- 1Institute of Plant and Microbial Biology, Academia Sinica, Taipei, Taiwan
- 2Institute of Plant Biology, National Taiwan University, Taipei, Taiwan
Meiosis, a specialized cell division to produce haploid cells, marks the transition from a sporophytic to a gametophytic generation in the life cycle of plants. In angiosperms, meiosis takes place in sporogenous cells that develop de novo from somatic cells in anthers or ovules. A successful transition from the mitotic cycle to the meiotic program in sporogenous cells is crucial for sexual reproduction. By contrast, when meiosis is bypassed or a mitosis-like division occurs to produce unreduced cells, followed by the development of an embryo sac, clonal seeds can be produced by apomixis, an asexual reproduction pathway found in 400 species of flowering plants. An understanding of the regulation of entry into meiosis and molecular mechanisms of apomictic pathway will provide vital insight into reproduction for plant breeding. Recent findings suggest that AM1/SWI1 may be the key gene for entry into meiosis, and increasing evidence has shown that the apomictic pathway is epigenetically controlled. However, the mechanism for the initiation of meiosis during sexual reproduction or for its omission in the apomictic pathway still remains largely unknown. Here we review the current understanding of meiosis initiation and the apomictic pathway and raised several questions that are awaiting further investigation.
Introduction
Meiosis is an extremely important step in sexual reproduction. It is widely accepted that it evolved from mitosis and shares certain features with mitosis (Maynard Smith, 1978). Yet at least three meiosis-specific events make meiosis a specialized cell division: meiotic recombination and pairing between homologous chromosomes during prophase I, the suppression of sister-chromatid separation during the first meiotic division, and the absence of chromosome replication at the start of the second division (Kleckner, 1996). While these meiosis-specific events have been studied extensively, the mechanisms that switch mitosis into meiosis are still puzzling. In multicellular organisms, meiosis initiation takes place within multicellular organs; consequently, mechanisms that initiate meiosis must integrate developmental cues. In plants, the decision to start meiosis may also be connected with reproductive cell fate specification since plants do not have pre-determined germ lines. Thus, the switch of somatic fate to germinal cell fate and the mitosis–meiosis cell cycle transition occur sequentially during the development of reproductive organs (i.e., anthers and ovules; Ma, 2005). Importantly, these sexual processes can be replaced by the asexual apomictic pathway in which meiosis is bypassed or a mitosis-like division occurs to produce unreduced daughter cells, followed by the development of an embryo without fertilization, apomictic plants can then produce diploid seeds with identical genetic content to their maternal genome. This phenomenon is called apomixis that occurs naturally in some flowering plants (Barcaccia and Albertini, 2013). If apomixis is engineered into crops to produce clonal seeds, its application on agriculture will be broad and profound. Here, we review the current understanding of the cell cycle transition that directs sporogenous cells to leave the mitotic cell cycle and enter the meiotic program in higher plants and additionally discuss advances in the apomictic pathway.
What Have We Learned From Other Model Species About Initiation of Meiosis?
The cellular events during meiosis are evolutionarily conserved among species; however, the mechanisms controlling the initiation of meiosis are diverse (Pawlowski et al., 2007). The molecular controls elucidated to date involve signaling pathways, transcriptional and translational regulations of meiotic genes, and cyclin-dependent kinase (CDK) circuits. Although different mechanisms are adopted, the final readout is likely the activation of a specific cyclin–CDK complex to initiate the meiotic S phase. It was also suggested from many studies that the decision to start meiosis is made before the onset of the pre-meiotic S phase (Watanabe et al., 2001). Here, we first summarize the discoveries from several model species, and then discuss recent advances in plants.
The meiosis decision in single-celled yeasts is often cued by environmental conditions. In the budding yeast Saccharomyces cerevisiae, starvation induces expression of the Initiator of Meiosis I (IME1) gene, which encodes a transcription factor responsible for activating early meiotic genes (Chu et al., 1998). One of these target genes, IME2, which encodes a Ser/Thr protein kinase, promotes meiotic DNA replication by directly phosphorylating Rfa2, a subunit of replication protein A (Foiani et al., 1996; Clifford et al., 2005). Sic1, an inhibitor of the CDK (CDC28), is also phosphorylated by Ime2p and then leads to its degradation. Subsequently, CDC28, in conjunction with the B-type S phase cyclins, Clb5, and Clb6, triggers the initiation of the premeiotic S phase (Dirick et al., 1998; Stuart and Wittenberg, 1998).
