterility system (Basunanda et al., 2010). Thus, no F1 heterosis was expected, except effects due to EMS mutations.
F1 hybrids were vernalized and grown in the greenhouse with the parental lines and Express 617. Both F1 hybrids showed differential effects. BnC6FTbG2009A hybrids showed no significant differences in seed number per plant and total seed weight in comparison to Express 617 as the best parent. In contrast, BnTFL1-2G750A F1 hybrids had significantly higher number of seeds/plant (20%) and total seed weight (40%) as compared to the best parent (Figure 6). Although the effects of background mutations cannot be ruled out, these results could indicate that BnTFL1 mutations impact heterosis in B. napus.
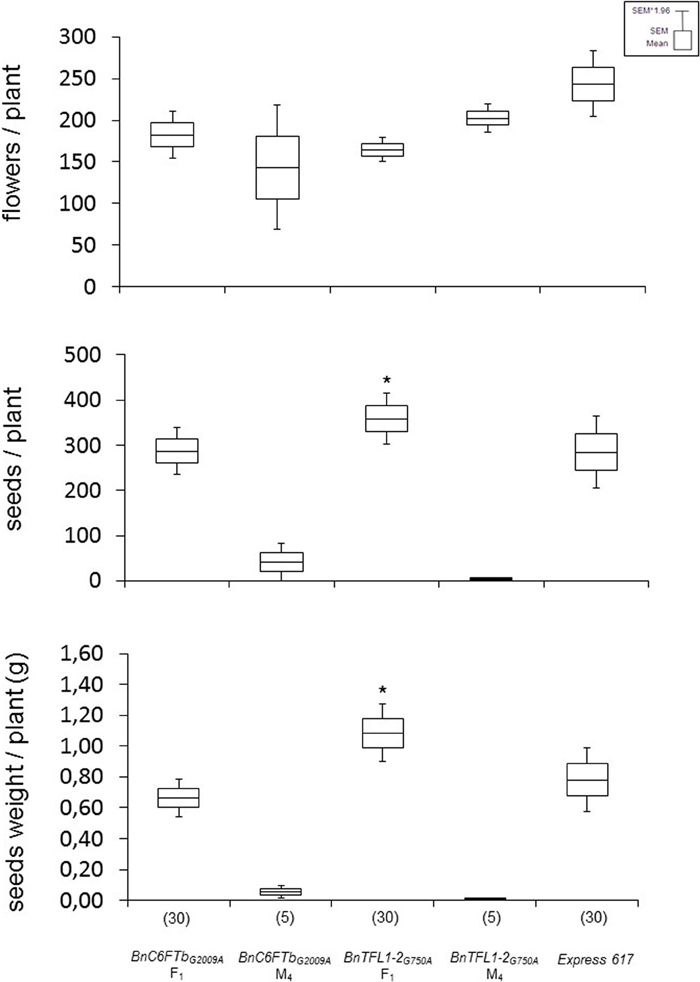
Figure 6. Growth experiments with vernalized F1 hybrids after crossing two mutants with a male sterile isogenic line (MSL007). Each F1 was obtained from two different crossing experiments. Mutant parents (M4 lines), and Express 617 were used as controls. Yield components were determined on single plants grown in the greenhouse. The number of plants analyzed is given in brackets. Differences in flowers, seed number and seed weight per plant against Express 617 were tested via t-test. Significant differences (P < 0.05) are depicted by asterisks. Growth conditions: 22°C, 16 h light, greenhouse. Vernalization: 4°C, 16 h light, 8 weeks.
Discussion
In the present study, more than 100 EMS-alleles have been found for three flowering time genes of B. napus. Based on previous reports, the average EMS-mutation frequencies are expected to be lower in diploid species (~1/380 kb) than in polyploids (~1/50 kb) (Till et al., 2007; Wang et al., 2012c). In this study, mutation frequencies ranged between 1/24 kb and 1/72 kb. Using the same EMS-population, mutation frequencies ranged between 1/12 and 1/22 kb for sinapine biosynthesis genes (Harloff et al., 2012). Although the observed BnFT /BnTFL1 mutation frequencies are slightly lower, our results are in the range expected for polyploid species. The present mutants will be a valuable resource to study flowering regulatory networks in polyploids and they can be introduced into breeding programs.
