- 1Plant Biotechnology, Department of Biology, ETH Zurich (Swiss Federal Institute of Technology), Zurich, Switzerland
- 2International Rice Research Institute, Los Banos, Philippines
Rice is a major cereal crop that contributes significantly to global food security. Biotic stresses, including the rice blast fungus, cause severe yield losses that significantly impair rice production worldwide. The rapid genetic evolution of the fungus often overcomes the resistance conferred by major genes after a few years of intensive agricultural use. Therefore, resistance breeding requires continuous efforts of enriching the reservoir of resistance genes/alleles to effectively tackle the disease. Seed banks represent a rich stock of genetic diversity, however, they are still under-explored for identifying novel genes and/or their functional alleles. We conducted a large-scale screen for new rice blast resistance sources in 4246 geographically diverse rice accessions originating from 13 major rice-growing countries. The accessions were selected from a total collection of over 120,000 accessions based on their annotated rice blast resistance information in the International Rice Genebank. A two-step resistance screening protocol was used involving natural infection in a rice uniform blast nursery and subsequent artificial infections with five single rice blast isolates. The nursery-resistant accessions showed varied disease responses when infected with single isolates, suggesting the presence of diverse resistance genes/alleles in this accession collection. In addition, 289 accessions showed broad-spectrum resistance against all five single rice blast isolates. The selected resistant accessions were genotyped for the presence of the Pi2 resistance gene, thereby identifying potential accessions for isolation of allelic variants of this blast resistance gene. Together, the accession collection with broad spectrum and isolate specific blast resistance represent the core material for isolation of previously unknown blast resistance genes and/or their allelic variants that can be deployed in rice breeding programs.
Introduction
Crop plant diseases caused by various pathogens such as viruses, bacteria, oomycetes and fungi pose major challenges to global crop production and food security. Global climate change is predicted to further increase the negative impact of biotic stresses. Higher temperatures and erratic weather pattern are likely to change the geographical pathogen distribution. This in turn might decrease the effectiveness of existing resistance genes in crop varieties (Garrett et al., 2006; Milus et al., 2009) by promoting more aggressive races of pathogens. For example, more frequent wheat rust disease outbreaks, during the last years (2009, 2010, and 2013) threaten production in almost all wheat-growing countries and have caused heavy annual yield losses, sometimes as high as 50% in case of an early infection (Huerta-Espino et al., 2011; FAO, 2014). The recently emerged virulent stem rust (Puccinia graminis) race Ug99 has overcome the major resistance gene Sr31 (Pretorius et al., 2000), which is widely used in India, China, Europe and South America. Similarly, rice blast caused by the fungus Magnaporthe oryzae is one of the most devastating rice pathogens because it can infect the plant during nearly all growth stages. Repeated epidemics and frequent breakdown of rice blast resistance causing yield losses of 20–100% have been reported over the last decades in India and Japan (Khush and Jena, 2009; Sharma et al., 2012). Effective management of such pathogens require constant breeding efforts for development of resistant cultivars.
Plant defense involves various mechanisms that range from physical barriers such as the waxy cuticle, release of phytochemicals (e.g., phenols and terpenoids), quantitative resistance triggered by pathogen-associated molecular pattern (PAMP) recognition receptors, as well as various minor and major resistance genes acting against specific pathogen races (Yang et al., 2013). PAMP-mediated immunity is a baseline defense system in plants during which transmembrane receptors activate resistance upon detection of conserved pathogen-associated molecules such as flagellins, lipopolysaccharides and chitin that are shared by all bacterial and fungal pathogens (DeYoung and Innes, 2006; Jacob et al., 2013). Pathogens can evade this basal defense by releasing effector molecules, which often are recognized by intracellular resistance proteins encoded by major resistance (R) genes thereby providing effector-triggered plant immunity (ETI) (Jones and Dangl, 2006; Jacob et al., 2013). Major R genes encode five different classes of proteins (Dangl and Jones, 2001), with additional subclasses defined on the basis of domain structures as well as their membrane topology (Jacob et al., 2013). The nucleotide binding site and leucine rich repeats (NBS-LRR) type form the largest class of R proteins that have either a coiled coil (CC) or a TOLL/interleukin 1 receptor (TIR) domain at the N-terminus. Each of these domains have their own structural and functional significance, for example the LRR domain that contains highly conserved segments (HCS) as well as variable segments (VS) is known to be involved in protein-protein or protein-ligand interactions (Matsushima and Miyashita, 2012). Approximately 500 NBS-LRR genes have been identified in the rice genome (Monosi et al., 2004) and about 150 genes in the Arabidopsis genome (Yang et al., 2013). The R genes are being widely used in crop breeding for protection against various diseases.
Advancements in molecular techniques, whole genome sequencing and functional genomics have facilitated the identification, fine mapping and cloning of several R genes in the recent years. Information on molecular markers linked to these genes is increasingly available and used in marker-assisted breeding programs to provide effective and broad resistance in the field. Pyramiding of major R genes that vary in their mode of action and confer resistance against various pathogens has been successfully deployed (Datta et al., 2002; Maruthasalam et al., 2007). For example, pyramiding of xa5, xa13, and Xa21 (Singh et al., 2001) has broadened resistance against bacterial blight (Xanthomonas oryzae) and of Rsv1, Rsv3, and Rsv4 against soybean mosaic virus (Shi et al., 2009) as compared to varieties carrying single R genes only. Additionally, mixed planting of crop varieties with different R genes has been recommended as an alternative for effective utilization of host plant resistance. Mixing varieties or multilines with varying levels of resistance and susceptibility reduce the disease pressure and severity in the field compared to planting of single resistant varieties that are more vulnerable to disease outbreaks (Zhu et al., 2000; Raboin et al., 2012). The usefulness of this strategy has been shown over the last decades in experiments for the control of fungal diseases (Mundt, 2002; Castilla et al., 2010). For example, Zhu et al. (2000) demonstrated its effectiveness in China for the control of rice blast, where susceptible sticky rice was planted as single rows interspersed between four to six rows of resistant hybrid rice. This greatly reduced rice blast infection (>90%) and increased grain production (>85%) in the mixture plots as compared to the monocultures. However, not all or random mixtures combinations will be functional because several controlling factors, including epidemiological conditions and varied degrees of susceptibility to different pathogens, can affect outcome. Therefore, the multi-variety strategy requires adaption to local agricultural priorities and available resistant genotypes. For example, in the uplands of Indonesia, farmers are willing to adopt variety mixture if these varieties have similar height, growth duration and grain quality (Vera Cruz et al., 2009).