In fission yeast S. pombe, a key transcription factor STE11, which is produced in response to environmental conditions, is responsible for early meiotic genes expression (Sugimoto et al., 1991). MEI2, an RRM-type RNA-binding protein, plays a crucial role in promoting entry into meiosis by regulating meiosis-specific mRNAs accumulation. During mitosis, MEI2 is inactivated by PAT1 kinase. Under meiosis-inducing conditions, this repression of MEI2 is released, allowing binding to and stabilization of meiosis-specific mRNAs at the G1 phase (Kitamura et al., 2001). In addition, this process reinforces stabilization also by sequestrating MMI1 protein, which function is to eliminate these meiotic mRNAs (Harigaya et al., 2006). Finally, CDC2 kinase binding with cyclin CIG2 is essential for entry into the pre-meiotic S phase (Borgne et al., 2002). Recently, protein S-palmitoylation, a lipid modification was also found to regulate the entry into meiosis (Zhang et al., 2013).
In mammals, meiosis is initiated at different stages of development in females and males (Bowles and Koopman, 2007). Mouse studies have revealed that retinoic acid (RA) produced during embryonic development can induce meiosis in both sexes. The level of RA is negatively regulated by the Cyp26b1 enzyme that has RA degradation activity (Bowles et al., 2006; Koubova et al., 2006). Stimulated by RA 8 (Stra8), a vertebrate-specific gene, is then induced by RA and is required for the transition to meiosis (Anderson et al., 2008). Stra8 plays no role in the mitotic phases of embryonic germ-cell development, but in females it is required for pre-meiotic DNA replication and the subsequent events of meiotic prophase. On the other hand, Dmrt1 represses Stra8 transcription in the mitotic phase, thereby preventing meiosis (Matson et al., 2010).
From these studies, the mechanisms that initiate meiosis are very different, and more importantly, the genes involved share no similarity. No doubt different strategies evolved because of the different reproductive requirements of diverse organisms.
The Decision of Mitosis–Meiosis Switch in Plants
In plants, meiosis is initiated in sporogenous cells that are differentiated in ovules and anthers (Bhatt et al., 2001). In each ovule, only a single megaspore mother cell (MMC) surrounded by the somatic nucellar cells is differentiated and then undergoes meiosis (Figure 1). During anther development, after primary sporogenous cells (i.e., the precursor of pollen mother cells, PMCs) are differentiated, they first undergo several rounds of mitosis to proliferate, and then meiosis occurs synchronously in all the PMCs of each anther (Figure 1; Palmer, 1971). Thus, the decision of mitosis–meiosis transition must coordinate with the developmental stages of anthers and ovules. For example, the signal that starts meiosis in an anther must be generated after complete development of the somatic layers of anthers (Kelliher and Walbot, 2011). Interestingly, the signal can also establish the synchronization of the meiotic cell cycle in an anther. On the other hand, only a single MMC in each ovule is specified to enter meiosis, which accompanies the development of ovule in parallel. Thus, the regulatory mechanism of meiosis initiation may be different between female and male in plants because of distinct development of sporogenesis.