Our aim was to provide data about the function of FT and TFL1 paralogs in rapeseed. We found that, despite the redundancy of mutations in a single gene, either non-sense or missense mutations in the BnC6FTb gene resulted in a marked flowering delay. This supports our hypothesis that BnFT paralogs contribute differently to flowering time regulation. A large plethora of Arabidopsis reports on FT loss-of-function mutants have established a robust correlation between FT mutations and flowering time delay in Arabidopsis (Andres and Coupland, 2012). Contrasting with expectations based on Arabidopsis TFL1-phenotypes, BnTFL1-2 mutants showed a slight delay in flowering time. In Arabidopsis, a single amino acid change in TFL1 (tfl1-1Gly105Asp) led to early flowering and limited the development of indeterminate inflorescence by promoting the formation of a terminal floral meristem (Bradley et al., 1997). On the other hand, F1 hybrids derived from crosses between BnTFL1-2 M3- and rapeseed MS lines showed increased seed yield compared to BnC6FTb F1 hybrids and Express 617 controls. Thus, although the role of BnTFL1-2 involving flowering time regulation is not likely to be conserved compared to its Arabidopsis ortholog, TFL1-2 appears to be involved in yield-related traits as reported for its tomato ortholog SP (Jiang et al., 2013b). As we mention in the following sections, confirming this hypothesis is a must for new research approaches.
During evolution, duplicated genes may undergo dosage adjustments (Papp et al., 2003; Conant and Wolfe, 2008), non-functionalization, or sub-/neo-functionalization (Force et al., 1999). We wanted to know whether different BnFT and BnTFL1 paralogs gained different function by studying their phenotypes and their transcriptional activities. Through digital gene expression analyses, differential expression within early generations of re-synthesized- (F1-F4) and natural B. napus accessions has been reported (Birchler and Veitia, 2010). Three highly similar genes encoding endoplasmic reticulum-bound sn-glycerol-3-phosphate acyltransferase-4 (BnGPAT4-C1, BnGPAT4-C2, and BnGPAT4-A1) showed different expression patterns and altered epigenetic features (Chen et al., 2011) which is in accordance with the assumption that in polyploids orthologous genes are frequently expressed in a non-additive manner (Jiang et al., 2013a). We have also observed marked differences in the expression of six BnFT paralogs in support of the non/sub-functionalisation hypothesis. In regard to their position within a major flowering time QTL (Wang et al., 2009), BnC6FTb paralogs seem to play the most important role as flowering time regulators in winter type B. napus. More evidence has been given by the expression analysis of putative FT downstream targets BnAP1 and BnSOC1 in late-flowering BnC6FTbG2009A mutants. SOC1 encodes a MADS-box transcription factor, acting as a floral integrator (Lee and Lee, 2010). The gene SOC1 gene is expressed in the shoot apical meristem, and SOC1 mutations lead to late flowering phenotype (Borner et al., 2000). However, SOC1 is also expressed in vegetative organs (leaves) (Hepworth et al., 2002). A reduction in SOC1 mRNA was detected in the meristem of late-flowering ft-7 (Trp138Stop) Arabidopsis Ler background (Searle et al., 2006). In the future, the activities of these genes shall also be studied in the shoot apical meristem.
The phenotypic studies presented here gave further support to our assumption that BnFT paralogs do not contribute equally to flowering time regulation. In sugar beet (Beta vulgaris), two FT paralogs (BvFT1 and BvFT2) were reported to antagonistically regulate flowering time (Pin et al., 2010). Knockdown of the FT potato paralog StSP3D resulted in a late flowering phenotype, while knockdown of the second paralog StSP6A had no effect on flowering time but on tuberization (Navarro et al., 2011). In line with these findings, we observed differential effects of BnC6FTa and BnC6FTb mutations. The strong effect of BnC6FTb as a flowering time regulator in rapeseed has been confirmed. To which extend BnC6FTa and other BnFT paralogs are involved in flowering time control needs to be investigated in the future (e.g., by using other TILLING mutants). As determined by our expression analyses, the strongest case of non-functionalization is shown by the lack of expression of the BnC2FT copy. This result is in full congruence with Wang et al. report (2012a) where this gene copy was neither expressed in B. napus nor in B. oleracea. A series of recent studies has demonstrated that beyond flowering time control, FT-like proteins act as mobile or cell-autonomous proteins that mediate other developmental processes, such as growth, plant architecture, and tuber formation (Carmona et al., 2007; Kinoshita et al., 2011; Navarro et al., 2011). In contrast to our BnFT mutant results, a previous analysis of sinapine biosynthesis mutants from the same EMS population as in our study, phenotypic or physiological effects had been observed only in double mutants (Harloff et al., 2012, Harloff, personal communication).