Most plant pathogens are fast evolving and can break down the resistance conferred by R genes, thus resulting in disease epidemics. These can be influenced by several factors, including weather conditions, disease pressure, as well as genome stability of the pathogen. The M. oryzae genome is rich in repetitive segments and retro-transposons (Dean et al., 2005), which allow the fungus to frequently change pathogenicity or escape from host recognition by altering the effector molecules. This requires the continuous identification of new sources of host disease resistance against continuously evolving and geographically diverse pathogen races. Crop germplasm collections maintained in genebanks at centers such as the International Rice Research Institute (IRRI) in the Philippines, the Leibniz Institute of Plant Genetics and Crop Plant Research in Germany, the International Maize and Wheat Improvement Center (CIMMYT) in Mexico, the International Potato Center (CIP) in Peru, Bioversity International in Belgium, or the Svalbard Global Seed Vault in Norway provide access to repertoires of favorable genes. These germplasm collections include several traditional cultivars and landraces as well as wild species from various geographical origins. The landraces and wild relatives may not possess favorable agronomic traits (such as plant height or yield) suitable for modern breeding programs, but they are highly relevant sources of genes for specific traits, including resistance or tolerance to biotic and abiotic stresses. Resistance to late blight (Phytophthora infestans) in potato derived from diverse wild Solanum species (Vossen et al., 2003; Foster et al., 2009), identification of broad spectrum rice blast resistance genes such as Pi9 from Oryza minuta (Qu et al., 2006) and the phosphorus uptake (Pup1) locus in rice (Gamuyao et al., 2012) from the traditional cultivar Kasalath are convincing examples of important contributions of wild species and traditional cultivars to our breeding programs.
Here we report the evaluation of 4246 rice gene bank accessions to identify new rice blast resistance sources. We selected rice accessions of diverse geographic origins on the basis of their annotated rice blast resistance in the IRRI rice germplasm collection. Systematic screening for rice blast resistance was carried out under both field and controlled environmental conditions. Using this approach we identified candidate accessions that can be deployed as donors in rice breeding programs and as starting material for the isolation of novel rice blast resistance genes and allelic variants of major rice blast resistance genes.
Materials and Methods
Plant Materials
The rice germplasm material was obtained from the International Rice Genebank (IRG) of the International Rice Research Institute (IRRI), Philippines. The International Rice Germplasm Collection (IRGC) of IRG holds more than 120,000 accessions from different geographical regions of the world (129 countries). The accessions for our study were selected on the basis of documented rice blast resistance information i.e., characterized as resistant, moderately resistant or susceptible, in the International Rice Germplasm Collection Information System (IRGCIS) of IRG. The rice cultivars IR72 and CO39 were used as susceptible control lines throughout the study. Blast monogenic resistant lines for Pi2 (IRBLz5-CA), Pi9 (IRBL9-W), Piz-t (IRBLzt-T), Pita (IRBLta-CP1), Pi54 (IRBLkh-K3), and Pib (IRBLb-B) were used as control lines for isolate specific rice blast resistance evaluation.
Magnaporthe Oryzae Isolates
For primary screening in the Uniform Blast Nursery (UBN), a mixture of naturally existing blast strains in the IRRI locality was used. Diseased plantlets of blast susceptible lines IR72 and CO39 were taken from IRRI pathology fields and were used as a source of inoculum for spreading the disease on control/disease-check lines in nursery screening beds.
For the second level screening under greenhouse conditions, the five highly virulent M. oryzae isolates (Supplementary Figure 1) M101-1-2-9-1, M39-1-2-21-2, JMB8401, M64-1-3-9-1, and Ca41 were selected based on their reported differential disease pattern/spectrum on blast monogenic lines carrying blast resistant genes Pi2, Pi9, Piz-t, Pi54, Pib, and Pita (Kobayashi et al., 2007; Telebanco-Yanoria et al., 2008). Protocols reported by Hayashi et al. (2009) were followed for the maintenance, culture and sporulation of individual rice blast isolates.
Inoculation and Disease Scoring
For UBN screening, test accessions (30 plants/test entry) were sown on nursery beds, along with a mixture of varieties with differing level of resistance to local M. oryzae strains sown as borderlines/spreader rows. Additionally the susceptible controls were sown as one line of IR72 for every 10 lines of test accessions and one line of CO39 for every 100 lines of test accessions. Inoculation was done on border lines (as described above) after 10 days from the date of sowing and disease was allowed to spread naturally via wind dispersal. Nursery beds were water sprayed 3–4 times per day and were covered during the night to maintain a high humidity until disease development and progression was observed in border lines.
In case of greenhouse screening, test accessions (15 plants/test entry) were sown in plastic trays (10 rows × 2 columns per tray) in five batches for inoculation with five individual blast isolates mentioned above. Plants were inoculated 10 days after sowing using 50 ml of spore suspension solution with a concentration of 1 × 105 spores per ml with 1% tween-20 per tray. After inoculation, the plants were maintained in moist chamber at 26–28°C for 24 h and after this period, the plants were transferred to the incubation chamber at 25°C ± 2 for 1 week before disease assessment.
Disease evaluations were performed for each test line according to the Standard Evaluation System of IRRI (1996), for screening leaf blast, with scores ranging from 0 to 9 (Supplementary Figure 2). Blast lesion type and the leaf area covered with infection were recorded. In this scoring system, score 0 represents no blast lesions observed and is graded as highly resistant, whereas score 9 marks as highly susceptible with more than 75% leaf area infected. The phenotypic scores of 0, 1, 2, and 3 are considered as blast resistant in the rice breeding programs and data were collected accordingly in the screening experiments.
Molecular Characterization of Candidate Accessions
Leaf samples were collected from the plants resistant to rice blast (scale 0–3) in UBN for genomic DNA isolation. Ground leaf samples were collected in 2 ml microcentrifuge tubes, 1.2 ml of 2 × CTAB-buffer was added, and the samples were incubated at 65°C for 30 min. After addition of 800 μl of phenol:chloroform:isoamylalcohol (25:24:1) and mixing, the sample tubes were centrifuged for 15 min at 13,000 rpm. The aqueous supernatant was collected in new 1.5 ml tubes and was treated with RNaseA at 37°C for 30 min. DNA was precipitated by adding 560 μl of isopropanol and pelleted by centrifugation at 13,000 rpm for 10 min. After washing twice with ethanol, the dried DNA pellet was dissolved in 50 μl of 1 × TE buffer and was used for polymerase chain reaction (PCR) analysis.