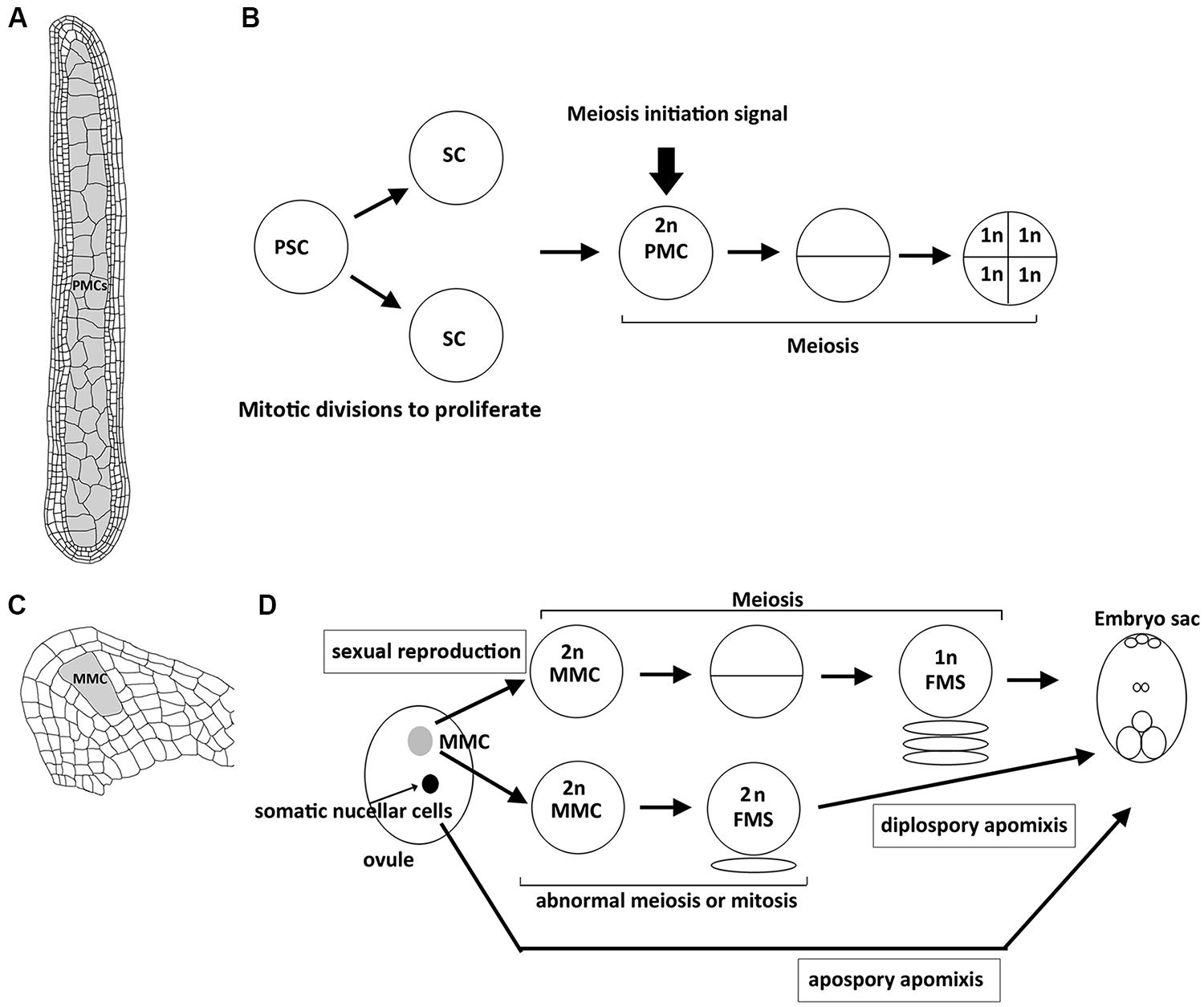
FIGURE 1. Structure of plant reproductive organs in maize and sequence of events leading to spore or gametophyte formation in anthers and ovules. (A) Longitudinal section of an anther with numerous pollen mother cells (PMCs, shown in gray) that are proliferated from primary sporogenous cells by mitosis, which accompanies the development of surrounding 4 layers of somatic cells. (B) After primary sporogenous cells (PSCs) are differentiated, they first undergo mitotic divisions to produce sporogenous cells (SCs) and further develop into PMCs. By the time when the development of surrounding somatic cells (shown in A) is complete, unknown meiosis initiation siganl is generated to start meisois synchronously in all PMCs of an anther. Each PMC enters meiosis to produce four haploid spore cells. (C) Longitudinal section of an ovule with a single megaspore mother cell (MMC, shown in gray). (D) Schematic illustration showing the sequential development of embryo sac through sexual reproduction or apomictic pathways. In sexual reproduction, the single MMC (shown in gray) is differentiated and then enters meiosis to produce a haploid functional megaspore (FMS), and then develops into an embryo sac. In diplospory apomixis, the specified MMC unergoes an abnormal meiosis or mitosis to produce a diploid FMS. In apospory apomixis, somatic nucellar cells develop into embryo sac without meiosis.
The first discovery about meiosis initiation was the isolation of the maize ameiotic1 (am1) mutant by Rhoades (1956). The original am1 mutant allele does not undergo meiosis; instead mitosis-like divisions take place in well-developed meiocytes in both female and male organs (Golubovskaya et al., 1993). Am1 encodes a plant specific coiled-coil protein with unknown functions (Pawlowski et al., 2009). All five null mutant alleles display identical phenotypes in male meiocytes in which mitosis replaces meiosis. However, female MMCs in the mutant may either undergo mitosis, or arrest at interphase. Interestingly, the am1-praI allele carrying a single amino acid substitution (R358W) can enter meiosis but cells arrest in the leptotene/zygotene stage, resembling the phenotype of the rice am1 mutant that also carries an amino acid substitution (R360W) in a conserved region (Golubovskaya et al., 1997; Pawlowski et al., 2009; Che et al., 2011). These results suggest that AM1 is required for meiosis initiation and may also regulate meiotic progression. In contrast to maize and rice, mutants in the closest homolog of Am1 in Arabidopsis, switch1/dyad (swi1), exhibit abnormal meiosis with sister chromatid cohesion defects in male meiocytes, and the mitosis-like division was only observed in female meiocytes (Mercier et al., 2001, 2003). These differences among species may indicate that the AM1-related genes have undergone species-specific diversification.