Although yield heterosis is regarded as a quantitative trait, single genes can contribute to heterotic effects through overdominance, such as the Arabidopsis Erecta locus (Moore and Lukens, 2011). As a first example of single gene overdominance, the yield of sft-4537/± heterozygous tomato plants was increased by up to 60% in comparison to their parents after crossing high yielding M82 inbred plants with low-yielding homozygous loss of function mutants (sft-4537) (Krieger et al., 2010). In our work, F1 hybrids carrying a BnTFL1-2 mutant allele had a higher seed yield as the Express 617 parent. Our study delivers the first insights about potential TFL1-related heterosis in B. napus. In the future, experimental data are needed to verify this hypothesis. We tested our initial hypothesis by crossing mutants with the non-mutated donor line. Although, the data point at BnTFL1 as a major gene for heterosis in rapeseed the possible impact of background mutations must be analyzed by additional hybrid combinations. Moreover, we will sequence the BnTFL1-2 loci from rapeseed lines with high and low combining ability (Qian et al., 2007). If our preliminary greenhouse data will be confirmed by field experiments, this study will offer new perspectives for a hybrid breeding strategy which make use of BnTFL1-2 sequence variations.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors gratefully acknowledge technical support by Verena Kowalewski, Bettina Rohardt, and Monika Bruisch. We thank the breeding company Norddeutsche Pflanzenzucht Hans-Georg Lembke KG (Hohenlieth) for providing us the Express inbred line and the male sterile material. We thank Professor Rod Snowdon and Mrs. Sarah Schiessl (University of Giessen, Germany) for providing the B. napus ASSYST materials. Yuan Guo acknowledges the China Scholarship Council for financial support. This work has been funded by the German research foundation (DFG) in the frame of the priority program PP1530 “Flowering Time Control—from Natural Variation to Crop Improvement” (grant No. JU205/19-1).
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/jouranl/10.3389/fpls.2014.00282/abstract
References
Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., et al. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056. doi: 10.1126/science.111598
Ahn, J. H., Miller, D., Winter, V. J., Banfield, M. J., Lee, J. H., Yoo, S. Y., et al. (2006). A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 25, 605–614. doi: 10.1038/sj.emboj.7600950
Amasino, R. (2010). Seasonal and developmental timing of flowering. Plant J. 61, 1001–1013. doi: 10.1111/j.1365-313X.2010.04148.x
Andres, F., and Coupland, G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639. doi: 10.1038/nrg3291
Basunanda, P., Radoev, M., Ecke, W., Friedt, W., Becker, H. C., and Snowdon, R. J. (2010). Comparative mapping of quantitative trait loci involved in heterosis for seedling and yield traits in oilseed rape (Brassica napus L.). Theor. Appl. Genet. 120, 271–281. doi: 10.1007/s00122-009-1133-z
Birchler, J. A., and Veitia, R. A. (2010). The gene balance hypothesis: implications for gene regulation, quantitative traits and evolution. New Phytol. 186, 54–62. doi: 10.1111/j.1469-8137.2009.03087.x
Böhlenius, H., Huang, T., Charbonnel-Campaa, L., Brunner, A. M., Jansson, S., Strauss, S. H., et al. (2006). CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312, 1040–1043. doi: 10.1126/science.1126038
Borner, R., Kampmann, G., Chandler, J., Gleissner, R., Wisman, E., Apel, K., et al. (2000). A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J. 24, 591–599. doi: 10.1046/j.1365-313x.2000.00906.x
Bradley, D., Ratcliffe, O., Vincent, C., Carpenter, R., and Coen, E. (1997). Inflorescence commitment and architecture in Arabidopsis. Science 275, 80–83. doi: 10.1126/science.275.5296.80
Bus, A., Körber, N., Snowdon, R., and Stich, B. (2011). Patterns of molecular variation in a species-wide germplasm set of Brassica napus. Theor. Appl. Genet. 123, 1413–1423. doi: 10.1007/s00122-011-1676-7
Carmona, M. J., Calonje, M., and Martinez-Zapater, J. M. (2007). The FT/TFL1 gene family in grapevine. Plant Mol. Biol. 63, 637–650. doi: 10.1007/s11103-006-9113-z
Chen, X., Truksa, M., Snyder, C. L., El-Mezawy, A., Shah, S., and Weselake, R. J. (2011). Three homologous genes encoding sn-glycerol-3-phosphate acyltransferase 4 exhibit different expression patterns and functional divergence in Brassica napus. Plant Physiol. 155, 851–865. doi: 10.1104/pp.110.169482
Conant, G. C., and Wolfe, K. H. (2008). Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9, 938–950. doi: 10.1038/nrg2482
Faure, S., Higgins, J., Turner, A., and Laurie, D. A. (2007). The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare). Genetics 176, 599–609. doi: 10.1534/genetics.106.069500
Force, A., Lynch, M., Pickett, F. B., Amores, A., Yan, Y. L., and Postlethwait, J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545.