PCR was carried out to determine the presence/absence of Pi2 locus using a Sequence Tagged Site (STS) marker (as reported in Zhou et al., 2006) that co-segregates with the Pi2 gene. Forward primer: 5′-TCTATAGAAGTGCAAACAGC and Reverse primer: 5′-TTAGGTACGAACATGAGTAG were used to amplify the targeted fragment of ~2 kb from the test accessions. The PCR program included initial denaturation at 95°C for 5 min; followed by additional denaturation at 95°C for 30 s, annealing at 51°C for 30 s, extension at 72°C for 1 min (these three steps repeated for 39 cycles); followed by final extension at 72°C for 7 min. Genomic DNA extracted from Pi2 monogenic line was used as the positive control and that from susceptible rice cultivar LTH was used as the negative control, in addition to a water control. In addition, rice actin gene was amplified within the same sample serving as an internal PCR control (actin forward primer: 5′-TTATGGTTGGGATGGGACA; actin reverse primer: 5′-AGCACGGCTTGAATAGCG).
Results
Selection of Diverse Rice Germplasm from IRGCIS Database
The IRGCIS online database contains 6338 rice accessions that are annotated as resistant to rice blast (excluding moderately resistant accessions) within the total rice germplasm collection at IRG. From these 6338 accessions, we selected genotypes of known varietal groups originating from 13 major rice growing countries, providing a total of 4246 rice genotypes as the starting material (excluding 96 accessions for which no seed was available or there was no germination). The 13 countries (and number of rice genotypes) chosen for this selection were India (1457), Indonesia (775), Bangladesh (613), Thailand (355), Philippines (333), Vietnam (248), China (116), Myanmar (92), Taiwan (76), Brazil (68), Japan (59), Nepal (47), and South Korea (7). These represent major rice producing and consuming countries. The accessions were assigned to three major varietal groups, i.e., indica (85.9%), javanica (9.2%), and japonica (4.9%). The 13 countries cover 66.9% of the annotated blast resistant genotypes within the full rice germplasm collection available at IRG while the remaining 33.1% is distributed over a total of 55 countries.
Screening of Candidate Accessions for Rice Blast Resistance in the Field Condition
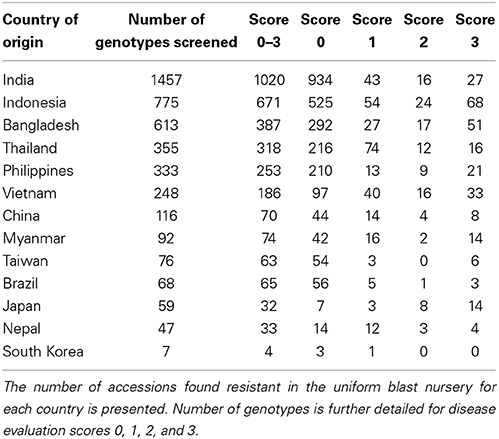
The 4246 selected rice accessions were screened for blast resistance in the uniform blast nursery (Figure 1). A large majority of the rice genotypes exhibited complete resistance when compared to the susceptible control lines. We found 3176 (74.8%) accessions to be blast resistant with scores between 0 and 3 (Figure 1 and Table 1). Most of these accessions scored as highly resistant with score 0, accounting for 78.5% of the 3176 resistant genotypes and 58.7% of the total 4246 genotypes screened. Although each country had a high number of resistant accessions, the percentages of blast resistant genotypes from different countries varied (Table 1), with Brazil containing the highest number of resistant accessions (95.6% of Brazilian accessions screened) while Japan had the lowest number (54.2% of Japanese accessions screened). For the other 11 countries, the percentage of resistant accessions ranged between 57.1 and 89.6% based on the number of genotypes screened from the respective country (South Korea 57.1, China 60.3, Bangladesh 63.1, India 70, Nepal 70.2, Vietnam 75, Philippines 75.9, Myanmar 80.4, Taiwan 82.9, Indonesia 86.6, and Thailand 89.6%). Within these resistant genotypes, 2726 (85.8%), 295 (9.2%), and 155 (4.9%) belong to indica, javanica and japonica varietal groups, respectively. Additionally, a relatively smaller set of 588 accessions exhibited moderate resistance with scores 4 and 5 (Supplementary Figure 3). The moderately resistant accessions were identified in all the 13 countries, with India, Bangladesh and Indonesia being the major contributors covering 75.5% of the total moderately resistant accessions.
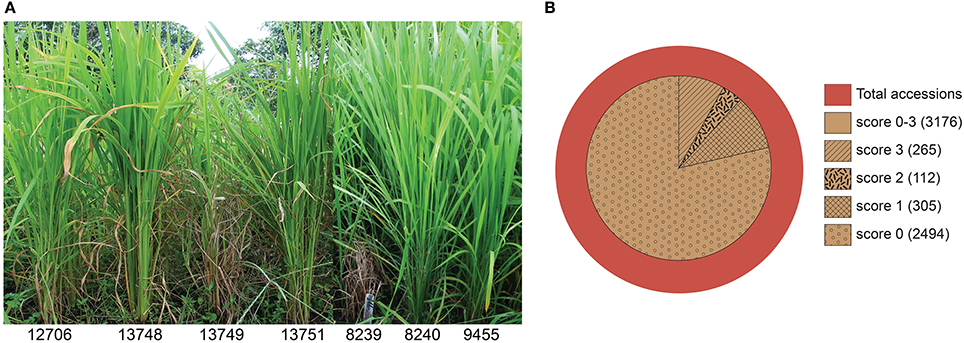
Figure 1. Rice blast resistance screening in the uniform blast nursery. (A) Sample images of rice genotypes showing full blast resistance phenotypes (IRGC 12706, 13748, 13751, 8240, and 9455) in contrast to susceptible genotypes (IRGC 13749 and 8239) in the uniform blast nursery. (B) Summary of rice blast resistance screening in the uniform blast nursery. Inner circle represent the number of accessions that scored as resistant (score 0–3) using the 0–9 scoring system. Majority of the accessions were observed with score 0 (78.5%) while accessions with score 1, 2, and 3 accounted for 9.6, 3.5, and 8.34% of the 3176 resistant accessions, respectively. The numbers of resistant accessions in each respective category are shown in brackets.