While the molecular functions of AM1/SWI1 are still unknown, microarray analyses showed that AM1 is required for normal expression of many meiotic genes (Nan et al., 2011). Using Agilent 44K microarrays, the authors compared transcriptomes in 1-mm and 1.5-mm anthers of am1-489 (null allele) and am1-praI (point mutant allele) and their fertile siblings. In 1-mm anthers when meiosis is about to start in the wild-type, 484 genes were missing and 1208 genes were ectopically expressed in am1-489 anthers. These genes are considered to contribute to the initiation of meiosis or the suppression of mitosis. In 1.5-mm anthers, during prophase I in the wild-type, 3700 transcripts were missing and another 3107 genes were differentially expressed in am1-489 anthers. Nearly 60% of transcriptome changes, regardless of stage, were genes enriched in PMCs and many putative meiosis-related genes were found among them. However, none of the meiosis-related genes were regulated in an absolute On/Off pattern on the am1-489 allele, somewhat surprising given that the am1-489 PMCs perform mitosis instead of abnormal meiosis. These results redefine the role of AM1 in the modulation of transcript accumulation for many meiotic genes rather than simply switching them on or off (Nan et al., 2011).
Recently, microarray analyses on laser-captured germinal and somatic initials from maize 0.3-mm anthers (right after sporogenous cells are differentiated) found about 2500 genes specific or enriched in germinal initials (Kelliher and Walbot, 2014). Surprisingly, more than 100 meiotic genes are expressed in the mitotic amplification period that is long before the onset of meiosis initiation. This finding raises a possibility that precocious expression of meiotic genes permits gradual dilution of mitotic chromatin components, a hypothesis recently proposed for the mouse germ-line (Hackett et al., 2013). Another possibility is that those PMC precursors are preparing for meiosis at the transcriptional level, and may store some meiotic transcripts for translation at later developmental stages (Zhang et al., 2014). Regardless, this finding suggests that the decision to start meiosis is a series of consecutive steps rather than a single switch. Perhaps, the expression of meiotic genes may be one of the earliest actions, and the following regulatory cascade finally governs the initiation and progression of meiosis. Thus, which transcription factors are responsible for early meiotic gene expression and whether meiotic genes are under translational control are interesting questions for further study. In addition, identification of components in the regulatory cascade will provide better understanding of this process.
Another gene that has been reported to be involved in meiosis initiation is rice MEL2, named for its “meiosis arrested at leptotene” phenotype. MEL2 encodes an RNA-recognition-motif (RRM) protein, and it is required for regulating the premeiotic G1/S-phase transition of male and female germ cells (Nonomura et al., 2011) as most germ cells fail to enter pre-meiotic S phase in mel2 mutant. A small proportion of PMCs can escape from the defects and undergo meiosis with a significant delay or continued mitotic cycles. How an RRM protein affects the initiation of meiosis is unclear at the molecular level, but this result implied a possible link between mRNA processing, transport or stability, and entry into meiosis in plants. Studies in yeast have shown that the final trigger to start meiosis is the activation of specific cyclin–CDK complexes to initiate the meiotic S phase. Arabidopsis has at least 50 cyclins and only a few of them are specifically expressed in the inflorescence (Bulankova et al., 2013). Mutant analyses revealed that some of these cyclins contribute to distinct meiosis-related processes, but none of cyclin mutants showed meiosis initiation defects, which was attributed to gene redundancy. Thus, it will be interesting to know which, if any, cyclin–CDK complex is responsible for the transition. Besides cyclin–CDK complexes, some meiosis-specific regulators, such as replication factor MUM2 and cohesion protein REC8, are involved at the meiotic S phase although much of the basic replication apparatus is employed (Strich, 2004). Therefore, what is special about the pre-meiotic S phase and which are the specific genes that differ from the mitotic S phase in plants? Understanding of these meiosis-specific components at meiotic S phase will help us to illustrate the molecular mechanisms of meiosis initiation. A proteomics study may offer valuable information on this aspect.