Frerichmann, S., Kirchhoff, M., Muller, A., Scheidig, A., Jung, C., and Kopisch-Obuch, F. (2013). EcoTILLING in beta vulgaris reveals polymorphisms in the FLC-like gene BvFL1 that are associated with annuality and winter hardiness. BMC Plant Biol. 13:52. doi: 10.1186/1471-2229-13-52
Girke, A., Schierholt, A., and Becker, H. C. (2012). Extending the rapeseed gene pool with resynthesized Brassica napus II: heterosis. Theor. Appl. Genet. 124, 1017–1026. doi: 10.1007/s00122-011-1765-7
Hanano, S., and Goto, K. (2011). Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell 23, 3172–3184. doi: 10.1105/tpc.111.088641
Hanzawa, Y., Money, T., and Bradley, D. (2005). A single amino acid converts a repressor to an activator of flowering. Proc. Natl. Acad. Sci. U.S.A. 102, 7748–7753. doi: 10.1073/pnas.0500932102
Harloff, H. J., Lemcke, S., Mittasch, J., Frolov, A., Wu, J. G., Dreyer, F., et al. (2012). A mutation screening platform for rapeseed (Brassica napus L.) and the detection of sinapine biosynthesis mutants. Theor. Appl. Genet. 124, 957–969. doi: 10.1007/s00122-011-1760-z
Hecht, V., Laurie, R. E., Vander Schoor, J. K., Ridge, S., Knowles, C. L., Liew, L. C., et al. (2011). The pea GIGAS gene is a FLOWERING LOCUS T homolog necessary for graft-transmissible specification of flowering but not for responsiveness to photoperiod. Plant Cell 23, 147–161. doi: 10.1105/tpc.110.081042
Hepworth, S. R., Valverde, F., Ravenscroft, D., Mouradov, A., and Coupland, G. (2002). Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J. 21, 4327–4337. doi: 10.1093/emboj/cdf432
Iniguez-Luy, F. L., and Federico, M. L. (2011). The Genetics of Brassica napus. Genet. Genomics Brassicaceae 9, 291–322. doi: 10.1007/978-1-4419-7118-0_10
Jiang, J., Shao, Y., Du, K., Ran, L., Fang, X., and Wang, Y. (2013a). Use of digital gene expression to discriminate gene expression differences in early generations of resynthesized Brassica napus and its diploid progenitors. BMC Genomics 14:72. doi: 10.1186/1471-2164-14-72
Jiang, K., Liberatore, K. L., Park, S. J., Alvarez, J. P., and Lippman, Z. B. (2013b). Tomato yield heterosis is triggered by a dosage sensitivity of the florigen pathway that fine-tunes shoot architecture. PLoS Genet. 9:e1004043. doi: 10.1371/journal.pgen.1004043
Kaufmann K., Nagasaki M., and Jauregui R. (2010). Modelling the molecular interactions in the flower developmental network of Arabidopsis thaliana. In Silico Biol. 10, 125–143. doi: 10.3233/ISB-2010-0414
Kardailsky, I., Shukla, V. K., Ahn, J. H., Dagenais, N., Christensen, S. K., Nguyen, J. T., et al. (1999). Activation tagging of the floral inducer FT. Science 286, 1962–1965. doi: 10.1126/science.286.5446.1962
Kinoshita, T., Ono, N., Hayashi, Y., Morimoto, S., Nakamura, S., Soda, M., et al. (2011). FLOWERING LOCUS T regulates stomatal opening. Curr. Biol. 21, 1232–1238. doi: 10.1016/j.cub.2011.06.025
Kojima, S., Takahashi, Y., Kobayashi, Y., Monna, L., Sasaki, T., Araki, T., et al. (2002). Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43, 1096–1105. doi: 10.1093/pcp/pcf156
Krieger, U., Lippman, Z. B., and Zamir, D. (2010). The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat. Genet. 42, 459–463. doi: 10.1038/ng.550