Out of the total 3176 resistant accessions (score 0–3), 1323 accessions (41.6% of total resistant) are annotated as traditional cultivars or landraces in the IRGCIS database (Figure 2), demonstrating the potential of landraces as promising sources of novel resistances. Considering that not all of the accessions in the database are fully annotated, it is likely that the number of resistant landraces is even higher. For example, in the case of India that contributes nearly one third (1457 accessions) of our study material, 88.7% of the accessions are annotated as “unknown sample status.” For the other 12 countries the number of resistant accessions with traditional cultivars/landraces status is 60% of the total blast resistant accessions. It should be noted, however that these countries also contain samples of unknown status (Figure 2).
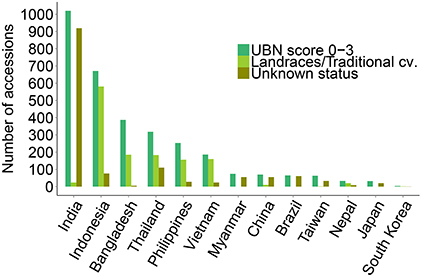
Figure 2. Landraces/traditional cultivars among identified resistant rice genotypes. Landraces/traditional cultivars among the blast resistant genotypes from the respective countries are shown, together with the resistant genotypes of unknown status. India contributes nearly one third of the total blast resistant genotypes but 90% of the accessions have an unknown sample status. Excluding India, 60% of the blast resistant rice genotypes identified in the 12 remaining countries are traditional cultivars/landraces. None of the resistant accessions from Myanmar, Brazil and Japan were landraces/traditional cultivars but a large numbers of identified resistant cultivars from these countries have an unknown status as well. A minor proportion of the UBN-resistant genotypes were inbred lines and improved varieties (data not shown).
Isolate Specific Screening of the UBN Resistant Accessions
The rice genotypes that were found to be resistant to rice blast (score 0, 1, 2, and 3) in the UBN screening were subjected to isolate-specific screening using five different M. oryzae isolates (M101-1-2-9-1, M39-1-2-21-2, JMB8401, M64-1-3-9-1, and Ca41). The isolates exhibit varied disease pattern, either unique or shared between different monogenic lines (see Materials and Methods). We expected that using these isolates for screening would facilitate the identification of potentially novel and broad-spectrum rice blast resistance sources. Out of the total 3176 UBN-resistant rice accessions, the genotypes resistant against the individual blast isolates (score 0–3) were 1093 for M101-1-2-9-1, 1444 for M39-1-2-21-2, 2253 for JMB8401, 1088 for M64-1-3-9-1, and 1298 for Ca41 (Figure 3). As expected, a varied degree of disease susceptibilities were observed among different rice genotypes when scoring against different M. oryzae isolates (Supplementary Figure 4, Supplementary Figure 5). For example, the variety DV109 (IRGC Nr. 8854) was highly resistant in the UBN screening and against the isolate M101-1-2-9-1, moderately resistant against Ca41, moderately susceptible against M39-1-2-21-2, and JMB8401 while it scored highly susceptible against M64-1-3-9-1. Such resistance and susceptibility pattern against different isolates identify these accessions as potential novel sources of race-specific resistance genes.
Figure 3. Isolate specific screening of UBN resistant rice genotypes. UBN-resistant accessions (score 0–3) were screened against five different single rice blast isolates under controlled conditions. Data for accessions exhibiting resistance against particular isolates is shown, with different color bars representing each isolate. It should be noted that several accessions exhibited resistance against two or more isolates. The screened rice accessions displayed varied patterns of disease symptoms, with accessions resistant or susceptible only to one blast isolate as well as accessions that exhibited resistance against two to all of the isolates.
We found a small set of 262 genotypes to be susceptible to all five M. oryzae isolates. However, these accessions might have resistance against isolates not included in our current study. It is also possible that these accessions have partial resistance but could become susceptible against a specific highly virulent isolate.
Identification of Genotypes Conferring Broad-Spectrum Resistance
We identified 289 genotypes that showed broad-spectrum resistance, of which 39.4% are landraces. These genotypes were found resistant (scale 0–3) to blast, both in the uniform blast nursery and when inoculated with the five individual rice blast isolates (Figure 4, Supplementary Table 1). The Philippines, India, Vietnam and Thailand contributed more broad-spectrum blast resistant genotypes (29.4, 22.8, 13.1, and 11.8%, respectively) among the 289 genotypes identified as broad-spectrum resistant (Figure 4). Nearly 30% of the 289 broad-spectrum resistant genotypes originated from the Philippines, which is high proportion considering that only 7.8% (333 accessions) of the total 4246 accessions screened were from Philippines (Table 1). This is expected because the rice accessions were challenged with M. oryzae isolates from the Philippines and these accessions likely have specific resistances against local isolates. Nevertheless, these accessions are promising donors for breeding local rice varieties in the Philippines and in other rice growing agricultural systems across Asia. It is also likely that these broad spectrum genotypes have multiple functional blast resistance genes and/or QTLs, which could be useful for detailed molecular analysis to identify the underlying blast resistance genes.
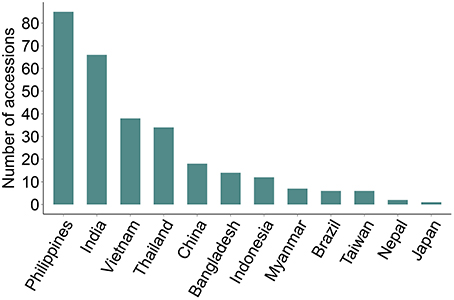
Figure 4. Broad spectrum resistant rice genotypes. Rice accessions showing broad spectrum UBN-resistance against the field mix inoculum as well as against all the five individual rice blast isolates are presented. 289 broad-spectrum resistance rice accessions were identified (scores 0–3), with Philippines, India, Vietnam, Thailand and China being the major contributors. None of accessions from South Korea was identified as broad spectrum resistant.
Molecular Screening for Identification of Accessions Carrying Pi2 Rice Blast Resistance Locus
We screened the rice accessions that were blast resistant in the UBN screen for the Pi2 locus using a STS marker (Zhou et al., 2006). The Pi2 locus represents a complex cluster of multiple NBS-LRR genes with different resistance specificities. Three genes (Pi2, Pi9, and Piz-t) have been cloned and another four genes (Pi40(t), Pigm, Pi26, and Piz) have been mapped to this locus. All of them confer broad spectrum resistance against rice blast, but at the same time these genes vary in their resistance pattern against different M. oryzae isolates (Qu et al., 2006; Liu et al., 2010; Wu et al., 2012). Among the 3176 rice accessions that were blast resistant in UBN screen, we identified 794 genotypes that carry the Pi2 locus (Figure 5, Supplementary Table 2). It is interesting to note that half of the 289 broad-spectrum blast resistant genotypes (50.2%) carry the Pi2 locus. These resistant accessions found to carry Pi2 locus are promising lines for the identification of allelic variants of the Pi2/Pi9/Piz-t genes and as well for the identification and cloning of other broad-spectrum resistant genes that were mapped to this locus.