To date, mutants directly affecting meiosis initiation showed similar phenotypes in that some of reproductive cells fail to enter meiosis in either female, male, or both sexes. Although some of these mutants produce unreduced daughter cells by mitosis-like division, there is no evidence that these resulting diploid cells in ovules would undergo the apomictic pathway without fertilization. However, an interesting study demonstrated that in the Arabidopsis swi1/dyad mutant, few seeds were produced when pollinated with wild-type pollens. Most of the progeny were triploid, suggesting that unreduced female daughter cells after mitosis-like division are able to develop further and be fertilized by haploid male gametes (Ravi et al., 2008).
Current Advances in the Apomictic Pathway
Apomixis is a type of asexual reproduction through seeds that avoid both meiosis and fertilization. In the apomictic pathway, differentiated MMCs or other somatic cells in ovules that gain germinal cell fate are able to bypass meiosis or undergo an abnormal meiosis to produce unreduced spores that further divide mitotically to form an embryo sac (Figure 1; Koltunow, 1993; Carman, 1997). Although apomixis is genetically regulated and occurs naturally in more than 400 species of flowering plants, its implementation at the molecular level is still unclear. Over the past few years, there has been increasing evidence to show that epigenetic control may regulate apomixis. In Arabidopsis, argonaute 9 (ago9) mutants exhibit multiple MMCs compared to a single MMC in the wild-type ovule, and additional MMCs in the mutant are able to initiate gametogenesis without undergoing meiosis, resembling apospory (Figure 1; Olmedo-Monfil et al., 2010). AGO9 preferentially interacts with 24-nucleotide small interfering RNAs (siRNA) derived from transposable elements to direct homolog-based RNA-dependent DNA methylation (RdDM). Moreover, mutations in SUPPRESSOR OF GENE SILENCING3 (SGS3) and RNA-DEPENDENT RNA POLYMERASE6 (RDR6), two genes required for siRNA biogenesis, also lead to an identical defect to that in ago9 mutants (Olmedo-Monfil et al., 2010). Similarly, maize AGO104, the homolog of Arabidopsis AGO9, is found to regulate reproductive fate despite some differences between maize ago104 and Arabidopsis ago9 phenotypes (Singh et al., 2011). The maize ago104 mutant has a single MMC; however, defective female meiosis with aberrant condensation results in functional female gametes with an unreduced chromosome set, resembling diplospory (Figure 1). Additionally, AGO104 is required for heterochromatic CHG and CHH methylation. Consistent with the idea that epigenetics regulates apomixis, mutations of two DNA methyltransferases, DMT102 and DMT103 in maize, also exhibit apomictic development (Garcia-Aguilar et al., 2010). Thus, loss of RdDM seems to direct somatic cells to distinct reproductive cells with an apomictic fate (seen in Arabidopsis ago9 mutant) or lead to apomixis in correctly specified MMCs (seen in maize ago104 mutant). Interestingly, both AGO9 in Arabidopsis and AGO104 in maize are specifically expressed in surrounding somatic nucellar cells, and not in the reproductive cells, implying that both genes control the apomictic pathway in a non-cell-autonomous manner. siRNAs produced from somatic cells may move to germinal cells to regulate the chromatin state by suppressing transposable elements. Indeed, many transposable elements are silenced in Arabidopsis wild-type ovules, in an AGO9-dependent manner (Durán-Figueroa and Vielle-Calzada, 2010). These results suggest a link between siRNA-dependent chromatin remodeling and the apomictic pathway (Garcia-Aguilar et al., 2010; Grimanelli, 2012).
Another gene belonging to the ARGONAUTE family with meiotic phenotypes is rice MEL1. It encodes an AGO5 protein that is required for maintaining germ cell identity and normal meiosis progression. Interestingly, the mel1 mutant also shows defective chromosome condensation with abnormal pericentromere histone modification (Nonomura et al., 2007). Recently, MEL1 has been shown to bind to 21-nucleotide phased small interfering RNA (Komiya et al., 2014). Further investigation is needed to understand the epigenetic regulation of plant reproduction.
Over the past few years, the identification of mutants has shed light on genetic control of epigenetic mechanisms involved in apomixis. However, it is still not clear how the RdDM-dependent process affects cell fate specification, meiosis, and gametophyte development? Why is there a need for transposes-derived siRNA in the germ line? Is it possible that RdDM resets cell fate in the germ line, a role also demonstrated for the animal PIWI pathway (Houwing et al., 2008; Juliano et al., 2011)? Perhaps, identifying the targets of the RdDM pathway at different stages will be essential for further definition of their roles. In addition, what is the relationship between AM1/SWI1-dependent meiosis initiation and the RdDM pathway? It is worth noting that alterations in histone modification were observed in the swi1 mutant (Boateng et al., 2008), raising the possibility that somehow AM1/SWI1 is involved in chromatin remodeling. Many exciting questions are awaiting further investigation.