Lee, J., and Lee, I. (2010). Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 61, 2247–2254. doi: 10.1093/jxb/erq098erq098
Long, Y., Shi, J., Qiu, D., Li, R., Zhang, C., Wang, J., et al. (2007). Flowering time quantitative trait loci analysis of oilseed brassica in multiple environments and genomewide alignment with Arabidopsis. Genetics 177, 2433–2444. doi: 10.1534/genetics.107.080705
Mimida, N., Goto, K., Kobayashi, Y., Araki, T., Ahn, J. H., Weigel, D., et al. (2001). Functional divergence of the TFL1-like gene family in Arabidopsis revealed by characterization of a novel homologue. Genes Cells 6, 327–336. doi: 10.1046/j.1365-2443.2001.00425.x
Mimida, N., Sakamoto, W., Murata, M., and Motoyoshi, F. (1999). TERMINAL FLOWER 1-like genes in Brassica species. Plant Sci. 142, 155–162. doi: 10.1016/S0168-9452(99)00020-5
Molinero-Rosales, N., Latorre, A., Jamilena, M., and Lozano, R. (2004). SINGLE FLOWER TRUSS regulates the transition and maintenance of flowering in tomato. Planta 218, 427–434. doi: 10.1007/s00425-003-1109-1
Moon, J., Suh, S. S., Lee, H., Choi, K. R., Hong, C. B., Paek, N. C., et al. (2003). The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 35, 613–623. doi: 10.1046/j.1365-313X.2003.01833.x
Moore, S., and Lukens, L. (2011). An evaluation of Arabidopsis thaliana hybrid traits and their genetic control. G3 (Bethesda) 1, 571–579. doi: 10.1534/g3.111.001156
Morjane, H., Geistlinger, J., Harrabi, M., Weising, K., and Kahl, G. (1994). Oligonucleotide fingerprinting detects genetic diversity among Ascochyta rabiei isolates from a single chickpea field in Tunisia. Curr. Genet. 26, 191–197. doi: 10.1007/BF00309547
Navarro, C., Abelenda, J. A., Cruz-Oró, E., Cuéllar, C. A., Tamaki, S., Silva, J., et al. (2011). Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 478, 119–122. doi: 10.1038/nature10431
Papp, B., Pal, C., and Hurst, L. D. (2003). Dosage sensitivity and the evolution of gene families in yeast. Nature 424, 194–197. doi: 10.1038/nature01771
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acid Res. 1, 29, e45. doi: 10.1093/nar/29.9.e45
Pin, P. A., Benlloch, R., Bonnet, D., Wremerth-Weich, E., Kraft, T., Gielen, J. J., et al. (2010). An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 330, 1397–1400. doi: 10.1126/science.1197004
Pin, P. A., and Nilsson, O. (2012). The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ. 35, 1742–1755. doi: 10.1111/j.1365-3040.2012.02558.x
Qian, W., Sass, O., Meng, J., Li, M., Frauen, M., and Jung, C. (2007). Heterotic patterns in rapeseed (Brassica napus L.): I. crosses between spring and Chinese semi-winter lines. Theor. Appl. Genet. 115, 27–34. doi: 10.1007/s00122-007-0537-x
Qiu, D., Morgan, C., Shi, J., Long, Y., Liu, J., Li, R., et al. (2006). A comparative linkage map of rapeseed and its use for QTL analysis of seed oil and erucic acid content. Theor. Appl. Genet. 114, 67–80. doi: 10.1007/s00122-006-0411-2
Schranz, M. E., Lysak, M. A., and Mitchell-Olds, T. (2006). The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 11, 535–542. doi: 10.1016/j.tplants.2006.09.002
Searle, I., He, Y., Turck, F., Vincent, C., Fornara, F., Krober, S., et al. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20, 898–912. doi: 10.1101/gad.373506