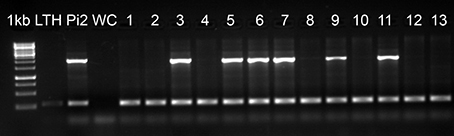
Figure 5. Molecular screening of UBN resistant genotypes for Pi2 locus. Molecular screening of 3176 UBN-resistant rice genotypes revealed 794 genotypes carrying the Pi2 locus. 1 kb, DNA marker; LTH, negative control; Pi2, positive control (Pi2-monogenic line); WC, water control; 1–13, test accessions. The accessions positive for Pi2 are 3 (IRGC 254), 5 (IRGC 257), 6 (IRGC 258), 7 (IRGC 840), 9 (IRGC 846), and 11 (IRGC 877). The upper band (~2 kb) indicates the presence of Pi2 locus and the lower band (~250 bp) shows the actin band, which is an internal control for the PCR.
Discussion
Identification of Genetic Variation is the Key to Future Crop Improvement
Domestication and modern breeding has reduced genetic diversity of crop plants (Tanksley and McCouch, 1997) by replacing landraces and traditional farmer cultivars with modern, high yielding varieties. Although this has allowed filtering out genes that cause detrimental traits, the reduction in genetic variation now limits the options of plant breeders to develop new varieties with the existing germplasm. New varieties are constantly needed to meet consumer demands and for protection of crops against highly unpredictable biotic and abiotic stresses that are encountered in agricultural systems. Breeding of improved varieties requires the identification of novel functional genes or alleles, and this calls for making effective use of our global crop genetic resources available in seed bank collections. These seed collections represent a wide range of genetic diversity that is critical for maintaining and enhancing the yield potential and other quality traits, because they can provide new sources of resistance and tolerance to various stresses. Many of the genes for highly relevant traits in modern crop cultivars have been transferred from their wild relatives and landraces, for example the Rht and sd-1 genes that confer dwarf stature in rice and wheat, respectively (Hedden, 2003; Hoisington et al., 1999). In the case of rice, the six wild species O. rufipogon, O. longistaminata, O. nivara, O. breviligulata, O. glumaepatula, and O. meridionalis together with the two cultivated species O. sativa and O. glaberrima have been identified as the primary gene pool for rice cultivars because genes can be easily transferred between these species (Khush, 1997). The wild species have been used as a source of many relevant genes for rice breeding, and O. rufipogon has been suggested as source of broad-spectrum rice blast resistance genes (Ram et al., 2007). Other examples include the bacterial blight resistance gene Xa21 that was first identified in O. longistaminata, the broad-spectrum blast resistance gene Pi-hk1 that was identified in the landrace Heikezijing (Wu et al., 2013), and the submergence tolerance gene SUB1 from the landrace FR13A (Bailey-Serres et al., 2010). Furthermore, Pi-Da(t) identified in an upland rice variety Dacca6 in Jin23B background has been used in an elite parental line (Shi et al., 2012), not only for its blast resistance but also for its good grain quality as well as for wide adoption for three-line hybrid rice breeding program over the past 20 years in China. In addition to traits for biotic and abiotic stress resistance or tolerance, rice landraces also served as an important source for yield traits such as SPIKE (an allele of NARROW LEAF1; NAL1) from an Indonesian rice landrace that increased rice grain yield by 18% (Fujita et al., 2013) and TGW6 (THOUSAND-GRAIN WEIGHT 6) from the Indian landrace Kasalath, which significantly increases grain yield as well (Ishimaru et al., 2013).
Despite the high genetic potential and efforts of identifying diversity from germplasm collections, the genetic base of our major crops remains narrow. Adaptation of pathogens and susceptibility to other stresses are continuous threats to existing elite crop varieties. Although there are demonstrated and valuable contributions of crop diversity to counter these threats, there is still a great potential hidden in available landraces, cultivars and wild species that remains under-explored. Large numbers of probably redundantly stored gene bank accessions and missing genotype × phenotype information make it difficult for modern breeding programs to select a feasible number of accessions for scoring traits of interest. Core collections have therefore been promoted as a means of exploiting large seed collections using smaller subsets of accessions that represent maximum diversity. However, while core collections aim to maximize genetic diversity in general, plant breeders and/or biologists are often interested in one or a few specific traits at a time. Other approaches such as the “Focused Identification of Germplasm Strategy (FIGS)” were therefore suggested to utilize eco-geographical information for identifying sets of gene bank accessions that are expected to have a maximum of functional diversity for one specific trait of interest (Bhullar et al., 2009).
We focused on potential rice accessions as donors of novel blast resistance alleles by selecting a test set based on annotated rice blast resistance information available in the IRGCIS database. Rice blast remains one of the most destructive diseases of rice, and resistance gene deployment has been suggested as most effective and environment friendly way of managing the disease. However, the continuously evolving genome of M. oryzae as well as existence of geographically diverse strains are challenging for the rice breeders. Genome studies of the rice blast fungus has revealed high probabilities of transposons mediated inactivation of genes involved in host specificity. Moreover, the high genetic variability in M. oryzae allows the fungus to broaden the host range and infect formerly resistant genotypes (Dean et al., 2005). It is therefore important to build a repertoire of resistant accessions/donors that could be screened and deployed in breeding programs according to needs of local agricultural systems. The rice accessions we selected from 13 major rice growing countries represent a broad geographic diversity. Nearly 75% of the UBN-screened accessions exhibited resistance to the local field mixture inoculum in the Philippines. We observed varied resistance patterns among these accessions when infected with five individual rice blast isolates. The severity of host plant disease phenotypes depends on interactions between various plant R genes or their allelic variants and corresponding pathogen effector molecules (DeYoung and Innes, 2006). Thus, the patterns of disease reactions against five differential isolates of rice blast varying from highly resistant to highly susceptible that we observed in these accessions indicate the presence of race specific genes/alleles and/or their combinations. Furthermore, 289 accessions conferred broad-spectrum resistance because they were resistant against the field-mixture inoculum as well as all the five tested blast isolates. Besides these completely resistant accessions, we also identified accessions conferring moderate resistance against rice blast. Considering the apparent lack of durability of major gene-conferred disease resistance, lines conferring such partial resistance could serve as potential donors for identifying and characterizing weaker yet more durable sources of resistance. Together, the identified resistant and moderately resistant accessions represent a rich stock of starting material that can be used directly as donor parents in breeding programs, as well as for identification/isolation of race specific, broad-spectrum resistance and/or durable resistance sources against rice blast. Further, there are evidences of quantitative trait loci (QTLs) conferring resistance to multiple pathogens. For example, the wheat leaf rust QTL Lr34 confers broad-spectrum and durable resistance to leaf rust, stripe rust and powdery mildew (Krattinger et al., 2009; Kou and Wang, 2010). Therefore, it would be highly interesting to test the lines showing broad-spectrum rice blast resistance in our study for their response to other economically important rice pathogens such as Xanthomonas oryzae pv. oryzae and Rhizoctonia solani.