Conclusion
Understanding the initiation of meiosis and apomixis in plants will be enlightening, and may have many potential applications for plant breeding and in agriculture including developing a strategy for acquiring apomixis in crops, and allowing manipulation of the meiotic cell cycle. It will be crucial to identify more participants in the mitosis–meiosis decision and the apomictic pathway and to explore their molecular functions.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the members of the Wang lab for insightful discussions and critical comments. This work was supported by the Career Development Award from Academia Sinica, Taiwan.
References
Anderson, E. L., Baltus, A. E., Roepers-Gajadien, H. L., Hassold, T. J., de Rooij, D. G., van Pelt, A. M.et al. (2008). Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc. Natl. Acad. Sci. U.S.A. 105, 14976–14980. doi: 10.1073/pnas.0807297105
Barcaccia, G., and Albertini, E. (2013). Apomixis in plant reproduction: a novel perspective on an old dilemma. Plant Reprod. 26, 159–179. doi: 10.1007/s00497-013-0222-y
Bhatt, A. M., Canales, C., and Dickinson, H. G. (2001). Plant meiosis: the means to 1N. Trends Plant Sci. 6, 114–121. doi: 10.1016/S1360-1385(00)01861-6
Boateng, K. A., Yang, X., Dong, F., Owen, H. A., and Makaroff, C. A. (2008). SWI1 is required for meiotic chromosome remodeling events. Mol. Plant. 4, 620–633. doi: 10.1093/mp/ssn030
Borgne, A., Murakami, H., Ayté, J., and Nurse, P. (2002). The G1/S cyclin Cig2p during meiosis in fission yeast. Mol. Biol. Cell. 13, 2080–2090. doi: 10.1091/mbc.01-10-0507
Bowles, J., Knight, D., Smith, C., Wilhelm, D., Richman, J., Mamiya, S.,et al. (2006). Retinoid signaling determines germ cell fate in mice. Science 312, 596–600. doi: 10.1126/science.1125691
Bowles, J., and Koopman, P. (2007). Retinoic acid, meiosis and germ cell fate in mammals. Development 134, 3401–3411. doi: 10.1242/dev.001107
Bulankova, P., Akimcheva, S., Fellner, B., and Riha, K. (2013). Identification of Arabidopsis meiotic cyclins reveals functional diversification among plant cyclin genes. PLoS Genet. 9:e1003508. doi: 10.1371/journal.pgen.1003508
Carman, J. G. (1997). Asynchronous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biol. J. Linn. Soc. 61, 51–94. doi: 10.1111/j.1095-8312.1997.tb01778.x
Che, L., Tang, D., Wang, K., Wang, M., Zhu, K., Yu, H.,et al. (2011). OsAM1 is required for leptotene-zygotene transition in rice. Cell Res. 21, 654–665. doi: 10.1038/cr.2011.7
Chu, S., DeRisi, J., Eisen, M., Mulholland, J., Botstein, D., Brown, P. O.,et al. (1998). The transcriptional program of sporulation in budding yeast. Science 282, 699–705. doi: 10.1126/science.282.5389.699
Clifford, D. M., Stark, K. E., Gardner, K. E., Hoffmann-Benning, S., and Brush, G. S. (2005). Mechanistic insight into the Cdc28-related protein kinase Ime2 through analysis of replication protein A phosphorylation. Cell Cycle 12, 1826–1833. doi: 10.4161/cc.4.12.2214
Dirick, L., Goetsch, L., Ammerer, G., and Byers, B. (1998). Regulation of meiotic S phase by Ime2 and a Clb5,6-associated kinase in Saccharomyces cerevisiae. Science 281, 1854–1857. doi: 10.1126/science.281.5384.1854
Durán-Figueroa, N., and Vielle-Calzada, J. P. (2010). ARGONAUTE9-dependent silencing of transposable elements in pericentromeric regions of Arabidopsis. Plant Signal. Behav. 5, 1476–1479. doi: 10.1038/nature08828
Foiani, M., Nadjar-Boger, E., Capone, R., Sagee, S., Hashimshoni, T., and Kassir, T. (1996). A meiosis-specific protein kinase, Ime2, is required for the correct timing of DNA replication and for spore formation in yeast meiosis. Mol. Gen. Genet. 253, 278–288. doi: 10.1007/s004380050323
Garcia-Aguilar, M., Michaud, C., Leblanc, O., and Grimanelli, D. (2010). Inactivation of a DNA methylation pathway in maize reproductive organs results in apomixis-like phenotypes. Plant Cell 22, 3249–3267. doi: 10.1105/tpc.109.072181
Golubovskaya, I., Avalkina, N., and Sheridan, W. F. (1997). New insights into the role of the maize ameiotic1 locus. Genetics 147, 1339–1350.