Shi, J., Li, R., Qiu, D., Jiang, C., Long, Y., Morgan, C., et al. (2009). Unraveling the complex trait of crop yield with quantitative trait loci mapping in Brassica napus. Genetics 182, 851–861. doi: 10.1534/genetics.109.101642
Till, B., Cooper, J., Tai, T., Colowit, P., Greene, E., Henikoff, S., et al. (2007). Discovery of chemically induced mutations in rice by TILLING. BMC Plant Biol. 7:19. doi: 10.1186/1471-2229-7-19
Turck, F., Fornara, F., and Coupland, G. (2008). Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59, 573–594. doi: 10.1146/annurev.arplant.59.032607.092755
Valverde, F. (2011). CONSTANS and the evolutionary origin of photoperiodic timing of flowering. J. Exp. Bot. 62, 2453–2463. doi: 10.1093/jxb/erq449
Wang, J., Hopkins, C. J., Hou, J., Zou, X., Wang, C., Long, Y., et al. (2012a). Promoter variation and transcript divergence in brassicaceae lineages of FLOWERING LOCUS T. PLoS ONE 7:e47127. doi: 10.1371/journal.pone.0047127
Wang, J., Long, Y., Wu, B., Liu, J., Jiang, C., Shi, L., et al. (2009). The evolution of Brassica napus FLOWERING LOCUS T paralogues in the context of inverted chromosomal duplication blocks. BMC Evol. Biol. 9:271. doi: 10.1186/1471-2148-9-271
Wang, T. L., Uauy, C., Robson, F., and Till, B. (2012c). TILLING in extremis. Plant Biotechnol. J. 10, 761–772. doi: 10.1111/j.1467-7652.2012.00708.x
Wang, X., Wang, H., Wang, J., Sun, R., Wu, J., Liu, S., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039. doi: 10.1038/ng.919
Wigge, P. A., Kim, M. C., Jaeger, K. E., Busch, W., Schmid, M., Lohmann, J. U., et al. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056–1059. doi: 10.1126/science.1114358
Yan, L., Fu, D., Li, C., Blechl, A., Tranquilli, G., Bonafede, M., et al. (2006). The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. U.S.A. 103, 19581–19586. doi: 10.1073/pnas.0607142103
Yoo, S. K., Chung, K. S., Kim, J., Lee, J. H., Hong, S. M., Yoo, S. J., et al. (2005). CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 139, 770–778. doi: 10.1104/pp.105.066928
Keywords: Flowering Locus-T, Terminal Flower-1, TILLING, rapeseed, allopolyploid, differential function
Citation: Guo Y, Hans H, Christian J and Molina C (2014) Mutations in single FT- and TFL1-paralogs of rapeseed (Brassica napus L.) and their impact on flowering time and yield components. Front. Plant Sci. 5:282. doi: 10.3389/fpls.2014.00282
Received: 04 February 2014; Accepted: 30 May 2014;
Published online: 17 June 2014.
Edited by:
Dorothee Staiger, Bielefeld University, GermanyReviewed by:
Mingsheng Chen, Chinese Academy of Sciences, ChinaFranziska Katharina Turck, Max Planck Society, Germany
Rod J. Snowdon, Justus Liebig University, Germany
Copyright © 2014 Guo, Hans, Christian and Molina. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carlos Molina, Plant Breeding Institute, University of Kiel, Olshausenstrasse 40, D-24098, Kiel, Germany e-mail: c.molina@plantbreeding.uni-kiel.de
†Present address: Carlos Molina, E-nema GmbH, Schwentinental, Germany
 Yuan Guo
Yuan Guo Harloff Hans
Harloff Hans Jung Christian
Jung Christian Carlos Molina
Carlos Molina