Approaches for Identification of High Value Genes from Crop Germplasm
Systematic evaluation of germplasm collections for agronomically relevant traits is necessary to facilitate strategies for efficient utilization of crop diversity. Conventional as well as high-throughput methods can be used to tap this diversity, including molecular marker screening, allele mining, genome wide association studies (GWAS), single nucleotide polymorphism (SNP) genotyping, as well as eco-TILLING (a variation of the “Targeting Induced Local Lesions In Genomes” method to identify natural mutations). Genome wide association studies help to discover and dissect functional variation among diverse genotypes for useful but complex traits such as grain yield (Huang et al., 2012). Allele mining and SNP genotyping could be particularly rewarding for (but not limited to) resistance traits (Bhullar et al., 2009, 2010; Wang et al., 2010) where even a small allelic variation can cause major changes in gene function (e.g., resistance vs. susceptibility) or in resistance spectrum (Bryan et al., 2000; Zhou et al., 2006). Large efforts are also being made to sequence genomes of diverse rice accessions. Using sequencing-by-synthesis technology for more than 500 Chinese rice landraces allowed the successful construction of a high-density haplotype map of the rice genome and GWAS for 14 agronomic traits in the population of O. sativa indica subspecies (Huang et al., 2010). Similarly, molecular analysis of blast resistant accessions identified in this study hold great potential for the characterization and isolation of novel rice blast resistance genes. Novel allelic forms of major R genes could be readily identified and cloned from this set using gene specific markers combined with PCR-based amplification and sequencing approaches. We found that 794 of the 3176 accessions that we screened and 50% of the 289 broad-spectrum resistance accessions carry Pi2/Pi2-like resistance genes. Pi2 shares high sequence homology with its other gene family members, including the cloned Pi9 and Piz-t broad spectrum resistance genes. It is therefore likely that these accessions not only contain allelic variants of Pi2 but also of other members within the gene cluster on chromosome 6 (Zhou and Wang, 2009; Wu et al., 2012). The accessions carrying Pi2 that we identified are the best candidates for isolating allelic variants and possibly characterizing additional broad-spectrum resistance genes from this locus. As reported here for Pi2, the approach we have used can be similarly extended to any major rice blast resistance gene of interest.
Together, increasing genome information and identification of candidate germplasm for novel traits represents an unprecedented resource for future rice breeding programs. Information gained from the systematic and standardized characterization of gene bank accessions using modern molecular tools must also be integrated into databases. Proper cataloging of genome and phenotypic information will enable breeders and plant biologists to use the existing genotypic diversity in seed banks for evaluating the biological significance of molecular and phenotypic diversity.
Author Contributions
Navreet K. Bhullar, Casiana M. Vera Cruz, Wilhelm Gruissem and Kumar Vasudevan designed the screen, Kumar Vasudevan carried out the experiments, Kumar Vasudevan and Navreet K. Bhullar analyzed the data, Kumar Vasudevan and Navreet K. Bhullar wrote the manuscript, Navreet K. Bhullar, Wilhelm Gruissem and Casiana M. Vera Cruz edited the manuscript. All authors have read the manuscript and agree with its content.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the International Rice Genebank of IRRI for providing seed materials. We thank International Rice Research Institute (IRRI), Philippines, for access to the uniform blast nursery, greenhouse facility and support of our rice biodiversity screen for blast resistance. The research was supported by funds from ETH Zurich to Wilhelm Gruissem.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00505/abstract
References
Bailey-Serres, J., Fukao, T., Ronald, P., Ismail, A., Heuer, S., and Mackill, D. (2010). Submergence tolerant rice: SUB1's journey from landrace to modern cultivar. Rice 3, 138–147. doi: 10.1007/s12284-010-9048-5
Bhullar, N. K., Street, K., Mackay, M., Yahiaoui, N., and Keller, B. (2009). Unlocking wheat genetic resources for the molecular identification of previously undescribed functional alleles at the Pm3 resistance locus. Proc. Natl. Acad. Sci. U.S.A. 106, 9519–9524. doi: 10.1073/pnas.0904152106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bhullar, N. K., Zhang, Z., Wicker, T., and Keller, B. (2010). Wheat gene bank accessions as a source of new alleles of the powdery mildew resistance gene Pm3: a large scale allele mining project. BMC Plant Biol. 10:88. doi: 10.1186/1471-2229-10-88
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bryan, G. T., Wu, K. S., Farrall, L., Jia, Y., Hershey, H. P., McAdams, S. A., et al. (2000). A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 12, 2033–2045. doi: 10.1105/tpc.12.11.2033
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Castilla, N. P., Willocquet, L., Suwarno, S., Santoso, S., Nasution, A., Sulaeman, Y., et al. (2010). Assessing the effect of resistant-susceptible associations and determining thresholds for associations in suppressing leaf and neck blast of rice. Crop Prot. 29, 390–400. doi: 10.1016/j.cropro.2009.11.016
Dangl, J. L., and Jones, J. D. G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. doi: 10.1038/35081161
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Datta, K., Baisakh, N., Maung Thet, K., Tu, J., and Datta, S. K. (2002). Pyramiding transgenes for multiple resistance in rice against bacterial blight, yellow stem borer and sheath blight. Theor. Appl. Genet. 106, 1–8. doi: 10.1007/s00122-002-1014-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dean, R. A., Talbot, N. J., Ebbole, D. J., Farman, M. L., Mitchell, T. K., Orbach, M. J., et al. (2005). The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434, 980–986. doi: 10.1038/nature03449
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
DeYoung, B. J., and Innes, R. W. (2006). Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol. 7, 1243–1249. doi: 10.1038/ni1410
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Food and Agriculture Organization of the United Nations (FAO). (2014). Wheat Rust Disease Global Programme. Available online at: www.fao.org/agriculture/crops/wheatrust/en/ (Accessed July, 2014).