Golubovskaya, I., Grebennikova, Z. K., Avalkina, N. A., and Sheridan, W. F. (1993). The role of the ameioticl gene in the initiation of meiosis and in subsequent meiotic events in maize. Genetics 135, 1151–1166.
Grimanelli, D. (2012). Epigenetic regulation of reproductive development and the emergence of apomixis in angiosperms. Curr. Opin. Plant Biol. 15, 57–62. doi: 10.1016/j.pbi.2011.10.002
Hackett, J. A., Sengupta, R., Zylicz, J. J., Murakami, K., Lee, C., Down, T. A.,et al. (2013). Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 339, 448–452. doi: 10.1126/science.1229277
Harigaya, Y., Tanaka, H., Yamanaka, S., Tanaka, K., Watanabe, Y., Tsutsumi, C.,et al. (2006). Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature 442, 45–50. doi: 10.1038/nature04881
Houwing, S., Berezikov, E., and Ketting, R. F. (2008). Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J. 27, 2702–2711. doi: 10.1038/emboj.2008.204
Juliano, C., Wang, J., and Lin, H. (2011). Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annu. Rev. Genet. 45, 447–469. doi: 10.1146/annurev-genet-110410-132541
Kelliher, T., and Walbot, V. (2011). Emergence and patterning of the five cell types of the Zea mays anther locule. Dev. Biol. 350, 32–49. doi: 10.1016/j.ydbio.2010.11.005
Kelliher, T., and Walbot, V. (2014). Maize germinal cell initials accommodate hypoxia and precociously express meiotic genes. Plant J. 77, 639–652. doi: 10.1111/tpj.12414
Kitamura, L., Katayama, S., Dhut, S., Sato, M., Watanabe, Y., Yamamoto, M.,et al. (2001). Phosphorylation of Mei2 and Ste11 by Pat1 kinase inhibits sexual differentiation via ubiquitin proteolysis and 14-3-3 protein in fission yeast. Dev. Cell. 1, 389–399. doi: 10.1016/S1534-5807(01)00037-5
Kleckner, N. (1996). Meiosis: how could it work. Proc. Natl. Acad. Sci. U.S.A. 93, 8167–8174. doi: 10.1073/pnas.93.16.8167
Koltunow, A. M. (1993). Apomixis: embryo sacs and embryos formed without meiosis or fertilization in ovules. Plant Cell 5, 1425–1437. doi: 10.1105/tpc.5.10.1425
Komiya, R., Ohyanagi, H., Niihama, M., Watanabe, T., Nakano, M., Kurata, N.,et al. (2014). Rice germline-specific Argonaute MEL1 protein binds to phasiRNAs generated from more than 700 lincRNAs. Plant J. 78, 385–397. doi: 10.1111/tpj.12483
Koubova, J., Menke, D. B., Zhou, Q., Capel, B., Griswold, M. D., and Page, D. C. (2006). Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proc. Natl. Acad. Sci. U.S.A. 103, 2474–2479. doi: 10.1073/pnas.0510813103
Ma, H. (2005). Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plant. Annu. Rev. Plant Biol. 56, 393–434. doi: 10.1146/annurev.arplant.55.031903.141717
Matson, C. K., Murphy, M. W., Griswold, M. D., Yoshida, S., Bardwell, V. J., and Zarkower, D. (2010). The mammalian doublesex homolog DMRT1 is a transcriptional gatekeeper that controls the mitosis versus meiosis decision in male germ cells. Dev. Cell. 19, 612–624. doi: 10.1016/j.devcel.2010.09.010
Mercier, R., Armstrong, S. J., Horlow, C., Jackson, N. P., Makaroff, C. A., Vezon, D.,et al. (2003). The meiotic protein SWI1 is required for axial element formation and recombination initiation in Arabidopsis. Development 130, 3309–3318. doi: 10.1242/dev.00550
Mercier, R., Vezon, D., Bullier, E., Motamayor J. C., Sellier A., Lefevre, F.,et al. (2001). SWITCH1 (SWI1): a novel protein required for the establishment of sister chromatid cohesion and for bivalent formation at meiosis. Gene Dev. 15, 1859–1871. doi: 10.1101/gad.203201
Nan, G. L., Ronceret, A., Wang, R. C., Fernandes, J. F., Cande, W. Z., and Walbot, V. (2011). Global transcriptome analysis of two ameiotic1 alleles in maize anthers: defining steps in meiotic entry and progression through prophase I. BMC Plant Biol. 11:120. doi: 10.1186/1471-2229-11-120