Foster, S. J., Park, T. H., Pel, M., Brigneti, G., Śliwka, J., Jagger, L., et al. (2009). Rpi-vnt1.1, a Tm-22 homolog from Solanum venturii, confers resistance to potato late blight. Mol. Plant Microbe Interact. 22, 589–600. doi: 10.1094/MPMI-22-5-0589
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fujita, D., Trijatmiko, K. R., Tagle, A. G., Sapasap, M. V., Koide, Y., Sasaki, K., et al. (2013). NAL1 allele from a rice landrace greatly increases yield in modern indica cultivars. Proc. Natl. Acad. Sci. U.S.A. 110, 20431–20436. doi: 10.1073/pnas.1310790110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gamuyao, R., Chin, J. H., Pariasca-Tanaka, J., Pesaresi, P., Catausan, S., Dalid, C., et al. (2012). The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature 488, 535–541. doi: 10.1038/nature11346
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Garrett, K. A., Dendy, S. P., Frank, E. E., Rouse, M. N., and Travers, S. E. (2006). Climate change effects on plant disease: genomes to ecosystems. Annu. Rev. Phytopathol. 44, 489–509. doi: 10.1146/annurev.phyto.44.070505.143420
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hayashi, N., Kobayashi, N., Vera Cruz, C. M., and Fukuta, Y. (2009). Protocols for sampling of disease specimens and evaluation of blast disease in rice. Japan International Research Center for Agricultural Sciences (JIRCAS), Tsukuba, Japan. Work. Rep. 63, 17–33.
Hedden, P. (2003). The genes of the green revolution. Trends Genet. 19, 5–9. doi: 10.1016/S0168-9525(02)00009-4
Hoisington, D., Khairallah, M., Reeves, T., Ribaut, J. M., Skovmand, B., Taba, S., et al. (1999). Plant genetic resources: what can they contribute toward increased crop productivity?. Proc. Natl. Acad. Sci. U.S.A. 96, 5937–5943. doi: 10.1073/pnas.96.11.5937
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huang, X., Wei, X., Sang, T., Zhao, Q., Feng, Q., Zhao, Y., et al. (2010). Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 42, 961–967. doi: 10.1038/ng.695
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Huang, X., Zhao, Y., Wei, X., Li, C., Wang, A., Zhao, Q., et al. (2012). Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat. Genet. 44, 32–39. doi: 10.1038/ng.1018
Huerta-Espino, J., Singh, R. P., Germán, S., McCallum, B. D., Park, R. F., Chen, W. Q., et al. (2011). Global status of wheat leaf rust caused by Puccinia triticina. Euphytica 179, 143–160. doi: 10.1007/s10681-011-0361-x
Ishimaru, K., Hirotsu, N., Madoka, Y., Murakami, N., Hara, N., Onodera, H., et al. (2013). Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 45, 707–711. doi: 10.1038/ng.2612
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jacob, F., Vernaldi, S., and Maekawa, T. (2013). Evolution and conservation of plant NLR functions. Front. Immunol. 4:297. doi: 10.3389/fimmu.2013.00297
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Khush, G. S. (1997). Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 35, 25–34. doi: 10.1023/A:1005810616885
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Khush, G. S., and Jena, K. K. (2009). “Current status and future prospects for research on blast resistance in rice (Oryza sativa L.),” in Advances in Genetics, Genomics and Control of Rice Blast Disease, eds G. L. Wang and B. Valent (Dordrecht: Springer), 1–10. doi: 10.1007/978-1-4020-9500-9_1
Kobayashi, N., Yanoria, M. J. T., and Fukuta, Y. (2007). Differential varieties bred at IRRI and virulence analysis of blast isolates from the Philippines. Japan International Research Center for Agricultural Sciences (JIRCAS), Tsukuba, Japan. Work. Rep. 53, 17–30.
Kou, Y., and Wang, S. (2010). Broad-spectrum and durability: understanding of quantitative disease resistance. Curr. Opin. Plant Biol. 13, 181–185. doi: 10.1016/j.pbi.2009.12.010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Krattinger, S., Lagudah, E., Spielmeyer, W., Singh, R. P., Huerta-Espino, J., McFadden, H., et al. (2009). A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323, 1360–1363. doi: 10.1126/science.1166453
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, J., Wang, X., Mitchell, T., Hu, Y., Liu, X., Dai, L., et al. (2010). Recent progress and understanding of the molecular mechanisms of the rice—Magnaporthe oryzae interaction. Mol. Plant Pathol. 11, 419–427. doi: 10.1111/j.1364-3703.2009.00607.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Maruthasalam, S., Kalpana, K., Kumar, K. K., Loganathan, M., Poovannan, K., Raja, J. A. J., et al. (2007). Pyramiding transgenic resistance in elite indica rice cultivars against the sheath blight and bacterial blight. Plant Cell Rep. 26, 791–804. doi: 10.1007/s00299-006-0292-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Matsushima, N., and Miyashita, H. (2012). Leucine-Rich Repeat (LRR) domains containing intervening motifs in plants. Biomolecules 2, 288–311. doi: 10.3390/biom2020288
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Milus, E. A., Kristensen, K., and Hovmoller, M. S. (2009). Evidence for increased aggressiveness in a recent widespread strain of Puccinia striiformis f. sp tritici causing stripe rust of wheat. Phytopathology 99, 89–94. doi: 10.1094/PHYTO-99-1-0089
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Monosi, B., Wisser, R. J., Pennill, L., and Hulbert, S. H. (2004). Full-genome analysis of resistance gene homologues in rice. Theor. Appl. Genet. 109, 1434–1447. doi: 10.1007/s00122-004-1758-x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mundt, C. C. (2002). Use of multiline cultivars and cultivar mixtures for disease management. Annu. Rev. Phytopathol. 40, 381–410. doi: 10.1146/annurev.phyto.40.011402.113723
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pretorius, Z. A., Singh, R. P., Wagoire, W. W., and Payne, T. S. (2000). Detection of virulence to wheat stem rust resistance gene Sr31 in Puccinia graminis. f. sp. tritici in Uganda. Plant Dis. 84:203. doi: 10.1094/PDIS.2000.84.2.203B
Qu, S., Liu, G., Zhou, B., Bellizzi, M., Zeng, L., Dai, L., et al. (2006). The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site–leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 172, 1901–1914. doi: 10.1534/genetics.105.044891
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Raboin, L. M., Ramanantsoanirina, A., Dusserre, J., Razasolofonanahary, F., Tharreau, D., Lannou, C., et al. (2012). Two-component cultivar mixtures reduce rice blast epidemics in an upland agrosystem. Plant Pathol. 61, 1103–1111. doi: 10.1111/j.1365-3059.2012.02602.x
Ram, T., Majumder, N. D., Mishra, B., Ansari, M. M., and Padmavathi, G. (2007). Introgression of broad-spectrum blast resistance gene(s) into cultivated rice (Oryza sativa ssp indica) from wild rice O. rufipogon. Curr. Sci. 92, 225–230.
Sharma, T. R., Rai, A. K., Gupta, S. K., Vijayan, J., Devanna, B. N., and Ray, S. (2012). Rice blast management through host-plant resistance: retrospect and prospects. Agric. Res. 1, 37–52. doi: 10.1007/s40003-011-0003-5
Shi, A., Chen, P., Li, D., Zheng, C., Zhang, B., and Hou, A. (2009). Pyramiding multiple genes for resistance to soybean mosaic virus in soybean using molecular markers. Mol. Breed. 23, 113–124. doi: 10.1007/s11032-008-9219-x
Shi, B. H., Zhang, J. H., Zheng, Y. M., Liu, Y. Q., Vera Cruz, C. M., Zheng, T. Q., et al. (2012). Identification of a new resistance gene Pi-Da(t) from Dacca6 against rice blast fungus (Magnaporthe oryzae) in Jin23B background. Mol. Breed. 30, 1089–1096. doi: 10.1007/s11032-011-9695-2
Singh, S., Sidhu, J. S., Huang, N., Vikal, Y., Li, Z., Brar, D. S., et al. (2001). Pyramiding three bacterial blight resistance genes (xa5, xa13, and Xa21) using marker-assisted selection into indica rice cultivar PR106. Theor. Appl. Genet. 102, 1011–1015. doi: 10.1007/s001220000495
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tanksley, S. D., and McCouch, S. R. (1997). Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277, 1063–1066. doi: 10.1126/science.277.5329.1063
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Telebanco-Yanoria, M. J., Imbe, T., Kato, H., Tsunematsu, H., Ebron, L. A., Vera Cruz, C. M., et al. (2008). A set of standard differential blast isolates (Magnaporthe gsrisea (Hebert) Barr.) from the Philippines for rice (Oryza sativa L.) resistance. Jap. Agric. Res. Q. 42, 23–34. doi: 10.6090/jarq.42.23
Vera Cruz, C. M., Castilla, N. P., Suwarno, S., Santoso, S., Hondrade, E., Hondrade, R. F., et al. (2009). “Rice disease management in the uplands of Indonesia and the Philippines,” in Natural Resource Management for Poverty Reduction and Environmental Sustainability in Fragile Rice-Based Systems, eds S. M. Haefele and A. M. Ismail (Los Baños: International Rice Research Institute), 10–18.
Vossen, E., Sikkema, A., Hekkert, B. L., Gros, J., Stevens, P., Muskens, M., et al. (2003). An ancient R gene from the wild potato species Solanum bulbocastanum confers broad-spectrum resistance to Phytophthora infestans in cultivated potato and tomato. Plant J. 36, 867–882. doi: 10.1046/j.1365-313X.2003.01934.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, X., Fjellstrom, R., Jia, Y., Yan, W. G., Jia, M. H., Scheffler, B. E., et al. (2010). Characterization of Pi-ta blast resistance gene in an international rice core collection. Plant Breed. 129, 491–501. doi: 10.1111/j.1439-0523.2009.01706.x
Wu, K., Xu, T., Guo, C., Zhang, X., and Yang, S. (2012). Heterogeneous evolutionary rates of Pi2/9 homologs in rice. BMC Genet. 13:73. doi: 10.1186/1471-2156-13-73
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wu, Y., Bao, Y., Xie, L., Su, Y., Chu, R., He, W., et al. (2013). Fine mapping and identification of blast resistance gene Pi-hk1 in a broad-spectrum resistant japonica rice landrace. Phytopathology 103, 1162–1168. doi: 10.1094/PHYTO-02-13-0044-R
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yang, S., Li, J., Zhang, X., Zhang, Q., Huang, J., Chen, J. Q., et al. (2013). Rapidly evolving R genes in diverse grass species confer resistance to rice blast disease. Proc. Natl. Acad. Sci. U.S.A. 110, 18572–18577. doi: 10.1073/pnas.1318211110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhou, B., Qu, S., Liu, G., Dolan, M., Sakai, H., Lu, G., et al. (2006). The eight amino-acid differences within three leucine-rich repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity to Magnaporthe grisea. Mol. Plant Microbe Interact. 19, 1216–1228. doi: 10.1094/MPMI-19-1216
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhou, B., and Wang, G. L. (2009). “Functional and evolutionary analysis of the Pi2/9 locus in rice,” in Advances in Genetics, Genomics and Control of Rice Blast Disease, eds G. L. Wang and B. Valent (Dordrecht: Springer), 127–135.
Zhu, Y., Chen, H., Fan, J., Wang, Y., Li, Y., Chen, J., et al. (2000). Genetic diversity and disease control in rice. Nature 406, 718–722. doi: 10.1038/35021046
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: disease resistance, rice blast, rice germplasm, genetic diversity, blast resistance, Pi2 locus
Citation: Vasudevan K, Vera Cruz CM, Gruissem W and Bhullar NK (2014) Large scale germplasm screening for identification of novel rice blast resistance sources. Front. Plant Sci. 5:505. doi: 10.3389/fpls.2014.00505
Received: 26 July 2014; Accepted: 09 September 2014;
Published online: 02 October 2014.
Edited by:
Benjamin Schwessinger, University of California, Davis, USAReviewed by:
Christian Staehelin, Sun Yat-sen University, ChinaDavid De Vleesschauwer, Ghent University, Belgium
Copyright © 2014 Vasudevan, Vera Cruz, Gruissem and Bhullar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Navreet K. Bhullar, Department of Biology, ETH Zurich (Swiss Federal Institute of Technology), Universitaetsstrasse 2, 8092 Zurich, Switzerland e-mail: bhullarn@ethz.ch
 Kumar Vasudevan
Kumar Vasudevan Casiana M. Vera Cruz2
Casiana M. Vera Cruz2 Wilhelm Gruissem
Wilhelm Gruissem Navreet K. Bhullar
Navreet K. Bhullar