Nonomura, K., Eiguchi, M., Nakano, M., Takashima, K., Komeda, N., Fukuchi, S.,et al. (2011). A novel RNA-recognition-motif protein is required for premeiotic G1/S-phase transition in rice (Oryza sativa L.). PLoS Genet. 7:e1001265. doi: 10.1371/journal.pgen.1001265
Nonomura, K., Morohoshi, A., Nakano, M., Eiguchi, M., Miyao, A., Hirochika, H.,et al. (2007). A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell 19, 2583–2594. doi: 10.1105/tpc.107.053199
Olmedo-Monfil, V., Durán-Figueroa, N., Arteaga-Vázquez, M., Demesa-Arévalo, E., Autran, D., Grimanelli, D.,et al. (2010). Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 464, 628–632. doi: 10.1038/nature08828
Palmer, R. G. (1971). Cytological studies of ameiotic and normal maize with reference to premeiotic pairing. Chromosoma 35, 233–246. doi: 10.1007/BF00326276
Pawlowski, W. P., Sheehan, M. J., and Ronceret, A. (2007). In the beginning: the initiation of meiosis. Bioessays 29, 511–514. doi: 10.1002/bies.20578
Pawlowski, W. P., Wang, C. R., Golubovskaya, I. N., Szymaniak, J. M., Shi, L., Hamant, O.,et al. (2009). Maize AMEIOTIC1 is essential for multiple early meiotic processes and likely required for the initiation of meiosis. Proc. Natl. Acad. Sci. U.S.A. 106, 3603–3608. doi: 10.1073/pnas.0810115106
Ravi, M., Marimuthu, M. P., and Siddiqi, I. (2008). Gamete formation without meiosis in Arabidopsis. Nature 451, 1121–1124. doi: 10.1038/nature06557
Rhoades, M. M. (1956). Genetic control of chromosomal behavior. Maize Genet. Coop. News L. 30, 38–42.
Singh, M., Goel, S., Meeley, R. B., Dantec, C., Parrinello, H., Michaud, C.,et al. (2011). Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell 23, 443–458. doi: 10.1105/tpc.110.079020
Strich, R. (2004). Meiotic DNA replication. Curr. Top. Dev. Biol. 61, 29–60. doi: 10.1016/S0070-2153(04)61002–61007
Stuart, D., and Wittenberg, C. (1998). CLB5 and CLB6 are required for premeiotic DNA replication and activation of the meiotic S/M checkpoint. Genes Dev. 12, 2698–2710. doi: 10.1101/gad.12.17.2698
Sugimoto, A., Iino, Y., Maeda, T., Watanabe, Y., and Yamamoto, M. (1991). Schizosaccharomyces pombe ste11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes Dev. 5, 1990–1999. doi: 10.1101/gad.5.11.1990
Watanabe, Y., Yokobayashi, S., Yamamoto, M., and Nurse, P. (2001). Pre-meiotic S phase is linked to reductional chromosome segregation and recombination. Nature 409, 359–363. doi: 10.1038/35053103
Zhang, H., Egger, R. L., Kelliher, T., Morrow, D., Fernandes, J., Nan, G. L.,et al. (2014). Transcriptomes and proteomes define gene expression progression in pre-meiotic maize anthers. G3 (Bethesda) 4, 993–1010. doi: 10.1534/g3.113.009738
Keywords: apomixis, meiosis initiation, RNA-directed DNA methylation, plant reproduction, ameiotic1
Citation: Wang C-JR and Tseng C-C (2014) Recent advances in understanding of meiosis initiation and the apomictic pathway in plants. Front. Plant Sci. 5:497. doi: 10.3389/fpls.2014.00497
Received: 17 July 2014; Paper pending published: 13 August 2014;
Accepted: 08 September 2014; Published online: 23 September 2014.
Edited by:
Ian Henderson, University of Cambridge, UKReviewed by:
Daphné Autran, Institut de Recherche pour le Développement, FranceGabriela Carolina Pagnussat, Universidad Nacional de Mar del Plata, Argentina
Copyright © 2014 Wang and Tseng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chung-Ju R. Wang, Institute of Plant and Microbial Biology, Academia Sinica, Room 120, Section 2, Academia Road, Taipei 11529, Taiwan e-mail: rwang@gate.sinica.edu.tw
 Chung-Ju R. Wang
Chung-Ju R. Wang Ching-Chih Tseng
Ching-Chih Tseng